Abstract
DNA methylation is responsive to various biotic and abiotic stresses. Heat stress is a serious threat to crop growth and development worldwide. Heat stress results in an array of morphological, physiological and biochemical changes in plants. The relationship between DNA methylation and heat stress in crops is relatively unknown. We investigated the differences in methylation levels and changes in the cytosine methylation patterns in seedlings of two rapeseed genotypes (heat-sensitive and heat-tolerant) under heat stress. Our results revealed that the methylation levels were different between a heat-tolerant genotype and a heat-sensitive one under control conditions. Under heat treatment, methylation increased more in the heat-sensitive genotype than in the heat-tolerant genotype. More DNA demethylation events occurred in the heat-tolerant genotype, while more DNA methylation occurred in the heat-sensitive genotype. A large and diverse set of genes were affected by heat stress via cytosine methylation changes, suggesting that these genes likely play important roles in the response and adaption to heat stress in Brassica napus L. This study indicated that the changes in DNA methylation differed between heat-tolerant and heat-sensitive genotypes of B. napus in response to heat stress, which further illuminates the molecular mechanisms of the adaption to heat stress in B. napus.
Keywords: rapeseed, heat stress, methylation, methylation-sensitive amplification polymorphism (MSAP)
Introduction
Global temperatures are rising and extreme temperature events have been observed in some areas of the world (Easterling et al. 2000). Extensive agricultural losses worldwide are attributed to heat, often in combination with drought (Mittler 2006). Heat stress disturbs cellular homeostasis and can lead to leaf etiolation, severe retardation in plant growth and development, and even death (Yu et al. 2012). B. napus is an important winter oilseed crop; early sowing of B. napus has several important advantages, such as the avoidance of disease infestation and aphid attacks during the flowering stage by early harvest and forced mature by high temperatures during the maturation period (Kaur et al. 2009). However, this crop encounters heat stress during the early sowing period and the maturation period, which affects plant growth and development and, ultimately, causes great losses in yield (Angadi et al. 2000).
Increasing evidence indicates that epigenetic mechanisms, such as DNA methylation and histone modification, play crucial roles in the regulation of gene expression in the plant response to environmental stresses, such as salinity, drought, extreme temperatures, heavy metals, and so on (Choi and Sano 2007, Dyachenko et al. 2006, Kou et al. 2011, Lukens and Zhan 2007, Wada et al. 2004). DNA methylation, an important epigenetic modification, may lead to environmentally induced phenotypic variations by regulating gene expression (Angers et al. 2010, Zhang et al. 2010). The levels of DNA methylation vary in different species. It has been reported that approximately 30–50% of all cytosine residues in the nuclear DNA are methylated in higher plants (Chan et al. 2005). Environmental stress can cause an increase or a decrease in the cytosine methylation levels throughout the genome and at specific loci (Aina et al. 2004, Labra et al. 2002, Li et al. 2009). In Brassicas, cadmium stress stimulated demethylation at specific loci (Filek et al. 2008); potassium dichromate induced extensive methylation changes in CCGG-sequences as revealed by the methylation-sensitive amplification polymorphism (MSAP) approach (Labra et al. 2004).
Several studies have shown that MSAP is an efficient and convenient method to detect DNA methylation (Ashikawa 2001, Cerveraet al. 2002, Xiong et al. 1999). MSAP profiling is an AFLP based method for the detection of DNA methylation (Xu et al. 2000) and has been applied to studies of drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.) (Wang et al. 2011), DNA methylation in Arabidopsis thaliana (Cervera et al. 2002), the association between the up-regulation of stress-responsive genes and the hypomethylation of genomic DNA in tobacco (Wada et al. 2004), epigenetic changes in cotton (Li et al. 2009) and DNA-methylation changes induced by salt stress in wheat (Triticum aestivum) (Zhong et al. 2009). MSAP has become one of the important methods for the investigation of methylation levels and pattern in the genome (Portis et al. 2004).
As sessile organisms, plants manifest numerous adaptive changes in response to heat stress. These responses include the induction of signaling cascades, the profound changes in the expression of specific genes (Xu et al. 2006) and the expression of heat shock proteins (HSPs) (Queitsch et al. 2000). However, little is known about the general pattern of DNA methylation in rapeseed under heat stress. The relationship between epigenetics and the regulation of gene expression under heat stress in different genotypes in rapeseed is also relatively unknown. In a previous study, we demonstrated that heat tolerance in different genotypes of B. napus were dissimilar; the genotypes Huyou 2 and Fengyou 1 were identified as a heat-tolerant genotype and a heat-sensitive genotype, respectively (Gao et al. 2010). Our goal was to investigate whether thermotolerance in seedlings was similar to that in seeds and whether DNA methylation was induced during seedling heat stress. We employed an electrolyte leakage analysis to determine the thermotolerance of B. napus seedlings. The MSAP technique was used to investigate the differences in methylation levels and changes in cytosine methylation patterns in seedlings of two rapeseed genotypes (heat-sensitive and heat-tolerant) under heat stress. The results obtained in this study revealed the effects of heat stress on DNA methylation in the B. napus genome; understanding these effects is important for the further analysis of the molecular mechanism of the adaptation to heat stress in rapeseed.
Materials and Methods
Plant material and treatment conditions
Two rapeseed genotypes (Huyou 2 and Fengyou 1) obtained from the Chinese National Mid-term Seed GeneBank of Oil Crops were used in this study. Previous studies demonstrated that the seeds of Huyou 2 were tolerant to high temperature and the seeds of Fengyou 1 were heat-sensitive (Gao et al. 2010). The seeds of both genotypes were germinated and grown in controlled growth conditions (21°C/23°C, 16 h/8 h light/dark photoperiod). Seedlings at the five-leaf stage were treated with a heat pretreatment at 37°C for 2 h and then heat stress treatment at 45°C for 3 h. The leaves were sampled and immediately frozen under liquid nitrogen for DNA and RNA extraction.
