Abstract
Microtubule-based motor proteins play key roles during mitosis to assemble the bipolar spindle, define the cell division axis, and align and segregate the chromosomes. The majority of mitotic motors are members of the kinesin superfamily. Despite sharing a conserved catalytic core, each kinesin has distinct functions and localization, and is uniquely regulated in time and space. These distinct behaviors and functional specificity are generated by variations in the enzymatic domain as well as the non-conserved regions outside of the kinesin motor domain and the stalk. These flanking regions can directly modulate the properties of the kinesin motor through dimerization or self-interactions, and can associate with extrinsic factors, such as microtubule or DNA binding proteins, to provide additional functional properties. This review discusses the recently identified molecular mechanisms that explain how the control and functional specification of mitotic kinesins is achieved. © 2013 Wiley Periodicals, Inc.
Keywords: mitosis, kinesin, microtubules, motors, regulation
Introduction
Microtubules are essential players in mitosis [Mitchison and Kirschner, 1984; Goshima and Scholey, 2010]. However, the dynamic properties and polarity of microtubules are not sufficient to organize microtubules into a spindle or align the chromosomes. Multiple microtubule-based motors, as well as non-motor microtubule-associated proteins, are key to mediating these processes. Most eukaryotic cells contain a single minus-end directed cytoplasmic dynein motor and multiple kinesin motor proteins that perform distinct microtubule-based processes, apart from higher plants that lack dynein. Kinesins have a conserved ATPase domain, but each kinesin family member utilizes the energy from ATP hydrolysis to perform different functions, relying on both its motor domain and its non-motor regions to create functional specificity. Although the motor domains of kinesins have been extensively studied, this domain represents only one part of the kinesin. Regions flanking the catalytic domain have divergent primary sequences and are essential for creating functional diversity. These divergent regions provide a molecular basis for unique interactions with cargo molecules, protein partners, and self-interaction that can also be uniquely regulated by post-translational modifications. Many of the early studies that revealed the molecular mechanism underlying kinesin diversity were conducted on kinesins involved in vesicular transport. Recently, there have been several reports revealing that mitotic kinesins require additional partners to target to the correct subcellular location. These regulatory domains and associated partners of the kinesin motors are thus key to understanding how mitotic kinesins target to and function at the correct place within the cell. Functional accuracy and specificity during mitosis is key to achieve correct chromosome segregation. Here, I review the intrinsic and extrinsic factors that provide specificity to mitotic kinesins and the underlying mechanisms for their correct targeting and function, focusing particularly on the kinesin proteins that play a role during mitosis.
Identification and Structural Characterization of the Kinesins
The first kinesin, Kinesin-1, was identified from squid giant axons [Vale et al., 1985] as a microtubule-based motor that was distinct from the minus end-directed cytoplasmic dynein motor. Subsequent work demonstrated that Kinesin-1 is conserved across species [Brady, 1985; Scholey et al., 1985; Neighbors et al., 1988; Saxton et al., 1988] and shares a common ancestor with the actin-based motor myosin [Kull et al., 1996]. Since the original discovery of the first kinesins, many additional kinesins have been identified due to the high degree of conservation of the kinesin motor domain that possesses the catalytic ATPase activity. In the early 1990s, it became apparent that a superfamily of kinesin motors existed with divergent motor-flanking regions and unique functions [Vale and Goldstein, 1990].
Biochemical and structural characterization of the Kinesin-1 established the tripartite architecture of this motor [Hirokawa et al., 1989; Yang et al., 1989]. The motor region containing the ATPase domain forms a large globular structure, also termed the kinesin “head.” Most kinesins have a coiled-coil region, the “stalk,” that flanks the motor domain and is often important for dimerization. Stalk length and flexibility can vary greatly between kinesins with the stalk in Kinesin-1 of 80 nm, but CENP-E's coiled-coil region being 230 nm long [Hirokawa et al., 1989; Kim et al., 2008]. The hinge region linking the motor domain to the stalk is termed the “neck.” At the other end of the stalk, the “tail” region is highly divergent, allowing kinesin specification. The kinesin superfamily can be subdivided into three groups depending on the position on the motor region within the polypeptide chain. The kinesin head is at the N-terminus in Kinesin-1 to -12 and at the C-terminus of Kinesin-14. Kinesin-13's are atypical with the motor domain flanked by additional regions on each side [Hirokawa, 1998]. In total, the kinesin superfamily shares common structural features such as the neck region and the presence or absence of a dimerization domain but the organization and position of these domains with respect to the ATPase region create functional diversity.
