Abstract
Rice fragrance is an important characteristic for Southeast Asian consumers, and fragrant landraces from Japan were first recorded in the 17th century. Principal component analysis clearly showed that Japanese fragrant landraces were genetically different from non-Japanese fragrant landraces. Japanese fragrant landraces were composed of six clades, none of which carried the most common fragrance mutation, an 8-bp deletion in exon 7 of Badh2. Fragrant landraces comprised two major groups carrying different Badh2 mutations. One group carried a known SNP at exon13 and the other a SNP at the exon1-intron1 junction as splicing donor site. The latter was considered to be a potential splicing mutant group as a novel allele at Badh2. Heterozygosity (He) scores in the two fragrant groups were not significantly different from non-fragrant landraces and modern cultivars. However, lower He scores were found around the Badh2 locus in the two groups. The potential splicing mutant group showed a more extended haplotype than the E13 SNP group. A likely causal factor responsible for loss of function is a novel splicing mutation allele that may have been generated quite recently. The fragrance allele has dispersed as a result of out-crossing under local environmental conditions.
Keywords: fragrance, landrace, 2-acetyl pyrroline, Badh2, diversity, selective sweep
Introduction
Rice consumers in Southeast and South Asian countries have a preference for fragrant rice, which has been selected from fragrant landraces that were initially improved by selection of pure lines from natural variants. Fragrant landraces have been considered a valuable resource in comparison with modern cultivars, and many genetic analyses of these landraces have been reported (Ahn et al. 1992, Amarawathi et al. 2008, Bradbury et al. 2005, Bourgis et al. 2008, Chen et al. 2006, Lanceras et al. 2000, Lorieux et al. 1996, Shi et al. 2008). Fragrance has been reported to be regulated by a single or multiple genes (Berner and Hoff 1986, Huang et al., 2008, Sood and Siddiq 1978) and although a major causal gene, Badh2, has been located on chromosome 8, other loci have also been reported to affect rice fragrance (Amarawathi et al. 2008, Lorieux et al. 1996). Loss of function of Badh2 results in accumulation of 2-actyl-1-pyrroline (2AP) (Bradbury et al. 2005, Vanavichit and Yoshihashi 2012), the major component of rice fragrance (Maga, 1984, Paule and Powers, 1989, Yajima et al. 1978). The most widely distributed polymorphism in Badh2 that causes 2AP accumulation is an 8-bp deletion in exon 7. Up to now, ten alleles including the major mutation have been reported (Kovach et al. 2009, Shi et al. 2008), all of which originate from japonica and have introgressed into other genetic backgrounds.
‘Kabashiko’ is the generic name for fragrant rice in Japan, and various fragrant Japanese landraces primarily distributed in mountainous areas, mentioned in 17th century historical records, Aizu-nousho, a document published in 1684 providing a record of agricultural information (Arashi 1975). By the end of the 19th century, application of stringent quality control had excluded red rice, slender grains, low-quality grains and so on. Most of the fragrant landraces also disappeared around that time. Why, then, was fragrance considered undesirable? Fragrant landraces may be exluded probably because of other undesirable traits such as a lower panicle yield (Arashi 1975). It is also true that the full undiluted smell of fragrant grains would probably have been unpopular in Japan. In general, consumers added a small portion of fragrant grains to aged grains to add a fresh flavor. Only restricted usage was applied in Japan. However, fragrant landraces were dispersed into Japan gradually during 18th to 19th centuries. These fragrant landraces show geographical divergence in their traits (Itani 2002a, Miyakawa and Nakamura 1984). Based on morphological and physiological traits, they can be roughly classified into two major groups: eastern and western. The eastern group distributed in Hokuriku, Kanto, and Tohoku, was characterized by early heading, short panicles, low yield, a fine culm and a propensity to lodge, relative to the western group distributed in Kinki, Shikoku, and Kyushu. Like red rice, they soon disappeared from the general market and the fields used for growing them were resown with modern cultivars over the last 100 years (Arashi 1975, Ishikawa et al. 2002a, 2002b, Itani 2002a, Tabuchi et al. 2007). Although landraces including fragrant landraces were displaced by modern cultivars they may retain a number of potentially valuable traits that have been lost in modern cultivars (Arashi 1975).
In the present study, we analyzed genetic diversity among Japanese fragrant landraces in comparison with Japanese non-fragrant landraces and modern cultivars. Genetic distance allowed us to specify particular groups of landraces that had adapted to local environmental conditions and carried a novel mutation related to fragrance. This novel mutation implied us different amount of fragrance based on the gene structure. Thus, it will allow us to design fragrance corresponding to needs of cunsumers’ preference.
Materials and Methods
Plant materials
Eighty-two cultivars, including landraces and modern varieties, were examined (Table 1). These landraces were collected at Hiroshima prefectural Hiroshima University and Hirosaki University. Most of those landraces were characterized for morphological and physiological traits (Itani 2002a, 2002b). Of these, 17 were non-Japanese fragrant landraces, 25 were Japanese non-fragrant cultivars including elite cultivars such as ‘Koshihikari’, and 40 were Japanese fragrant landraces included five non-scented ones. Overseas category included Meragome originated in Kagoshima which is an indica cultivars known as Daitoumai-group introduced into Japan in the middle age (Ishikawa 2002a, Ogawa and Itani 2008). Thus, this landrace was categorized as a non-Japanese landrace. ‘Masshigura’ and ‘Koihonoka’ were used to confirm allelic relations among various fragrant landraces. Both of them are improved varieties in Aomori prefecture. The former one is non-fragrant leading variety. The latter one is a fragrant rice inherited 8bp deletion in Badh2 gene from ‘Sari Queen’. ‘Sari Queen’ was generated from the cross between ‘Basmati370’ and ‘Nipponbare’. Then, the variety was crossed with other Japanese varieties to obtain ‘Koihonoka’ (Ando et al. 2004, Mikami et al. 2007).
Table 1.