Electrolyte leakage analysis
An electrolyte leakage (EC) analysis was carried out using a modified version of the procedure described by March et al. (1982). Leaf discs (1.4 cm) were cut using cork borers, placed in 15-mL polypropylene tubes with 5.0 mL of distilled water and incubated in 37°C and 45°C water baths. The EC was measured using a conductivity meter (METTLER TOLEDO FE30) every 30 min (EC1). The total leakage (EC2) was determined after the samples were frozen and thawed. The ratio of EC1/EC2 × 100 was used to determine relative damage by heat stress. The experiment was performed three times.
DNA isolation
Genomic DNA was extracted using a modified hexadecyl trimethyl ammonium bromide (CTAB) protocol. DNA quality was verified by separation on 0.8% agar gels and analysis with an UV-1800 spectrophotometer (SHIMADZU, JAPAN). The DNA concentration and purity were determined spectrophotometrically. Aliquots were diluted to a final concentration of 10–15 ng/μl.
MSAP analysis
The MSAP marker was used to assess the stability/alteration in the cytosine methylation patterns at the 5′-CCGG sites. MSAP analysis was performed as described by Xiong et al. (1999), with minor modifications; a pair of methylation-sensitive restriction enzymes, MspI and HpaII, were used in combination with EcoRI. HpaII and MspI are isoschizomers that recognize the same restriction site (5-CCGG) with a differential sensitivity to methylation modifications at the two cytosines. HpaII will not cut if either of the cytosines is fully methylated (double-strand), whereas MspI will not cut if the external cytosine is hemi-methylated (Dong et al. 2006).
The DNA was digested with EcoRI/HpaII and EcoRI/MspI at 37°C for 4 h. Digested aliquots were ligated with EcoRI-and MspI-or HpaII-specific adopters at 20°C for 5 h. After the ligation reaction, the samples were incubated for 10 min at 60°C to inactivate the enzymes. The ligated DNA was diluted 1 : 10 and pre-amplified. The pre-amplified product was diluted 1 : 10 and amplified using different combinations of EcoRI and MspI or HpaII primers. Each primer contained three selective nucleotides at the 5′ and 3′ ends. The sequences of the adapters and the primers used for pre-amplification and selective amplification are provided in Supplemental Tables 1, 2. The MSAP PCR products were separated by 6% polyacrylamide gels electrophoresis (PAGE) and visualized after silver staining.
The MSAP patterns of the DNA fragments resulting from the digestion with the isoschizomers were divided into the following four types. Type I: two bands presenting for both enzyme combinations indicated that the corresponding CCGG site is unmethylated. Type II: One band representing only EcoRI/HpaII indicated a hemimethylation state of DNA due to the methylation of one DNA strand but not its complementary strand. Type III: one band appearing for EcoRI/MspI reflects the case of a full CG (internal cytosine) methylation. Type IV: the absence of bands for both enzyme combinations indicating that full methylation occurred at both cytosines (McClelland et al. 1994). The MSAP bands were scored “1” or “0” to indicate the presence or absence, respectively, of a band at a particular position.
Isolation and characterization of the amplified fragments
The polymorphic fragments were excised from the gel, hydrated in 50 μl of water, and incubated in boiling water for 5 min. The supernatant was recovered by centrifugation and used for re-amplification using the same primers under the conditions described above for the selective amplification. The re-amplified DNA fragments were purified and cloned into a T-vector (TransGen Biotech, China) for sequencing. The sequences obtained were analyzed using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BRASSICA (http://brassicadb.org/brad/blastPage.php) search algorithms.
Quantitative RT-PCR analysis
To test the effect of heat stress, seedlings of Huyou 2 and Fengyou 1 were treated at 45°C for 3 h. Seedlings grown in normal conditions were used as control. The leaves were sampled and immediately frozen under liquid nitrogen for RNA extraction. We used RNeasy Plant Mini Kit (QIAGEN, Germany) to extract the total RNA from each sample. Quantitative RT-PCR analysis was performed by LightCycler® 480 SYBR Green I Master using a LightCycler®480II real-time PCR machine (Roche, http://www.roche-applied-science.com). Each sample was analyzed at least three times and three replicates were used in each case. The method of reverse transcription-polymerase chain reaction (RT-PCR) was performed as described by Li et al. (2013). The relative expression levels were analyzed as described by Yuan et al. (2008). Primers were designed to amplify the genes that were homologous to M7, M9 and M16, which are genes differentially methylated, using GeneSript (https://www.genscript.com/ssl-bin/app/primer). The primers used in this study are listed in Supplemental Table 3.
