Abstract
Amylose content is one of the most important factors influencing the physical and chemical properties of starch in rice. Analysis of 352 Vietnamese rice cultivars revealed a wide range of variation in apparent amylose content and the expression level of granule-bound starch synthase. On the basis of single-nucleotide polymorphisms (SNP) at the splicing donor site of the first intron and in the coding region of the granule-bound starch synthase I gene, Waxy gene, alleles can be classified into seven groups that reflect differences in apparent amylose content. The very low and low apparent amylose content levels were tightly associated with a G to T in the first intron whereas intermediate and high amylose was associated with a T genotype at SNP in exon 10. The correlation between the combination of T genotype at SNP in the first intron, C in exon 6, or C in exon 10 was predominant among low amylose rice varieties. Our analysis confirmed the existence of Wxop allele in Vietnamese rice germplasm. The results of this study suggest that the low amylose properties of Vietnamese local rice germplasm are attributable to spontaneous mutations at exons, and not at the splicing donor site.
Keywords: amylose, granule-bound starch synthase I, rice germplasm, single-nucleotide polymorphism, Waxy gene
Introduction
Starch consists of two kinds of glucan polymers, amylose and amylopectin. Amylose is predominantly a linear molecule of α-1,4-linked D-glucose, although some of the molecules are slightly branched by α-1,6-glucosidic linkages (Takeda and Hizukuri 1987). Amylopectin is composed of highly branched α-1,4-polyglucans, which are short, linear α-1,4-glucan chains that are regularly branched by α-1,6-glucosidic linkages (Takeda et al. 1987). The physicochemical properties of rice starch are affected by the ratio of amylose to amylopectin and their molecular structures (Nakamura et al. 2006).
Amylose is synthesized by the granule-bound starch synthase (GBSS), whereas amylopectin is synthesized by the concerted action of four classes of enzymes: ADP-glucose pyrophosphorylase, starch synthase, branching enzyme, and debranching enzyme (Denyer et al. 2001, Hannah and James 2008). Granule-bound starch synthase I (GBSSI) in rice is encoded by the Waxy (Wx) gene (Hirano and Sano 1991, Hirose and Terao 2004, Okagaki 1992). In rice cultivars, there are three functional alleles at the Waxy locus: Wxa, Wxb, and Wxop. Wxa and Wxb are found mainly in Indica and Japonica rice, respectively. The expression level of Wxa and Wxb are associated with amylose content (Sano 1984, Sano et al. 1985, 1986). Expression level of mRNA and accumulation of waxy protein in Wxa cultivars is 10-fold higher than that of Wxb cultivars (Isshiki et al. 1998). The Waxy opaque (Wxop) allele is the modified form of the Wxa gene and endosperms with the Wxop allele have 10% amylose content (Mikami et al. 1999, 2008).
The association between amylose content (AC) and single-nucleotide polymorphisms (SNPs) in the rice Wx gene has been described at the splicing donor sites of the first intron (Isshiki et al. 1998, Sano et al. 1985), exon 4 (Larkin and Park 1999, Mikami et al. 1999, 2008), exon 6 (Cai et al. 1998, Larkin and Park 2003, Mikami et al. 2008, Wang et al. 1995), and exon 10 (Cai et al. 1998, Hirano et al. 1996, Mikami et al. 2008, Wang et al. 1995). The cytosine and thymidine (CTn) dinucleotide repeats in the 5′-untranslated region (UTR) of the Wx gene were reported to be a factor associated with AC (Ayres et al. 1997, Bergman et al. 2001, Bligh et al. 1995). However, the relationship between these polymorphisms and amylose contents is not clear.
Subsequent studies demonstrated that the SNP at the splicing donor site of the first intron reduces the efficiency of GBSS prior to processing of mRNA and causes the low levels of the mature Waxy transcript, GBSS, and apparent amylose content (AAC) (Cai et al. 1998, Hirano et al. 1996, 1998, Larkin and Park 1999, 2003, Wang et al. 1995). Moreover, recent reports show a tight correlation between SNP in the first intron, coding regions, and AAC (Chen et al. 2008a, 2008b, Liu et al. 2009, Dobo et al. 2010). Two SNPs in exons 6 and 10, the A/C and C/T polymorphisms, respectively, resulted in nonsynonymous amino acid changes (Chen et al. 2008a, 2008b, Dobo et al. 2010). The combination of two SNPs in the Waxy gene, including a single G/T polymorphism at the splicing donor site of the first intron and an SNP in exon 6, can effectively differentiate all three classes of low, intermediate, and high AAC (Chen et al. 2008a). The combination of three SNPs (the single G/T polymorphism at the splicing donor site of the first intron, A/C in exon 6, and C/T in exon 10) was associated with AAC among US rice varieties and in European rice germplasm (Chen et al. 2008b, Dobo et al. 2010).
Vietnamese rice cultivars display a wide range of variation in amylose content (Suu et al. 2012). According to Nakagahra et al. (1986), the center of diversity for amylose content is the hilly area of southeast Asia. Indeed, wide variation in apparent amylose content has been reported in rice germplasm from many Asian countries, including Myanmar, northeastern India, China, and Vietnam (Aung et al. 2002, Heu 1986, Jahan et al. 2002, Nakagahra et al. 1986, Suu et al. 2012). Vietnam is one of the centers of diversity of cultivated rice, as shown in previous studies that confirmed the great diversity of Vietnamese rice landraces (Fukuoka et al. 2003, Suu et al. 2012).