List of materials characterized genotypes and phenotypic traits of landraces (LR) and modern varieties (MV)
| Overseas fragrant landraces | Japanese fragrant landraces | Japanese non-fragrant landraces/modern varieties (MV) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||
| Variety name | Origin | Criteria | Fragrance | Clade* | Variety name | Origin | Criteria | Fragrance | Clade | Variety name | Origin | Criteria | Fragrance | Clade |
| Carolina | USA | LR | + | 4 | Nioi-wase1 | Iwate | LR | − | 1 | Aichiasahi | Aichi | MV | − | 6 |
| Karalath | India | LR | + | 4 | Kaori-wase | Iwate | LR | − | 1 | Aikoku | Shizuoka | LR | − | 6 |
| Ikkikoku | China | LR | + | 4 | Jakou-ine 3 | Yamagata | LR | − | 1 | Akage | Hokkaido | LR | − | 6 |
| Meragome | Kagoshima** | LR | + | 4 | Kaori-ine 5 | – | LR | − | 1 | Akamai 3 | Unknown | LR | − | 6 |
| Muskakudanti | India | LR | + | 4 | Kaori-wase 2 | Hokkaido | LR | + | 2 | Akihikari | Aomori | MV | − | 6 |
| Surjamkhi | India | LR | + | 4 | Kaori-wase 3 | Hokkaido | LR | + | 2 | Akitakomachi | Akita | MV | − | 6 |
| Choukokou | China | LR | + | 4 | Kaori-ine 4 | – | LR | + | 2 | Asahi | Kyoto | LR | − | 6 |
| Azucena | Philippines | LR | + | 5 | Hokkaido-40nichi-wase | Iwate | LR | + | 2 | Fujisaka 5 | Aomori | MV | − | 6 |
| Basmati 370 | India, Pakistan | LR | + | 5 | Jakou (Niigata) | Niigata | LR | + | 2 | Ginbozu | Toyama | MV | − | 6 |
| Binicol | Philippines | LR | + | 5 | Nioi-wase2 | Akita | LR | + | 2 | Gohyaku-mangoku | Niigata | LR | − | 6 |
| Beijing-koutou | China | LR | + | 5 | Iwaka | Miyagi | LR | + | 3 | Hyogoomachi | Hyogo | MV | − | 6 |
| DawDam | Thai | LR | + | 5 | Jakou-ine 2 | Tohoku | LR | + | 3 | Jukkoku | Unknown | LR | − | 6 |
| Della | USA | LR | + | 5 | Jakou (Ibaraki) | Ibaraki | LR | + | 3 | Jyoshu | Yamagata | LR | − | 6 |
| KhaoPickLaos | Laos | LR | + | 5 | Jakou-wase | Niigta | LR | + | 3 | Kameji | Shimane | LR | − | 6 |
| KU 701 | Thai | LR | + | 5 | Kaori-ine 1 | Ibaraki | LR | + | 3 | Kamenoo | Yamagata | LR | − | 6 |
| Mangareez | Philippines | LR | + | 5 | Kaori-ine 2 | Kanagawa | LR | + | 3 | Koshihikari | Fukui | MV | − | 6 |
| Sabarmati | India | LR | + | 5 | Nioi-akako | Miyagi | LR | + | 3 | Nipponbare | Aichi | MV | − | 6 |
| Nioi-mai | Miyagi | LR | + | 3 | Oba | Ishikawa | LR | − | 6 | |||||
| Narukosan-koutou | Miyagi | LR | − | 3 | Rikuu132 | Aomori | MV | − | 6 | |||||
| Kumamoto | Kumamoto | LR | + | 7 | Sasanishiki | Miyagi | MV | − | 6 | |||||
| Kabashiko A | Miyazaki | LR | + | 7 | Senichi | Tochigi | LR | − | 6 | |||||
| Kabashiko B | Kumamoto | LR | + | 7 | Shinriki | Hyogo | LR | − | 6 | |||||
| Kabashiko C | Yamaguchi | LR | + | 7 | Takenari | Mie | LR | − | 6 | |||||
| Kaori-ine 3 | Nara | LR | + | 7 | Tsushima | Nagasaki | LR | − | 6 | |||||
| Keitoku | Kumamoto | LR | + | 7 | Tsushima-akamai | Nagasaki | LR | − | 6 | |||||
| Kumamoto | Kumamoto | LR | + | 7 | ||||||||||
| Mangoku | Kumamoto | LR | + | 7 | ||||||||||
| Nioi-yoshi 1 | Kochi | LR | + | 7 | ||||||||||
| Nioi-yoshi 2 | Kochi | LR | + | 7 | ||||||||||
| Seigoku-kawachi | Nagasaki | LR | + | 7 | ||||||||||
| Shiro-wase | Wakayama | LR | + | 7 | ||||||||||
| Jakou-ine 1 | Yamagata | LR | + | 8 | ||||||||||
| Jakou-ine 4 | Yamagata | LR | + | 8 | ||||||||||
| Jakou-mochi | Touhoku | LR | + | 8 | ||||||||||
| Kamari | Ibaraki | LR | + | 8 | ||||||||||
| Nioi-mochi 1 | Fukuhsima | LR | + | 8 | ||||||||||
| Nioi-mochi 2 | Fukushima | LR | + | 8 | ||||||||||
| Hieri | Kouchi | LR | + | 9 | ||||||||||
| Kaisen | Kouchi | LR | + | 9 | ||||||||||
| Wase-hieri | Kouchi | LR | + | 9 | ||||||||||
Clade was based on genetic distance shown in Fig. 2.
Landrace introduced from overseas was classified into oversea origin.
Fragrance testing by treatment with KOH
The fragrance (2AP) status of cultivars was confirmed by soaking small pieces of leaves in 2% KOH solution heated to 60°C for 10 minutes. Sensory analysis was performed by multiple persons at multiple times to confirm phenotypes. The varieties were also classified as tropical or temperate japonica (Tr or Tm) using the method described by Sato (1991).
Molecular markers
Thirty SSR loci dispersed across the genome (Supplemental Table 1) were amplified with Ampliqon-Taq (Ampliqon Co., Denmark). PCR conditions were 94°C for 3 min, 30 cycles of 94°C for 10 s, 55°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min for post-heating. Amplified products were genotyped using 6% denatured sequencing gels with a general silver staining method based on the SILVER SEQUENCE™ DNA Sequencing System (Promega Co., USA). Additional SSRs were analyzed in Japanese landraces and elite cultivars to screen for a selective sweep around Badh2 (Supplemental Table 2). INDEL markers were developed for Badh2 from representatives of fragrant landraces (Supplemental Table 3). The PCR protocol was the same as that for SSRs. A CAPS marker including the splice donor site mutation at the exon1-intron 1 junction was developed to confirm whether the SNP was cleaved by the restriction enzyme KpnI (New England BioLabs, Japan, Inc.) using PCR fragments amplified with the 5′-UTR forward primer and I-986 reverse primer shown in Supplemental Table 3.
Badh2 sequences
Parts of Badh2 were amplified with primer pairs listed in Supplemental Table 4. Amplified fragments were purified with a QIAquick PCR purification kit (QIAGEN Co., Japan). These fragments were then prepared for sequencing using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems Co., Japan) to obtain sequences including the ABI310 sequence. SNP mutation found at the exon-intron junction has been registered (AB821274).