Phylogenetic Organization and Functional Diversity of Kinesins
After the initial discovery of Kinesin-1 in diverse species, additional kinesin-like molecules were identified in various organisms [Brady, 1985; Scholey et al., 1985; Vale et al., 1985; Yang et al., 1988]. The minus bend directed motor Kar3 and the Eg5 homologues Cut7 and BimC were identified in fungi [Enos and Morris, 1990; Hagan and Yanagida, 1990; Meluh and Rose, 1990]. This marked the birth of the kinesin superfamily [Vale and Goldstein, 1990]. The sequence of the Drosophila Khc gene, encoding the heavy chain of Kinesin-1, allowed the identification of multiple kinesin genes, which were initially termed kinesin-like proteins (KLP) based on sequence homology of the ATPase domain [Yang et al., 1989; McDonald and Goldstein, 1990; McDonald et al., 1990]. A phylogeny of the kinesin superfamily was established to classify the kinesins according to their sequences [Moore and Endow, 1996; Hirokawa, 1998]. Kinesins are classified into 14 families according to their sequence and function [Lawrence et al., 2004]. Large-scale genome sequencing further refined this kinesin phylogeny and helped identify new kinesins [Dagenbach and Endow, 2004; Miki et al., 2005; Wickstead and Gull, 2006]. The large scale sequence data on eukaryotic kinesins however challenged the proposed classification and stated that Kinesin-4 and -10 should be united into one class, while Kinesin-12 members Kif12 and Kif15 should be in separate classes [Wickstead and Gull, 2006]. In this review, we follow the classification nomenclature proposed by Lawrence et al. [2004]. The Kinesin-1 to -12 families are plus-end directed motors that move towards the rapidly growing end of microtubules, whereas Kinesin-13 are microtubule depolymerases. There is only one minus end directed family, Kinesin-14, most likely due to dynein acting as a main contributor to minus-end directed transport along microtubules. In species lacking dynein, motors may adopt bidirectional properties. For example, budding yeast Cin8 single motors are minus-end directed on individual microtubules. However, teams of Cin8 become plus-end directed motors [Roostalu et al., 2011]. All eukaryotes contain multiple kinesin motors with conserved function, suggesting the last common ancestor also possessed multiple kinesins [Wickstead et al., 2010]. The number of kinesins within a species can vary widely due to gene loss (e.g., loss of Kinesin-14 in Apicomplexa), gene duplications and subsequent divergence, generating diversity, and versatility within each kinesin motor family [Vale, 2003; Wickstead and Gull, 2006]. Budding yeast contain six different kinesins belonging to five different subfamilies, Cin8 and Kip1 being partially redundant, whereas human cells encode 45 different kinesin family members [Hirokawa et al., 2009bb], most of which are plus end directed motors. Flowering plants such as Arabidopsis have at least 61 kinesins, of which 21 are minus-end directed motors perhaps to compensate for the lack of dynein in plants [Lawrence et al., 2001; Reddy and Day, 2001; Lee and Liu, 2004]. Overall, each kinesin fulfills functions distinct from its related family members and has unique properties. Here, I will focus on the diverse kinesin proteins that play essential roles during mitosis to organize the spindle and direct chromosome alignment and segregation.
Most kinesins appear to have non-overlapping functions, but these can be masked by the existence of multiple parallel pathways. For example, RNAi-based screens in Drosophila S2 cells have defined groups of kinesin motors that play synergistic roles in chromosome congression or bipolar assembly [Goshima and Vale, 2003b; Goshima et al., 2005b]. In metazoans, kinesin motors with distinct molecular functions act synergistically in parallel pathways to ensure the robustness of bipolar spindle assembly [Mountain et al., 1999; Sharp et al., 1999; Tanenbaum et al. 2008, 2009]. In general, the kinesins that participate in related functional processes do not belong to the same family, and have different domain structures such that they act together through distinct molecular mechanisms. For example, the Kinesin-12 Kif15 and the Kinesin-13 MCAK are important for maintaining a bipolar spindle when the activity of the Kinesin-5 Eg5 is compromised [Tanenbaum et al., 2009]. Overall, mitotic kinesins have distinct cellular functions that allow fine-tuning and robustness of mitotic microtubule-based processes.
