Abstract
Influences of allelic variations in starch synthesis-related genes (SSRGs) on rice grain quality were examined. A total of 187 nonglutinous Korean rice varieties, consisting of 170 Japonica and 17 Tongil-type varieties, were grown in the field and in two greenhouse conditions. The percentages of head rice and chalky grains, amylose content, alkali digestion value, and rapid visco-analysis characteristics were evaluated in the three different environments. Among the 10 previously reported SSRG markers used in this study, seven were polymorphic, and four of those showed subspecies-specific allele distributions. Six out of the seven polymorphic SSRG markers were significantly associated with at least one grain quality trait (R2 > 0.1) across the three different environments. However, the association level and significance were markedly lower when the analysis was repeated using only the 170 Japonica varieties. Similarly, the significant associations between SSRG allelic variations and changes in grain quality traits under increased temperature were largely attributable to the biased allele frequency between the two subpopulations. Our results suggest that within Korean Japonica varieties, these 10 major SSRG loci have been highly fixed during breeding history and variations in grain quality traits might be influenced by other genetic factors.
Keywords: rice, grain quality, starch synthesis-related genes, high temperature
Introduction
The increase in temperature as a result of global warming is threatening stable crop production in many countries. The grain yield and quality are degenerating in rice, a staple for half of the world’s population. The global average temperature rose by 0.85°C during 1880–2012 and an additional increase of 1.0°C–3.7°C is expected for 2081–2100 relative to 1986–2005 (IPCC 2013). At the flowering stage, excessive heat causes spikelet sterility and yield reduction in rice. When panicles are exposed to temperature higher than 35°C for more than 1 hour, spikelet fertility is significantly reduced in most rice varieties because of deterioration in pollen viability and anther dehiscence (Jagadish et al. 2007, 2010, Prasad et al. 2006, Rang et al. 2011, Ye et al. 2012). During the grain-filling stage, temperature higher than the optimum causes lower grain weights and decreases the percentage of ripened grains because of accelerated respiration and delayed dry matter production and endosperm cell enlargement (Mohammed and Tarpley 2009, Morita et al. 2005, She et al. 2010, Zakaria et al. 2002). Furthermore, the appearance and palatability of rice are diminished under high temperature because of loose and irregular starch granule formation (Chun et al. 2009, Ishimaru et al. 2009, Tanaka et al. 2009). Starch biosynthesis is hindered under high temperature, as the expressions of starch-synthesizing genes are suppressed, while those of starch-degrading genes are enhanced (Yamakawa et al. 2007). Therefore, maintaining stable yield and quality under high temperature is one of the most important goals in rice breeding to cope with climate change.
Starch is the main component of rice, constituting 90% of the milled grain. Thus, the physicochemical properties of starch, such as amylose content (AC), gel consistency, gelatinization temperature (GT), and rapid visco-analysis (RVA) characteristics, greatly affect the appearance and the processing, cooking, and eating qualities of rice grains (Bao et al. 2006c, Cruz et al. 1989, Fitzgerald et al. 2009, Tian et al. 2009). Starch biosynthesis is mainly catalyzed by the enzymes ADP-glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), starch synthase (SS), starch branching enzyme (SBE), and starch debranching enzyme (SDBE) (Jeon et al. 2010, Tian et al. 2009). Naturally, allelic variations in starch synthesis-related genes (SSRGs) affect phenotypic variations in grain quality traits. For example, the G/T single nucleotide polymorphism (SNP) in the 5′ leader intron of GBSSI, which influences the splicing efficiency of the gene, explains AC variations in rice along with the (CT)n microsatellite in the first exon of the same gene (Ayres et al. 1997, Bao et al. 2006a, Bligh et al. 1995, Dobo et al. 2010, Larkin and Park 2003). Also, the TT/GC SNPs in SSIIa that result in a leucine-to-phenylalanine substitution affect the GT and alkali digestion value (ADV) of rice (Bao et al. 2006b, Nakamura et al. 2005, Umemoto et al. 2004, Umemoto and Aoki 2005). Allelic effects of SBE and SDBE on grain quality traits have been documented as well (Bao et al. 2006a, Han et al. 2004, Larkin et al. 2003).
Recently, advanced sequencing technology has allowed large-scale identification of sequence polymorphisms in SSRGs, and their associations with the variations in grain quality traits have been widely investigated. By sequencing the entire loci of 18 SSRGs from 16 core entries of diverse rice germplasms and analyzing the allelic effects of SSRGs on starch physicochemical properties, Tian et al. (2009) identified the roles of different SSRGs affecting rice cooking and eating qualities. Lee et al. (2009) sequenced complete or partial genic regions of eight SSRGs from 27 rice varieties with diverse genetic backgrounds and developed 25 cleaved amplified polymorphic sequence (CAPs) or derived CAPs markers, among which SNPs in GBSSI and AGPase-S were strongly associated with AC. Yan et al. (2011) developed 43 sequence tagged site (STS) and CAPs markers for genotyping glutinous rice germplasms by sequencing 17 SSRGs from 13 representative varieties and studied the roles of SSRGs in controlling the RVA characteristics of 118 glutinous rice varieties. Through massive parallel sequencing, Kharabian-Masouleh et al. (2011) found 501 SNPs and 113 insertion/deletion (InDels) markers in 17 SSRGs from 233 Australian rice breeding lines. Among those, 66 functional SNPs were chosen for association analysis; their results reaffirmed that the GBSSI G/T SNP and SSIIa TT/GC SNPs are the major SSRG polymorphisms explaining variations in AC and GT, respectively (Kharabian-Masouleh et al. 2012).
While extensive studies have been conducted on the effects of SSRG allelic variations on grain quality traits, few have explored the relationships between SSRG allelic variations and the changes in grain quality traits under increased temperature. One of the few examples is the study of Inukai and Hirayama (2010), which showed that the reduction in AC under high temperature was greater in the genotypes with the Wxb allele of GBSSI than in those with the Wxa allele. Hence, more research is required to identify SSRG alleles that are useful for maintaining stable grain quality under high temperature. In this study, we investigated the effects of allelic variations in 10 previously reported SSRG markers on the phenotypic variations in grain quality traits of 187 nonglutinous Korean rice varieties under ambient (field) and two greenhouse conditions. Further analyses were carried out to identify relationships between SSRG allelic variations and the changes in grain quality traits under different temperatures.
Materials and Methods
Plant materials and growth conditions
A total of 187 nonglutinous Korean rice varieties consisting of 170 Japonica and 17 Tongil-type (derived from an Indica-Japonica cross) were used in this study. These varieties have been developed since 1979, when ‘Taebaeg’ was released, while most of the other varieties have been released to farmers since the late 1980’s. The seeds were sown in a seedling nursery at the Rice Experimental Station of the Department of Rice and Winter Cereal Crop (35.9343°N, 126.9259°E), National Institute of Crop Science, Iksan, South Korea, on May 4, 2010. On June 7, 20 plants of each variety were transplanted in a row in the field and two greenhouses with a planting density of 30 cm × 15 cm, one plant per hill. The greenhouses were covered with the ‘F-clean® Clear film’ (60 μm thickness with 94% light transmission; AGC GREEN-TECH Co., LTD., Tokyo, Japan) to mitigate sun light shadiness by covering film. The greenhouses were also equipped with temperature sensor-controlled ventilation fans at the top and manual side windows to avoid over-heating. To create different temperature conditions in the two greenhouses, the ventilation fans were set to operate when the temperature reached 32°C in greenhouse I and 34°C in greenhouse II. Side windows were opened to two-thirds of the full height in greenhouse I and half of the full height in greenhouse II. Temperatures in the field and the two greenhouses were recorded every 30 minutes from transplanting to harvest with SK-L200T II data loggers (SATO, Tokyo, Japan). N, P2O5, and K2O fertilizers were applied at 90 kg/ha, 45 kg/ha, and 57 kg/ha, respectively. The fertilization split ratio was as follows: 50% as basal fertilizer, 20% at the tillering stage, and 30% at the panicle initiation stage for N, 100% as basal fertilizer for P2O5, and 70% as basal fertilizer and 30% at the panicle initiation stage for K2O.