Tr-japonica, Tm-japonica classification
Hull size, mesocotyl length, and KOH endosperm degradation were tested using a protocol described by Sato (1991) (Supplemental Table 5). Hull size was measured using Nikon Digital Sight (NIKON, Japan) and expressed as the average for ten hulls. Mesocotyl length (mm) was measured after two weeks of germination in total darkness. Ten young seedlings were subjected to 1st internode measurement and the values averaged. KOH degradation was measured as the loss of weight after soaking in 1.5% KOH solution, ten grains being thus treated. Z2 score was determined according to Sato (1991):
where R: ratio of hull length to width, M: mesocotyl length (mm), E: reduction of endosperm weight after soaking in 1.5% KOH. Cultivars showing a Z2 score of <0 were identified as Tm-japonica (Tm), and those showing a score of >0 as Tr-japonica (Tr).
Data analysis
The GenAlEx 6.2 software package (Peakall and Smouse 2006) was used to evaluate genetic variation in loci and populations, including the number of different alleles per locus (Na), Fst, observed heterozygosity (Ho), and expected heterozygosity (He). Dendrograms were constructed using the neighbor-joining method based on Nei’s unbiased genetic distances using the Populations 1.2.30 beta2 program (http://bioinformatics.org/~tryphon/populations/#ancre_bibliographie). All dendrograms were drawn by TreeExplorer (Tamura et al. 2011). Bootstrap values were assessed with 1000 replicates.
Results
Diversity in fragrant rice
Confirmation of fragrance found five Japanese fragrant landraces, ‘Nioi-wase 1’, ‘Kaori-wase’, ‘Jakou-ine 3’, ‘Kaori-ine 5’, and ‘Narukosan-koutou’, revealed that they were not fragrant, even though they shared similar names implying fragrance (Table 1). All other Japanese and non-Japanese fragrant landraces were fragrant. Including these landraces, genetic divergence among landraces and modern cultivars was genotyped using 30 SSRs.
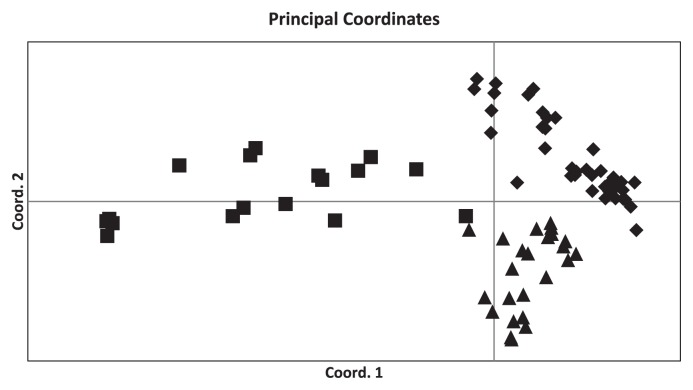
Principle component analysis (PCA) clearly identified three major groups: overseas (non-Japanese) fragrant landraces, Japanese fragrant landraces, and Japanese non-fragrant landraces/varieties (Fig. 1). The non-Japanese fragrant landraces were the most diverse, with He = 0.652 (Table 2). These were followed by Japanese landraces and cultivars in combination, He = 0.397, Japanese non-fragrant landraces and modern cultivars with He = 0.349, and Japanese fragrant landraces with He = 0.336. There was no significant difference between the Japanese fragrant and non-fragrant groups.
Fig. 1.
Principal component analysis of fragrant rice by using 30 SSR loci. Overseas fragrant landraces (squares), Japanese fragrant landraces (diamonds), and Japanese non-fragrant landraces/elite varieties (Triangles) were separated clearly.
Table 2.
Heterozygosity among groups with different genetic background such as overseas fragrant group carrying 8bp deletion or no-deletion at exon 8, non-scented group, E13 SNP group carrying a SNP in exon 13, splicing donor site mutant (splic-mut) group, and non-fragrant group
| Group | No. of cultivars | He (all loci) | He around Badh2 | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Mean ± SE | 544kup (RM8264) | 45kup | 12kup | SSRat7 | 106kdwn (RM1109) | 142kdwn (RM3459) | ||
| Total | 82 | 0.516 ± 0.043 | 0.736 | – | – | 0.510 | 0.668 | 0.775 |
| (Total—Non-scented) | 77 | 0.523 ± 0.043 | 0.741 | – | – | 0.528 | 0.676 | 0.771 |
| Oversea | 17 | 0.652 ± 0.028 | 0.761 | – | – | 0.734 | 0.699 | 0.747 |
| 8 bp-del. | 7 | 0.467 ± 0.044 | 0.612 | – | – | 0.490 | 0.408 | 0.694 |
| non 8 bp-del. | 10 | 0.523 ± 0.042 | 0.740 | – | – | 0.480 | 0.580 | 0.660 |
| Japan | 65 | 0.397 ± 0.052 | 0.631 | – | – | 0.334 | 0.631 | 0.766 |
| (Japan—Non-scented) | 60 | 0.397 ± 0.053 | 0.623 | – | – | 0.352 | 0.639 | 0.759 |
| Fragrant landraces scented | 35 | 0.336 ± 0.055 | 0.483 | – | – | 0.000 | 0.633 | 0.738 |
| E13 SNP group | 29 | 0.325 ± 0.055 | 0.328 | 0.000 | 0.000 | 0.000 | 0.523 | 0.709 |
| Splic-mutant group | 6 | 0.217 ± 0.047 | 0.444 | 0.000 | 0.000 | 0.000 | 0.000 | 0.500 |
| Fragrant landraces non-scented | ||||||||
| Non-scented group | 5 | 0.253 ± 0.044 | 0.000 | – | – | 0.320 | 0.480 | 0.640 |
| Non-fragrant group | 25 | 0.349 ± 0.048 | 0.435 | 0.480 | 0.499 | 0.570 | 0.557 | 0.509 |
–: not genotyped.
