Abstract
Self-incompatibility (SI) is a genetic system that prevents self-fertilization in many Angiosperms. Although plants from the Brassicaceae family present an apparently unique SI system that is ancestral to the family, investigations at the S-locus responsible for SI have been mostly limited to two distinct lineages (Brassica and Arabidopsis-Capsella, respectively). Here, we investigated SI in a third deep-branching lineage of Brassicaceae: the tribe Biscutelleae. By coupling sequencing of the SI gene responsible for pollen recognition (SRK) with phenotypic analyses based on controlled pollinations, we identified 20 SRK-like sequences functionally linked to 13 S-haplotypes in 21 individuals of Biscutella neustriaca and 220 seedlings. We found two genetic and phylogenetic features of SI in Biscutelleae that depart from patterns observed in the reference Arabidopsis clade: (1) SRK-like sequences cluster into two main phylogenetic lineages interspersed within the many SRK lineages of Arabidopsis; and (2) some SRK-like sequences are transmitted by linked pairs, suggesting local duplication within the S-locus. Strikingly, these features also were observed in the Brassica clade but probably evolved independently, as the two main SRK clusters in Biscutella are distinct from those in Brassica. In the light of our results and of what has been previously observed in other Brassicaceae, we discuss the ecological and evolutionary implications on SI plant populations of the high diversity and the complex dominance relationships we found at the S-locus in Biscutelleae.
Keywords: self-incompatibility, Biscutelleae, SRK, controlled crosses, dominance/recessivity, genetics of sex
Self-incompatibility (SI) is a genetic system that prevents self-fertilization in plants (reviewed in Charlesworth 1987) and is present in approximately 40% of Angiosperm species (reviewed in Igic et al. 2008). SI is generally controlled by a single locus (S-locus) at which tightly linked genes forming coadapted haplotypic combinations encode pollen and pistil specificities. Pollen is rejected after pollination if its specificity is encoded by the same haplotype as that of the pistil. The determination of pollen phenotype occurs in two distinct flavors in SI systems. In gametophytic systems, the pollen phenotype is defined by its haploid genotype (reviewed in Franklin-Tong and Franklin 2003), whereas in sporophytic systems (SSIs), pollen phenotypes are controlled by the diploid genotype of the paternal plant (reviewed in Hiscock and Tabah 2003).
The molecular mechanism of pollen rejection in SSI has been described only in Brassicaceae and involves the interaction of a cysteine-rich protein (SCR) deposited on the pollen surface (Schopfer et al. 1999) with a transmembrane receptor kinase (SRK) of the stigma (Takasaki et al. 2000). Knowledge about the genes involved in SI has allowed comparative investigations of molecular diversity of both genes in different groups of Brassicaceae. Functional alleles of SRK have been found in several genera of Brassicaceae, including Brassica (Kusaba et al. 1997; Sato et al. 2002), Raphanus (Lim et al. 2002), Capsella (Guo et al. 2009; Paetsch et al. 2006), Arabidopsis (Castric and Vekemans 2007; Kusaba et al. 2001; Schierup et al. 2001), Arabis (Tedder et al. 2011), as well as functional alleles of a SRK ortholog in Leavenworthia (Busch et al. 2008), suggesting that the SCR/SRK SSI system is ancestral in this family. SRK alleles in these genera share the property of trans-specific or even trans-generic polymorphisms, as expected due to the strong negative frequency-dependent selection occurring at the S-locus (Schierup et al. 1998). However, phylogenetic relationships among alleles differ in different groups. SI species of Arabidopsis and Capsella have SRK alleles distributed into many lineages, with highly diverged sequences, whereas SRK alleles in Brassica and Raphanus, taken together, cluster into only two sequence clades, called classes I and II, evolutionary distinct since they are intermingled with SRK lineages from Arabidopsis and Capsella (Figure 1 and Supporting information, Table S1) (Castric and Vekemans 2007; Edh et al. 2009; Schierup et al. 2001). In the genus Leavenworthia, the S alleles clustered into a single clade that is highly divergent from SRK alleles from the other genera (Busch et al. 2008). This seems to be due to independent evolution of the pollen and pistil genes involved in SI, from different members of the same gene families as SCR and SRK (Chantha et al. 2013).
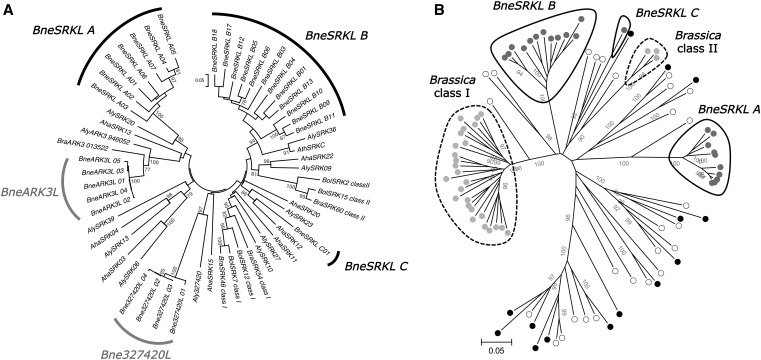
Figure 1.
Phylogenetic relationships between the 29 SRK-related sequences found in Biscutella neustriaca and sequences from other Brassicaceae. The evolutionary trees were built using Neighbor-Joining with a Poisson corrected distance based on amino acid sequences. The proportion of replicate trees in which branches clustered together is indicated close to nodes (1000 bootstrap replicates). (A) The 29 SRK-related sequences found in B. neustriaca cluster in three monophyletic groups similar in sequence to SRK (BneSRKL_A, B, and C) and paralogs ARK3 and Aly327420 found in other other Brassicaceae. Aha, Arabidopsis halleri; Aly, A. lyrata; Ath, A. thaliana; Bol, Brassica oleracera; Bra, B. rapa; Bne, Biscutella neustriaca). (B) Phylogenetic relationships among SRK and SRK-like (SRKL) sequences in four species of Brassicaceae: Capsella grandiflora (black); Brassica oleracera (gray), Arabidopsis lyrata (white), and B. neustriaca (dark gray). The three clades in Biscutella (continuous line) are distinct to the two clades of Brassica (dotted line). Accession numbers of SRK-related sequences are detailed in Table S1.
In SI species of Arabidopsis, only two functional genes, SRK and SCR, have been found within the S-locus (Goubet et al. 2012; Guo et al. 2011), whereas most Brassica haplotypes have a third gene (SLG), which is a paralog of SRK lacking its transmembrane and kinase domains (Nishio and Kusaba 2000; Takasaki et al. 2000). Although SLG is not required for the SI reaction (Nishio and Kusaba 2000), the tightly intermingled phylogeny of SRK and SLG alleles observed in Brassica suggests frequent gene conversion events between these two genes (Sato et al. 2002; Takuno et al. 2008).