Results
Relative thermotolerance in two rapeseed genotypes
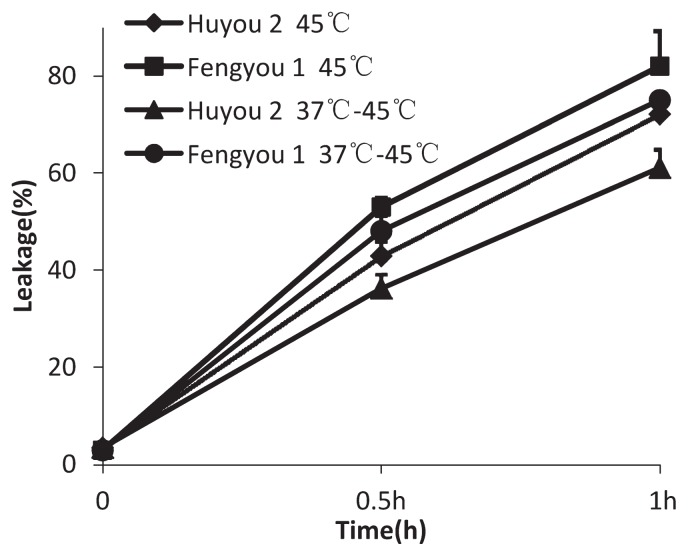
To examine the heat stress response in rapeseed seedlings, we analyzed two genotypes. In response to high temperature storage, the seeds of the first genotype (Huyou 2) have been described as heat-tolerant and the seeds of the second genotype (Fengyou 1) have been described as heat-sensitive (Gao et al. 2010). Electrolyte leakage analysis has been shown to be a reliable, quantitative and reproducible assay to predict thermotolerance in a variety of plants (Coria et al. 1998, Keeler et al. 2000). To observe thermotolerance in the green leaf tissue of seedlings, we measured electrolyte leakage from leaf discs as an indicator of heat injury. The leaves of Fengyou 1 seedlings reached 82% injury; this observed injury was greater than observed in Huyou 2 after exposure to a test temperature of 45°C for 90 min (Fig. 1). When seedlings were pretreated at 37°C for 2 h before the 45°C heat treatment, the thermotolerance of both Huyou 2 and Fengyou 1 increased. These results strongly suggest that pretreatment at 37°C increases the thermotolerance of rapeseed seedlings. The Fengyou 1 seedlings were more sensitive to heat stress than Huyou 2 seedlings; these results correlate with those observed in seeds.
Fig. 1.
Electrolyte leakage of heat-tolerance and heat-sensitive rapeseed under heat stress.
The dynamic level of DNA methylation induced in heat-tolerant and heat-sensitive genotypes by heat stress
A total of 47 primer pair combinations (listed in Supplemental Table 2) were used to assess changes in the methylation of DNA and the polymorphisms in methylated DNA from the leaves of seedlings of two rapeseed genotypes (Huyou 2 and Fengyou 1). The seedlings were subjected to different levels of heat stress. Between 675 and 794 clear bands were amplified in seedlings of both genotypes under control conditions and high temperature conditions. Of these bands, 196–297 (1/0 and 0/1) were methylated (hemi-or full-methylation), of which 138–223 were full-methylated. The percentages of the total methylated bands and the full-methylated bands were 26.8%–37.4% and 18.5%–28.3%, respectively. Overall, the number of methylated DNA bands observed in Huyou 2 (199) was lower than that observed in Fengyou 1 (295) after a 45°C heat stress treatment preceded by either a 37°C pretreatment or no pretreatment (Table 3).
Table 3.
Blast results of 17 polymorphic methylated DNA fragments
| MSAP fragment | Size (bp) | Accession No./locus | E value | Nuclear identity (%) | Score | chromosome Position | Gene Position | Sequence homology |
|---|---|---|---|---|---|---|---|---|
| M1 | 243 | Bol030657 | 4.00E-71 | 90 | 268 | C03 | Exon | CSDP1; CSDP1 (cold shock domain protein 1) |
| M2 | 338 | Bra016894 | 6.00E-11 | 95 | 68 | A04 | Exon | POLD2; DNA binding/DNA-directed DNA polymerase |
| M3 | 245 | Bra013050 | 2.00E-19 | 98 | 96 | A04 | Exon | GTP binding / GTPase |
| M4 | 153 | FR715249.1 | 1.00E-55 | 98 | 224 | mitochondria | Intergenic regions | Brassica napus complete mitochondrial genome |
| M5 | 137 | AP006444.1 | 2.00E-15 | 98 | 92 | mitochondria | Intergenic regions | Brassica napus mitochondrial DNA |
| M6 | 222 | DQ321812.1 | 5.00E-94 | 99 | 352 | mitochondria | Intergenic regions | Brassica napus AFLP marker M5 sequence |
| M7 | 287 | Bol023401 | 1.00E-139 | 98 | 496 | C08 | Exon | calcium-transporting ATPase |
| M8 | 256 | Bol029195 | 1.00E-124 | 99 | 448 | C08 | Promotor | FUT8 (FUCOSYLTRANSFERASE 8) |
| M9 | 256 | Bra002340 | 1.00E-111 | 97 | 400 | A10 | Exon | protein kinase family protein |
| M10 | 180 | Bra036388 | 3.00E-45 | 96 | 180 | A07 | Exon | zinc ion binding |
| M11 | 433 | AC241103.1 | 6.00E-34 | 78 | 154 | A06 | Intergenic regions | Brassica rapa subsp. pekinensis clone KBrH015L02 |
| M12 | 437 | Bol030652 | 0.00E + 00 | 97 | 761 | C03 | Intron | WD-40 repeat family protein/beigerelated |
| M13 | 299 | AK228893.1 | 2.00E-18 | 70 | 102 | C02 | Intergenic regions | Arabidopsis thaliana mRNA for hypothetical protein |
| M14 | 300 | AC189501.2 | 6.00E-57 | 79 | 230 | A10 | Intergenic regions | Brassica rapa subsp. pekinensis clone KBrB013O20 |
| M15 | 246 | AC189495.2 | 2.00E-05 | 78 | 99 | C09 | Intergenic regions | Brassica rapa subsp. pekinensis clone KBrB085G17 |
| M16 | 225 | Bra002219 | 7.00E-99 | 97 | 359 | A10 | Exon | ECR1; ECR1 (E1 C-terminal related 1) |
| M17 | 169 | AC189233.2 | 9.00E-14 | 74 | 86 | A03 | Intergenic regions | Brassica rapa subsp. pekinensis clone KBrB013O20 |
Under control conditions, the overall methylation levels in the DNA isolated from the leaves of Huyou 2 were 2.2% greater than that observed in the leaves of Fengyou 1 (Table 1). Compared with the control conditions, heat stress induced a greater number of observed methylated bands (both hemi- and full-methylation) at the expense of unmethylated bands in Fengyou 1. However, no obvious changes in the overall methylation level were observed in the Huyou 2 seedlings following heat stress. However, the overall level of DNA methylation was approximately 26.8% in the leaves of Fengyou 1 under the control condition and 37.4% after the 45°C heat treatment; The percentage of full-methylated bands increased from 18.5% to 28.3%. This result indicated that there was a great difference in the overall level of genomic DNA methylation in Fengyou 1 under control conditions and heat stress. These results also indicated that in both rapeseed genotypes, no obvious changes were detected when the seedlings underwent pretreatment at 37°C before direct heat stress.