Genetic relationships among fragrant landraces
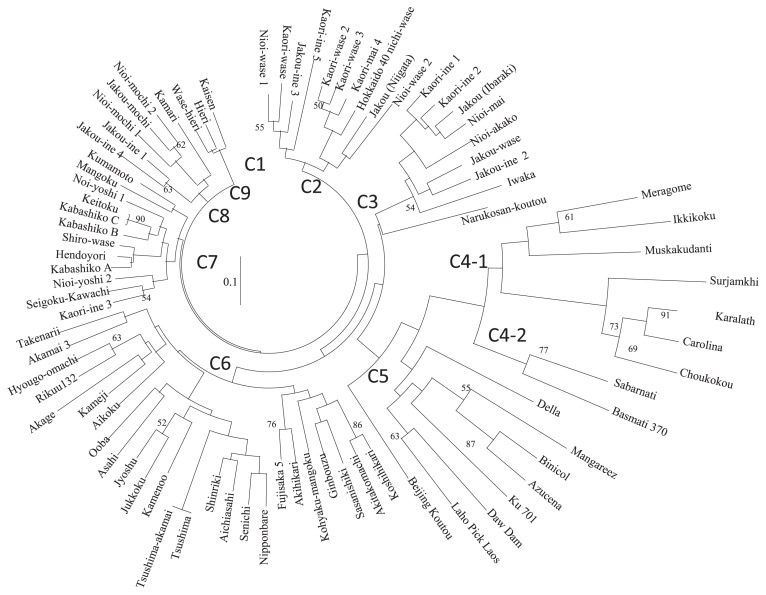
A phylogenetic tree was constructed based on genetic distance (Fig. 2). Overseas cultivars were grouped into Clades 4 and 5. Landraces in Clades 4-2 and 5 carried an 8-bp deletion, which is known to be the major causal mutation in Badh2, but not in Clades 4-1 or in other Japanese fragrant landraces. Japanese non-fragrant landraces and modern varieties were included in Clade 6. Japanese fragrant landraces were classed into six groups (Clade 1, 2, 3, 7, 8, and 9). Clade 1 comprised fragrant landraces giving no aroma in the sensory test, but which had names related to fragrance and originated from Iwate and Yamagata in Tohoku district. Varietal names similar to them, such as ‘Kaori-wase’ and ‘Nioi-wase’, were also found in Clade 2, and were phenotyped as fragrant in the sensory test. Landraces in Clade 2 originated from Eastern Japan, extending from Niigata to Hokkaido. Clade 3 included one fragrant landrace, ‘Narukosan-koutou’, which gave no aroma in the sensory test. In addition, it was most distantly separated from the other members of the clade. Fragrant landraces in Clade 3 originated from Tohoku area to Kanto area (Ibaraki Prefecture). Clade 7 comprised landraces from Western Japan. Clade 8 included three glutinous and three non-glutinous landraces, all of which originated from northern Kanto up to the southern areas of Tohoku. Clade 9 comprised three landraces that originated from the same prefecture, Kochi, in Western Japan.
Fig. 2.
Phylogenetic tree based on NJ method. Bar showed genetic distance measured by 30 SSR genotypes. Nine clades (Clade 1 to 9) were shown as C1 to C9. Bootstrap values were assessed with 1000 replicates and the scores were shown beside each clade.
Novel fragrant allele at the Badh2 locus
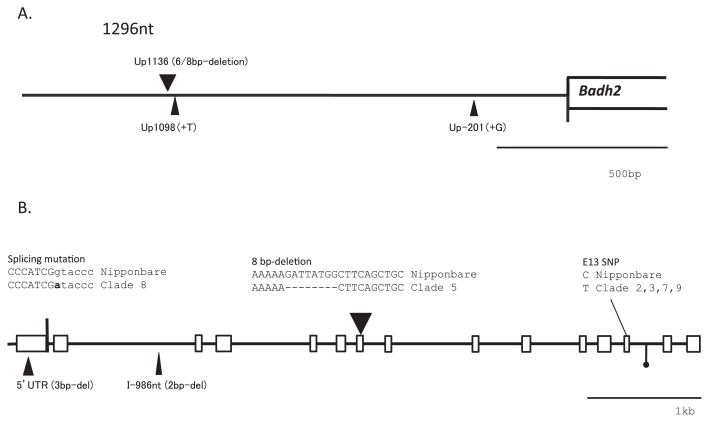
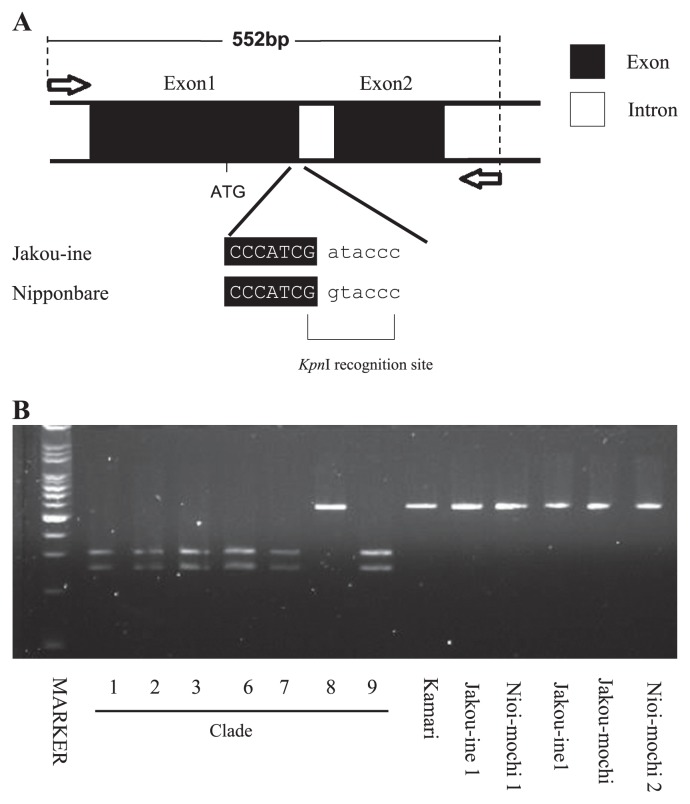
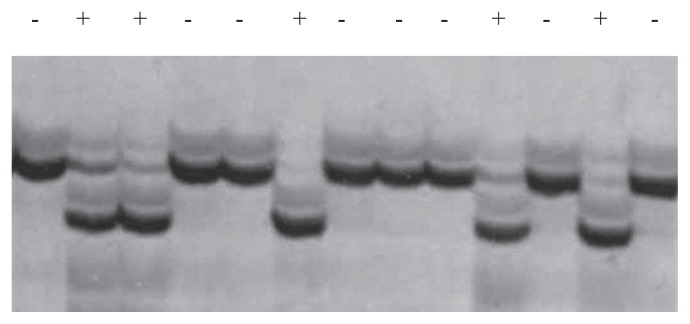
In order to confirm molecular variation in Badh2, the full-length Badh2 gene including the 1296-nt upstream region was sequenced from representatives of each clade (Fig. 3). A C to T (C/T) substitution as a single nucleotide polymorphism (SNP) in exon 13, leading to substitution of a single amino acid, has already been reported in a single fragrant landrace, ‘Shiratama’, originating from Japan. ‘Hokkaido-40-nichi-wase’, ‘Iwaka’, ‘Kabashiko A’, and ‘Hieri’ belonging to Clades 2, 3, 7, and 9, respectively, were sequenced and showed the C/T substitution, which results in a single amino acid change (alanine to valine) (Table 3). However, ‘Nioi-mochi 2’ in Clade 8 did not carry the mutation. Exon 13 was sequenced in all Japanese landraces, and all fragrant landraces carried the substitution at exon 13 except for those in Clade 8. Another significant nucleotide change was detected only in Clade 8, which carried the same G to T (G/T) substitution in the splice donor site at the exon 1-intron 1 junction in ‘Nioi-mochi 2’ (Fig. 4 and Table 3). The G/T substitution involves to a recognition site of KpnI restriction enzyme. Non-fragrant rice such as ‘Nipponbare’ and general fragrant rice can be digested with Kpn I, whereas fragrant landraces in Clade 8 can not be digested because of the substitution at the splicing donor site. Thus, a CAPS marker was developed to confirm the sequence substitution. All fragrant landraces were re-confirmed by Kpn I restriction digestion of amplicons including the relevant site (Fig. 4). As the SNP has not yet been reported, it was concluded that six varieties in Clade 8 carry the novel allele at the Badh2 locus. This has now been registered at DDBJ (AB821274).