Molecular marker analysis
Genomic DNA of the 187 rice varieties was extracted from the fresh young leaves of 2-week-old seedlings using a BioSprint 96 DNA Plant Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. PCR was carried out using AccuPower PCR Premix (Bioneer, Daejeon, Korea) with 10 pmol forward and reverse primers and 50–100 ng template DNA in a 20-μl total reaction volume. The primer sequences of 10 previously reported SSRG markers are listed in Table 1. In a MyGenie 96 thermal block (Bioneer), amplifications were carried out with the following conditions: initial denaturation at 95°C for 3 min followed by 35 cycles of 45 s at 94°C, 45 s at 55–60°C, and 1 min at 72°C, and a final extension at 72°C for 5 min. Banding patterns were visualized by electrophoretic separation of the PCR products on 1.0% agarose (AGPase-L InDel, SSIIa TT/GC SNPs, and SBEI InDel) or 8.0% polyacrylamide gels (GBSSI InDel and SDBE InDel). For other SSRG markers, PCR product was digested with an appropriate restriction enzyme before the electrophoresis on 1.0% agarose gel (SBEIII C/G SNP) or 8.0% polyacrylamide gel (AGPase-S C/T and A/T SNPs, and GBSSI T/G and T/C SNPs). Detailed PCR and electrophoresis procedures for each marker are described in the references provided in Table 1. Based on the detected banding patterns of SSRG markers, allele types were assigned to each rice variety.
Table 1.
Starch synthesis-related gene (SSRG) markers used for genotyping 187 nonglutinous Korean rice varieties Gene Chromosome
| Gene | Chromosome | Marker | Sequence (5′→3′)c | REd | PCR product size (bp) | Reference |
|---|---|---|---|---|---|---|
| AGPase-La | 3 | InDel (TEb) | F: GCTCCATTGAGTTGAATTGGC | – | In: 1104 | Mo et al. 2012b |
| R: CCGCATGGACTACATGGAC | Del: 704 | |||||
|
| ||||||
| AGPase-Sa | 8 | SNP (C/T) | F: ATGTAAACTGTTAGAATCGAAGA | EcoR V | C: 305 | Lee et al. 2009 |
| R: CAAGATCTATGTGCTGACACAAT | T: 281 + 24 | |||||
| SNP (A/T) | F: TGTGTGTTTGTGGTAGGGTGAAGC | Alu I | A: 338 | Lee et al. 2009 | ||
| R: CGATGCAAATTGGATTTGGATAG | T: 315 + 23 | |||||
|
| ||||||
| GBSSI (Wx) | 6 | SNP (T/G) | F: CTTTGTCTATCTCAAGACAC | Acc I | T: 253 | Ayres et al. 1997, Bao et al. 2006a |
| R: TTTCCAGCCCAACACCTTAC | G: 127 + 126 | |||||
| InDel | F: TGCAGAGATCTTCCACAGCA | – | In: 196 | Wanchana et al. 2003 | ||
| R: GCTGGTCGTCACGCTGAG | Del: 173 | |||||
| SNP (T/C) | F: AGAAGGGGTGAGGCTTTGAACCCGG | Nhe I | T: 295 | Lee et al. 2009 | ||
| R: CTGGATGAGTCCACAGGGCTCGAAG | C: 244 + 51 | |||||
|
| ||||||
| SSIIa | 6 | SNPs (TT/GC)e | F1: CTGGATCACTTCAAGCTGTACGAC | – | TT: 1336 + 831 | Bao et al. 2006b |
| F2: CAAGGAGAGCTGGAGGGGGC | GC: 1336 + 542 | |||||
| R1: GCCGGCCGTGCAGATCTTAAC | ||||||
| R2: ACATGCCGCGCACCTGGAAA | ||||||
|
| ||||||
| SBEI | 6 | InDel (TEb) | F: GAGTTGAGTTGCGTCAGATC | – | In: 882 | Bao et al. 2006a, |
| R: AATGAGGTTGCTTGCTGCTG | Del: 547 | Han et al. 2004 | ||||
|
| ||||||
| SBEIII | 2 | SNP (C/G) | F: GTCTTGGACTCAGATGCTGGACTC | Spe I | C: 510 | Bao et al. 2006a, |
| R: ATGTATAACTGGCAGTTCGAACGG | G: 295 + 215 | Han et al. 2004 | ||||
|
| ||||||
| SDBE | 4 | InDel | F: TGGCCTATCGATTGATGAGA | – | In: 299 | Larkin et al. 2003 |
| R: AAAATTTTGCTGGTAATAGAAAAA | Del: 280 | |||||
L and S, large and small subunits, respectively.
Insertion and deletion polymorphism of a transposable element.
F and R, forward and reverse primers, respectively.
Restriction enzymes used for PCR product digestion.
TT/GC SNPs are two contiguous SNPs in SSIIa (Bao et al. 2006b).
Grain quality evaluation
The 187 varieties were divided into six maturity groups according to their heading dates, and were harvested 112–129 days after transplanting, with 3–4 day intervals between adjacent maturity groups (Table 2). For each variety, 10 plants with relatively uniform growth were harvested in the field and from the two greenhouses. The panicles were hand-threshed, dehulled with SY88-TH (Ssangyong, Incheon, Korea), and milled with VP-32T (Yamamoto, Yamagata, Japan) to investigate head rice percentage (HRP), chalky grains percentage (CGP), AC, ADV, and RVA characteristics. HRP and CGP were examined by scanning milled rice grain with an RN-300 Grain Inspector (Kett, Tokyo, Japan). AC was estimated indirectly by scanning the milled rice samples with near infrared spectroscopy using an Infratec 1241 Grain Analyzer (Foss, Hillerod, Denmark). ADV was measured visually by soaking the milled rice grains in 1.4% KOH solution for 24 h at 30°C as described by Little et al. (1958). Rice flour (3.0 g) stirred in 25 ml water was used to measure RVA characteristics including pasting temperature (PT), peak viscosity (PKV), hot paste viscosity (HPV), cool paste viscosity (CPV), setback viscosity (SBV), and breakdown viscosity (BDV) with a Rapid Visco Analyzer (Newport Scientific Pty, Warriewood, NSW, Australia).
Table 2.
Average daily temperature during grain filling (from heading to harvest) in the field and two greenhouses for each maturity group
| Maturity groupa | No. of varieties | Average daily temperature during grain-filling | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Field | Greenhouse I | Greenhouse II | ||||||||
|
|
|
|
||||||||
| Min | Mean | Max | Min | Mean | Max | Min | Mean | Max | ||
| I | 36 | 21.5 | 25.7 | 33.4 | 22.3 | 26.1 | 32.5 | 22.8 | 26.5 | 33.0 |
| II | 27 | 20.6 | 24.9 | 32.7 | 21.5 | 25.3 | 31.8 | 22.1 | 25.9 | 32.3 |
| III | 26 | 19.5 | 23.9 | 31.6 | 20.5 | 24.4 | 30.8 | 21.1 | 25.0 | 31.3 |
| IV | 25 | 18.8 | 23.4 | 31.2 | 19.8 | 23.9 | 30.5 | 20.4 | 24.5 | 30.8 |
| V | 46 | 18.1 | 22.8 | 30.8 | 19.1 | 23.3 | 30.2 | 19.8 | 23.9 | 30.6 |
| VI | 27 | 17.5 | 22.3 | 30.4 | 18.7 | 22.9 | 29.9 | 19.3 | 23.5 | 30.2 |
Plants were harvested 112–129 days after transplanting according to their maturity, with 3- to 4-day intervals between adjacent groups.