Another distinctive feature of SSI systems is the occurrence of dominance relationships between S-haplotypes in heterozygous genotypes. SI Brassica species have two main dominance classes of S-haplotypes, which correspond to the two phylogenetic clusters of haplotypes (Nasrallah et al. 1991). In Arabidopsis, dominance relationships appear to be more complex with more than two dominance levels, showing only partial congruence with phylogenetic clustering of alleles (Llaurens et al. 2008; Prigoda et al. 2005). Dominance relationships are expected to affect S-haplotypes frequencies because the negative frequency-dependent selection typical for SI systems (Wright 1939) will be more intense on dominant than on recessive haplotypes (reviewed in Billiard et al. 2007), resulting in greater frequencies of recessive haplotypes in natural populations (Schierup et al. 1997). This prediction has been validated empirically in Arabidopsis halleri (Llaurens et al. 2008), A. lyrata (Mable et al. 2003; Schierup et al. 2006), and Brassica insularis (Glemin et al. 2005).
Although SI is well described and documented in Brassicaceae, there is still little clue about the evolutionary mechanisms responsible for the differences between species just outlined, e.g., regarding the patterns of phylogenetic clustering of alleles, the co-occurrence of a SRK paralog at the S-locus, and the patterns of dominance relationships. Because most studies focused on two divergent model groups (Brassica-Raphanus and Arabidopsis-Capsella, respectively) we investigates SSI in Biscutella neustriaca, a self-incompatible member of the distantly related Biscutelleae tribe (Franzke et al. 2011). In a previous study, we defined eight phenotypic incompatibility groups by using a full diallele crossing design between 21 individuals of B. neustriaca, which implies that the species has at least eight functional S-haplotypes (Leducq et al. 2010).
We used molecular approaches to identify SRK-like sequences in B. neustriaca individuals, together with segregation analyses, controlled pollinations and paternity assignment from open pollinations to relate the sequences to SI phenotypes and to investigate dominance among haplotypes. Two striking features of the SRK-like sequences were observed: (1) most haplotypes appear to carry two linked SRK-like sequences; and (2) all but one of the SRK-like sequences grouped into distinct phylogenetic clusters whose sequences differ from Brassica ones.
Materials and Methods
Identification of SRK-like sequences
We used 21 B. neustriaca individuals (collection F0) from a collection of plants maintained in the Conservatoire Botanique National de Bailleul (France) that we previously studied by controlled pollinations to identify incompatibility groups (Leducq et al. 2010). We used these plants to identify putative SRK sequences and validate their SI function by investigating association with the incompatibility groups.
DNA was extracted from leaf material as described in Leducq et al. (2010). Candidate SRK sequences were initially obtained by polymerase chain reaction (PCR) amplification with primers 13SEQ2 and SLGR as described in Schierup et al. (2001). The PCR mixture (15 µL) contained 20 ng of DNA, 1X buffer (Applied Biosystems, Foster City, CA), 2.5mM MgCl2, 400 μM Fermentas dNTP mix (Fermentas Canada, Burlington, ON, Canada), 150 µg/mL BSA, 0.5 mΜ each primer, and 0.05 U/μL Taq polymerase (Amplitaq DNA polymerase; Applied Biosystems). Amplifications were performed on a Mastercycler EpGradient Eppendorf thermocycler with the following conditions: 15 min at 95°, 40 cycles of three steps: (1) 45 sec at 95°, (2) 1 min at 50° and (3) 1 min at 72°, followed by 10 min at 72°. PCR products were purified using NucleoSpin Extract II kit from Macherey-Nagel and ligated and transformed into chemically competent bacteria as described in Castric and Vekemans (2007). At least eight clones per individual were sequenced with the BigDye3.1 sequencing kit (Applied Biosystems) and loaded on a 3130 capillary sequencer. Sequences were edited using MEGA5 (Tamura et al. 2011) and validated when identical copies were found in at least two clones.
Validated sequences were checked for similarity with previously described sequences of SRK and SRK-related genes from other Brassicaceae species using blast searches against the nucleotide database in Genbank, and against the genome of A. lyrata at the Phytozome website (http://www.phytozome.net/). In all positive clones sequenced in a first set of 16 individuals (135 clones), sequences from one type (BneARK3L) were found in all individuals (60% of all clones). This sequence is closely related to that of ARK3, a paralog of SRK (also called Aly8 in A. lyrata, and corresponding to NCBI gene ID 946052), which is not involved in pollen-pistil recognition (Charlesworth et al. 2003; Kusaba et al. 2001). We therefore developed a method to minimize amplification of this gene before the cloning step intended to obtain SRK sequences. We identified a restriction site that was present only in this sequence and digested the DNA (purified PCR product) with the HindIII restriction enzyme (A-AGCTT). The restriction was carried out overnight at 37° in a mixture (20 µL) containing 15 µL of fresh purified PCR product, 0.2 mM Spermidin Sigma, 2 µL of 1X Fermentas R buffer, and 0.05 U/μL HindIII Fermentas restriction enzyme. Restriction products were mixed with loading dye and run at 110 V on 2% agarose gels in TBE buffer for 45 min. Fragments were visualized by ethidium bromide under ultraviolet light and compared with a 100-bp DNA ladder. Digested PCR products of BneARK3L gave two fragments (150 and 450 pb size). Undigested PCR fragments (≈600 pb size) were extracted from the agarose gel, cloned, and sequenced as described previously. Of the 269 positive clones obtained, only 6% contained BneARK3L, but 37% contained another sequence type (Bne327420L) that is similar to an A. lyrata gene with unknown function (Hu et al. 2011). We designed specific primers for Bne327420L to test whether it segregates at the S-locus and it proved to be unlinked (Table S2). The reaction mixture for PCR amplification (15 µL) contained 20 ng of DNA, 1X buffer (Applied Biosystems), 2.5mM MgCl2, 200 μM Fermentas dNTP mix, 200 µg/mL bovine serum albumin, 0.5 mm of each primer, and 0.025 U/μL Taq polymerase (Applied Biosystems). Amplifications were performed on Eppendorf thermocycler with the following conditions: 15 min at 95°, 35 cycles of three steps: (1) 40 sec at 95°, (2) 40 sec at 60° and (3) 40 sec at 72°, and finally 10 min at 72°. PCR products were visualized as described previously.