In this report, we describe the variation in AAC, GBSS levels, and the SNP of the GBSSI gene in the germplasm of Vietnamese rice cultivars. We also examine the association between the variation in AAC and the SNP distribution at the splicing donor site of the first intron or in the coding region of the GBSSI gene.
Materials and Methods
Plant materials
Of the 352 Vietnamese local rice cultivars examined in this study, 98 cultivars were previously examined for AAC by Suu et al. (2012). Three rice cultivars, ‘Taichung65’ (Japonica), ‘IR36’ (Indica), and ‘EM21’ (waxy mutant) were used as a control. Out of 352 rice cultivars, 87 non waxy rice cultivars were used for EcoTILLING SNP analysis. For sequencing analysis of the GBSSI gene, 23 were chosen from 87 non waxy rice cultivars. All Vietnamese local rice cultivars are preserved in National seed genebank of Plant Resources Center, Vietnam.
Estimation of the AAC
Apparent amylose content was estimated by using the DU 7500 Spectrophotometer (Beckman), following Satoh et al. (1990). A single seed was cut into two pieces, gelatinized by treatment with 2 ml of 1 N NaOH, and kept at room temperature for 24 h. Next, 4 ml of 1 N CH3COOH and 4 ml of distilled water were added and the solution was homogenized. A 0.8-ml volume of this solution was transferred into a small tube, to which 0.2 ml of 0.2% (w/v) I2, 2% (w/v) KI, and 4 ml of distilled water were added. The AAC was determined by measuring the blue value at 680 nm and λ-max.
Western blotting
To each 20 mg of seed powder, 700 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffers was added. The material was vortexed for 3 h at room temperature and then centrifuged at 20°C, 6,000–7,000 rpm for 10 min. SDS-PAGE was performed using 10% SDS polyacrylamide gel. A polyvinylidene difluoride (PVDF) membrane was used to blot the band of proteins, and the membrane was incubated with the primary antibody (rabbit anti-wheat GBSS waxy protein). Immunoreactive proteins were detected by incubation with the secondary antibody (goat anti-rabbit IgG H + L). An equal amount of the enhanced chemiluminescence substrate (GE Healthcare) was mixed, and the membrane was incubated for 1 min in this mixture before use. The x-ray film was developed, washed, and fixed.
DNA extraction
A set of 87 nonwaxy rice cultivars was selected to represent a wide range of AACs from 352 rice cultivars. Total genomic DNAs were extracted from young leaves of 10–14-day-old seedlings using the hexadecyl trimethylammonium bromide (CTAB) method (Doyle 1991). DNAs from all samples were adjusted to a concentration of 15 ng/μl.
EcoTILLING SNP analysis
We used the EcoTILLING method to investigate the GBSSI gene in germplasm from Vietnamese rice cultivars, following the method of Comai et al. (2004) and Till et al. (2006). The Primer3 software was used to design primers for the Wx gene (http://frodo.wi.mit.edu/cgi-bin/primer3). GBSSI (AccNo Os06t0133000-01 of the ‘Nipponbare’ Wx genome sequence), which was acquired from the DNA Data Bank of Japan (DDBJ), was used to design the primer.
Heteroduplexes between the wild-type (one of the control cultivars, ‘IR36’ - Indica or ‘Taichung65’ - Japonica) and the DNA samples were formed by denaturation followed by reanneaning of PCR products. PCR was performed in a 10-μl final volume by using 0.4 U/reaction of Taq DNA polymerase (TaKaRa Ex Taq™). The forward and reverse primers were used for PCR amplification (Table 1). PCR was performed as follows: one cycle at 94°C for 4 min; followed by 29 cycles at 94°C for 30 s, 61°C for 30 s, and 72°C for 1 min 30 s. The subsequent denaturation and reannealing steps followed the method described by Suzuki et al. (2008). The digestion was carried out at 37°C for 20 min by using a crude celery juice extract (CEL I enzyme) (Anai et al. 2008). The CEL I reaction was stopped by 10 mM EDTA (final conc.). Then, 10 μl of digested product was resolved on 1.5% agarose gel in TAE buffer, run at 5 V/cm. The gel was stained with ethidium bromide and visualized under a UV transilluminator.
Table 1.