Statistical analyses
Descriptive statistics were calculated for grain quality traits under each temperature condition. The changes in grain quality traits under increased temperature were calculated by subtracting the values obtained in the field from those obtained in greenhouse II, which had the highest temperature. The percentage of phenotypic variation explained (R2) was obtained to estimate the relative contribution of SSRG loci to grain quality traits by single-locus ANOVA. For F-tests, a locus with a P-value less than 0.05 was considered to be significant. Then the least significant difference (LSD) test was conducted to compare the means of the grain quality traits according to the different allele types for each marker. Calculations and analyses were performed using PROC MEANS and PROC GLM of the SAS statistical software package (version 9.1, SAS Institute, NC, USA). Nei’s (1973) gene diversity value for each SSRG marker was calculated using PopGen version 1.31 (http://www.ualberta.ca/~fyeh/). To identify pair-wise genetic relationships between rice varieties according to SSRG alleles, a genetic similarity matrix was calculated using simple matching coefficients. The similarity matrix was used to construct a dendrogram by the unweighted pair-group method with the arithmetic mean (UPGMA) algorithm and the sequential, hierarchical, and nested clustering (SHAN) routine in the NTSYS program (version 2.0; Rohlf 1998).
Results
Changes in grain quality traits under increased temperature
The daily mean temperature from transplanting to harvest in the field and two greenhouses is shown in Table 2. Compared to the field, the average daily mean temperature during grain filling was 0.4–0.6°C higher in greenhouse I and 0.8–1.2°C higher in greenhouse II. Also, the average daily mean temperature during grain filling was higher for early maturity groups than for late maturity groups. For example, the average daily mean temperature during grain filling in the field was 25.7°C for Group I and 22.3°C for Group VI. Similarly, the average daily mean temperature during grain filling in greenhouse II was 26.5°C for Group I and 23.5°C for Group VI.
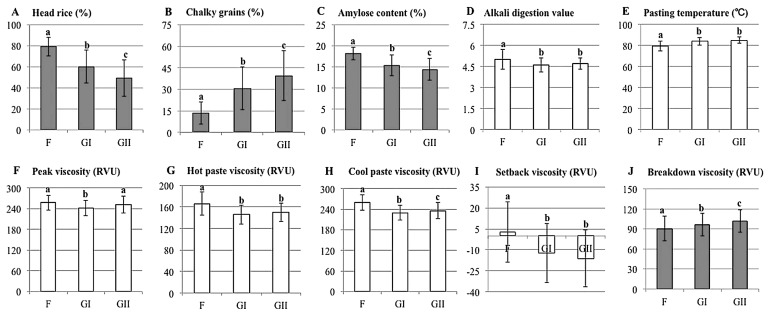
The changes in the average values of 10 grain quality traits under increasing temperature in 187 nonglutinous Korean rice varieties are shown in Fig. 1 (see Supplemental Table 1 for descriptive statistics). The average HRP significantly decreased under high temperature; it was 79.4% in the field, 60.2% in greenhouse I, and 49.1% in greenhouse II (Fig. 1A). There were significant increases under high temperature for the average CGP, which was 13.4%, 30.7% and 39.4% in the field, greenhouse I, and greenhouse II, respectively (Fig. 1B). Average AC showed a significant decrease with increasing temperature, decreasing from 18.2% in the field to 15.4% and 14.4% in greenhouse I and II, respectively (Fig. 1C). There was no significant difference in average ADV between the rice grown in the two greenhouses, but that of rice grown in the field was higher than those of rice grown in the greenhouses (Fig. 1D). Among the six RVA characteristics, only average BDV showed significant changes under increasing temperature (Fig. 1E–1J). The average BDV was 90.8 in the field and significantly higher in greenhouse I (96.1) and greenhouse II (102.0) (Fig. 1J). The PT was significantly higher in the greenhouses than in the field, but there was no significant difference between the two greenhouses (Fig. 1E). Average HPV, CPV, and SBV were higher in the field than in the greenhouses (Fig. 1G–1I). Average PKV did not change significantly under high temperature (Fig. 1F).
Fig. 1.
Changes in the grain quality traits of 187 nonglutinous Korean rice varieties under increased temperature. (A) head rice percentage (HRP); (B) chalky grains percentage (CGP); (C) amylose content (AC); (D) alkali digestion value (ADV); (E) pasting temperature (PT); (F) peak viscosity (PKV); (G) hot paste viscosity (HPV); (H) cool paste viscosity (CPV); (I) setback viscosity (SBV); (J) breakdown viscosity (BDV). Traits showing significant (P < 0.01) changes under increased temperature both between F&GI and GI&GII are indicated with gray bars, while the others are displayed with white bars. Error bars show standard deviation. Different letters above error bars indicate significant difference at P < 0.01. F, GI, and GII represent the field, greenhouse I, and greenhouse II, respectively.
Temperature during grain filling was higher in early-maturing group compared to late-maturing group (Table 2). Therefore, we conducted analysis of variance (ANOVA) with the maturity group (I–VI) as an independent variable to estimate the effect of maturity on the variations in grain quality traits under different temperature conditions (Table 3). There were significant associations between the maturity group and the variations in HRP, CGP, AC, ADV, and CPV, in the field, greenhouse I, and greenhouse II. The maturity group explained >49% of the variations in HRP, CGP, and AC in greenhouse II. In greenhouse II, there were notable differences in the three grain quality traits among maturity groups I–IV, V, and VI (Supplemental Table 2). The average HRP in greenhouse II was 37.6%–43.0% in group I–IV, 59.2% in group V, and 71.8% in group VI, while the average CGP in greenhouse II was 42.8%–51.0% in group I–IV, 30.2% in group V, and 17.2% in group VI. Similarly, the average AC in greenhouse II was 12.5%–13.5% in group I–IV, 15.9% in group V, and 17.6% in group VI. The maturity group also explained >31% variations in the changes in HRP, CGP, and AC under increased temperature (Δ in Table 3). The effect of the maturity group on grain quality traits was also detected when the ANOVA was conducted with 170 Japonica varieties, after excluding 17 Tongil-type varieties (Table 3). From this result, we could infer that the deterioration in grain appearance and the decrease in AC under high temperature were more severe in the varieties with early heading dates because they were exposed to higher temperature during the grain filling period, compared with those with late heading dates (Table 2).
Table 3.
Effect (R2) of maturity groups on the grain quality traits of 187 nonglutinous Korean rice varieties under different temperature conditionsa
| Traitb | All (n = 187)c | Japonica (n = 170)c | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| F | GI | GII | Δ | F | GI | GII | Δ | |
| HRP | 0.178*** | 0.396*** | 0.534*** | 0.450*** | 0.218*** | 0.430*** | 0.581*** | 0.485*** |
| CGP | 0.142*** | 0.386*** | 0.491*** | 0.413*** | 0.170*** | 0.427*** | 0.575*** | 0.464*** |
| AC | 0.440*** | 0.474*** | 0.515*** | 0.319*** | 0.436*** | 0.478*** | 0.517*** | 0.329*** |
| ADV | 0.211*** | 0.182*** | 0.107*** | 0.118*** | 0.187*** | 0.105** | 0.067* | 0.140*** |
| PT | NS | NS | 0.078* | NS | NS | NS | 0.087* | NS |
| PKV | 0.210*** | 0.094** | NS | 0.204*** | 0.318*** | 0.151*** | NS | 0.206*** |
| HPV | 0.354*** | 0.230*** | NS | 0.306*** | 0.349*** | 0.239*** | NS | 0.297*** |
| CPV | 0.241*** | 0.148*** | 0.095** | 0.291*** | 0.252*** | 0.148*** | 0.092** | 0.289*** |
| SBV | NS | 0.083** | NS | NS | NS | 0.129*** | 0.081* | NS |
| BDV | 0.059* | 0.067* | NS | NS | 0.069* | 0.076* | 0.074* | NS |
R2 was calculated from ANOVA results in which the maturity group (Group I–Group VI; see Table 2) was used as an independent variable. R2 values are significant at *P < 0.05, **P < 0.01, and ***P < 0.001; NS no significance.