We performed a phylogenetic analysis of all our B. neustriaca sequences, together with a sample of SRK alleles and SRK-related genes from Arabidopsis and Brassica species (Table S1). We used PhyML 3.0 (Guindon et al. 2010) on amino acid sequences with the LG model of substitution and 100 bootstraps. We identified several clades of SRK-related sequences from Biscutella by characterizing the largest monophyletic groups containing exclusively sequences from B. neustriaca. We estimated synonymous (πS) and nonsynonymous (πN) nucleotide divergence between sequences identified as belonging to the different clades, using DNAsp v.5 (Librado and Rozas 2009). For putative SRK alleles, we checked for signatures of positive selection at the codon level using CODEML from the PAML package (Yang 2007) by comparing the likelihood of the nested models M7 and M8, which differ by the existence in M8 of a category of codon sites under diversifying selection (ω = dN/dS > 1). Putatively selected codons were determined by the Bayes empirical Bayes inference procedure (Yang et al. 2005). Based on annotation of hypervariable regions previously identified in Brassica SRK alleles (Kusaba et al. 1997), we determined whether putatively selected codons were located within hypervariable regions.
Based on the SRK-like sequences, we developed a typing strategy by designing primers with Primer3 (http://primer3.sourceforge.net/) to specifically target each candidate SRK sequence identified in collection F0 (Table S2). PCR amplifications were performed as described previously for Bne327420L, and individuals were genotyped by absence/presence of a band on agarose gel. Because of the low divergence between some sequences, some nonspecific amplifications occurred (revealed by frequent or systematic amplification in all genotyped individuals, regardless of their incompatibility type). In these cases, PCR products were systematically sequenced, as described previously, using the primers used for the amplifications.
Segregation of SRK-like sequences and association with the incompatibility phenotype
Our second aim was to verify that SRK-like sequences are indeed allelic and that they segregate with the SI phenotype. For this we used segregation patterns in F1 offspring to test for associations between the SI phenotype and the occurrence of particular SRK-like sequences in the F0 collection. We also tested whether dominance occurs between the B. neustriaca S-haplotypes, as we previously inferred (Leducq et al. 2010). For this purpose, we used two collections of seedlings obtained from 13 individuals of the F0 collection (Table S3), representing the eight previously identified incompatibility groups (Leducq et al. 2010). The first collection of 82 seedlings (F1o) was obtained from open pollinations among seven F0 individuals (four pollen donors and three pollen receptors) and paternities were assigned a posteriori with parentage analysis (Leducq et al. 2010). This collection included eight groups of verified full sibs. The second collection consisted of five sets of 138 seedlings (F1c) obtained from controlled pollinations among five pairs of compatible F0 individuals. These controlled crosses were performed as described in Leducq et al. (2010).
To test whether observed genotype frequencies at the S-locus followed Mendelian expectations in the 13 cohorts of F1 individuals, we defined n as cohort size (which range from 5 to 50 in collection F1, see Table S3) and randomly generated 100,000 hypothetical cohorts of individuals for each n value. Each simulated set of progeny (which we denote as a ‘cohort’) was assumed to be produced by a cross between two diploid individuals A and B with known genotype at the S-locus (or binary combinations of four possible S-haplotypes S1, S2, S3, and S4). We performed the analysis for two cases, depending on the number of different genotypes at the S-locus observed in the cohort: (1) individuals A and B both heterozygous (e.g., genotypes S1S2 and S3S4); (2) parent A homozygous (S1S1) and parent B heterozygous (S3S4; considered only when only one or two genotypes were present in the sibship). Each sibship from a cross could thus include either (1) genotypes S1S3, S1S4, S2S3, and S2S4 with expected frequency of 0.25 each, or (2) genotypes S1S3 and S1S4 with expected frequency of 0.5 each. For each cohort size n and each possible genotype, we used the distribution of simulated genotype frequencies to determine whether observed genotype frequencies differed significantly from those expected.
To test for associations between the putative S-haplotypes and the SI phenotypes in the offspring, we performed 1472 controlled pollinations between individuals of collection F0, F1o, and F1c. A total of 1229 pollinations were performed between individuals putatively sharing at least one S-haplotype, and the others were between individuals not thought to share S-haplotypes, to control for pollen viability and stigma receptivity. In collection F0, we had 18 different combinations between 13 putative S-haplotypes. We made crosses between different individuals with the same S-locus genotype for 21 out of 35 genotypes tested. Controlled pollinations and determination of compatibility were performed as described in Leducq et al. (2010).
Results
Identification of SRK-like sequences in B. neustriaca
Overall, our approach yielded 29 distinct sequences in the F0 individuals (accession numbers KF905295−KF905324; Table S4). These sequences clustered into five clades, defined as the largest monophyletic groups containing exclusively sequences from B. neustriaca. Two clades were closer to sequences of SRK paralogs than to SRK alleles from Arabidopsis or Brassica (Figure 1A). One of these clades includes five sequences (BneARK3L) that are similar to the ARK3 gene sequence in B. rapa (gene Bra 013522) and A. lyrata (Aly8 or NCBI gene ID 946052). A second clade of four sequences (Bne327420L) is similar to a SRK paralog present in the A. lyrata genome (gene 327420) that lack a kinase domain, unlike ARK3. Specific PCR primers for Bne327420L yielded amplification products in all individuals from collection F0 (data not shown), confirming that these do not correspond to S-locus alleles. The 20 remaining sequences were similar to those from SRK alleles from several Arabidopsis species, and we will refer to those as SRK-like (SRKL) sequences (Figure 1A). B. neustriaca SRKL sequences formed three distinct clades, which we call classes A, B, and C. The seven class A sequences are similar to AlySRK20, a functional SRK allele of A. lyrata (also called SRKb in Kusaba et al. 2001). There were 12 class B sequences, which are closest to AthSRKC, one of the three alleles still segregating at the S-locus in A. thaliana (Shimizu et al. 2004), and to AlySRK36, a functional allele in A. lyrata (Bechsgaard et al. 2006). The third clade, class C, is represented by a single sequence similar to AhSRK12, a functional A. halleri SRK allele (Castric and Vekemans 2007). All three clades of SRK-like sequences in B. neustriaca are distinct from those of Brassica (Figure 1B).
Segregation of SRK-like sequences from F0, F1C, and F1O collections
Patterns or presence and absence results for the 20 SRKL sequences among the 21 individuals of collection F0 are presented in Table 1. Despite the fact that B. neustriaca is diploid (Leducq et al. 2013; Tremetsberger et al. 2002), we observed up to four SRKL sequences per individual, sometimes including A, B, and C classes. However, we observed consistent associations between certain sequences. Hereafter, we denote a putative SRK sequence in a simpler form, for example using A01 as the abbreviation for BneSRKL_A01, where A refers to class A. In this notation, we found, for instance, that A01 is associated with either A02 or A03, and B01 was always associated with B13, B04 with C01, B10 with B11, and B09 with B12 (Table 1). Genotyping of the sibships confirmed these associations and provided two more associations (A06 with A07 and B17 with B18). These associations suggest linkage disequilibrium between eight pairs of sequences, forming the following haplotypes at a single locus: A01−A02, A01−A03, A06−A07, B01−B13, B04−C01, B10−B11, B09−B12, and B17−B18 (Table 2). Most of the associations involve sequence pairs from the same class (A or B). However, one haplotype (B04-C01) did not. In addition, several sequences identified in the F0 collection, A04, A05, B03, B05, and B06, were not transmitted in pair. Using information about the eight haplotypes described previously, plus the five distinct haplotypes involving the five sequences just mentioned, allowed us to infer the genotype of each of the 21 F0 individuals (Table 1, Table 2, and Table 3).