List of primers used for EcoTILLING and sequencing
| Primer | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| 1 | CCATTCCTTCAGTTCTTTGTCT | CCTTTGACCAACTCCGGCTAC |
| 2 | TAGCCGAGTTGGTCAAAGGA | TGGATTGGGGATTAGAATTTGA |
| 3 | TGCAGAGATCTTCCACAGCA | CTCCTAGCCACAACG |
| 4 | TCGCATTGGATGGATGGATGTGTA | CACTGACCTGGCAAAGAAGG |
| 5 | TTCAGGTTTGGGGAAAGACC | TGTTGTGGATGCAGAAAGCA |
| 6 | TGCACACTGCATTCTGTTCA | GTCGTACTTGGCGGTGATGT |
| 7 | ACATCACCGCCAAGTACGAC | GGTCTTCCGGCTAACTCCAC |
| 8 | TCAGAACAAATTCAGTGGCAAA | AAGCACAGGCTGGAGAAAT |
| 9 | ATTTCTCCAGCCCTGTGCTT | TTGACCGTTCGTCTTGTTCA |
| 10 | TGAACAAGACGAACGGTCAA | GCATAAAACAAAAATGGCATGG |
| 11 | TCTTATCGGACCCTGAATTTATGT | TCCTGAGTCAAACTACTGCTCCTT |
Sequencing analysis
We constructed a PCR primer set 1 including a forward primer (5′-CCATTCCTTCAGTTCTTTGTCT-3′) and a reverse primer (5′-CACTGACCTGGCAAAGAAGG-3′), which amplified the fragment containing the first exon–intron junction of the Waxy gene. To amplify the fragment containing exons 4–10, we designed the following PCR primer sets: primer set 2, containing the forward primer (5′-TAGCCGAGTTGGTCAAAGGA-3′) and reserve primer (5′-AAGCACAGGCTGGAGAAAT-3′), and primer set 3, containing the forward primer (5′-TCGCATTGGATGGATGTGTA) and reserve primer (5′-GCATAAAACAAAAATGGCATGG-3′) (Fig. 1). PCR amplification was performed using KOD_FX (TOYOBO). PCR was performed as follows: one cycle at 94°C for 2 min, followed by 29 cycles of 94°C for 15 s, 61°C for 30 s, and 68°C for 5 min. PCR products were purified using Microcon Centrifugal Filter Devices (GE Healthcare). Purified PCR products were directly sequenced from both strands using 11 primers (Table 1) with a BigDye Terminator Cycle Sequencing Kit using an ABI 3130xl Genetic Analyzer (Applied Biosystems). Data were analyzed using ClustalW (http://clustalw.ddbj.nig.ac.jp).
Fig. 1.
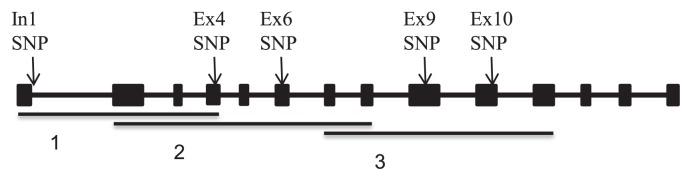
Schematic representation of GBSSI gene structure and the position of single-nucleotide polymorphism in GBSSI gene. The boxes and the lines between boxes indicate the exon and the intron, respectively. Lines under the gene and number from 1 to 3 indicate the region amplified by PCR and the primer set used for amplification, respectively.
Results
Variation in GBSS levels and AAC
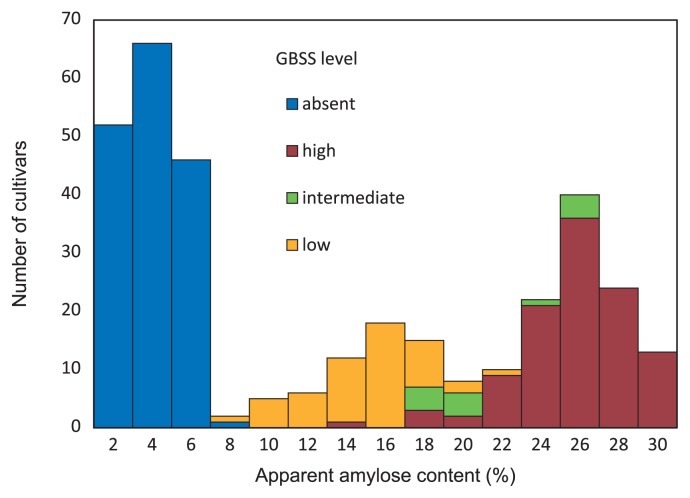
Amylose is synthesized by GBSSI protein. Three control cultivars, ‘IR36,’ ‘Taichung65,’ and ‘EM21,’ showed high, low, and absent GBSS levels, respectively, according to the intensity of 60 kDa bands by Western blot analysis (Fig. 2). Based on the amount of GBSS protein, 352 rice cultivars were divided into four classes, high in 122 cultivars (34.6%), intermediate in 13 cultivars (3.7%), low in 52 cultivars (14.8%), and absent in 165 cultivars (46.9%) (Figs. 2, 3). Intermediate between ‘IR36’ and ‘Taichung65’ was defined as intermediate (lane 5 in Fig. 2)
Fig. 2.
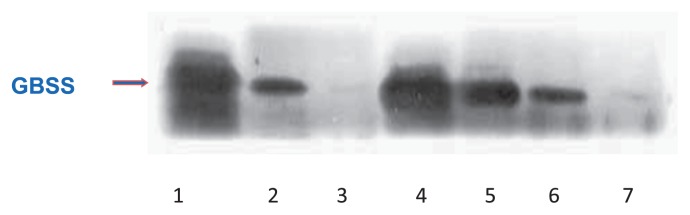
The expression level of granule-bound starch synthase I by Western blotting analysis in Vietnamese rice cultivars. Lane 1 to lane 3: Control cultivars: Lane 1: ‘IR36’; Lane 2: ‘Taichung65’; Lane 3: ‘EM21’; Lane 4 to lane 7: The expression level of granule-bound starch synthase I of local rice cultivars were classified into four types (high, intermediate, low and absent) as compared with that of ‘IR36’. Lane 4: ‘Accession number 2479: Te Moc chau’; Lane 5: ‘Accession number 2217: Me noll’; Lane 6: ‘Accession number 2596: Khau se tao’; Lane 7: ‘Accession number 2468: Khau pe’.