Table 1.
DNA methylation changes in leaves of Huyou 2 and Fengyou 1 under control and heat stress
| MSAP band type | H | M | Huyou 2 | Fengyou 1 | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| CK | 45°C | 37–45°C | CK | 45°C | 37–45°C | |||
| Type I | 1 | 1 | 479 | 479 | 479 | 574 | 494 | 497 |
| Type II | 1 | 0 | 58 | 61 | 60 | 65 | 72 | 74 |
| Type III | 0 | 1 | 138 | 138 | 140 | 145 | 223 | 223 |
| Total amplified bands | 675 | 678 | 679 | 784 | 789 | 794 | ||
| Total methylated bands | 196 | 199 | 200 | 210 | 295 | 297 | ||
| MSAP (%) | 29.0 | 29.4 | 29.5 | 26.8 | 37.4 | 37.4 | ||
| Fully methylated ratio (%) | 20.4 | 20.4 | 20.6 | 18.5 | 28.3 | 28.1 | ||
CK indicates the control check.
A score of 1 and 0 represents the presence and absence of bands, respectively.
H and M represent digestion with HpaII/EcoRI and MspI/EcoRI, respectively.
Differences in the DNA methylation patterns in heat-tolerant and heat-sensitive genotypes under heat stress
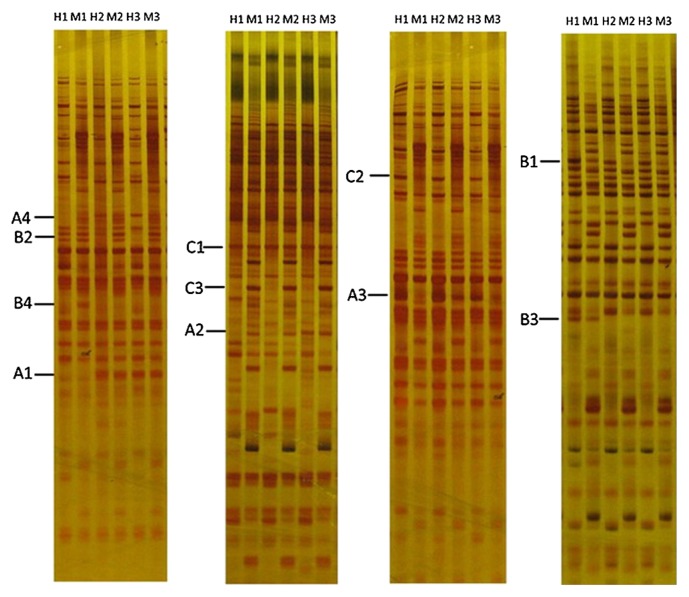
Twelve methylation patterns, organized into polymorphism and monomorphism groups, were observed in the products of the MSAP assay (Table 2 and Fig. 2). In the polymorphism group, the CCGG/GGCC sites were methylated differently between the control and the heat stress treatments; such a result indicates that the methylation patterns of the genomic DNA changed under heat stress. The methylation banding patterns were divided into the following three major types: A, B and C (Table 2). Type A indicates cytosine demethylation patterns under heat stress, showing that demethylation can occur in both internal cytosines (A2) and external cytosines (A3). Contrary to type A patterns, type B indicates cytosine methylated patterns in the treatment that corresponds with the control treatment; heat induced methylation can occur in both the external cytosine (B1) and internal cytosine (B2). Type C indicates patterns of a monomorphic classification; the same CCGG sites were detected following both the heat stress treatment and the control treatment (Fig. 2).
Table 2.
Changes of DNA methylation pattern under control and heat stress in Huyou 2 and Fengyou 1
| Banding pattern | CK | T | Methylation status | 45°C | 37–45°C | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| H | M | H | M | Huyou 2 | Fengyou 1 | Huyou 2 | Fengyou 1 | |||
| A1 | 0 | 0 | 1 | 1 | CCGG GGCC |
CCGG GGCC |
2 | 5 | 4 | 2 |
| A2 | 0 | 1 | 1 | 1 |
CCGG GGCC |
CCGG GGCC |
1 | 0 | 0 | 1 |
| A3 | 1 | 0 | 1 | 1 |
CCGG GGCC |
CCGG GGCC |
0 | 0 | 2 | 0 |
| A4 | 0 | 0 | 1 | 0 |
CCGG GGCC |
CCGG GGCC |
2 | 6 | 2 | 9 |
| A5 | 0 | 0 | 0 | 1 | CCGG GGCC |
CCGG GGCC |
1 | 4 | 3 | 4 |
| Demethylation | 6 (0.9%) | 15 (1.9%) | 11 (1.6%) | 16 (2.0%) | ||||||
| B1 | 1 | 1 | 1 | 0 | CCGG GGCC |
CCGG GGCC |
1 | 1 | 1 | 0 |
| B2 | 1 | 1 | 0 | 1 | CCGG GGCC |
CCGG GGCC |
1 | 74 | 0 | 75 |
| B3 | 1 | 1 | 0 | 0 | CCGG GGCC |
CCGG GGCC |
1 | 11 | 2 | 12 |
| B4 | 0 | 1 | 0 | 0 | CCGG GGCC |
CCGG GGCC |
1 | 0 | 1 | 0 |
| Methylation | 4 (0.6%) | 86 (10.7%) | 4 (0.6%) | 87 (10.9%) | ||||||
| C1 | 1 | 1 | 1 | 1 | CCGG GGCC |
CCGG GGCC |
476 | 489 | 476 | 488 |
| C2 | 1 | 0 | 1 | 0 |
CCGG GGCC |
CCGG GGCC |
58 | 66 | 57 | 65 |
| C3 | 0 | 1 | 0 | 1 | CCGG GGCC |
CCGG GGCC |
136 | 145 | 137 | 145 |
| No change | 670 (98.5%) | 700 (87.4%) | 670 (97.8%) | 698 (87.1%) | ||||||
CK and T indicate the control and heat stress conditions, respectively. Methylated cytosine is underlined, A score of 1 and 0 represents the presence and absence of bands, respectively. H and M represent digestion with HpaII/EcoRI and MspI/EcoRI, respectively.