Table 1. SRK-like sequences identified in the F0 collection.
| Individual | Sequences Identified | Putative Genotypes |
|---|---|---|
| 1 | B09, B10, B11a, B12 | B09B12 / B10B11 |
| 2 | B09, B10, B11a, B12 | B09B12 / B10B11 |
| 3 | B10, B11 | B10B11 / B10B11c |
| 4 | A01a, A03, B10, B11 | A01aA03 / B10B11 |
| 5 | A01b, A03, B09, B12 | A01bA03 / B09B12 |
| 6 | A01a, A02, A01a, A03a | A01aA02 / A01aA03 |
| 7 | A01a, A03, B06 | A01aA03 / B06 |
| 8 | A06, A07, B06 | A06A07 / B06 |
| 9 | B06, B18 | B06 / B17bB18 |
| 10 | B01, B06 | B01B13b / B06 |
| 11 | B01, B13, B04, C01 | B01B13 / B04C01 |
| 12 | A05, B01, B13 | A05 / B01B13 |
| 13 | A05, B04, C01 | A05 / B04C01 |
| 14 | A05, B04, C01 | A05 / B04C01 |
| 15 | A05, B05 | A05 / B05 |
| 16 | A05, B05 | A05 / B05 |
| 17 | B04, C01 | B04C01 / B04C01c |
| 18 | A01a, A02, B04, C01a | A01aA02 / B04C01 |
| 19 | B03, B06 | B03 / B06 |
| 20 | B03, B05 | B03 / B05 |
| 21 | A04, B01, B13 | A04 / B01B13 |
Putative genotypes are deduced from patterns of segregation in the F1c and F1o collections (see Table 3).
Typed but not sequenced.
Supposed by association.
Homozygote genotypes.
Table 2. Pattern of SRK-like sequences segregation observed in 13 cohorts of seedlings issued from crosses between individuals of collection F0.
| Seedling Collection | na | Parentsb | Seedlings Genotype (Observed Frequency)c | Deduced Linkage Groups | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parent 1 | Parent 2 | ||||||||||
| 1 | 2 | 1 | 2 | 3 | 4 | 1 | 2 | 1 | 2 | ||
| F1o | 7 | 3 | 6 | A01A02 | A01A03 | – | – | B10 | B10 | A01 | A01 |
| B10B11 | B10B11 | B11 | B11 | A02 | A03 | ||||||
| (4) | (3) | ||||||||||
| 13 | 3 | 7 | A01A03 | B06B10 | – | – | B10 | B10 | A01 | B06 | |
| B10B11 | B11 | B11 | B11 | A03 | |||||||
| (7) | (6) | ||||||||||
| 5 | 8 | 4 | A01A03 | A06A07 | A01A03 | – | A06 | B06 | A01 | B10 | |
| A06A07 | B10B11 | B06 | A07 | A03 | B11 | ||||||
| (2) | (1) | (2) | |||||||||
| 7 | 8 | 6 | A01A03 | A01A02 | A01A03 | – | A06 | B06 | A01 | A01 | |
| A06A07 | B06 | B06 | A07 | A02 | A03 | ||||||
| (1) | (3) | (3) | |||||||||
| 21 | 9 | 4 | A01A03 | B10B11 | A01A03 | B06B10 | B06 | –d | A01 | B10 | |
| (4) | (1) | B06 | B11 | A03 | B11 | ||||||
| (10) | (6) | ||||||||||
| 7 | 9 | 6 | A01A02 | A01A03 | A01A02 | – | B06 | –d | A01 | A01 | |
| (3) | (2) | B06 | A02 | A03 | |||||||
| (2) | |||||||||||
| 6 | 10 | 4 | A01A03 | B01B10 | A01A03 | – | B01d | B06 | A01 | B10 | |
| B01 | B11 | B06 | A03 | B11 | |||||||
| (1) | (4) | (1) | |||||||||
| 16 | 10 | 6 | A01A02 | A01A03 | A01A02 | A01A03 | B01d | B06 | A01 | A01 | |
| B01 | B01 | B06 | B06 | A02 | A03 | ||||||
| (2) | (6) | (6) | (2) | ||||||||
| F1c | 14 | 4 | 20 | A01A03 | A01A03 | B03B10 | B05B10 | A01 | B10 | B03 | B05 |
| B03 | B05 | B11 | B11 | A03 | B11 | ||||||
| (3) | (2) | (4) | (5) | ||||||||
| 30 | 12 | 8 | A05A06 | A05B06 | A06A07 | B01B06 | A05 | B01 | A06 | B06 | |
| A07 | (6) | B01 | (11) | A07 | |||||||
| (7) | (6) | ||||||||||
| 30 | 17 | 18 | A01A02 | B04C01 | – | – | B04 | B04 | A01 | B04 | |
| B04C01 | (17) | C01 | C01 | A02 | C01 | ||||||
| (13) | |||||||||||
| 50 | 18 | 1 | A01A02 | A01A02 | B04B09 | B04B10 | A01 | B04 | B10 | B09 | |
| B09B12 | B10B11 | B12C01 | B11C01 | A02 | C01 | B11 | B12 | ||||
| (11) | (15) | (9) | (15) | ||||||||
| 14 | 21 | 8 | A06A07 | A04A06 | A04 | B01d | A06 | B06 | |||
| B01 | B01B06 | A07 | A04B06 | A07 | |||||||
| (4) | (1) | (5) | (4) | ||||||||
Collection F1o (eight cohorts) was obtained from open pollinations and seedlings were assigned by paternity analyses (Leducq et al. 2010). Collection F1c was obtained by controlled crosses. For each cohort, the following information is shown: number of seedlings, name of F0 parental individuals, SRK-like sequences found in parents, associations of SRK-like sequences found in progeny with frequency of association and deduced SRK-like linkage groups in parents
Cohort size.
F0 collection; see Table 2 for genotypes.
F1 collection.
B13, B17, and B18 could not be typed in progeny.