Fig. 3.
The relationship between apparent amylose content and the expression level of granule-bound starch synthase in Vietnamese rice cultivars.
The AAC of Vietnamese rice cultivars varied from 0 to 32% (Fig. 3). Based on varied range of AAC, rice cultivars can be grouped into five groups: glutinous (0–6%), very low (6–12%), low (12–20%), intermediate (20–25%), and high (25–32%) (Suu et al. 2012) (Table 2 and Fig. 3). Nearly half (46.9%) of the cultivars belonged to the glutinous group (0–6%), while the other 53.1% belonged to nonglutinous groups (>6%). The nonglutinous group was divided into four groups on the basis of AAC: very low (3.4% of cultivars), low (15.1%), intermediate (12.2%), and high (22.4%).
Table 2.
Variation in apparent amylose content (%) in Vietnamese rice cultivars
| Apparent amylose content (%) | Amylose class | Number of cultivars | Frequency (%) |
|---|---|---|---|
| 0–6 | Glutinous | 165 | 46.9 |
| 7–12 | Very low | 12 | 3.4 |
| 12–20 | Low | 53 | 15.1 |
| 20–25 | Intermediate | 43 | 12.2 |
| 25–32 | High | 79 | 22.4 |
The relationship between GBSS level and AAC in Vietnamese rice cultivars is shown in Fig. 3. Cultivars with different GBSS levels showed continuous variation in AAC. All the cultivars with no GBSS exhibited the glutinous phenotype (waxy endosperm), and the AAC of the varieties was distributed from 0% to 6%. In the nonglutinous group, the varieties with low, intermediate, and high levels of GBSS varied in AAC from 8% to 26%, from 14% to 26%, and from 18% to 32%, respectively (Fig. 3).
SNPs of the GBSS1 gene
The SNPs in GBSSI gene of 23 nonglutinous rice cultivars were determined by the EcoTILLING and the sequencing of the genomic DNA. The G/T polymorphism at the splicing donor site of the first intron and four SNPs in GBSSI gene were found in the 23 rice cultivars (Table 3). Based on the combination of the G/T polymorphism and SNPs in exon 4 (Ex4), exon 6 (Ex6), exon 9 (Ex9), and exon 10 (Ex10), the cultivars were classified into seven types.
Table 3.
Waxy gene haplotypes and geographical distribution of Wx alleles in Vietnamese local rice cultivars
| Wx alleles | G/T in first intron | Exon 4 | Amino acid | Exon 6 | Amino acid | Exon 9 | Amino acid | Exon 10 | Amino acid | GBSS level | Apparent amylose content (%) | Geographical distribution |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TC65 | T | GAC | Asp | TAT | Tyr | CCT | Pro | CCT | Pro | Low | 20 | |
| IR36 | G | GAC | Asp | TAT | Tyr | CCC | Pro | TCT | Ser | High | 23 | |
| Wxg1 (5*) | G | GAC | Asp | TAT | Tyr | CCC | Pro | TCT | Ser | High | 20–32 | Northwest, Northeast |
| Wxg2 (2) | G | GAC | Asp | TAT | Tyr | CCC | Pro | CCT | Pro | High, low | 13–26 | Northeast |
| Wxg3 (3) | G | GGC | Gly | TAT | Tyr | CCC | Pro | CCT | Pro | High | 13–18 | Northwest |
| Wxg4 (2) | G | GAC | Asp | TCT | Ser | CCT | Pro | CCT | Pro | High | 17–20 | Northwest, South central coast |
| Wxt1 (1) | T | GAC | Asp | TAT | Tyr | CCT | Pro | CCT | Pro | Low | 17 | Northwest |
| Wxt2 (2) | T | GAC | Asp | TAT | Tyr | CCC | Pro | CCT | Pro | Low | 15–16 | Northeast, Central highland |
| Wxt3 (8) | T | GAC | Asp | TCT | Ser | CCT | Pro | CCT | Pro | Low | 9–16 | Northwest, Northeast, Central highland |
The number in parenthesis is the number of rice cultivars in this group.
The 23 cultivars were divided into two groups based on the G/T polymorphism at the splicing donor site of the first intron. In the first group (‘IR36’ type), containing 12 cultivars, the splicing donor site of the first intron was AGGTATA. The AAC of this group ranged from 13 to 30. In the second group (‘Taichung65’ type), containing 11 cultivars, the splicing donor site of the first intron was AGTTATA. The AAC of the second group varied from 9 to 20.