Fig. 3.
Gene structure of BADH2 with upstream region. Sequence differences found in fragrant landraces and INDELs compared to reference sequence in Nipponbare and others were noted as solid triangles. The size of triangles corresponded to relative size of the INDELs. A. Three INDELs were recognized in upstream regions. B. Inside Badh 2, two functional SNP and deletions were recognized. Splicing junction at exon1-intron1, intron sequences were shown as small letters and g at splicing donor site was substituted into a showing bold letter. One known mutation was C to T substitution at exon 13. Three INDELs (solid triangles) and SSR (flag with solid circle) were detected. One of INDELs, 8bp deletion at exon 7 was carried by Koihonoka and some oversea landraces.
Table 3.
Sequence substitution, insertion (+), and deletion (−) compared to Nipponbare sequence among representatives of different clades in Japanese fragrant landraces
| Variety/landrace | Clade | Upstream | 5′-UTR | Intron 1 | Intron 2 | Intron 4 | Intron 7 | Exon 13 | Intron 13 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
||||||||
| up1136 | up1098 | up962 | up201 | up160 | 257 | 986 | 1045 | 2291–2304 repeat of (at) motif | 3434 | 5391 | 5678 | |||
| Nipponbare | 0 | 0 | G | 0 | C | 0 | G | 0 | T | 7 | C | C | A | |
| Hokkaido-40-nichi-wase | Clade-2 | +AAATCTAG | +T | A | −G | T | 0 | G | −TT | C | 11 | T | T | G |
| Iwaka | Clade-3 | +AAATCTAG | +T | A | −G | T | 0 | G | −TT | C | 11 | T | T | G |
| Kabashiko A | Clade-7 | +AAATCTAG | +T | A | −G | T | 0 | G | −TT | C | 11 | T | T | G |
| Nioi-mochi 2 | Clade-8 | +AAATCTAG | +T | A | −G | T | 0 | A | −TT | C | 11 | C | C | G |
| Hieri | Clade-9 | +AAATCTAG | +T | A | −G | T | 0 | G | −TT | C | 11 | T | T | G |
Fig. 4.
Novel allele at Badh 2 locus. A. A single nucleotide substitution was identified at exon-splicing junction, G to A, which is Kpn I resctriction enzyme recogniton site. Jakou-ine in Clade 8 carried the mutation type and reference genome Nipponbare, non-fragrant elite varieties carried a general type. B. PCR amplicon in Nioi-mochi 2 was not digested by enzyme digestion but others were. All fragrant landraces were applied and only six landraces in Clade 8 showed that their amplicons were not able to digest.
Since the six landraces belonging to Clade 8 shared the splicing donor site mutation, their morphological and physiological characteristics were compared. Three of them were non-glutinous and the remainder were glutinous (Fig. 5). Grain shape, presence or absence of awns, and colors in parts of the spikelets were polymorphic.
Fig. 5.
Spikelets of six landraces in Clade 8. Nioi-mochi 1, Nioi-mochi 2, and Jakou-mochi represented glutinous endosperm and others were non-glutinous. Jakou-ine 1, Jakou-ine 2, Nioi-mochi 2, and Kamari were awned spiekelets. Nioi-mochi 1, Nioi-mochi 2, Kamari, and Jakou-mochi carried C and A alleles at C locus and A locus, respectively. Thus, their awns or apiculus part (top of the spikelet) and sterile glumes were colored.
INDEL polymorphism
Based on the sequence data, six INDELs were detected among representatives of fragrant landraces (Fig. 3). These INDELs were detected through sequencing of the entire coding region including the 5′-upstream region with Japanese fragrant landraces shown in Table 3, ‘Della’, and ‘Meragome’. Only Della showed the 3-bp deletion at the 5′-UTR, and subsequent screening of the deletion also indicated that it is unique in ‘Della’. The 8-bp deletion at exon 7 is known to cause loss of function and is found only in overseas landraces in the C4-1 Clade and ‘Della’ in the C4-2 Clade (Table 4).
Table 4.
Haplotypes evaluated with inserts (+) and deletions (−) compared to ‘Nipponbare’ sequence and the frequencies of landraces and varieties among overseas fragrant carrying 8bp deletion or no-deletionat exon 8, E13 SNP, splicing donor site mutant (Splic-mut), and non-fragrant groups
| Haplotype | DNA variation in each haplotype | No. of landraces/varieties | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Upstream | Inside BADH2 | Overseas | Japanese groups | ||||||
|
|
|
|
|
||||||
| Up1136 | Up1098 | 5′UTR | I2-986nt | 8 bp del | non-8 bp | E13 SNP | Splic-mut | Non-fragrant | |
| Indel-Hap1* | – | – | – | −2 bp | 0 | 0 | 0 | 0 | 8 |
| Indel-Hap2 | – | +T | – | – | 0 | 0 | 0 | 0 | 7 |
| Indel-Hap3 | – | +T | – | −2 bp | 0 | 0 | 0 | 0 | 3 |
| Indel-Hap4 | −6 bp | – | – | −2 bp | 0 | 3 | 0 | 0 | 0 |
| Indel-Hap5 | −8 bp | +T | +3bp | −2 bp | 1 | 0 | 0 | 0 | 0 |
| Indel-Hap6 | −8 bp | +T | – | – | 2 | 0 | 3 | 0 | 7 |
| Indel-Hap7 | −8 bp | +T | – | −2 bp | 7 | 4 | 26 | 6 | 0 |
| Total | 10 | 7 | 29 | 6 | 25 | ||||
‘Nipponbare’ was included in Hap 1.
In total, seven haplotypes were detected among overseas and Japanese groups (Table 4). Three INDELs, Up201, Up1098 and Up1136, were detected in the upstream region. Up1136 was a 6-bp or 8-bp deletion without reference to ‘Nipponbare’. However, seven non-fragrant varieties carried the 8-bp deletion for Up1136. Two single-nucleotide insertions were found in the Japanese fragrant group. One (Up201) could not be converted to an INDEL marker, but another (Up1098) could, and this INDEL was also found in the non-fragrant group.