Table 3. Occurrence of putative S-haplotypes identified in F0 collection (sequencing).
| S-Haplotype | Sequence | Incompatibility Group | Haplotype Co-occurrence in F0 | Cohorts With the Haplotypea | |
|---|---|---|---|---|---|
| 1 | 2 | ||||
| S01 | B10 | B11 | I | 5 | 7 |
| S02 | A01 | A03 | II | 4 | 9 |
| S03 | B06 | – | III | 5 | 9 |
| S04 | B01 | B13b | IV | 4 | 2b |
| S05 | A05 | – | V | 5 | 1 |
| S06 | B04 | C01 | VI | 6 | 2 |
| S07 | A01 | A02 | VII | 2 | 4 |
| S08 | B03 | – | VIII | 2 | 1 |
| S09 | A06 | A07 | – | 1 | 4 |
| S10 | B05 | – | – | 3 | 1 |
| S11 | B09 | B12 | – | 3 | 1 |
| S12 | B17b | B18b | – | 1 | 0b |
| S13 | A04 | – | – | 1 | 1 |
Incompatibility groups previously identified in F0 collection by Leducq et al. (2010) are indicated with associated S-haplotypes.
B13, B17, and B18 were not typed in the progeny.
These results indicate that B. neustriaca has two SRKL genes in most of its S-locus haplotypes, and that most individuals we analyzed are heterozygotes. In two individuals (3 and 17) we found only one haplotype (haplotypes B10−B11 and B04−C01, respectively), so we hypothesize that these individuals could either be homozygotes for those haplotypes or heterozygotes with another haplotype, so far undetected (Table 1 and Table 2).
We tested the 13 putative S-haplotypes inferred from SRKL sequences in sibships F1c and F1o. In eight of ten sibships the genotype frequencies followed Mendelian expectations from crossing two individuals heterozygous at the S-locus (case 1), but two F1o sibships had significant deviations frequencies (one for one genotype, and one for two). In three other sibships from crossing one heterozygous individual and individuals 3 or 17, for which only a single haplotype was identified, genotype frequencies of 0.5 were found (case 2) and the alternative of more than two progeny genotypes was rejected (Table S3). This confirms that these two individuals are homozygotes for their respective haplotypes. Thus, all S-locus genotypes were inferred in the F0 collection.
Associations between SRK-like sequences and SI phenotypes
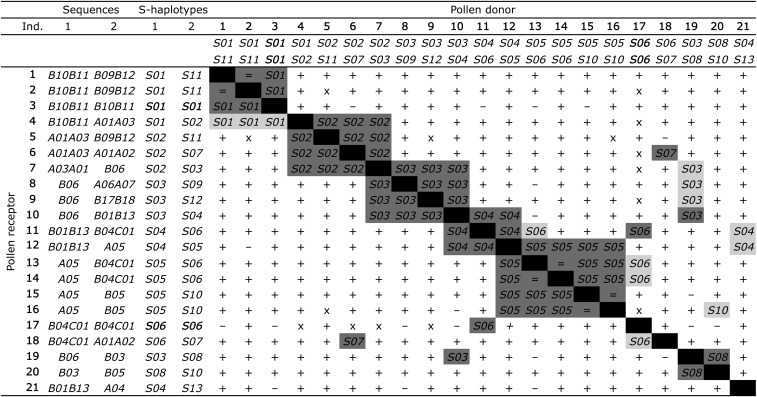
As shown in Figure 3, the eight incompatibility groups (groups I−VIII) identified from S phenotypes in the F0 collection (Leducq et al. 2010) correspond to groups of individuals sharing at least one SRKL sequence (putative SRK allele) and we will now refer to these as S-haplotypes S01 to S08 (Table 3).
Figure 3.
Results from controlled crosses realized among 21 individuals of collection F0 (Leducq et al. 2010) related with S-genotypes determined in this study. Linked groups of SRK-like sequences found in each individual are indicated in second and third columns; corresponding S-haplotypes are indicated in fourth and fifth columns. Black boxes represent unsuccessful self-pollinations; gray boxes represent reciprocal unsuccessful cross-pollinations, filled with the expressed S-haplotype when shared between crossed individuals (sign equal when to crossed individuals have identical genotypes). Light gray box represent nonreciprocal unsuccessful cross-pollinations. Successful cross-pollinations are represented by plus signs (+). Unsuccessful cross pollinations realized between individuals sharing no S-haplotypes are indicated by minus signs (-). Unrealized pollinations are represented by crosses (x).
To confirm the link between these haplotypes and the SI phenotypes in the offspring and to validate it for other putative S-haplotypes, we performed 1472 additional controlled pollinations between individuals of collection F1 (Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, Figure S10, Figure S11, Figure S12, Figure S13, and Figure S14). Figure 4 summarizes our results from all crosses between individuals of collections F0 and F1. A total of 71% of positive controls (243 crosses between individuals sharing no putative S-haplotype) yielded fruits vs. 9% of crosses between individuals sharing one putative S-haplotype (1229 crosses; Figure 4, A−D). These results confirmed the functional incompatibility types of the S-haplotypes previously identified as S01−S08 (Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, and Figure S8) and validate the function of four other putative S-haplotypes S09 (A06/A07; Figure S9), S10 (B05; Figure S10), S12 (B17/B18; Figure S12) and S13 (A04; Figure S13), but not S11 (B09/B12; Figure S11). Successful positive controls between individuals sharing no common sequence attested that these aborted pollinations most probably resulted from incompatibility reactions (Figure 4, A and B). Compatibility results were almost the same for different plants with identical genotypes (but see Figure S10 for an exception). Most crosses between individuals carrying S07 (A01/A02) and S02 (A01/A03) were fully compatible, indicating that, despite sharing the A01 sequence, these two haplotypes confer distinct incompatibility types (Figure S14). The low sequence diversity we found within A01 sequences (one nonsynonymous nucleotide substitution separating sequences A01a and A01b) was not associated with whether the other sequence in the haplotype was A02 or A03 (Table 1 and Table S4).
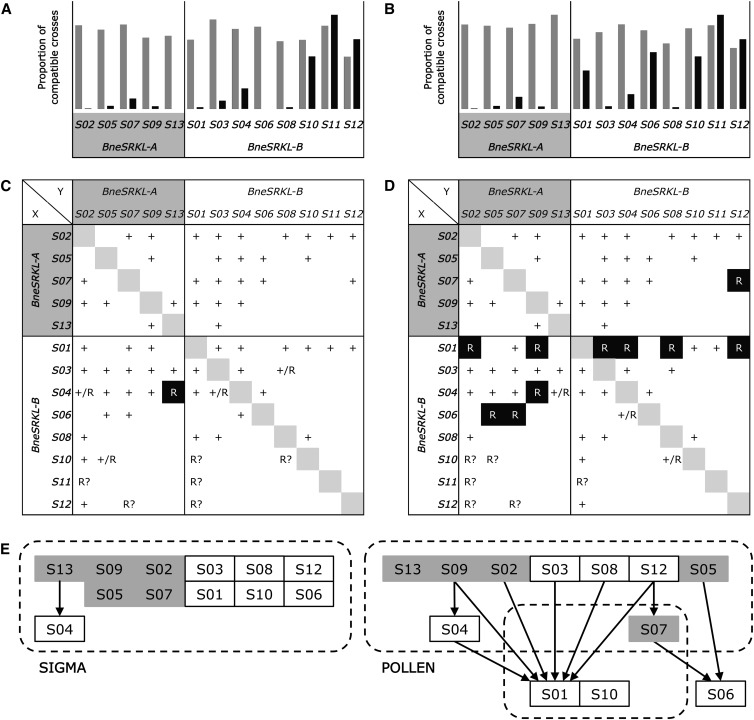
Figure 4.