A sequence analysis of 23 rice cultivars indicated seven allelic patterns: Wxg1 (G-A-A-C-T), Wxg2 (G-A-A-C-C), Wxg3 (G-G-A-C-C), Wxg4 (G-A-C-T-C), Wxt1 (T-A-A-T-C), Wxt2 (T-A-A-C-C), and Wxt3 (T-A-C-T-C) (Table 3). The first letter in the allelic pattern corresponds to the G/T at the splicing donor site of the first intron, the second letter to the A/G polymorphism in Ex4, the third letter to the A/C polymorphism in Ex6, the fourth letter to the C/T polymorphism in Ex9, and the fifth letter to the C/T polymorphism in Ex10. The Wxg1 pattern (G-A-A-C-T - ‘IR36’ type) was found in five cultivars with intermediate and high AAC, whereas the Wxg4 pattern (G-A-C-T-C) was found in two cultivars with low AAC. The Wxg3 pattern (G-G-A-C-C) allele was associated with the low AAC class, while the Wxg2 pattern (G-A-A-C-C) was associated with both low and high AAC. As the cultivars in Wxg2 contained both low and high levels of GBSS, Wxg2 cultivars were divided into subgroups, Wxg2-1 and Wxg2-2 (Fig. 4). The Wxt1 pattern (T-A-AT-C-‘Taichung65’ type) was associated with low AAC. The Wxt2 pattern (T-A-A-C-C) was found in two low AAC cultivars. The Wxt3 pattern (T-A-C-T-C) was found in eight cultivars with low AAC.
Fig. 4.
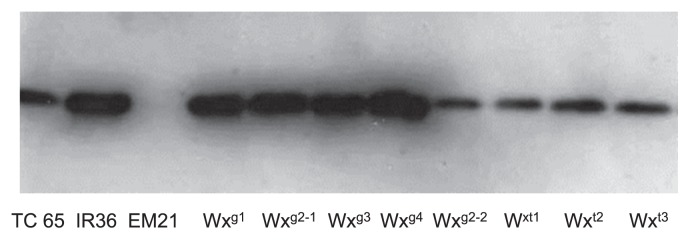
The expression level of granule-bound starch synthase in Vietnamese rice cultivars by Western blotting analysis, grouped according to the single-nucleotide polymorphism at the 5′ splicing donor site of the first intron and at the coding region of the granule-bound starch synthase I gene.
We found four SNPs on four exons (Ex4, Ex6, Ex9, Ex10) of the GBSSI gene. The SNP at the position of 3,013 bp from ATG starting site was a synonymous C to T substitution (Ex9). On the other hand, A to C substitution at 2,016 bp (Ex4), A to C substitution at 2,385 bp (Ex6), and C to T substitution at 3,377 bp (Ex10) were responsible for non-synonymous changes from Asp to Gly, from Tyr to Ser, and from Pro to Ser, respectively (Table 3).
Waxy gene haplotypes and geographical distribution
Table 3 showed the geographical distribution of Waxy allele haplotypes. Northwest regions is richness diversity of Waxy allele with the distribution of all Waxy haplotypes were found except Wxg2. Most of Wxt2 and Wxt3 cultivars with low AAC were detected in Northeast and Central highland of Vietnam. The Wxg3 and Wxt1 were distributed only in Northwest, Wxg1 was found in Northeast regions.
Discussion
One of the useful tools for identifying the GBSS allele and explaining variations in AC is the CT dinucleotide repeats in Ex1 (Ayres et al. 1997, Bergman et al. 2001, Bligh et al. 1995). For example, the CT dinucleotide repeats can explain 75.6% of the variation in AAC in international rice germplasm (Chen et al. 2008a) and 81.2% of the variation in AAC in European rice germplasm (Dobo et al. 2010), However, recent studies indicate that the combination of an SNP in the splicing donor site of the first intron, Ex6, or Ex10 can more effectively distinguish all three classes of apparent amylose than the CT dinucleotide repeats can (Chen et al. 2008a, Dobo et al. 2010). The present study identified a diversity of nucleotides in the GBSSI gene sequence among 23 rice landrace cultivars. As the SNP C-to-T change in Ex9 was a silent mutation, we present here our finding of an association between the AAC and the combination of an SNP in the first intron and an SNP in Ex4, Ex6, or Ex10 of the GBSSI gene in Vietnamese rice germplasm.
The G/T SNP at the splicing donor site of the first intron was observed in several of the Vietnamese rice cultivars. The very low and low AAC levels were tightly associated with the T SNP in the first intron in the rice cultivars we investigated. This finding was consistent with previous studies that showed that the non waxy cultivars, which carried T in the first intron, decreased levels of AAC (Bligh et al. 1998, Cai et al. 1998, Hirano et al. 1998, Isshiki et al. 1998, Olsen and Purugganan 2002, Wang et al. 1995, Yamanaka et al. 2004). Thus, T SNP in the first intron may be used to discriminate between the low AAC and the intermediate to high AAC classes (Ayres et al. 1997, Hirano et al. 1998, Isshiki et al. 1998, Larkin and Park 1999, Liu et al. 2009). Those studies assumed that the T SNP in the first intron was attributable to control the processing and steady levels of GBSS mRNA and the subsequent low amylose in these cultivars.
In the present study, three classes of low, intermediate and high AAC were found in cultivars with the presence of G in the first intron. This finding contrasted with the conclusion of some other studies (e.g., Ayres et al. 1997, Dobo et al. 2010, Larkin and Park 1999). Those studies concluded that intermediate or high AAC of cultivars from US and European rice germplasms was associated with G genotype.
The association between the combination of SNPs in the first intron, Ex6, or Ex10 and low amylose content in US, European, Asian rice germplasms was reported (Chen et al. 2008b, Dobo et al. 2010). However, whereas most rice varieties from Europe had the combination of TAC, the combination of TCC was predominant in US rice germplasm. In the present study, more cultivars had the TCC combination (Wxt3) than the TAC combination (Wxt1 and Wxt2), suggesting that the Wxt3 allele was predominant among low amylose rice varieties from Vietnam.