There were two INDELs in transcribed regions, a 3-bp deletion in the 5′-UTR and a 2-bp deletion within intron 2 (I-986). The former deletion was found only in non-Japanese fragrant groups while the latter (I-986) was detected in both non-fragrant and fragrant groups. These INDELs did not lead to loss of gene function.
Allelism test
‘Koihonoka’ is a recently released cultivar from Aomori prefecture, Japan, and carries the 8-bp deletion of exon 7 in Badh2 as a single causal mutation of fragrance (Fig. 6 and Table 5). This was confirmed using F7 recombinant inbred lines (RILs) from the cross between the non-fragrant modern variety, ‘Masshigura’, and the fragrant variety, ‘Koihonoka’. One hundred sixty-five individuals were phenotyped by sensory testing to confirm fragrance, and they segregated as 85 non-fragrant : 80 fragrant. This was compatible with a 1 : 1 ratio (chi-squared test = 0.15), suggesting that the fragrance was controlled by a single gene. All progeny individuals were genotyped using a INDEL marker to show the 8-bp deletion. Complete linkage was obtained between fragrance and genotypes of Badh2.
Fig. 6.
A 8 bp deletion of exon 7 in BADH2 corresponding to phenotypes of fragrance in recombinant inbred lines of the F7 progeny of ‘Masshigura’ × ‘Koihonoka’. Deletion type in ‘Koihonokara’ was inherited from Basmati370. Phenotypes were segregated as a single gene. + and − meaning fragrance and non-fragrance by using leaf test with 2% KOH treatment.
Table 5.
Allelism test of fragrant mutations among different landraces and ‘Koihonoka’ (modern variety) carrying the 8bp deletion in Badh2 inherited from Basmati 370
| F2/F7 populations | No. of plants | No. of plants scored for the fragrance | Chi-test against a single gene segregation (3 : 1 or 1 : 1) | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Non-fragrance | Fragrance | |||||
| F2 populations | ||||||
| Koihonoka | × Clade 3: Iwaka | 10 | 0 | 10 | (All recessive type) | |
| Koihonoka | × Clade 3: Kaori-ine | 10 | 0 | 10 | (All recessive type) | |
| Koihonoka | × Clade 8: Jakou-mochi | 10 | 0 | 10 | (All recessive type) | |
| Clade 3 : Iwaka | × Clade 8: Jakou-mochi | 10 | 0 | 10 | (All recessive type) | |
| Clade 3 : Kaori-ine | × Clade 8: Jakou-mochi | 10 | 0 | 10 | (All recessive type) | |
| Koihonoka | × Masshigura | 50 | 39 | : | 11 | NS (0.5 < p) |
| Clade 4: Narukosan-koutou | × Clade 3: Iwaka | 50 | 39 | : | 11 | NS (0.5 < p) |
| Clade 3 : Kaori-ine | × Clade 1: Nioi-wase 1 | 18 | 42 | : | 8 | NS (0.1 < p) |
| Masshigura | × Clade 8: Jakou-ine 4 | 50 | 38 | : | 12 | NS (0.5 < p) (test for 3 : 1 ratio) |
| F7 RIL | ||||||
| Masshigura | × Koihonoka | 165 | 85 | : | 80 | NS (0.5 < p) (test for 1 : 1 ratio) |
‘Masshigura’ was crossed with ‘Koihonoka’ and ‘Jakou-ine 4’ (Clade 8). In addition, other non-fragrant cultivars were crossed to ‘Iwaka’ (Clade 3), ‘Kaori-ine’ (Clade 3), and ‘Jakou-mochi’ (Clade 8) in order to test Mendelian segregation as a single gene. These F2 populations segregated into non-fragrant and fragrant individuals in a 3 : 1 ratio. Thus, the fragrant trait was controlled by a single gene.
F1 individuals of ‘Koihonoka’ × ‘Iwaka’, ‘Koihonoka’ × ‘Kaori-ine’, ‘Koihonoka’ × ‘Jakou-mochi’, ‘Iwaka’ × ‘Jakou-mochi’, and ‘Kaori-ine’ × ‘Jakou-mochi’ were subjected to an allelism test. These F1 individuals were genotyped using SSR markers to confirm whether they were heterozygous as F1. Sensory testing suggested that all the F1 individuals and the self-pollinated progenies were fragrant. It was concluded that their fragrant alleles were controlled by the same single gene, Badh2, as recessive multiple alleles. There were at least two groups as Japanese landraces, 1) the E13 SNP group with the SNP in exon 13, as described above, and 2) the splicing mutant (Splic-mut) group carrying the G/T substitution in the splicing donor site at the exon 1-intron 1 junction without the SNP at exon 13. Additionally, there is another group included ‘Koihonoka’ and ‘Meragome’ inherited the 8-bp deletion in Badh2 from overseas fragrant landraces.
Genetic diversity around the Badh2 locus
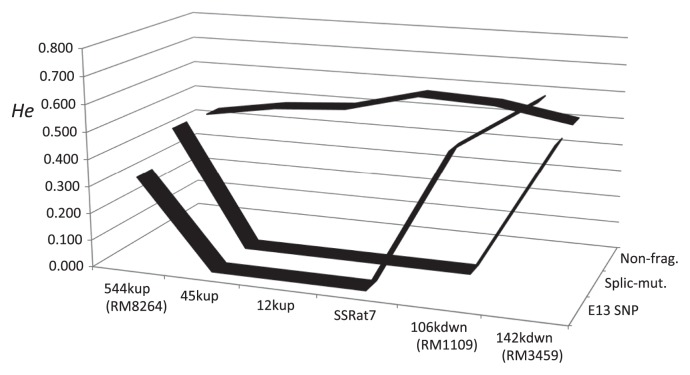
Heterozygosity was measured as the average score for 30 SSR loci distributed over the whole genome (Fig. 7 and Supplemental Table 1). An additional 16 SSR markers were assayed within a 543-kb stretch of Badh2 in order to identify selective sweeps around the Badh2 locus (Supplemental Table 2). Fourteen of the additional SSRs were monomorphic or nearly monomorphic as only one cultivar showed an allele different from the others. Such rare alleles are probably due to a recent spontaneous mutation. Two additional loci, 45kup and 12kup, were relatively highly heterozygous, He ranging from 0.480 to 0.499, respectively, in the non-fragrant group. He scores at four loci, RM8264, SSRat7, RM1109, and RM3459, were compared with the data obtained at the two polymorphic loci. The splicing mutant group was monomorphic within at least a 151-kb region of Badh2 and the E13 SNP group was monomorphic within at least a 45-kb region. Splicing mutants were cultivated at the restricted area carried more wider selective sweep region. It may result from the recent origin of the novel mutation.
Fig. 7.