Fig. 2.
MSAP fingerprints of rapeseed seedling between control and heat stress. Lanes H1 and M1 are MSAP patterns of control, lanes H2 and M2 are MSAP of 45°C heat stress, lanes H3 and M3 are MSAP of 45°C heat stress followed 37°C pretreatment. H represent digestion with EcoRI/HpaII, M represent digestion with EcoRI/MspI. Band patterns of A, B, and C can be seen in Table 2.
By using 23 pairs of primer combinations which illustrated polymorphisms in the MSAP analysis, banding patterns were detected between control conditions and the heat stress treatments in the two genotypes. Approximately 87.1%–97.8% of the CCGG sites in the two genotypes remain unchanged under heat stress (Table 2). The percentage of demethylated bands in Huyou 2 were 0.9% and 1.6% for the 45°C treatment without pretreatment and the 45°C treatment with a 37°C pretreatment, respectively. In Fengyou 1, the percentage of demethylated bands were 1.9% and 2.0% for the 45°C treatment without pretreatment and the 45°C treatment with a 37°C pretreatment, respectively. Under heat stress, the percentage of methylated bands was 0.6% in Huyou 2 and 10.7% in Fengyou 1. These results indicate that more DNA demethylation events occurred in Huyou 2 than methylation and relatively more DNA methylation occurred in Fengyou 1 than demethylation under heat stress. In general, DNA methylation and demethylation events caused by heat stress were different in the two genotypes, which may be related to the genotype-specific differences in thermotolerance.
Cloning and sequence analysis of the methylated DNA bands
Of the polymorphic fragments described above, a random set of 17 were cloned and sequenced to identify the nature of the DNA sequences involved in methylation and demethylation. The resulting sequences were subjected to BLAST searches against the NCBI and Brassica databases. The length of the cloned bands varied between 137 to 437 bp, with an average length of 258 bp (Table 3). Among these 17 DNA sequences, seven sequences (M11–M17) differed between genotypes under non-stress conditions. These seven sequences were homologous to a WD-40 repeat family protein/beige-related; an A. thaliana mRNA for a hypothetical protein; ECR1; and four Brassica rapa subsp. pekinensis clones (Table 3). These results indicate an epigenetic difference in these sequences between the two genotypes.
Three sequences were demethylated in both lines in response to heat stress (M6–M8); these were found to be homologous to a calcium-transporting ATPase, an AFLP marker M5 sequence of B. napus mitochondrial DNA, and a galactoside 2-alpha-L-fucosyltransferase. Five sequences were methylated or demethylated in response to heat stress in Fengyou 1 (M1–M3, M9–M10). Two sequences were demethylated under heat stress in Huyou 2 (M4–M5) (Table 3). These results indicate that heat stress affects a large number of diverse genes in rapeseed via DNA-specific changes in cytosine methylation.
Expression analysis of polymorphic fragments
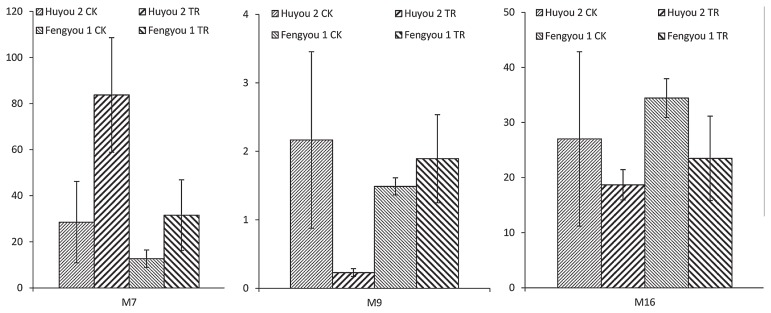
The expression analysis of three MSAP polymorphic fragments was performed using quantitative RT-PCR. The expression results of heat treated leaves and untreated leaves were showed in Fig. 3. M7 was demethylated in heat treated leaves contrasted to untreated leaves, M9 was demethylated in Fengyou 1 and M16 was methylated in Fengyou 1 but the two genes were not changed in Huyou 2 under heat stress. The expression level of the M7 demethylated gene was up-regulated obviously, but the M9 was up-regulated slightly, and the M16 methylated gene was down-regulated in heat treated leaves.
Fig. 3.
Expression of genes homologous to M7, M9 and M16 in leaves of rapeseed under control and heat stress condition. CK and TR indicate the control and heat stress conditions, respectively.