Regarding the relationship between the combination of three SNPs in In1, Ex6 and Ex10 and AAC, we found that the Wxt1 (TAC), Wxt2 (TAC) and Wxt3 (TCC) were associated with low AAC. This finding supposed that the change of A to C in Ex6 and T to C in Ex10 may affect the enzyme activity, reducing the amylose content. Our results suggest that the combination between three SNPs in the first intron, Ex6, or Ex10 (TAC and TCC) may be used as a tool to distinguish low amylose content from other classes of amylose content (intermediate and high). In Chen’s study and Dobo’s study, Waxy-H (G in In1, A in Ex6, C in Ex10) had intermediate or high ACC, whereas in this study, Wxg2 and Wxg3 (Waxy-H) had both low and high AAC. Thus, this finding contrasts with the results of previous studies (Chen et al. 2008b, Dobo et al. 2010). It is interesting to note that in the present study, all Wxg1 cultivars (Waxy-HH) having the combination of three SNPs (G in In1, A in Ex6 and T in Ex10) showed intermediate or high AAC. Our finding supports the previous studies that the combination of three SNPs in In1, Ex6 and Ex10 (GAT) could be associated with intermediate or high amylose (Chen et al. 2008b, Dobo et al. 2010, Mikami et al. 2008).
In the present study, we also found that ranges in AAC of Wxt2 and Wgt3, which differ only by Exon 6 and 9, are also overlapping with each other (Table 3), suggesting the influence of other genes or genetic background. Similar observation was recorded in Wxg2 and Wxg4. Ranges in AAC of Wxg2 and Wxg4, which differ only by Ex6, are overlapping with each other (Table 3), suggesting the influence of other genes or genetic background.
In our study of 23 rice cultivars, most of the cultivars with low amylose had the C SNP in Ex10, whereas all the cultivars with intermediate and high amylose had the T SNP in Ex10. The results suggested that the low amylose content observed in Vietnamese local rice germplasm might be attributable to the SNP in Ex10, not by the SNP at the splicing donor site of the first intron.
Recently, Teng et al. (2012) demonstrated that the Ex10 SNP was the cause of the AAC phenotypic diversity between the high amylose sub-class I (24.36–25.20%) and high amylose sub-class II (25.81–26.19%) lines, and used this SNP to subdivide the Wxa allele into two subgroups demonstrating high AAC. The lines with serine residue from the amino acid substitution had higher GBSS activity, suggesting that the Wx protein might be phosphorylated. This SNP was responsible for amino acid change to Ser, which might play an important role in regulating GBSS activity through phosphorylation. Rice pasting viscosity determined by Rapid Visco Analyzer (RVA), is used to estimate cooking and eating quality characteristic. The association of SNP in Waxy gene with viscosity characteristics was reported in some studies (Chen et al. 2008b, Larkin et al. 2003, Larkin and Park 2003). Larkin et al. (2003) reported that the Waxy locus has significant effects on viscosity characteristics such as peak viscosity, hot paste viscosity, cool paste viscosity, breakdown and setback viscosity. Chen et al. (2008b) demonstrated that the SNP in Ex10 associated with RVA profiles differ between the two high AAC types. Recently, Traore et al. (2011) reported that the Waxy Ex10 SNP marker was associated with most RVA profiles. Furthermore, Tran et al. (2011) also found that the SNP on Ex10 of the Wx gene explained a significant component of the differences in gel consistency. The results from previous studies suggested that the SNP in Ex10 can be used in molecular breeding program focused on quality improvement. In the present study, we assume that this polymorphism results in the substitution of the polar amino acid serine for the non-polar proline in Ex10, which might lead to a change in GBSS activity and thus reduce amylose content. Our results suggested that the T genotype at SNP in Ex10 could be used as a marker for discriminating the high amylose class. The one exception to this rule was the Wxg2 group, which included both low and high AAC levels. The expression of GBSS in this group suggested that other factors might contribute to the variation in AAC among these rice varieties.
Our research confirmed that the combination of the G/T SNP in the first intron and SNPs in Ex4, Ex6, Ex9, and Ex10 were related to the AAC. The Wxg3 cultivars had opaque endosperm types and low AAC. Two SNPs, G in the first intron and G in Ex4, were observed in Wxop cultivars, and the C SNP in Ex10 was also found in Wxg3 cultivars. Thus, the Wxg3 in this study was similar to the Wxop and Wxhp types reported by other studies (Liu et al. 2009, Mikami et al. 1999, 2008). Mikami et al. (1999) reported that the level of gene product bound to starch granules was markedly reduced in the Wxop line, resulting in a lower amylose content. Apparently the combination of SNPs at the splicing donor site of the first intron, Ex4, and Ex10 affects the binding of GBSS to starch granules, causes a low activity level in GBSS, and results in low AC of these cultivars.
In nonwaxy rice, the presence of all amylose classes in Vietnamese rice germplasm support the fact that the Southeast Asian countries is the center of genetic diversity for amylose content in rice (Aung et al. 2002, Fukuoka et al. 2003, Heu 1986, Liu et al. 2009, Nakagahra et al. 1986, Suu et al. 2012). Those results imply that the local rice resources in Vietnam are rich in spontaneous mutations that play a vital role in rice quality breeding programs.