Selective sweep found around Badh 2 locus. Vertical line showed Heterozygosity measured by 30 SSR genotypes. Horizontal axis corresponded to each SSR locus.
Discussion
Fragrant rice is an economically important crop in terms of global trade. The FAO All Rice Price Index (2002–04 = 100) indicates that among all types, fragrant rice varieties were priced more highly than others, as of January 2013. The causal gene for fragrance has been reported as Badh2 (Kovachi et al. 2009). Ten multiple alleles have been found. One of them, Badh2.10, is shared between Taiwanese fragrant landraces and one Japanese landrace (Shiratama). As we have various Japanese fragrant landraces except for Shiratama, we characterized them at the molecular level in the present study to screen for novel mutations. When data of all SSR markers were analyzed together, we recognized nine different clades among modern cultivars and landraces, including non-fragrant landrace/varieties. The Japanese fragrant landraces were relatively highly diverse and formed Clades 1, 2, 3, 7, 8 and 9, which reflected their regional divergence. However, based on the Badh2 haplotypes, fragrant landraces comprised only two groups, one corresponding to Clade 8 and the other to Clades 2, 3, 7, and 9. The latter group carried the SNP in exon 13 (E13 SNP group), which was reported by Kovach et al. (2009) as badh2.10. Although te E13 SNP group shared the single allele, they showed higher structure which would result from adaptation to each environmental condition through outcrossing. The original lineage carying the SNP was spontaneously outcrossed with other landraces for exchange of the genetic background. Although landraces in Clade 8 were fragrant, they did not carry the exon 13 SNP. Like the low-amylose-type Wxb allele at the Wx locus (Sano 1984) they shared a single mutation at the splicing donor site, which was the only mutation within the gene and upstream of it. INDEL haplotyping suggested that there were no INDELs related to the function of Badh2. Our allelism test also suggested that the splicing mutation was allelic to the 8-bp deletion allele shared with ‘Basmati 370’ and ‘Koihonoka’, suggesting that the splicing donor site mutation results in fragrance. The exon 13 SNP mutation was the most common among Japanese fragrant landraces while the remainder (17%) were found to carry the splicing donor site mutation whih has never been reported.
Whole-genome genetic diversity estimated in terms of SSRs showed that the two groups of Japaneses fragrant landraces were not significantly different from the non-fragrant group, probably due to genetic exchange with the non-fragrant group. In contrast, estimation of genetic diversity with SSR markers flanking Badh2 showed that the non-fragrant group had higher genetic diversity in this region relative to the two groups of fragrant landraces, although the splicing mutant group carried a longer stretch of sequences without any polymorphisms than the E13 SNP group. Outside this region, genetic diversity increased dramatically. The high diversity in fragrant landraces were also visible as morphological and physiological traits. Although the splicing donor site mutants shared a single mutation that had originated recently, they showed differences in endosperm traits, awns, and organ coloration, which had probably arisen due to outcrossing with different landraces. The lack of polymorphism around the Badh2 locus can be explained by the presence of a selective sweep similar to the situation in glutinous rice (Olsen and Purugannan 2002) where a reduction of nucleotide variation spans a stretch of ~250 kb centered on the Waxy locus (Olsen et al. 2006).
Except for one Japanese fragrant landrace, Jakou-ine, variety names of the novel mutants were not listed in historical records of Japanese fragrant landraces of the 17th to 19th centuries, but do appear in records published in 1970 (Arashi 1975). A number of landraces carrying the E13 SNP have been named independently, which have a single origin for the fragrant gene known as badh2.10. It was also cultivated at Taiwan. However, the novel splicing mutation has not been found outside Japan. The distribution of the mutants is partly overlapped with that of fragrant soybean landraces in the same southern Tohoku area, one of which is known as the fragrant soybean, ‘Dadacha-mame’, a well known product that was recently found to carry a mutation in the Badh2 homolog, GmAMADH2 (Arikit et al. 2011). Fukushima prefecture, which neighbors Yamagata, also has a fragrant soybean landrace. The origin of the splicing mutation in rice plants may be a recent event happened around the area. No variety sharing the splicing mutation was found in other areas. Preference of local people allowed the selection as an independent lineage to create novel fragarnt crops. Probable conscious selection such as this has been noted for Indian corn, which contrasts with more recent negative selection for traits such as red pericarp in Japanese rice (Arashi 1976, Ishikawa et al. 2002a). Because the locality, landraces in the splicing mutant group was only composed from Tm-japonica, but not from Tr-japonica due to the local origin and genetic background cultivated at the area (Supplemental Table 5).
The quantity of 2AP is influenced by environmental conditions and may be affected by seasonal and storage factors (Itani 2004). Multiple alleles may also alter the activity of enzymes that pass precursors down the biosynthetic pathway. The amino acid change originating from the E13 SNP group does not lead to complete loss of function in the enzyme BADH2, and probably explains the lower concentration of 2AP (Kovach et al. 2009) in this group or fragrant landraces relative to other groups or fragrant rice. Variation in the level of fragrance is a useful breeding target, as fragrant varieties differing in this trait can be offered to consumers. Furthermore, these landraces have acquired various genetic constituents through outcrossing. Fragrant landraces without any aroma would be segregating descendants of such spontaenous crossing. In addition, higher structure of fragrant landraces sharing the single gene, badh2.10 in E13 SNP group, resulted from the spontaenous outcrossing. This lead them to adapt to various environmental condition. Such spontaenous outcrossing, however, has never been allowed in modern varieties. Because original seeds of modern varieties are strictly controlled to exclude outcrossed seeds from nursery fields in which seeds are propagated for farmers. Captured genetic constituents from different landraces might be lost from current genetic resources. In this sence, fragrant landraces are a genetic reservoir that could allow rice breeders to find unexpected traits such as tolerance to biotic and abiotic stress, which had been lost in elite varieties. Some presumed tolerance could be recognized under varietal names (Arashi 1975). Parts of that would be beneficial for modern varieties to acquire further valuable traits.