AC, amylose content; ADV, alkali digestion value; BDV, breakdown viscosity; CGP, chalky grains percentage; CPV, cool paste viscosity; HPV, hot paste viscosity; HRP, head rice percentage; PKV, peak viscosity; PT, pasting temperature; SBV, setback viscosity.
F, GI, and GII represent the field, greenhouse I, and greenhouse II, respectively. Δ changes in grain quality traits under increased temperature (calculated by subtracting the field values from their corresponding values in greenhouse II).
Allele distribution of 10 SSRG markers in 187 nonglutinous Korean rice varieties
Although the maturity group (based on heading date) explained large portion of the variations in grain quality traits under different temperature conditions (Table 3), there were also high level of variations in grain quality traits among different varieties within the same maturity group (Supplemental Table 2). To examine the effect of SSRG alleles on the variations in grain quality traits under different temperature conditions, we genotyped 187 nonglutinous Korean rice varieties with 10 previously reported SSRG markers (Table 1).
Among the 10 SSRG markers, seven were polymorphic (Table 4). The SSIIa TT/GC SNPs showed the highest gene diversity (0.499) among the 10 SSRG markers. Although all Tongil-type varieties contained the TT allele, the Japonica varieties showed even allele distribution with 80 varieties carrying the TT allele and 88 varieties carrying the GC allele. AGPase-S C/T and A/T SNPs also had relatively high gene diversities (0.402 for C/T SNP and 0.415 for A/T SNP) with relatively even allele distribution among both Japonica and Tongil-type varieties. It should be noticed that two additional allele types, unknown and absent, were identified by the DNA marker A/T SNP in AGPase-S. Lee et al. (2009) reported that, among 55 rice germplasm with different ecotypes (Japonica, Japonica/Indica hybrid, and Indica), the A allele produces a single band (338 bp) while the T allele cleaves into two fragments (315 bp and 23 bp) by application of an endonuclease, Alu I. However, when we genotyped the Korean Japonica rice varieties, 25 varieties did not generate PCR products. We also observed bands with the size obviously smaller than 300 bp, and designated them as unknown (U) allele which is clearly distinguished from the T allele. The U allele was actually derived from a 50 bp deletion within the amplicon (data not shown). The results indicate there is sequence variations within the AGPase-S locus, which was not identified in the 55 rice germplasm evaluated by Lee et al. (2009).
Table 4.
Allele distribution and gene diversity of 10 SSRG markers among 187 nonglutinous Korean rice varieties according to the two subpopulations— Japonica and Tongil
| Gene | Marker | Allele | No. of varieties | Nei’s gene diversitya | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| All | Japonica | Tongil | All | Japonica | Tongil | |||
| AGPase-L | InDel | In | 169 | 169 | 0 | 0.174 | 0.012 | 0 |
| Del | 18 | 1 | 17 | |||||
|
| ||||||||
| AGPase-S | SNP | C | 135 | 127 | 8 | 0.402 | 0.378 | 0.498 |
| T | 52 | 43 | 9 | |||||
|
|
||||||||
| SNP | A | 119 | 109 | 10 | 0.415 | 0.383 | 0.484 | |
| T | 10 | 3 | 7 | |||||
| Ub | 33 | 33 | 0 | |||||
| absent | 25 | 25 | 0 | |||||
|
| ||||||||
| GBSSI | SNP | T | 187 | 170 | 17 | 0 | 0 | 0 |
| G | 0 | 0 | 0 | |||||
|
|
||||||||
| InDel | In | 0 | 0 | 0 | 0 | 0 | 0 | |
| Del | 187 | 170 | 17 | |||||
|
|
||||||||
| SNP | T | 187 | 170 | 17 | 0 | 0 | 0 | |
| C | 0 | 0 | 0 | |||||
|
| ||||||||
| SSIIa | SNP | TT | 97 | 80 | 17 | 0.499 | 0.499 | 0 |
| GC | 88 | 88 | 0 | |||||
| absent | 2 | 2 | 0 | |||||
|
| ||||||||
| SBEI | InDel | In | 165 | 165 | 0 | 0.193 | 0.035 | 0 |
| Del | 20 | 3 | 17 | |||||
| absent | 2 | 2 | 0 | |||||
|
| ||||||||
| SBEIII | SNP | C | 18 | 2 | 16 | 0.174 | 0.023 | 0.111 |
| G | 169 | 168 | 1 | |||||
|
| ||||||||
| SDBE | InDel | In | 13 | 0 | 13 | 0.129 | 0 | 0.360 |
| Del | 174 | 170 | 4 | |||||
A previously unknown allele different from A and T SNPs was identified in 33 varieties and was designated as the unknown (U) allele.
For the AGPase-L InDel, 169 out of 170 Japonica varieties had the insertion allele, while all of the 17 Tongil-type varieties had the deletion allele, indicating subspecies-specific allele distribution. This type of biased allele frequency in the two subpopulations was also observed for the SBEI InDel, the SBEIII C/G SNP, and the SDBE InDel, all of which had genetic diversity values of less than 0.2. The three markers in GBSSI were monomorphic; all of the 187 varieties had the T alleles for both T/G and T/C SNPs in GBSSI, and the deletion allele for the GBSSI InDel marker.
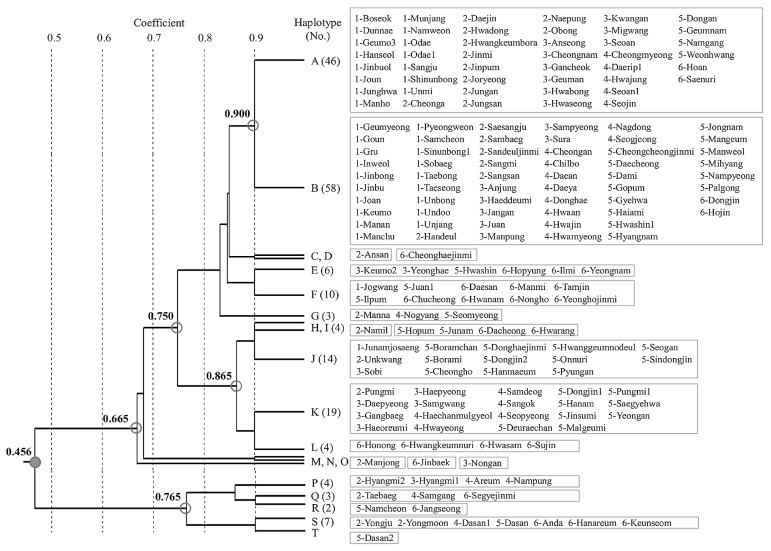
A dendrogram generated from the genotyping results revealed 20 haplotypes for the 10 SSRG markers among the 187 nonglutinous Korean rice varieties (Fig. 2). The two most common haplotypes were Haplotypes A and B, containing 25% and 31% of the tested varieties, respectively. The Haplotypes C, D, H, M, N, O, and T were variety-specific for ‘Ansan’, ‘Cheonghaejinmi’, ‘Namil’, ‘Manjong’, ‘Jinbaek’, ‘Nongan’, and ‘Dasan2’, respectively. Both Haplotypes A and B contained varieties from all of the maturity groups (I–VI in Table 2). Most of the other haplotypes did not show maturity group-dependent variety composition either, indicating that the allele distributions of most SSRG markers tested in this study are likely to be independent from heading dates. In contrast, the haplotypes of the 170 Japonica varieties (A–O) were clearly distinguished from those of the 17 Tongil-type varieties (P–T) in the dendrogram, reflecting the subspecies-specific allele distribution of many SSRG markers, i.e., AGPase-L InDel, SBEI InDel, SBEIII C/G SNP, and SDBE InDel (Fig. 2 and Table 4).