Discussion
Many studies have indicated that primary DNA methylation plays important roles in the responses to environment stresses and may contribute to environmentally inducing phenotypic variation by modifying gene expression (Angers et al. 2010, Pecinka et al. 2010). In this study, we analyzed the relative thermotolerance of seedlings in two genotypes of B. napus using electrolyte leakage analysis. The seeds of these two genotypes were previously considered to be heat-sensitive (Fengyou 1) or heat-tolerant (Huyou 2) during seed preservation at high temperatures (Gao et al. 2010). We found that the response to heat stress in seeds of both genotypes correlated with that of the seedlings. Furthermore, our results demonstrated that high temperature (45°C) affects leaf membrane permeability and a pretreatment at 37°C increases leaf membrane permeability. Our results are consistent with previous reports that demonstrated that pretreatment at 37°C results in an observable decrease in leaf cell damage at 45°C (Hikosaka et al. 2006, Keeler et al. 2000, Larkindale et al. 2005, Senthil-Kumar et al. 2007).
Some studies have revealed that environmental stimuli can result in an increase or decrease in cytosine methylation throughout the genome and at specific loci (Lukens and Zhan 2007, Mastan et al. 2012, Tan 2010, Wang et al. 2011). In this study, we found that the percentage of MSAP sites in the Huyou 2 and Fengyou 1 genotypes were approximately 29.0% and 26.8%, respectively; these percentages were lower than those observed in A. thaliana (35–43%) (Cervera et al. 2002). Our results indicated that differential DNA methylation occurs among distinct genotypes; this finding is consistent with studies in rice (Karan et al. 2012, Takata et al. 2005). In addition, heat stress tended to increase the overall DNA methylation levels in the leaves of seedlings of both rapeseed genotypes. Similar results were observed in rice (Oryza sativa L.) and wheat (Triticum aestivum L.) under heavy metal stress (Ge et al. 2002), in pea (Pisum sativum L.) under water deficit stress (Labra et al. 2002), and in Arabidopsis and Pinus silvestris in response to irradiation (Kovalchuk et al. 2003, 2004). DNA methylation was found to be specific to the genotype, the tissue and the developmental stage (Mastan et al. 2012, Tan 2010, Wang et al. 2011). Further studies are needed to identify the tissue and developmental specificity of these epigenetic changes in the rapeseed genome under heat stress. According to our study, the heat-sensitive genotype Fengyou 1 showed a greater increase in cytosine methylation under heat stress compared to the heat-tolerant genotype Huyou 2. Rapp and Wendel (2005) stated that methylation changes may generate nonspecific (random) differences between individuals, which may have adaptive significance during times of stress; such differences in methylation patterns increase the range of variation that natural selection can act upon (Verhoeven et al. 2010). Several studies have also suggested that crop genotypes that avoid cytosine methylation may be agriculturally superior to genotypes that are sensitive to methylation (Guo et al. 2006). A study by Tani et al. (2005) supported the hypothesis that hybrids perform better than inbred lines because they resist alterations in methylation under stress. A greater number of methylation/demethylation loci were observed in the heat-sensitive genotype compared to the heat-tolerant genotype. Our results are in agreement with previous global gene expression analyses revealing that the expressions of a strikingly large number of genes are induced by salinity stress in sensitive genotypes (Walia et al. 2005, 2007). The methylation and demethylation of DNA cytosines in specific regions of these genes, their promoters or in the neighboring sequences play an important role in regulating gene expression during plant development (Filek et al. 2008, Portis et al. 2004).
Heat stress induced extensive alterations at multiple genomic loci. In total, 17 MSAP fragments that were methylated or demethylated were identified and represent both responses to stress and genotype-specific methylation patterns. Our results indicate that changes in DNA methylation occur throughout the entire genome in response to abiotic stress, as previously observed in rice under salt stress (Wang et al. 2011). Recent genomic studies in A. thaliana revealed that many endogenous genes are methylated either within their promoters or within their transcribed regions and that gene methylation is highly correlated with the transcription level (Cokus et al. 2008). A close correlation exists between methylation and gene expression in response to abiotic stress (Choi and Sano 2007). In rapeseed seedlings, an increase in the Cd concentration in the medium stimulated demethylation at specific loci (Filek et al. 2008). In tobacco, viral infections (Wada et al. 2004) as well as several abiotic stresses (Choi and Sano 2007) resulted in demethylation and the associated up-regulation of stress-related genes (Verhoeven et al. 2010). In this study, we found that demethylated M7 was homologous to calcium-transporting ATPase. Through RT-PCR analysis, the expression of the gene was up-regulated in two genotypes under heat stress. Generally, the plasma membrane and ER of plant cells have been shown to possess primary calcium-transporting ATPase that use ATP hydrolysis to drive the direct transport of calcium ions (Thomson et al. 1993), and it has been suggested that a stress-induced change in [Ca2+]cyt might be one of the primary transduction mechanisms whereby gene expression and biochemical events are altered to adapt plant cells to environmental stresses under the influence of various stress signals (Gong et al. 1998). Our results suggest that genotype-specific epigenetic changes may be an important regulatory mechanism in response to heat stress through the modification of the expression network of responsive genes. In sum, the changes in DNA methylation patterns induced by heat stress were different in two rapeseed genotypes. In addition, the methylation/demethylation induced by heat stress involved a wide range of genes. Our results help to clarify the molecular mechanisms of the adaption to heat stress in rapeseed.
Supplementary Material
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31100236), Chen Guang Program for Young Scientists of Wuhan Municipal Government (201271031402, 2013070104010031), and National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAD35B09), Strategic Japanese-Chinese Cooperative Program on “Climate Change” (2012DFG90290).