Supplementary Material
Literature Cited
- Ahn, S.N., Bollich, C.N. and Tanksley, S.D. (1992) RFLP tagging of a gene for aroma in rice. Theor. Appl. Genet. 84: 825–828 [DOI] [PubMed] [Google Scholar]
- Amarawathi, Y., Singh, R., Singh, A.K., Singh, V.P., Mohapatra, T., Sharma, T.R. and Singh, N.K. (2008) Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 21: 49–65 [Google Scholar]
- Ando, I., Kaneda, C., Yokoo, M., Nemoto, H., Hata, T., Ise, K., Ikeda, R., Akama, Y., Nakane, A., Shimura, E.et al. (2004) “Sari Queen”, a new aromatic rice cultivar with Basmati rice grain character. Bull. Natl. Inst. Crop Sci. 5: 53–66 [Google Scholar]
- Arashi, K. (1975) Kinsei inasaku gijutsu shi: sono ritchi seitaiteki kaiseki (In Japanese: History of rice cultivation in modern times: Geological and ecological view point). Nobunkyo, Tokyo, Japan, pp. 468–490 [Google Scholar]
- Arashi, K. (1976) Nihon Akamai kou (In Japanese: Thought of Japanese red rice). Yuuzankaku, Tokyo, Japan, pp. 296 [Google Scholar]
- Arikit, S., Yoshihashi, T., Wanchana, S., Uyen, T.T., Huong, N.T., Wongpornchai, S. and Vanavichit, A. (2011) Deficiency in the amino aldehyde dehydrogenase encoded by GmAMADH2, the homologue of rice Os2AP, enhances 2-acetyl-1-pyrroline biosynthesis in soybeans (Glycine max L.). Plant Biotechnol. J. 9: 75–87 [DOI] [PubMed] [Google Scholar]
- Berner, D.K. and Hoff, B.J. (1986) Inheritance of scent in American long grain rice. Crop Sci. 26: 876–878 [Google Scholar]
- Bradbury, L.M.T., Fitzgerald, T.L., Henry, R.J., Jin, Q. and Waters, D.L.E. (2005) The gene for fragrance in rice. Plant Biotech. J. 3: 363–370 [DOI] [PubMed] [Google Scholar]
- Bourgis, F., Guyot, R., Gherbi, H., Tailliez, E., Amabile, I., Salse, J., Lorieux, M., Delseny, M. and Ghesquière, A. (2008) Characterization of the major fragrance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor. Appl. Genet. 117: 353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Wu, J., Yang, Y., Shi, W. and Xu, M. (2006) The fgr gene responsible for rice fragrance was restricted within 69 kb. Plant Sci. 171: 505–514 [DOI] [PubMed] [Google Scholar]
- Huang, T.C., Teng, C.S., Chang, J.L., Chuang, H.S., Ho, C.T. and Wu, M.L. (2008) Biosynthetic mechanism of 2-acetyl-1-pyrroline and its relationship with pyrroline-5-carboxylic acid and methylglyoxal in aromatic rice (Oryza sativa L.) callus. J. Agric. Food Chem. 56: 7399–7404 [DOI] [PubMed] [Google Scholar]
- Ishikawa, R., Nakamura, I., Nishihara, T., Kikuchi, M., Senda, M., Akada, S., Harada, T. and Niizeki, M. (2002a) Origin of cytoplasm substituted rice cultivars found in Japan. Theor. Appl. Genet. 105: 608–613 [DOI] [PubMed] [Google Scholar]
- Ishikawa, R., Sato, Y.-I., Tang, T. and Nakamura, I. (2002b) Different maternal origins of Japanese lowland and upland rice populations. Theor. Appl. Genet. 104: 976–980 [DOI] [PubMed] [Google Scholar]
- Itani, T. (2002a) Agronomic characteristics of aromatic rice cultivars collected from Japan and other countries. Jpn. J. Crop Sci. 71: 68–75 [Google Scholar]
- Itani, T. (2002b) Apperance and alkali slubility of the brown rice of aromatic rice cultivars collected in Japan and other countries. Jpn. J. Crop Sci. 71: 178–185 [Google Scholar]
- Itani, T. (2004) Variation of 2-acetyl-pyrroline concentration in aromatic rice grains collected in the same region in Japan and factors affecting its concentration. Plant Prod. Sci. 7: 178–183 [Google Scholar]
- Kovach, M.J., Calingacion, M.N., Fitzgerald, M.A. and McCouch, S.R. (2009) The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 106: 14444–14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceras, J.C., Huang, Z.L., Naivikul, O., Vanavichit, A., Ruanjaichon, V. and Tragoonrung, S. (2000) Mapping of genes for cooking and eating qualitiesin Thai jasmine rice (KDML 105). DNA Res. 7: 93–101 [DOI] [PubMed] [Google Scholar]
- Lorieux, M., Petrov, M., Huang, N., Guiderdoni, E. and Ghesquiere, A. (1996) Aroma in rice: Genetic analysis of a quantitative trait. Theor. Appl. Genet. 93: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Maga, J.A. (1984) Rice product volatile: A review. J. Agric. Food Chem. 32: 924–970 [Google Scholar]
- Mikami, T., Takadate, M., Yokoyama, H., Kawamura, Y., Kobayashi, W., Tateyama, M., Maeda, K., Kudo, R., Nakahori, T., Oyamada, Z.et al. (2007) A new aromatic rice variety ‘Koihonoka’. Bulletin of Aomori Pref. Agric. And For. Res. Cent. 41: 45–62 [Google Scholar]
- Miyakawa, S. and Nakamura, S. (1984) Regional differences in varietal characteristics of scentedrice. Jpn. J. Crop Sci. 53: 494–502 [Google Scholar]
- Ogawa, M. and Itani, T. (2008) Natural history of red rice in Japan (In Japanese: Akamai-no-hakubutsushi). Daigaku Kyouiku Shuppan, Okayama, Japan, pp. 183 [Google Scholar]
- Olsen, K.M. and Purugganan, M.D. (2002) Molecular evidence on the origin and evolution of glutinous rice. Genetics 162: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.M., Caicedo, A.L., Polato, N., McClung, A., McCouch, S. and Purugganan, M.D. (2006) Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule, C.M. and Powers, J.J. (1989) Sensory and chemical examination of aromatic and nonaromatic rices. J. Food Sci. 54: 343–346 [Google Scholar]
- Sano, Y. (1984) Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68: 467–473 [DOI] [PubMed] [Google Scholar]
- Sato, Y.-I. (1991) Variation in spikelet shape of the indica and japonica rice cultivars in Asian origin. Jpn. J. Breed. 3: 33–43 [Google Scholar]
- Shi, W., Yang, Y., Chen, S. and Xu, M. (2008) Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 22: 185–192 [Google Scholar]
- Sood, B.C. and Siddq, E.A. (1978) A rapid techinique for scent determination in rice. Indian J. Genet. Plant Breed. 38: 268–271 [Google Scholar]
- Tabuchi, H., Sato, Y.-I. and Ashikawa, I. (2007) Mosaic structure of Japanese rice genome composed mainly of two distinct genotypes. Breed. Sci. 57: 213–221 [Google Scholar]
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanavichit, A. and Yoshihashi, T. (2012) Molecular aspects of fragrance and aroma in rice. Advances in Botanical Research 56: 50–73 [Google Scholar]
- Yajima, I., Yanai, T., Nakamura, M., Sakakibara, H. and Habu, T. (1978) Volatile flavor components of cooked rice. Agricultural and Biological Chemistry 42: 1229–1233 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.