Fig. 2.
Dendrogram of 187 nonglutinous Korean rice varieties by UPGMA cluster analysis based on 10 SSRG loci. A total of 20 unique SSRG haplotype groups were identified within the 187 rice varieties. For each haplotype group, the number of rice varieties is shown in parentheses, except for haplotype groups with only one entry. Variety names are shown beside the corresponding haplotype group; Arabic numeral prefixes show their designated maturity groups (see Table 2). Note that the node in gray (0.456) split 187 nonglutinous Korean rice varieties into two subspecies— 170 Japonica varieties and 17 Tongil-type varieties, in which 15 (A–O) and 5 (P–T) SSRG haplotypes were identified, respectively.
Effects of SSRG allelic variations on grain quality traits under different temperature conditions
Table 5 shows the significant associations between the allelic variations in SSRG markers and the phenotypic variations in grain quality traits of 187 nonglutinous Korean rice varieties under three different temperature conditions (the field, greenhouse I, and greenhouse II). Among the seven polymorphic SSRG markers, six showed a relatively high level of association (R2 > 0.1) with variations in one or more grain quality traits of the 187 varieties in the three different environments; i.e., AGPase-L InDel (PKV, SBV, BDV), AGPase-S A/T SNP (SBV, BDV), SSIIa TT/GC SNPs (ADV), SBEI InDel (SBV, BDV), SBEIII C/G SNP (PKV, SBV, BDV), and SDBE InDel (PKV, SBV, BDV).
Table 5.
Summary of association analyses (R2) between SSRG marker alleles and grain quality traits among 187 nonglutinous Korean rice varieties under different temperature conditions
| Gene (Markera) | Traitb | All (n = 187)c | Japonica (n = 170)c | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| F | GI | GII | F | GI | GII | ||
| AGPase-L (InDel) | CGP | 0.021* | 0.078*** | 0.138*** | NS | NS | NS |
| AC | 0.023* | 0.057*** | 0.024* | NS | NS | NS | |
| ADV | 0.045** | 0.038** | 0.069*** | NS | NS | NS | |
| PKV | 0.189*** | 0.291*** | 0.131*** | NS | NS | NS | |
| SBV | 0.315*** | 0.313*** | 0.297*** | 0.030* | 0.038* | 0.037* | |
| BDV | 0.432*** | 0.418*** | 0.344*** | 0.035* | 0.035* | 0.029* | |
|
| |||||||
| AGPase-S (C/T) | SBV | 0.074*** | 0.035* | 0.034* | 0.046** | NS | NS |
| BDV | 0.081*** | 0.104*** | 0.074*** | 0.044** | 0.078*** | 0.038* | |
|
| |||||||
| AGPase-S (A/T) | SBV | 0.152*** | 0.123*** | 0.119*** | NS | NS | NS |
| BDV | 0.178*** | 0.144*** | 0.134*** | NS | 0.065** | NS | |
|
| |||||||
| SSIIa (TT/GC) | ADV | 0.287*** | 0.294*** | 0.141*** | 0.277*** | 0.342*** | 0.126*** |
| PT | 0.110*** | 0.043** | 0.050** | 0.104*** | 0.054** | 0.041** | |
|
| |||||||
| SBEI (InDel) | ADV | 0.078*** | 0.049** | 0.093*** | 0.037* | NS | 0.033* |
| PKV | 0.135*** | 0.188*** | 0.070*** | NS | 0.028* | 0.032* | |
| SBV | 0.215*** | 0.205*** | 0.198*** | NS | NS | NS | |
| BDV | 0.313*** | 0.286*** | 0.236*** | NS | NS | NS | |
|
| |||||||
| SBEIII (C/G) | PKV | 0.156*** | 0.224*** | 0.100*** | NS | NS | NS |
| SBV | 0.284*** | 0.275*** | 0.261*** | 0.029* | NS | NS | |
| BDV | 0.372*** | 0.352*** | 0.292*** | NS | NS | NS | |
|
| |||||||
| SDBE (InDel) | AC | 0.026* | 0.063*** | 0.033* | – | – | – |
| ADV | 0.076*** | 0.110*** | 0.110*** | – | – | – | |
| PKV | 0.157*** | 0.258*** | 0.127*** | – | – | – | |
| SBV | 0.223*** | 0.214*** | 0.213*** | – | – | – | |
| BDV | 0.312*** | 0.320*** | 0.269*** | – | – | – | |
Number of allele types for each marker is listed in Table 4.
Traits listed had significant R2 values in 187 rice varieties under the three different conditions. Traits showing relatively high level of association (R2 > 0.1) with the corresponding SSRG marker in the three different conditions are highlighted in bold. AC, amylose content; ADV, alkali digestion value; BDV, breakdown viscosity; CGP, chalky grains percentage; PKV, peak viscosity; PT, pasting temperature; SBV, setback viscosity; R2 values are significant at *P < 0.05, **P < 0.01, and ***P < 0.001; NS no significance; – monomorphism.
F, GI, and GII represent the field, greenhouse I, and greenhouse II, respectively.
The data were reanalyzed after excluding 17 Tongil-type varieties to rule out the effect of population structure. In this analysis, significant associations were not observed in most markers, or the R2 value decreased to a large extent (Table 5). For example, although allelic variations in the AGPase-L InDel significantly explained some of the phenotypic variations in CGP (2.1–13.8%), AC (2.3–5.7%), ADV (3.8–6.9%), and PKV (13.1–29.1%) of the 187 varieties in the three different environments, there was no significant relationship when the 17 Tongil-type varieties were excluded from the analysis. Also, while more than 29% of the variations in SBV and BDV of the 187 varieties was explained by the AGPase-L InDel polymorphism in the three different environments, less than 4% was explained after excluding the 17 Tongil-type varieties. Similar trends were observed for the AGPase-S C/T SNP (SBV, BDV), the AGPase-S A/T SNP (SBV, BDV), the SBEI InDel (ADV, PKV, SBV, BDV), the SBEIII C/G SNP (PKV, SBV, BDV), and the SDBE InDel (AC, SBV, BDV).
The SSIIa TT/GC SNP marker was the only SSRG marker showing a relatively high level of association with certain grain quality traits in the field and two greenhouse conditions when the analysis included only the 170 Japonica varieties (Table 5). It explained 12.6%–34.2% of the ADV variation in 170 Japonica varieties, which was similar to the association analysis conducted without considering the population structure comprising Japonica and Tongil-type varieties.
Relationship between SSRG allelic variations and changes in grain quality traits under increased temperature
To evaluate the allelic effects of SSRGs on maintaining stable grain quality traits under high temperature, we analyzed the relationships among SSRG allelic variations and the changes in grain quality traits under increased temperature (Table 6). The changes in grain quality traits under increased temperature were calculated by subtracting the field values from their corresponding values in greenhouse II, which had the highest temperature (Table 2).
Table 6.