Literature Cited
- Aina, R., Sgorbati, S., Santagostino, A., Labra, M., Ghiani, A. and Citterio, S. (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plant. 121: 472–480 [Google Scholar]
- Angadi, S.V., Cutforth, H.W., Miller, P.R., McConkey, B.G., Entz, M.H., Brandt, S.A. and Volkmar, K.M. (2000) Response of three Brassica species to high temperature stress during reproductive growth. Can. J. Plant Sci. 80: 693–701 [Google Scholar]
- Angers, B., Castonguay, E. and Massicotte, R. (2010) Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19: 1283–1295 [DOI] [PubMed] [Google Scholar]
- Ashikawa, I. (2001) Surveying CpG methylation at 5′-CCGG in the genomes of rice cultivars. Plant Mol. Biol. 45: 31–39 [DOI] [PubMed] [Google Scholar]
- Cervera, M.T., Ruiz-Garcia, L. and Martinez-Zapater, J.M. (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol. Genet. Genomics 268: 543–552 [DOI] [PubMed] [Google Scholar]
- Chan, S.W.L., Hendeson, I.R. and Jacobsen, S.E. (2005) Gardening the genome DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Choi, C.S. and Sano, H. (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genomics 277: 589–600 [DOI] [PubMed] [Google Scholar]
- Cokus, S.J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C.D., Pradhan, S., Nelson, S.F., Pellegrini, M. and Jacobsen, S.E. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coria, N.A., Sarquis, J.I., Peñalosa, I. and Urzua, M. (1998) Heat-induced damage in potato (Solanum tuberosum) tubers: membrane stability, tissue viability, and accumulation of glycoalkaloids. J. Agric. Food Chem. 46: 4524–4528 [Google Scholar]
- Dong, Z.Y., Wang, Y.M., Zhang, Z.J., Shen, Y., Lin, X.Y., Ou, X.F., Han, F.P. and Liu, B. (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor. Appl. Genet. 113: 196–205 [DOI] [PubMed] [Google Scholar]
- Dyachenko, O.V., Zakharchenko, N.S., Shevchuk, T.V., Bohnert, H.J., Cushman, J.C. and Buryanov, Y.I. (2006) Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biochemistry (Mosc) 71: 461–465 [DOI] [PubMed] [Google Scholar]
- Easterling, D.R., Meehl, G.A., Parmesan, C., Changnon, S.A., Karl, T.R. and Mearns, L.O. (2000) Climate extremes: observations, modelling, and impacts. Science 289: 2068–2074 [DOI] [PubMed] [Google Scholar]
- Filek, M., Keskinen, R., Hartikainen, H., Szarejko, I., Janiak, A., Miszalski, Z. and Golda, A. (2008) The protective role of selenium in rape seedlings subjected to cadmium stress. J. Plant Physiol. 165: 833–844 [DOI] [PubMed] [Google Scholar]
- Gao, G.Z., Wu, X.M., Lv, X.D., Chen, B.Y., Xu, K. and Yan, G.X. (2010) Genotype different of seed viability in rapeseed during storage at different temperature. Zhongguo You Liao Xue Bao 32: 495–499 [Google Scholar]
- Ge, C.L., Yang, X.Y., Liu, X.N., Sun, J.H., Luo, S.S. and Wang, Z.G. (2002) Effects of heavy metal on the DNA methylation level in rice and wheat. J. Plant Physiol. Mol. Biol. 28: 363–368 [Google Scholar]
- Gong, M., van der Luit, A.H., Knight, M.R. and Trewavas, A.J. (1998) Heat-shock-induced changes in intracellular Ca2+ level in tobacco seedlings in relation to thermotolerance. Plant Physiol. 116: 429–437 [Google Scholar]
- Guo, M., Rupe, M.A., Yang, X., Crasta, O., Zinselmeier, C., Smith, O.S. and Bowen, B. (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 113: 831–845 [DOI] [PubMed] [Google Scholar]
- Hikosaka, K., Ishikawa, K., Borjigidai, A., Muller, O. and Onoda, Y. (2006) Temperature acclimation of photosynthesis: mechanisms involved in the changes in temperature dependence of photosynthetic rate. J. Exp. Bot. 57: 291–302 [DOI] [PubMed] [Google Scholar]
- Karan, R., DeLeon, T., Biradar, H. and Subudhi, P.K. (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS One 7: e40203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, P., Ghai, N. and Sangha, M.K. (2009) Induction of thermotolerance through heat acclimation and salicylic acid in Brassica species. Afr. J. Biotechnol. 8: 619–625 [Google Scholar]
- Keeler, S.J., Boettger, C.M., Haynes, J.G., Kuches, K.A., Johnson, M.M., Thureen, D.L., Keeler, C.L. and Kitto, S.L. (2000) Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 123: 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou, H.P., Li, Y., Song, X.X., Ou, X.F., Xing, S.C., Ma, J., Wettstein, D.V. and Liu, B. (2011) Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.). J. Plant Physiol. 168: 1685–1693 [DOI] [PubMed] [Google Scholar]
- Kovalchuk, O., Burke, P., Arkhipov, A., Kuchma, N., James, S.J., Kovalchuk, I. and Pogribny, I. (2003) Genome hypermethylation in Pinus silvestris of Chernobyl—a mechanism for radiation adaptation? Mutat. Res. 529: 13–20 [DOI] [PubMed] [Google Scholar]
- Kovalchuk, I., Abramov, V., Pogribny, I. and Kovalchuk, O. (2004) Molecular aspects of plant adaptation to life in the Chernobyl zone. Plant Physiol. 135: 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labra, M., Ghiani, A., Citterio, S., Sgorbati, S., Sala, F., Vannini, C., Ruffini-Castiglione, M. and Bracale, M. (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol. 4: 694–699 [Google Scholar]