The findings of this research will contribute to our understanding of the molecular mechanisms of amylose biosynthesis that are controlled by naturally occurring alleles at the Wx locus in rice.
Acknowledgments
This work was partially supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (grant no. 21380008 and 25257413 to T.K.) and by JSPS Ronpaku (Dissertation Ph.D) Program (VAST-10933).
Literature Cited
- Anai, T., Yamada, T., Hideshima, R., Kinoshita, T., Rahman, S.M. and Takagi, Y. (2008) Two high-oleic-acid soybean mutants, M23 and KK21, have disrupted microsomal omega-6 fatty acid desaturase, encoded by GmFAD2-1a. Breed. Sci. 58: 447–452 [Google Scholar]
- Aung, P.P., Nishi, A., Kumamaru, T. and Satoh, H. (2002) Genetic variation of granule-bound starch synthase (GBSS) level in Myanmar local rice cultivars. Rice Genet. Newslett. 19: 67–69 [Google Scholar]
- Ayres, N.M., McClung, A.M., Larkin, P.D., Bligh, H.F.J., Jones, C.A. and Park, W.D. (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor. Appl. Genet. 94: 773–781 [Google Scholar]
- Bergman, C.J., Delgado, J.T., McClung, A.M. and Fjellstrom, R.G. (2001) An improved method for using a microsatellite in the rice waxy gene to determine amylose class. Cereal Chem. 78: 257–260 [Google Scholar]
- Bligh, H.F.J., Till, R.I. and Jones, C.A. (1995) A microsatellite sequence closely linked to the Waxy gene of Oryza sativa. Euphytica 86: 83–85 [Google Scholar]
- Cai, X.L., Wang, Z.Y., Xing, Y.Y., Zhang, J.L. and Hong, M.M. (1998) Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14: 459–465 [DOI] [PubMed] [Google Scholar]
- Chen, M.H., Bergman, C., Pinson, S. and Fjellstrom, R. (2008a) Waxy gene haplotypes: Associations with apparent amylose content and the effect by the environment in an international rice germplasm collection. J. Cereal Sci. 47: 536–545 [Google Scholar]
- Chen, M.H., Bergman, C.J., Pinson, S.R.M. and Fjellstrom, R.G. (2008b) Waxy gene haplotypes: Associations with pasting properties in an international rice germplasm collection. J. Cereal Sci. 48: 781–788 [Google Scholar]
- Comai, L., Young, K., Till, B.J., Reynolds, S.H., Greene, E.A., Codomo, C.A., Enns, L.C., Johnson, J.E., Burtner, C., Odden, A.R.et al. (2004) Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 37: 778–786 [DOI] [PubMed] [Google Scholar]
- Denyer, K.A.Y., Johnson, P., Zeeman, S. and Smith, A.M. (2001) The control of amylose synthesis. J. Plant Physiol. 158: 479–487 [Google Scholar]
- Dobo, M., Ayres, N., Walker, G. and Park, W.D. (2010) Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J. Cereal Sci. 52: 450–456 [Google Scholar]
- Doyle, J.J. (1991) DNA protocols for plants-CTAB total DNA isolation. In: Hewitt, G.M. and Johnston, A. (eds.) Molecular Techniques in Taxonomy, Springer, Berlin Heidelberg New York, pp. 283–293 [Google Scholar]
- Fukuoka, S., Alpatyeva, N.V., Ebana, K., Luu, N.T. and Nagamine, T. (2003) Analysis of Vietnamese rice germplasm provides an insight into Japonica rice differentiation. Plant Breed. 122: 497–502 [Google Scholar]
- Hannah, L.C. and James, M. (2008) The complexities of starch biosynthesis in cereal endosperms. Curr. Opin. Biotechnol. 19: 160–165 [DOI] [PubMed] [Google Scholar]
- Heu, M.H. (1986) Inheritance of chalkiness of brown rice found in non-glutinous rice cultivar “Pokhareli Mashino”. Korean J. Breed. 18: 162–166 [Google Scholar]
- Hirano, H.Y. and Sano, Y. (1991) Molecular characterization of the waxy locus of rice (Oryza sativa). Plant Cell Physiol. 32: 989–997 [Google Scholar]
- Hirano, H.Y., Eiguchi, M. and Sano, Y. (1996) A point mutation, G to T, causes the differentiation of the Wxb allele from Wxa allele, which is specific to Japonica rice. Rice Genet. Newslett. 13: 148–149 [Google Scholar]
- Hirano, H.Y., Eiguchi, M. and Sano, Y. (1998) A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 15: 978–987 [DOI] [PubMed] [Google Scholar]
- Hirose, T. and Terao, T. (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220: 9–16 [DOI] [PubMed] [Google Scholar]
- Isshiki, M., Morino, K., Nakajima, M., Okagaki, R.J., Wessler, S.R., Izawa, T. and Shimamoto, K. (1998) A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 15: 133–138 [DOI] [PubMed] [Google Scholar]
- Jahan, M.S., Kumamaru, T., Hamid, A. and Satoh, H. (2002) Diversity of granule-bound starch synthase (GBSS) level in Bangladesh rice cultivars. Rice Genet. Newslett. 19: 69–71 [Google Scholar]