Dominance relationships determined by controlled pollinations between 13 S-haplotypes in B. neustriaca. Proportion of successful pollinations calculated for all crosses involving stigma (A) or pollen (B) of an individual with a given S-haplotype (x-axis). Crosses between individuals sharing or not sharing the haplotype are filled in black and gray, respectively. Pattern of SI expression for S-haplotypes involved in 35 possible genotypes in stigma (C) and pollen (D). Results are indicated for the haplotype X (in rows) as compared to haplotype Y (in columns) and report whether X was expressed (+) or not (R). Uncertain cases resulted from either the lack of evidence for X function (R?) or from inconsistencies between different individuals with the same genotype (+/R). (E) Dominance relationships among 12 S-haplotypes (S11 removed) in stigma (left) and pollen (right). Only nonambiguous cases were reported here. Dotted frames regroup haplotypes that are mostly codominant. Arrows indicate dominance relationships. Haplotypes from classes A and B are filed in gray and white respectively. See Figure 3, Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, Figure S10, Figure S11, Figure S12, Figure S13, and Figure S14 for detailed crosses.
Patterns of dominance among S-haplotypes
In controlled pollinations among F0 individuals, some pairs sharing one S-haplotype were fully compatible, indicating the occurrence of dominance relationships between S-haplotypes, as typical for sporophytic SI (Figure 3). Dominance relationships were not identical between pollen and stigmas. For instance, pollen from individual 4, carrying alleles S01 and S02, was compatible with individual 3, carrying two copies of allele S01 (whose expression in stigmas was validated by unsuccessful pollination by other individuals having S01), but these individuals were incompatible when individual 3 was the pollen donor (Figure 3 and Figure 4, A−D). Thus, allele S01 is expressed in stigma of individual 4 (and is thus either dominant or codominant with allele S02) but recessive to S02 in pollen. The varied compatibility in pollen and stigmas among the S-haplotypes in crosses between individuals sharing one S-haplotype (combining F0 and F1 crosses) is shown in Figure 4, A−B. The average proportion of compatible crosses was lower for S-haplotypes of class BneSRK-A (3%) than for BneSRK-B (23%), and the difference was less pronounced in stigmas (3 and 18% for A and B classes, respectively; Figure 4A) than in pollen (3 and 28% for A and B classes, respectively; Figure 4B). In stigmas, S-haplotypes of class A were expressed in all genotypes, whereas there was at least one case of recessivity for a class B haplotype (Figure 4C). We observed many more cases of recessivity in pollen, at least 10 cases, mostly involving two class B S-haplotypes (S01 and S06), which were not expressed in most genotypes tested (Figure 4D). Overall, codominance was more frequent in stigmas than in pollen and recessive haplotypes generally belonged to class B (Figure 4E).
Analysis of polymorphism among SRKL sequences and test of diversifying selection
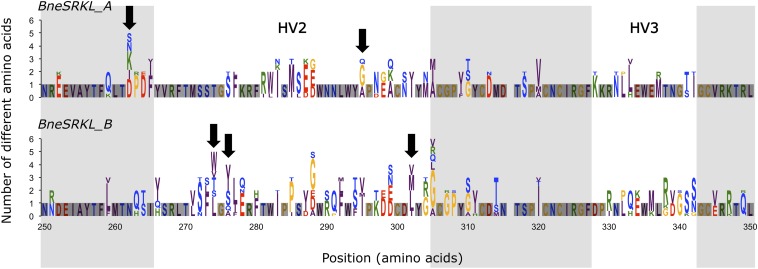
To test for positive selection resulting from negative frequency-dependent selection, we analyzed polymorphisms in the SRKL sequences. Diversity was found to be high among sequences of BneSRKL classes A and B at both synonymous (πS = 9.9–17.0%) and non-synonymous sites (πN = 7.8–9.8%; 528−465 nucleotide sites; Table 4, Figure 2); class C has a single sequence. PAML analysis yielded evidence for a class of sites evolving under diversifying selection for both classes A and B (Table 4). Using a cut-off at P = 0.95 in the Bayes empirical Bayes analysis, we detected 2 codons evolving under positive selection for class A and 3 codons for class B (Table 4 and Figure 2). All three positively selected codons of class B were within regions corresponding to the hypervariable regions of Brassica sequences (Nishio and Kusaba 2000), and the same was true for one of the two class A selected codons (Figure 2). Three of these sites were also previously detected as positively selected in other Brassicaceae (Table 4). The posterior mean dN/dS for positively selected sites in classes A and B were 4.110 and 2.915, respectively (Table 4). Only one sequence, BneSRKL_B12, had a premature stop codon; this sequence has a 1bp deletion that resulted in a reading frame shift.
Table 4. Analyses of nucleotide polymorphism and detection of positive selection in sequences within phylogenetic groups BneSRKL_A and BneSRKL_B.
| Log Likelihood | Positively Selected Sites | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| nseq | nsites | πS | πN | πN/πS | M7 | M8 | -2ΔLn | p-Value | Sitesa | Average dN/dS | |
| BneSRKL_A | 7 | 528 | 0.0993 | 0.0777 | 0.78 | −1432 | −1429 | 6.37 | 0.041 | 262N 295Sb | 4.110 |
| BneSRKL_B | 10 | 465 | 0.1705 | 0.0984 | 0.58 | −1714 | −1708 | 11.71 | 0.003 | 274Db,c | 2.915 |
| 276b,d | |||||||||||
| 303Vb,c | |||||||||||
Abbreviations: nseq, number of sequences analyzed; nsites, number of nucleotide sites; πS and πS, nucleotide diversity at synonymous and non-synonymomus sites, respectively.
BoSRK60 used as reference.
Sites in hypervariable regions.
Sites also detected in Arabidopsis and Brassica.
Site also detected in Arabidopsis.
Figure 2.