Although the kinesin motors domains all bind to microtubules and hydrolyze ATP, the way in which this enzymatic activity is harnessed to direct intracellular functions varies dramatically. The unique roles that kinesins play in a given process arise primarily from the functional contributions of the divergent non-motor regions, through both their intrinsic features and extrinsic associated factors. Here, I will discuss the primary molecular mechanisms that appear to create this kinesin functional specification:
Functional diversity of the kinesin motor domain.
Oligomerization and self-interactions.
Non-conserved regions that modify the properties or targeting of the motor domain, or alter motor activity.
Divergent regions that interact with microtubules, thereby providing a second microtubule binding site to the kinesin.
Interactions with proteins that target kinesins to a particular sub-cellular location, for example by providing a microtubule or DNA binding activity.
Post-translational modifications of kinesins.
Functional Diversity of the Motor and Neck Domains
A significant focus of work on kinesin motor proteins has involved the analysis of the molecular and catalytic mechanisms underlying the ATP hydrolysis cycle of the motor and how this chemical energy is coupled to produce mechanical force [Cross, 2004; Carter and Cross, 2006; Block, 2007]. Molecular snapshots of kinesin motor domains in distinct nucleotide-bound states by X-ray crystallography or kinesins bound to microtubules visualized using cryo-electron microscopy have provided important insights into the catalytic mechanism by which kinesins utilize ATP to produce a conformational change that can be converted into a mechanical force [Vale et al., 2000; Marx et al., 2009]. Depending on the unique features of the kinesin motor domain, this force manifests in different ways. For processive kinesins, this force can be converted into movement and stepping along the microtubule lattice using a hand-over-hand mechanism [Asbury et al., 2003; Kaseda et al., 2003; Yildiz et al., 2004]. In contrast, non-processive kinesins such as Ncd, adopt a microtubule hold-and-release mechanism that does not involve stepping. Finally, microtubule depolymerases utilize force to induce or stabilize a curved conformation of single protofilaments thereby causing microtubule catastrophe and disassembly [Niederstrasser et al., 2002; Ogawa et al., 2004; Varga et al., 2006]. Some kinesins can act both as processive motors and as microtubule depolymerases. For example, Kar3/Cik1 is a S. Cerevisiae Kinesin-14 that has minus end directed activity and microtubule directed plus end depolymerase activity [Endow et al., 1994; Sproul et al., 2005]. It is under debate whether the Kinesin-8 members also act both as depolymerases and processive motors. The yeast Kinesin-8 Kip3 is a microtubule depolymerase, proposed to depolymerize microtubules in an age-dependent manner [Varga et al., 2006; Gardner et al., 2011; Su et al., 2011]. Yet, mammalian Kinesin-8 members do not show intrinsic microtubule depolymerase activity in vitro, but rather causes pausing in microtubule dynamics [Stumpff et al., 2008; Peters et al., 2010; Stumpff et al., 2012]. Kif18a depletion results in longer spindles which could correlate with microtubule depolymerase activity [Mayr et al., 2007]. However, spindle length also increases in absence of kinetochore proteins mediated-pulling forces [DeLuca et al., 2002; Toso et al., 2009]. It is possible that Kinesin-8 motors, while processive in most species, have acquired new additional depolymerase properties to compensate for the lack of Kinesin-13 in S. cerevisiae.