- Larkin, P.D. and Park, W.D. (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 40: 719–727 [DOI] [PubMed] [Google Scholar]
- Larkin, P.D., McClung, A.M., Ayres, N.M. and Park, W.D. (2003) The effect of the Waxy locus (Granule Bound Starch Synthase) on pasting curve characteristics in specialty rice (Oryza sativa L.). Euphytica 131: 243–253 [Google Scholar]
- Larkin, P.D. and Park, W.D. (2003) Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol. Breed. 12: 335–339 [Google Scholar]
- Liu, L., Ma, X., Liu, S., Zhu, C., Jiang, L., Wang, Y., Shen, Y., Ren, Y., Dong, H., Chen, L.et al. (2009) Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71: 609–626 [DOI] [PubMed] [Google Scholar]
- Mikami, I., Aikawa, M., Hirano, H.Y. and Sano, Y. (1999) Altered tissue-specific expression at the Wx gene of opaque mutants in rice. Euphytica 105: 91–97 [Google Scholar]
- Mikami, I., Uwatoko, N., Ikeda, Y., Yamaguchi, J., Hirano, H.Y., Suzuki, Y. and Sano, Y. (2008) Allelic diversification at the Wx locus in landraces of Asian rice. Theor. Appl. Genet. 116: 979–989 [DOI] [PubMed] [Google Scholar]
- Nakagahra, M., Nagamine, T. and Okuno, K. (1986) Spontaneous occurrence of low amylose genes and geographical distribution of amylose content in Asian rice. Rice Genet. Newslett. 3: 46–48 [Google Scholar]
- Nakamura, Y., Sato, A. and Juliano, B.O. (2006) Short-chain-length distribution in debranched rice starches differing in gelatinization temperature or cooked rice hardness. Starch/Stärke 58: 155–160 [Google Scholar]
- Okagaki, R.J. (1992) Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol. Biol. 19: 513–516 [DOI] [PubMed] [Google Scholar]
- Olsen, K.M. and Purugganan, M.D. (2002) Molecular evidence on the origin and evolution of glutinous rice. Genetics 162: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y. (1984) Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68: 467–473 [DOI] [PubMed] [Google Scholar]
- Sano, Y., Katsumata, M. and Amano, E. (1985) Correlation between the amount of amylose and Wx protein in rice endosperm. SABRAO J. 17: 121–127 [Google Scholar]
- Sano, Y., Katsumata, M. and Okuno, K. (1986) Genetic studies of speciation in cultivated rice. 5. Inter- and intraspecific differentiation in the waxy gene expression of rice. Euphytica 35: 1–9 [Google Scholar]
- Satoh, H., Ronald, R.X. and Katayama, T.C. (1990) On amylose content of cultivated rice collected in Madagascar, 1988. Kagoshima University Research Center South Pacific, Occasional Papers 18: 83–91 [Google Scholar]
- Suu, T.D., Hoai, T.T.T., Hoa, N.T.L., Loan, H.M., Yen, D.B., Kumamaru, T. and Satoh, H. (2012) Variation on grain quality in Vietnamese rice cultivars collected from Central Vietnam. J. Fac. Agr., Kyushu Univ. 57: 365–371 [Google Scholar]
- Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H., Moriguchi, K., Nagato, Y. and Kurata, N. (2008) MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279: 213–223 [DOI] [PubMed] [Google Scholar]
- Takeda, Y. and Hizukuri, S. (1987) Structures of branched molecules of amyloses of various origins, and molar fractions of branched and unbranched molecules. Carbohydr. Res. 165: 139–145 [Google Scholar]
- Takeda, Y., Hizukuri, S. and Juliano, B.O. (1987) Structure of rice amylopectins with low and high affinities for iodine. Carbohydr. Res. 168: 79–88 [Google Scholar]
- Teng, B., Zeng, R., Wang, Y., Liu, Z., Zhang, Z., Zhu, H., Ding, X., Li, W. and Zhang, G. (2012) Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol. Breed. 30: 583–595 [Google Scholar]
- Till, B.J., Zerr, T., Conai, L. and Henikoff, S. (2006) A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 1: 2465–2477 [DOI] [PubMed] [Google Scholar]
- Tran, N.A., Daygon, V.D., Resurreccion, A.P., Cuevas, R.P., Corpuz, H.M. and Fitzgerald, M.A. (2011) A single nucleotide polymorphism in the Waxy gene explains a significant component of gel consistency. Theor. Appl. Genet. 123: 519–525 [DOI] [PubMed] [Google Scholar]
- Traore, K., McClung, A.M., Chen, M-H. and Fjellstrom, R. (2011) Inheritance of flour paste viscosity is associated with a rice Waxy gene exon 10 SNP marker. J. Cereal Sci. 53: 37–44 [Google Scholar]
- Wang, Z.Y., Zheng, F.Q., Shen, G.Z., Gao, J.P., Snustad, D.P., Li, M.G., Zhang, J.L. and Hong, M.M. (1995) The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7: 613–622 [DOI] [PubMed] [Google Scholar]
- Yamanaka, S., Nakamura, I., Watanabe, K.N. and Sato, Y.-I. (2004) Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 108: 1200–1204 [DOI] [PubMed] [Google Scholar]