Amino acid variation in two SRK-related sequence groups found in Biscutella neustriaca. Height of bars is proportional to the number of different amino-acids at each position within a group. Relative frequency of each amino acid (symbolized by its letter) is proportional to its height in the bar. Black arrows indicate positions under positive selection in groups A and B (see Table 4) that fall into or close to hypervariable regions known to be involved in SRK-SCR protein recognition (HV2 and HV3 indicated by white frames). The alignment (100 amino-acids positions) was based on eight BneSRKL-A and 11 BneSRKL-B sequences in MEGA5.
Discussion
Molecular identification of SRK-like sequences linked to the SI phenotype in the tribe Biscutelleae
Here, we identified SRK-like sequences in B. neustriaca and demonstrated that these sequences segregate at a single locus and are tightly linked with the SI phenotype. Our results suggest that these SRK-like sequences are functional SRK alleles similar to those in the Brassicaceae SSI system previously described in Brassica (Kusaba et al. 1997; Sato et al. 2002), Capsella (Guo et al. 2009; Paetsch et al. 2006), Arabidopsis (Castric and Vekemans 2007; Kusaba et al. 2001; Schierup et al. 2001), and Arabis (Tedder et al. 2011) and not those of Leavenworthia, where the gene involved in stigma SI is not homologous to the SRK of Arabidopsis and Brassica (Chantha et al. 2013).
SRK sequences in the Biscutella and Brassica lineages have several features in common
The two large monophyletic groups, A and B, of B. neustriaca SRKL sequences resemble the situation observed in Brassica outlined in the Introduction, where the two sequence groups represent dominant and recessive alleles. A demographic event such as a strong bottleneck may have caused the low SRK diversity in Brassica (Castric and Vekemans 2007; Edh et al. 2009). The B. neustriaca clusters, including the single sequence of group C, do not cluster with those of Brassica. Moreover, the reduction in the number of lineages in B. neustriaca appears to have been followed by allelic diversification of two of the three surviving SRK lineages, with evidence of strong positive selection (dN/dS in the range 2.92−4.11) of the same order of magnitude as inferred in Brassica (2.98; Castric and Vekemans 2007) and in Leavenworthia (3.49; Herman et al. 2012) and contrasting with lower estimates for A. lyrata and A. halleri (1.49 and 1.44, respectively) whose S-locus diversification seems to be much more ancient.
The duplicated SRKL sequences within B. neustriaca S-haplotypes also resemble the situation in Brassica, where SRK and SLG co-segregate at the S-locus (Nishio and Kusaba 2000; Takuno et al. 2008). Cosegregation of SI-related sequences also was recently observed in some genera of Papaveraceae, which have a gametophytic system (Paape et al. 2011), and in Solaneaceae a set of genetically linked F-box genes collectively determines the pollen phenotype (Kubo et al. 2010). We cannot currently determine whether the two B. neustriaca SRKL sequences together determine the pistil phenotype, or, a single SRK sequence, as in Brassica. In one case, however, we have evidence that a sequence is probably not involved in pistil recognition, since BneSRKL-A01 is shared by two functionally distinct S-haplotypes, S02 and S07. Hence, further functional investigations should focus on the other BneSRKL sequences, A02 and A03, also carried by these haplotypes. Genomic studies are now needed to determine the structure and genomic localization of the S-locus, and to obtain the full coding sequences of both SRK-like copies within S-haplotypes, to determine whether both sequences have a functional kinase domain, or whether one of them consistently lacks this like Brassica SLG.
Number of S-alleles and patterns of dominance
B. neustriaca is a narrow endemic species whose populations are highly disconnected, with restricted gene flow (Leducq et al. 2013). Within natural populations, individuals typically aggregate in high-density patches (J.-B. Leducq, personal observations). In our previous study, we showed that highly reduced diversity at the S-locus and low local density could greatly reduce maternal reproductive success (Leducq et al. 2010), i.e., there is a S-Allee effect (Wagenius et al. 2007). Given that B. neustriaca has at least 13 functionally distinct S-alleles in the 21 individuals sampled in collection F0, it might appear unlikely that ecological events could reduce allelic diversity enough to cause an overall reduced reproductive success in natural populations.
Recessivity of the SI phenotype in B. neustriaca is commoner in pollen than in stigmas, where codominance is most frequent, as was also found in Brassica and Arabidopsis. The difference between dominance in pollen and stigmas is often interpreted as a consequence of the more intense sexual selection on the male than on the female function, because dominance in pollen allows a plant to minimize the chances of its pollen being rejected by stigmas from other plants (Schoen and Busch 2009). Interestingly, all recessive S-haplotypes except for one appear to belong to class B, suggesting that different dominance is associated with different S-haplotype lineages, as observed in Brassica (Nasrallah et al. 1991) and Arabidopsis (Llaurens et al. 2008; Prigoda et al. 2005).
Because negative frequency-dependent selection acting on S-allele diversity is expected to be partially relaxed for recessive S-haplotypes, they are expected to reach greater frequencies in populations than dominant ones (Billiard et al. 2007; Llaurens et al. 2008; Schierup et al. 1997). Consistent with this prediction, the two most recessive S-haplotypes found in our sample of 21 individuals (S01 and S06) were the most frequent. It should now be tested whether this is also true in natural populations of B. neustriaca.
Supplementary Material
Acknowledgments
We address special thanks to Sophie Vauquier and Damien Drubet for help in phenotyping, Matthieu Poiret, Aline Courseaux, Anne-Catherine Holl, Nathalie Faure, and Benoit Leducq for technical help and two anonymous reviewers and Deborah Charlesworth for comments that greatly improved the manuscript. Yves Piquot and Nina Hautekèete helped us to get access to plant material. J.-B.L. was supported by a fellowship from the Fonds de Recherche en Santé du Québec (FRSQ). We acknowledge financial support from the life science department of the CNRS (ATIP-plus research grant to X.V.), from ANR Jeunes Chercheurs BRASSIDOM (ANR 11 JSV7-008-01), and from the European Union through the Life Program ‘‘Rescue of Viola hispida and Biscutella neustriaca on the Seine Valley.”