Changes in grain quality traits in 187 nonglutinous Korean rice varieties under increased temperature according to allelic differences in SSRG markersa
| Gene | Allele | No. of variety | ΔHRP (%) | ΔCGP (%) | ΔAC (%) | ΔBDV (RVU) |
|---|---|---|---|---|---|---|
| AGPase-L | In | 169 | −32.2a | 27.4a | −3.9a | 12.0a |
| Del | 18 | −15.9b | 9.7b | −3.3a | 3.8b | |
| AGPase-S | C | 135 | −32.9a | 28.2a | −4.1a | 11.6a |
| T | 52 | −24.7b | 19.3b | −3.3b | 10.0a | |
| AGPase-S | A | 119 | −34.3a | 30.0a | −4.3a | 10.8a |
| T | 10 | −20.7b | 12.6b | −3.7a | 3.7a | |
| U | 33 | −29.7a | 24.7a | −3.6a | 11.9a | |
| SSIIa | TT | 97 | −28.9a | 24.0a | −4.0a | 11.2a |
| GC | 88 | −32.4a | 27.6a | −3.8a | 10.8a | |
| SBEI | In | 165 | −32.1a | 27.5a | −3.9a | 11.8a |
| Del | 20 | −17.6b | 11.2b | −3.6a | 4.6b | |
| SBEIII | C | 18 | −18.4a | 7.0a | −3.9a | 4.1a |
| G | 169 | −31.9b | 27.7b | −3.9a | 11.9b | |
| SDBE | In | 13 | −12.1a | 6.6a | −3.1a | 4.9a |
| Del | 174 | −32.0b | 27.2b | −3.9a | 11.7a |
Changes in grain quality traits under high temperature were calculated by subtracting the field values from their corresponding values in greenhouse II. Only traits showing significant changes under increased temperature are listed (see Fig. 1). Values with a different letter are significantly different at P < 0.05 (highlighted in bold). Traits showing significant allelic effect in Japonica varieties (n = 170) are underlined.
AC, amylose content; BDV, breakdown viscosity; CGP, chalky grains percentage; HRP, head rice percentage.
There were significant differences in the changes in HRP and CGP under increased temperature (ΔHRP and ΔCGP, respectively) among the varieties with different alleles of six SSRGs (Table 6). ΔHRP and ΔCGP were significantly lower in the varieties containing the deletion allele for AGPase-L InDel, the T alleles for both C/T and A/T SNPs in AGPase-S, the deletion allele for SBEI InDel, the C allele for SBEIII C/G SNP, and the insertion allele for SDBE InDel. The AGPase-S C/T SNP was the only locus showing significant relationship with the change in AC under increased temperature (ΔAC); ΔAC was significantly lower in the varieties carrying the T allele than in those carrying the C allele. The AGPase-L InDel, SBEI InDel, and SBEIII C/G SNP had significant associations with the change in BDV under increased temperature (ΔBDV); rice varieties with deletion, deletion, and C alleles for the three markers had significantly lower ΔBDV than those with the other alleles. When the analyses were carried out only for the Japonica group (n = 170), however, the significant associations disappeared or were markedly decreased for the AGPase-L InDel, the SBEI InDel, the SBEIII C/G SNP, and the SDBE InDel (Tables 6, 7). Meanwhile AGPase-S C/T SNP showed significant association with ΔHRP, ΔCGP, and ΔAC, and AGPase-S A/T SNP exhibited significant association with ΔHRP and ΔCGP in the 170 Japonica varieties, which was similar to the association analyzed in the total of 187 varieties. However, their effects on the grain quality traits were minor (R2 < 0.07, Table 7).
Table 7.
Association (R2) of SSRG allelic variations with changes in grain quality traits of nonglutinous Korean rice varieties under increased temperature
| Markera | Traitb | All (n = 187) | Japonica (n = 170) |
|---|---|---|---|
| AGPase-L InDel | ΔHRP | 0.100*** | NS |
| ΔCGP | 0.106*** | NS | |
| ΔBDV | 0.029* | NS | |
|
| |||
| AGPase-S C/T | ΔHRP | 0.059*** | 0.052** |
| ΔCGP | 0.061*** | 0.043** | |
| ΔAC | 0.031* | 0.042** | |
|
| |||
| AGPase-S A/T | ΔHRP | 0.056* | 0.063** |
| ΔCGP | 0.079** | 0.052* | |
|
| |||
| SBEI InDel | ΔHRP | 0.088*** | NS |
| ΔCGP | 0.098*** | NS | |
| ΔBDV | 0.025* | NS | |
|
| |||
| SBEIII C/G | ΔHRP | 0.069*** | NS |
| ΔCGP | 0.145*** | 0.066*** | |
| ΔBDV | 0.027* | NS | |
|
| |||
| SDBE InDel | ΔHRP | 0.111*** | – |
| ΔCGP | 0.106*** | – | |
Only the markers significantly associated with the changes in grain quality traits under increased temperature were analyzed. The number of allele types for each marker is listed in Table 4.
AC, amylose content; BDV, breakdown viscosity; CGP, chalky grains percentage; HRP, head rice percentage. R2 values are significant at *P < 0.05, **P < 0.01, and ***P < 0.001; NS no significance, – no polymorphism.
Discussion
Among the 10 grain quality traits investigated in this study, four showed significant changes with increasing temperature. Grain appearance deteriorated as the average HRP decreased, while the average CGP increased under higher temperatures. This was similar to the previous findings that the deterioration in grain appearance under high temperature was mainly because of the increase in chalkiness (Mo et al. 2012a). It was reported that AC decreased under high temperature in most rice varieties, although a few varieties showed increase or no significant change (Cruz et al. 1989, Tanaka et al. 2009, Zhong et al. 2005). Similarly, the average AC of the 187 nonglutinous Korean rice varieties decreased significantly as temperature increased. It is assumed that suppression of starch biosynthesis genes such as GBSSI and SBE under high temperature might explain the decrease in AC (Yamakawa et al. 2007). Among the six RVA characteristics, only BDV showed significant change under increased temperature. Tanaka et al. (2009) reported that PKV and BDV increased while SBV decreased significantly under high temperature in three Japonica rice varieties. Zhong et al. (2005) showed that CPV and SBV decreased significantly under high temperature in four Indica rice varieties. In this study, the average BDV of the 187 varieties increased significantly as the temperature rose while the other RVA characteristics and ADV did not change significantly in response to higher temperature. Therefore, we decided to trace the changes in HRP, CGP, AC, and BDV to estimate the levels of stable grain quality traits under increased temperature in Korean nonglutinous rice varieties.
The early maturing varieties were exposed to higher temperature during the grain-filling period compared to the late-maturing varieties. Therefore, a large proportion of the variations in grain quality traits under different temperature conditions was explained by the maturity group (Table 3). However, large variations in grain quality traits were observed even among the varieties in the same maturity group (Supplemental Table 2). Therefore, we investigated the effect of previously reported SSRG loci on grain quality traits under different temperature conditions to identify SSRG alleles favorable for maintaining grain quality under high temperature.
Allelic variations in GBSSI are well-known for determining starch physicochemical properties such as AC and RVA characteristics. These characteristics affect the processing, cooking, and eating qualities of rice (Dobo et al. 2010, Kharabian-Masouleh et al. 2012, Larkin et al. 2003, Larkin and Park 2003, Tian et al. 2009, Yamanaka et al. 2004). However, there was no polymorphism for the three markers in GBSSI (T/G SNP, T/C SNP, and InDel) among the 187 nonglutinous Korean rice varieties used in this study. As for the GBSSI T/G SNP, Bao et al. (2006a) revealed that the Wxa (G allele) and Wxb (T allele) are evenly distributed among Chinese nonglutinous rice breeding lines, reflecting the breeders’ selections for varieties with various eating and cooking qualities. In contrast, all of the nonglutinous Korean rice varieties used in this study had only the Wxb allele. Considering that Wxa and Wxb are the predominant alleles for Indica and Japonica subspecies, respectively (Hirano et al. 1998, Sano et al. 1986), it was notable that all of the 17 Tongil-type (derived from Indica-Japonica cross) varieties contained only the Wxb allele. This implied that the Korean consumers’ preference for a soft and sticky texture of Japonica cooked rice has been reflected in the breeders’ selections in developing Tongil-type varieties. According to previous reports, GBSSI expression and AC decreased under high temperature to a greater extent in varieties with the Wxb allele than in those with the Wxa allele (Hirano and Sano 1998, Inukai and Hirayama 2010, Larkin and Park 1999). Further research is underway to introduce the Wxa allele into elite Korean Japonica varieties by marker-assisted backcrosses. This will allow us to evaluate and exploit the effect of Wxa to retain stable grain quality under high temperature, to cope with global warming.