High structural conservation of the motor domain is necessary to preserve the ATPase properties and microtubule binding properties of the kinesin. Overall, a layer of β-strands sandwiched by α-helices form the highly conserved core of the kinesin motor domain, which also contains the microtubule binding region [Hirokawa et al., 1998a]. However, comparative analyses of known kinesin structures have identified unique features within the ATPase domain that create functional specificity of the motor. Variations in the primary and tertiary sequences of kinesins are mapped primarily to the loop regions within the motor domain [Kozielski et al., 1999; Ogawa et al., 2004; Neumann et al., 2006; Cochran et al., 2009; Peters et al., 2010]. These structural variations impact the affinity of the motor for the microtubule, its processivity, the nucleotide-gated switch to modulate the ATP cycle, and its kinetic parameters. Kinesins can be classified according to these kinetic parameters and properties [Friel and Howard, 2012]. The adjacent neck region is also conserved across kinesin families, although less so than the motor domain. The neck is essential for specifying the directionality of kinesin movement along the microtubule and gives rise to unique properties within kinesin families [Case et al., 1997; Henningsen and Schliwa, 1997; Endow and Waligora, 1998; Sablin et al., 1998; Endow, 1999]. In addition, comparative structural analysis of kinesin motors in different nucleotide-bound states and conformations allows the evaluation of individual mitotic kinesins as cancer drug targets [Rath and Kozielski, 2012]. In total, while being conserved, the motor domains of kinesins show structural and biophysical differences translate into unique functional properties.
Oligomerization and Self-interactions
In many cases, kinesins homo-multimerize through a coiled-coil region called the stalk, which allows them to step along the length of the microtubule in a hand-over-hand manner with one head remaining associated with the microtubule at all times (Fig. 1A). The Kinesin-5 family forms tetramers and functions to slide antiparallel microtubules away from each other, especially during formation of a bipolar spindle [Cole et al., 1994; Kashina et al. 1996a, 1996b; Kapitein et al., 2005; van den Wildenberg et al., 2008]. This tetramerization property is essential to allow each pair of motor heads to crosslink and slide along the length of proximally-associated antiparallel microtubules. One long coiled-coil adjacent to the N-terminal motor domain forms the central stalk and dimerization interface of the motor pair. Three additional downstream coiled-coils mediate the tetramerization of the Kinesin-5 complex [Weinger et al., 2011]. While many kinesins multimerize to allow them to walk along microtubules, some kinesins can even display highly processive motility acting as monomers. The first kinesins to be identified as monomeric are Kif1a and Kif1b, both of which are involved in axonal transport of synaptic vesicles and mitochondria in neurons [Nangaku et al., 1994; Okada et al., 1995]. Kif1a and Kif1b lack a dimerization domain and form a globular compact structure, yet are highly processive in vitro [Hirokawa and Noda, 2008; Hirokawa et al., 2009a]. However, Kif1a is also found in a dimeric state in vivo that is essential for its motility [Tomishige et al., 2002; Lee et al., 2004; Hammond et al., 2009]. The controlled transition of a kinesin monomeric to dimeric state might regulate the activity of such kinesins in cells. Subsequently, the Kinesin-10 chromokinesins have also been shown to be functionally monomeric [Matthies et al., 2001; Shiroguchi et al., 2003]. Kid displays plus-end directed motility [Yajima et al., 2003; Stumpff et al., 2012], whereas its meiotic counterpart NOD in Drosophila has been proposed to lack motility [Matthies et al., 2001]. Recent work suggests that Nod may be able to track the plus ends of microtubules [Cane et al., 2013]. For non-canonical kinesins that do not display motility, oligomerization can also be important. Dimerization of the Kinesin-13 MCAK/Kif2c increases the efficiency of the microtubule depolymerase activity relative to a monomeric depolymerase domain of MCAK in vitro [Maney et al., 2001; Hertzer et al., 2006]. Overall, kinesins can form higher order structures through coiled-coil regions, such as dimers or tetramers that further define their properties.
Fig. 1.