- Labra, M., Grassi, F., Imazio, S., Fabio, T.D., Citterio, S., Sgorbati, S. and Agradi, E. (2004) Genetic and DNA-methylation changes induced by potassium dichromate in Brassica napus L. Chemosphere 54: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Larkindale, J., Hall, J.D., Knight, M.R. and Vierling, E. (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Gao, G.Z., Xu, K., Chen, B.Y., Yan, G.X., Li, F., Qiao, J.W., Zhang, T.Y. and Wu, X.M. (2013) Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS One 9: e84556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.L., Lin, Z.X., Nie, Y.C., Guo, X.P. and Zhang, X.L. (2009) MSAP analysis of epigenetic changes in cotton (Gossypium hirsutum L.) under salt stress. Acta Agron. Sin. 35: 588–596 [Google Scholar]
- Lukens, L.N. and Zhan, S. (2007) The plant genome’s methylation status and response to stress: implications for plant improvement. Curr. Opin. Plant Biol. 10: 317–322 [DOI] [PubMed] [Google Scholar]
- March, L.E., Davis, D.W., Li, P.H. and Silbernagel, M.J. (1982) Two methods of evaluating genotypes of Phaseolus vulgaris under high temperature stress. Annu. Rep. Bean Improv. Coop. 25: 55–56 [Google Scholar]
- Mastan, S.G., Rathore, M.S., Bhatt, V.D., Yadav, P. and Chikara, J. (2012) Assessment of changes in DNA methylation by methylation-sensitive amplification polymorphism in Jatropha curcas L. subjected to salinity stress. Gene 508: 125–129 [DOI] [PubMed] [Google Scholar]
- McClelland, M., Nelson, M. and Raschke, E. (1994) Effect of site specific modification restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22: 3640–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Pecinka, A., Dinh, H.Q., Banbec, T., Rosa, M., Lettner, N. and Scheid, O.M. (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis, E., Acquadro, A., Cominio, C. and Lanteri, S. (2004) Analysis of DNA methylation during germination of pepper (Capsicum annuum L.) seeds using methylation-sensitive amplification polymorphism (MSAP). Plant Sci. 166: 169–178 [Google Scholar]
- Queitsch, C., Hong, S.W., Vierling, E. and Lindquist, S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, R.A. and Wendel, J.F. (2005) Epigenetics and plant evolution. New Phytol. 168: 81–91 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar, M., Kumar, G., Srikanthbabu, V. and Udayakumar, M. (2007) Assessment of variability in acquired thermotolerance: potential option to study genotypic response and the relevance of stress genes. J. Plant Physiol. 164: 111–125 [DOI] [PubMed] [Google Scholar]
- Takata, M., Yuji, K. and Yoshio, S. (2005) DNA methylation polymorphisms in rice and wild rice strains: detection of epigenetic markers. Breed. Sci. 55: 57–63 [Google Scholar]
- Tan, M.P. (2010) Analysis of DNA methylation of maize in response to osmotic and salt stress based on methylation-sensitive amplified polymorphism. Plant Physiol. Biochem. 48: 21–26 [DOI] [PubMed] [Google Scholar]
- Tani, E., Polidoros, A.N., Nianiou-Obeidat, I. and Tsaftaris, A.S. (2005) DNA methylation patterns are differentially affected by planting density in maize inbreds and their hybrids. Maydica 50: 19–23 [Google Scholar]
- Thomson, L.J., Xing, T., Hail, J.L. and Williams, L.E. (1993) Investigation of the calcium-transporting ATPases at the endoplasmic reticulum and plasma membrane of red beet (Beta vulgaris). Plant Physiol. 102: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven, K.J., Jansen, J.J., van Dijk, P.J. and Biere, A. (2010) Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Wada, Y., Miyamoto, K., Kusano, T. and Sano, H. (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol. Genet. Genomics 271: 658–666 [DOI] [PubMed] [Google Scholar]
- Walia, H., Wilson, C., Condamine, P., Liu, X., Ismail, A.M., Zeng, L., Wanamaker, S.I., Mandal, J., Xu, J., Cui, X.et al. (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 139: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia, H., Wilson, C., Zeng, L., Ismail, A.M., Condamine, P. and Close, T.J. (2007) Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol. Biol. 63: 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.S., Pan, Y.J., Zhao, X.Q., Dwivedi, D., Zhu, L.H., Ali, J., Fu, B.Y. and Li, Z.K. (2011) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 62: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L.Z., Xu, C.G., Saghai Maroof, M.A. and Zhang, Q. (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261: 439–446 [DOI] [PubMed] [Google Scholar]
- Xu, M.L., Li, X.Q. and Korban, S.S. (2000) AFLP-based detection of DNA methylation. Plant Mol. Biol. Rep. 18, 361–368 [Google Scholar]
- Xu, S., Li, J., Zhang, X., Wei, H. and Cui, L. (2006) Effects of heat acclimation pretreatment on changes of membrane lipid peroxidation, antioxidant metabolites, and ultrastructure of chloroplasts in two cool-season turfgrass species under heat stress. Environ. Exp. Bot. 56: 274–285 [Google Scholar]
- Yu, X., Wang, H., Lu, Y., de Ruiter, M., Cariaso, M., Prins, M., van Tunen, A. and He, Y. (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 63: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Chen, D., Ren, Y., Zhang, X. and Zhao, J. (2008) Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 146: 1637–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Kimatu, J.N., Xu, K. and Liu, B. (2010) DNA cytosine methylation in plant development. J. Genet. Genomics 37: 1–12 [DOI] [PubMed] [Google Scholar]
- Zhong, L., Xu, Y. and Wang, J. (2009) DNA-methylation changes induced by salt stress in wheat Triticum aestivum. Afr. J. Biotechnol. 8: 6201–6207 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.