Among the seven polymorphic SSRG markers, four showed subspecies-specific allele distribution in the 187 nonglutinous Korean rice varieties. The SBEI InDel had significant associations with ADV, PKV, SBV, and BDV in the field and two greenhouse conditions and SBEIII C/G SNP had significant associations with PKV, SBV, and BDV in the three different environments. This finding was similar to the previous reports that the SBEI InDel and SBEIII C/G SNP markers were significantly associated with variations in RVA characteristics (Bao et al. 2006a, Han et al. 2004). The AGPase-L InDel and SDBE InDel were also relatively strongly associated (R2 > 0.1) with PKV, SBV, and BDV in the field and two greenhouse conditions. However, when the association analysis was conducted after separating Japonica and Tongil-type varieties, the significance level was highly reduced or disappeared because of the biased allele frequency between the two subpopulations (Table 5).
Only three out of 10 SSRG markers showed relatively even allele distributions among the Japonica varieties. The C/T and A/T SNPs in AGPase-S were relatively evenly distributed among both Japonica and Tongil-type varieties (Table 4). Lee et al. (2009) reported that the two SNPs in AGPase-S explained >40% of the AC variation in 55 rice accessions with diverse geographical origins; however, no significant association with AC was found in the present study. Instead, the AGPase-S C/T SNP was significantly associated with SBV and BDV and the A/T SNP was significantly associated with SBV and BDV in the field and two greenhouse conditions. Nevertheless, when the analysis was conducted on the 170 Japonica varieties, the significant associations were markedly decreased. The SSIIa TT/GC SNPs were also evenly distributed among the Japonica varieties, with 80 varieties carrying the TT allele and 88 carrying the GC allele. This was consistent with the study by Bao et al. (2006b), in which 334 Chinese rice varieties also showed even allele distributions of SSIIa TT/GC SNPs. Also, the strong associations between SSIIa TT/GC SNPs and ADV and PT reported in other studies were verified in this study in all three environments (Bao et al. 2006b, Nakamura et al. 2005, Umemoto and Aoki 2005). In particular, we observed a significant association (R2 > 0.126) between SSIIa TT/GC SNPs and the variation in ADV in the Japonica group, which was similar to the association analyzed in the total of 187 varieties. In summary, out of the 10 SSRG markers investigated in this study, only the two SNPs (C/T and A/T) in AGPase-S showed relatively even allele distributions in both Japonica and Tongil-type nonglutinous Korean rice varieties. The SSIIa TT/GC SNPs were the only marker showing significant associations (R2 > 0.1) with a grain quality trait in the three different environments for the 170 Japonica varieties.
In the association analysis between the SSRG allelic variations and the changes in grain quality traits under increased temperatures, all polymorphic SSRG markers except for SSIIa TT/GC SNPs showed a significant allelic effect on certain grain quality traits (Table 6). However, when the analysis was conducted only on Japonica varieties, only the C/T and A/T SNPs in AGPase-S showed significant allelic effects on the variations in certain grain quality traits by increased temperature, which was similar to the association analysis conducted without excluding the 17 Tongil-type varieties (Table 7). Since the varieties with the T alleles for both C/T and A/T SNPs in AGPase-S had significantly less decrease in HRP and less increase in CGP under increased temperature, these alleles could be used in a breeding program to develop rice varieties that retain stable grain appearance under high temperature. However, it is likely that their effect would be limited because the two markers explained only 4.3–6.3% of the variations in ΔHRP and ΔCGP. As for the four subspecies-specific SSRG markers, the changes in the grain quality traits under increased temperature were significantly smaller in the varieties with Indica-specific alleles than in those with Japonica-specific alleles. This is reasonable considering that Indica subspecies originated from the tropics while the Japonica subspecies are more adapted to a temperate climate. Significant allelic effects of the four subspecies-specific SSRG markers largely disappeared when the analysis was conducted only on the 170 Japonica varieties (Table 7). Jin et al. (2010) mentioned that functional loci with subspecies-specific allele distribution would not be detected if population structure is considered in the association analysis. Therefore, further research is needed to confirm whether the effect of the subspecies-specific SSRG alleles on the changes in grain quality traits under increased temperature is merely because of the population structure or not. Overall, the level of polymorphism in the 10 SSRG markers was highly limited or subspecies-specific among the 187 nonglutinous Korean rice varieties. Accordingly, the polymorphisms had limited abilities to explain variations in grain quality traits under different temperature conditions. It is assumed that the 10 SSRG loci have been highly fixed with optimum alleles according to market demands during the history of Korean rice breeding. Therefore, there may be other genetic factors influencing the grain quality variations among the Korean Japonica varieties under increasing temperature. Another association study is currently being carried out with genome-wide SNP and SSR markers. We hope that this will allow us to identify other genetic factors related to the variations in grain quality traits among Korean nonglutinous Japonica rice varieties under different temperature conditions. Efforts to improve genetic diversity in terms of grain quality under high temperature are also being made using germplasm search and mutagenesis approaches.
Supplementary Material
Acknowledgements
This research was supported by grants from the Rural Development Administration, Korea.
Literature Cited
- Ayres, N.M., McClung, A.M., Larkin, P.D., Bligh, H.F.J., Jones, C.A. and Park, W.D. (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germ plasm. Theor. Appl. Genet. 94: 773–781 [Google Scholar]
- Bao, J.S., Corke, H. and Sun, M. (2006a) Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in nonwaxy rice (Oryza sativa L.). Theor. Appl. Genet. 113: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Bao, J.S., Corke, H. and Sun, M. (2006b) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 113: 1171–1183 [DOI] [PubMed] [Google Scholar]
- Bao, J., Shen, S., Sun, M. and Corke, H. (2006c) Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch-Stärke 58: 259–267 [Google Scholar]
- Bligh, H.F.J., Till, R.I. and Jones, C.A. (1995) A microsatellite sequence closely linked to the Waxy gene of Oryza sativa. Euphytica 86: 83–85 [Google Scholar]
- Chun, A., Song, J., Kim, K.J. and Lee, H.J. (2009) Quality of head and chalky rice and deterioration of eating quality by chalky rice. J. Crop Sci. Biotechnol. 12: 239–244 [Google Scholar]
- Cruz, N., Kumar, I., Kaushik, R.P. and Khush, G.S. (1989) Effect of temperature during grain development on stability of cooking quality components in rice. Jpn. J. Breed. 39: 299–306 [Google Scholar]
- Dobo, M., Ayres, N., Walker, G. and Park, W.D. (2010) Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J. Cereal Sci. 52: 450–456 [Google Scholar]
- Fitzgerald, M.A., McCouch, S.R. and Hall, R.D. (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci. 14: 133–139 [DOI] [PubMed] [Google Scholar]
- Han, Y., Xu, M., Liu, X., Yan, C., Korban, S.S., Chen, X. and Gu, M. (2004) Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci. 166: 357–364 [Google Scholar]