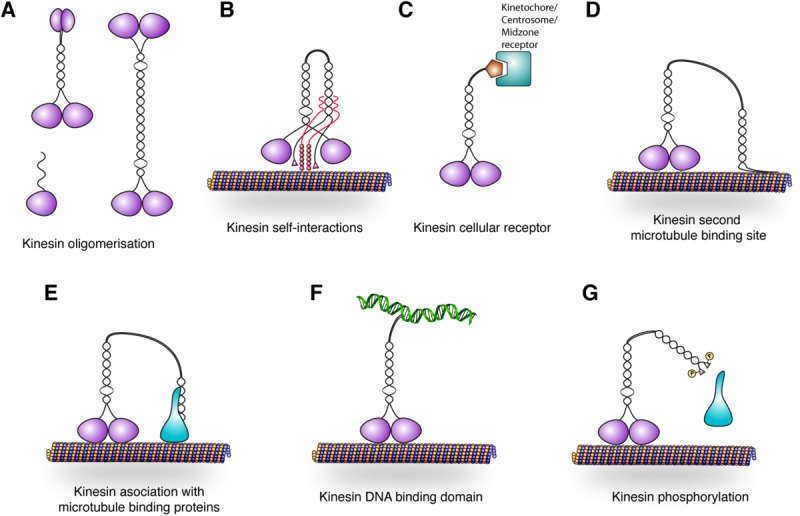
Schematic diagram showing how the non-motor regions of kinesins may acquire properties that would specify unique function to the catalytic motor domain. (A) The non-motor domains can support the monomeric or oligomeric state of kinesins. (B) The non-motor regions can self interact or interact with the ATPase domain of kinesins, most of the time to inhibit kinesin function. (C) The non-motor regions can direct the targeting of the kinesin to a specific localization that acts as a kinesin receptor. (D) Non-motor regions may have a microtubule binding domain or (E) may associate with a microtubule binding protein. (F) Chromokinesins have a DNA binding region. (G) All of these interactions can be regulated by post-translational modifications to add an additional layer of regulation to kinesin function. The schematic diagram of kinesins was adapted from the review Verhey and Hammond [2009].
Kinesins can also use self-interactions to regulate their activity and prevent non-productive ATP hydrolysis. Binding of non-motor regions to the kinesin ATPase domain traps the ATPase domain in an auto-inhibited state (Fig. 1B). Auto-inhibition is particularly well-documented with motors involved in the transport of cargos [Verhey and Hammond, 2009]. The most abundant kinesin, Kinesin-1, has been used as a paradigm for studies on auto-inhibition of kinesins. Kinesin-1 transports cargos throughout the cell. To avoid the unnecessary use of ATP, Kinesin-1 remains in an autoinhibited state until cargo binding occurs, which activates the motor [Cai et al., 2007]. Early electron microscopy and biophysical studies revealed that kinesin-1 adopts a folded conformation where the motor domain is in close proximity with the tail region [Hackney et al., 1991, 1992]. Full-length Kinesin-1 has only limited ATPase activity due to the tail region binding to and inhibiting the ATPase motor domains, with one tail binding to the two heads [Coy et al., 1999; Friedman and Vale, 1999; Stock et al., 1999; Hackney et al., 2009]. The inhibitory tail binds at the interface of a motor domain dimer and makes a second point of attachment between the two motors, thereby preventing movement of one motor with respect to the other (Fig. 2) [Kaan et al., 2011]. Similarly, an auto-inhibitory mechanism via the C-terminal tail of the Kinesin-7, CENP-E, and the Kinesin-2, Kif17 has been proposed to regulate their motility [Espeut et al., 2008; Hammond et al., 2010]. The inhibition of CENP-E is relieved by CDK1 or Mps1-dependent phosphorylation in the tail region. Finally, the extreme C-terminal region of MCAK is thought to influence its catalytic activity and affinity of MCAK for microtubules [Moore and Wordeman, 2004]. This auto-inhibition mechanism does not appear to be conserved across species, as the removal of the C-terminal tail of Xenopus MCAK results in a decrease in depolymerase activity [Hertzer et al., 2006]. Typically, the self-interaction of a non-motor region with the motor domain of the kinesin results in inhibition that is relieved by an active mechanism. This auto-inhibition mechanism represents an interesting paradigm for the temporal or spatial control of a kinesin.
Fig. 2.
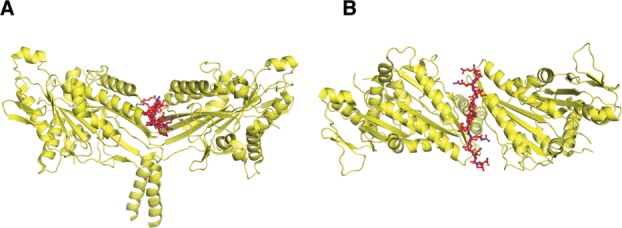
Structure of Kinesin-1 bound to its tail. (A) Side and (B) top view of Drosophila melanogaster kinesin motor domain dimer complexed with tail domain. This is the only structure to date to show interactions of a kinesin motor domain with other regions (based on the structure published in Kaan et al. [2011], PDB: 2Y65).