Footnotes
Communicating editor: D. Charlesworth
Literature Cited
- Bechsgaard J. S., Castric V., Vekemans X., Schierup M. H., Schierup M. H., 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750 [DOI] [PubMed] [Google Scholar]
- Billiard S., Castric V., Vekemans X., 2007. A general model to explore complex dominance patterns in plant sporophytic self-incompatibility systems. Genetics 175: 1351–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J. W., Sharma J., Schoen D. J., 2008. Molecular characterization of Lal2, an SRK-like gene linked to the S-locus in the wild mustard Leavenworthia alabamica. Genetics 178: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V., Vekemans X., 2007. Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol. Biol. 7:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantha S. C., Herman A. C., Platts A. E., Vekemans X., Schoen D. J., 2013. Secondary evolution of a self-incompatibility locus in the Brassicaceae genus Leavenworthia. PLoS Biol. 11: e1001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 1987. Self-incompatibility systems in the flowering plants. Perspect. Biol. Med. 30: 263–277 [Google Scholar]
- Charlesworth D., Mable B. K., Schierup M. H., Bartolome C., Awadalla P., 2003. Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164: 1519–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edh K., Widen B., Ceplitis A., 2009. Molecular population genetics of the SRK and SCR self-incompatibility genes in the wild plant species Brassica cretica (Brassicaceae). Genetics 181: 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong N. V., Franklin F. C. H., 2003. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 8: 598–605 [DOI] [PubMed] [Google Scholar]
- Franzke A., Lysak M. A., Al-Shehbaz I. A., Koch M. A., Mummenhoff K., 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Glemin S., Gaude T., Guillemin M. L., Lourmas M., Olivieri I., et al. , 2005. Balancing selection in the wild: testing population genetics theory of self-incompatibility in the rare species Brassica insularis. Genetics 171: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet P. M., Berges H., Bellec A., Prat E., Helmstetter N., et al. , 2012. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 8: e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., et al. , 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guo Y. L., Bechsgaard J. S., Slotte T., Neuffer B., Lascoux M., et al. , 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. L., Zhao X., Lanz C., Weigel D., 2011. Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 157: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A. C., Busch J. W., Schoen D. J., 2012. Phylogeny of Leavenworthia S-alleles suggests unidirectional mating system evolution and enhanced positive selection following an ancient population bottleneck. Evolution 66: 1849–1861 [DOI] [PubMed] [Google Scholar]
- Hiscock S. J., Tabah D. A., 2003. The different mechanisms of sporophytic self-incompatibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J. F., et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B., Lande R., Kohn J. R., 2008. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 169: 93–104 [Google Scholar]
- Kubo K., Entani T., Takara A., Wang N., Fields A. M., et al. , 2010. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799 [DOI] [PubMed] [Google Scholar]
- Kusaba M., Nishio T., Satta Y., Hinata K., Ockendon D., 1997. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: Implications for the evolution and recognition mechanism. Proc. Natl. Acad. Sci. USA 94: 7673–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J. B., et al. , 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A-lyrata and its autogamous relative A-thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Leducq J. B., Gosset C. C., Poiret M., Hendoux F., Vekemans X., et al. , 2010. An experimental study of the S-Allee effect in the self-incompatible plant Biscutella neustriaca. Conserv. Genet. 11: 497–508 [Google Scholar]
- Leducq J.-B., Siniarsky C., Gosset C., Godé C., Poiret M., et al. , 2013. Intriguing small-scale spatial distribution of chloropastic and nuclear diversity in the endangered plant Biscutella neustriaca (Brassicaceae). Conserv. Genet. 14: 65–77 [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lim S. H., Cho J., Lee J., Cho Y. H., Kim B. D., 2002. Identification and classification of S haplotypes in Raphanus sativus by PCR-RFLP of the S locus glycoprotein (SLG) gene and the S locus receptor kinase (SRK) gene. Theor. Appl. Genet. 104: 1253–1262 [DOI] [PubMed] [Google Scholar]
- Llaurens V., Billiard S., Leducq J. B., Castric V., Klein E. K., et al. , 2008. Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution 62: 2545–2557 [DOI] [PubMed] [Google Scholar]
- Mable B. K., Schierup M. H., Charlesworth D., 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90: 422–431 [DOI] [PubMed] [Google Scholar]
- Nasrallah J. B., Nishio T., Nasrallah M. E., 1991. The self-incompatibility genes of brassica—expression and use in genetic ablation of floral tissues. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 393–422 [Google Scholar]
- Nishio T., Kusaba M., 2000. Sequence diversity of SLG and SRK in Brassica oleracea L. Ann. Bot. (Lond.) 85: 141–146 [Google Scholar]
- Paape T., Miyake T., Takebayashi N., Wolf D., Kohn J. R., 2011. Evolutionary genetics of an S-like polymorphism in Papaveraceae with putative function in self-incompatibility. PLoS ONE 6: e23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetsch M., Mayland-Quellhorst S., Neuffer B., 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97: 283–290 [DOI] [PubMed] [Google Scholar]
- Prigoda N. L., Nassuth A., Mable B. K., 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22: 1609–1620 [DOI] [PubMed] [Google Scholar]
- Sato K., Nishio T., Kimura R., Kusaba M., Suzuki T., et al. , 2002. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M. H., Vekemans X., Christiansen F. B., 1997. Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics 147: 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M. H., Vekemans X., Christiansen F. B., 1998. Allelic genealogies in sporophytic self-incompatibility systems in plants. Genetics 150: 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M. H., Mable B. K., Awadalla P., Charlesworth D., 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M. H., Bechsgaard J. S., Nielsen L. H., Christiansen F. B., 2006. Selection at work in self-incompatible Arabidopsis lyrata: mating patterns in a natural population. Genetics 172: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen D. J., Busch J. W., 2009. The evolution of dominance in sporophytic self-incompatibility systems. II. Mate availability and recombination. Evolution 63: 2099–2113 [DOI] [PubMed] [Google Scholar]
- Schopfer C. R., Nasrallah M. E., Nasrallah J. B., 1999. The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Shimizu K. K., Cork J. M., Caicedo A. L., Mays C. A., Moore R. C., et al. , 2004. Darwinian selection on a selfing locus (retracted article. See vol 320, pg 176, 2008). Science 306: 2081–2084 [DOI] [PubMed] [Google Scholar]
- Takasaki T., Hatakeyama K., Suzuki G., Watanabe M., Isogai A., et al. , 2000. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916 [DOI] [PubMed] [Google Scholar]
- Takuno S., Nishio T., Satta Y., Innan H., 2008. Preservation of a pseudogene by gene conversion and diversifying selection. Genetics 180: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedder A., Ansell S. W., Lao X., Vogel J. C., Mable B. K., 2011. Sporophytic self-incompatibility genes and mating system variation in Arabis alpina. Ann. Bot. (Lond.) 108: 699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremetsberger K., Konig C., Samuel R., Pinsker W., Stuessy T. F., 2002. Infraspecific genetic variation in Biscutella laevigata (Brassicaceae): new focus on Irene Manton’s hypothesis. Plant Syst. Evol. 233: 163–181 [Google Scholar]
- Wagenius S., Lonsdorf E., Neuhauser C., 2007. Patch aging and the S-Allee effect: breeding system effects on the demographic response of plants to habitat fragmentation. Am. Nat. 169: 383–397 [DOI] [PubMed] [Google Scholar]
- Wright S., 1939. The distribution of self-sterility alleles in populations. Genetics 24: 538–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W. S., Nielsen R., 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Yang Z. H., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.