- Hirano, H.Y., Eiguchi, M. and Sano, Y. (1998) A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 15: 978–987 [DOI] [PubMed] [Google Scholar]
- Hirano, H.Y. and Sano, Y. (1998) Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol. 39: 807–812 [Google Scholar]
- Inukai, T. and Hirayama, Y. (2010) Comparison of starch levels reduced by high temperature during ripening in japonica rice lines near-isogenic for the Wx locus. J. Agron. Crop Sci. 196: 296–301 [Google Scholar]
- IPCC (2013) Summary for Policymakers. In: Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V. and Midgley, P.M. (eds.) Climate Change 2013: The Physical Science Basis. Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA [Google Scholar]
- Ishimaru, T., Horigane, A.K., Ida, M., Iwasawa, N., San-oh, Y.A., Nakazono, M., Nishizawa, N.K., Masumura, T., Kondo, M. and Yoshida, M. (2009) Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under high-temperature stress. J. Cereal Sci. 50: 166–174 [Google Scholar]
- Jagadish, S.V.K., Craufurd, P.Q. and Wheeler, T.R. (2007) High temperature stress and spikelet fertility in rice (Oryza sativa L.). J. Exp. Bot. 58: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Jagadish, S.V.K., Muthurajan, R., Oane, R., Wheeler, T.R., Heuer, S., Bennett, J. and Craufurd, P.Q. (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J. Exp. Bot. 61: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J.S., Ryoo, N., Hahn, T.R., Walia, H. and Nakamura, Y. (2010) Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48: 383–392 [DOI] [PubMed] [Google Scholar]
- Jin, L., Lu, Y., Xiao, P., Sun, M., Corke, H. and Bao, J. (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor. Appl. Genet. 121: 475–487 [DOI] [PubMed] [Google Scholar]
- Kharabian-Masouleh, A., Waters, D.L.E., Reinke, R.F. and Henry, R.J. (2011) Discovery of polymorphisms in starch-related genes in rice germplasm by amplification of pooled DNA and deeply parallel sequencing. Plant Biotechnol. J. 9: 1074–1085 [DOI] [PubMed] [Google Scholar]
- Kharabian-Masouleh, A., Waters, D.L.E., Reinke, R.F., Ward, R. and Henry, R.J. (2012) SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci. Rep. 2: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, P.D. and Park, W.D. (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 40: 719–727 [DOI] [PubMed] [Google Scholar]
- Larkin, P.D., McClung, A.M., Ayres, N.M. and Park, W.D. (2003) The effect of the Waxy locus (Granule Bound Starch Synthase) on pasting curve characteristics in specialty rices (Oryza sativa L.). Euphytica 131: 243–253 [Google Scholar]
- Larkin, P.D. and Park, W.D. (2003) Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol. Breed. 12: 335–339 [Google Scholar]
- Lee, G.A., Koh, H.J., Chung, H.K., Dixit, A., Chung, J.W., Ma, K.H., Lee, S.Y., Lee, J.R., Lee, G.S., Gwag, J.G.et al. (2009) Development of SNP-based CAPS and dCAPS markers in eight different genes involved in starch biosynthesis in rice. Mol. Breed. 24: 93–101 [Google Scholar]
- Little, R.R., Hilder, G.B. and Dawson, E.H. (1958) Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 35: 111–126 [Google Scholar]
- Mo, Y.J., Kim, K.Y., Park, H.S., Ko, J.C., Shin, W.C., Nam, J.K., Kim, B.K. and Ko, J.K. (2012a) Changes in the panicle-related traits of different rice varieties under high temperature condition. Aust. J. Crop Sci. 6: 436–443 [Google Scholar]
- Mo, Y.J., Kim, K.Y., Shin, W.C., Lee, G.M., Ko, J.C., Nam, J.K., Kim, B.K., Ko, J.K., Yu, Y. and Yang, T.J. (2012b) Characterization of Imcrop, a Mutator-like MITE family in the rice genome. Genes Genom. 34: 189–198 [Google Scholar]
- Mohammed, A.R. and Tarpley, L. (2009) Impact of high nighttime temperature on respiration, membrane stability, antioxidant capacity, and yield of rice plants. Crop Sci. 49: 313–322 [Google Scholar]
- Morita, S., Yonemaru, J.I. and Takanashi, J.I. (2005) Grain growth and endosperm cell size under high night temperatures in rice (Oryza sativa L.). Ann. Bot. 95: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Francisco, P.B., Hosaka, Y., Sato, A., Sawada, T., Kubo, A. and Fujita, N. (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58: 213–227 [DOI] [PubMed] [Google Scholar]
- Nei, M. (1973) Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, P.V.V., Boote, K.J., Allen, L.H., Sheehy, J.E. and Thomas, J.M.G. (2006) Species, ecotype and cultivar differences in spikelet fertility and harvest index of rice in response to high temperature stress. Field Crop Res. 95: 398–411 [Google Scholar]
- Rang, Z.W., Jagadish, S.V.K., Zhou, Q.M., Craufurd, P.Q. and Heuer, S. (2011) Effect of high temperature and water stress on pollen germination and spikelet fertility in rice. Environ. Exp. Bot. 70: 58–65 [Google Scholar]
- Rohlf, F.J. (1998) NTSYS-PC: Numerical taxonomy and multivariate analysis system, version 2.00. Exter Software, Setauket, New York, USA [Google Scholar]
- Sano, Y., Katsumata, M. and Okuno, K. (1986) Genetic studies of speciation in cultivated rice. 5. Inter- and intraspecific differentiation in the Waxy gene expression of rice. Euphytica 35: 1–9 [Google Scholar]
- She, K.C., Kusano, H., Yaeshima, M., Sasaki, T., Satoh, H. and Shimada, H. (2010) Reduced rice grain production under high-temperature stress closely correlates with ATP shortage during seed development. Plant Biotechnol. 27: 67–73 [Google Scholar]
- Tanaka, K., Onishi, R., Miyazaki, M., Ishibashi, Y., Yuasa, T. and Iwaya-Inoue, M. (2009) Changes in NMR relaxation of rice grains, kernel quality and physicochemical properties in response to a high temperature after flowering in heat-tolerant and heat-sensitive rice cultivars. Plant Prod. Sci. 12: 185–192 [Google Scholar]
- Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., Liu, G., Gao, Z., Tang, S., Zeng, D.et al. (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 106: 21760–21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto, T., Aoki, N., Lin, H., Nakamura, Y., Inouchi, N., Sato, Y., Yano, M., Hirabayashi, H. and Maruyama, S. (2004) Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 31: 671–684 [DOI] [PubMed] [Google Scholar]
- Umemoto, T. and Aoki, N. (2005) Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 32: 763–768 [DOI] [PubMed] [Google Scholar]
- Wanchana, S., Toojinda, T., Tragoonrung, S. and Vanavichit, A. (2003) Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 165: 1193–1199 [Google Scholar]
- Yamakawa, H., Hirose, T., Kuroda, M. and Yamaguchi, T. (2007) Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144: 258–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, S., Nakamura, I., Watanabe, K.N. and Sato, Y.I. (2004) Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 108: 1200–1204 [DOI] [PubMed] [Google Scholar]
- Yan, C.J., Tian, Z.X., Fang, Y.W., Yang, Y.C., Li, J., Zeng, S.Y., Gu, S.L., Xu, C.W., Tang, S.Z. and Gu, M.H. (2011) Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor. Appl. Genet. 122: 63–76 [DOI] [PubMed] [Google Scholar]
- Ye, C., Argayoso, M.A., Redoña, E.D., Sierra, S.N., Laza, M.A., Dilla, C.J., Mo, Y., Thomson, M.J., Chin, J., Delaviña, C.B.et al. (2012) Mapping QTL for heat tolerance at flowering stage in rice using SNP markers. Plant Breed. 131: 33–41 [Google Scholar]
- Zakaria, S., Matsuda, T., Tajima, S. and Nitta, Y. (2002) Effect of high temperature at ripening stage on the reserve accumulation in seed in some rice cultivars. Plant Prod. Sci. 5: 160–168 [Google Scholar]
- Zhong, L.J., Cheng, F.M., Wen, X., Sun, Z.X. and Zhang, G.P. (2005) The deterioration of eating and cooking quality caused by high temperature during grain filling in early-season indica rice cultivars. J. Agron. Crop Sci. 191: 218–225 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.