Specification of Kinesin Function by Non-motor Regions
Kinesins are large molecules and contain multiple divergent domains flanking the motor ATPase domain. These regions may associate with distinct kinesin “receptors” within a cell, thereby creating specificity that targets a given kinesin to a defined subcellular location (Fig. 1C). The role of non-motor regions in specific targeting was proposed once the extent of the functional diversity within the kinesin superfamily was realized [Vale and Goldstein, 1990]. Extensive work on kinesins involved in trafficking, particularly in neurons, has provided information on how kinesin diversity of the non-motor regions is essential for targeting to specific subcellular locations [Goldstein and Yang, 2000; Hirokawa and Noda, 2008; Hirokawa et al., 2009b].
The non-motor regions of mitotic kinesins also direct the targeting of the motor to the correct subcellular location. The highly conserved microtubule cross-linking protein PRC1 (Ase1p in yeast and MAP65 in plants) is important for specifying the spindle mid-zone, a region of overlapping microtubules that determine the cleavage plane in anaphase and telophase [Jiang et al., 1998; Smertenko et al., 2000; Mollinari et al., 2002; Schuyler et al., 2003; Verbrugghe and White, 2004; Verni et al., 2004]. In addition, PRC1 crosslinks antiparallel microtubules through a spectrin-fold domain and bundles microtubules in vitro [Subramanian et al., 2010]. In cells, PRC1 acts as a receptor for multiple mitotic kinesins in the spindle mid-zone during anaphase. The N-terminus of Kif14 flanking the motor region associates with PRC1 and localizes to the mid-zone, independently of the motor domain [Gruneberg et al., 2006]. PRC1 also associates with the C-terminal region of the chromokinesin Kif4a to control the length of the mid-zone microtubules [Kurasawa et al., 2004; Bieling et al., 2010b; Subramanian et al., 2010]. The Kif4a/PRC1 complex accumulates at microtubule ends in a microtubule-length dependent manner [Subramanian et al., 2013]. Phosphorylation of PRC1 by Cdk1/cyclinB prevents its association with Kif4a until anaphase. Regulation of the PRC1-Kif4a interaction thus provides a spatial and temporal context to restrict this interaction until after the metaphase-anaphase transition [Zhu and Jiang, 2005; Zhu et al., 2006]. PRC1 also associates with additional kinesins such as MKLP1/Kif23 and MKLP2/Kif20 to target them to the central spindle and mid-zone during the late stages of mitosis [Verni et al. 2004; Gruneberg et al., 2006; Bassi et al., 2013]. Disruption of PRC1 localization leads to altered localization of these kinesins and multiple defects in cytokinesis. Therefore PRC1 acts as a receptor to recruit mitotic kinesins to the mid-zone and allow correct cytokinesis.
Within a kinesin family, the divergent N-termini can provide diversity in targeting the conserved catalytic activity. When expressed alone, the motor domains of the kinesins-13 family proteins localize uniformly throughout the mitotic spindle [Welburn and Cheeseman, 2012]. However, in the context of the full-length protein, each of the three human family members (Kif2a, Kif2b, and Kif2c/MCAK) displays a distinct localization within the cell where they play different functions (Figs. 3A and 3B). The divergent non-motor regions provide targeting specificity. The N-terminus of MCAK/Kif2c directs the kinesin to centromeres [Maney et al., 1998; Walczak et al., 2002]. At kinetochores, MCAK associates with Sgo2 in an Aurora B dependent manner to promote kinetochore alignment [Tanno et al., 2010; Rivera et al., 2012]. The N-terminus of Kif2a is also responsible for targeting the Kif2a microtubule depolymerase activity to the centrosome, but its interaction partners that target Kif2a to centrosomes are unknown [Welburn and Cheeseman, 2012]. Interestingly, the Kinesin-13 member Kif24 utilizes its motor domain to target to the centrosome [Kobayashi et al., 2011]. Thus comparative analysis highlights the differences between closely related proteins within a kinesin family and identifies both intrinsic and extrinsic factors that contribute to creating kinesin specificity and targeting.
Fig. 3.
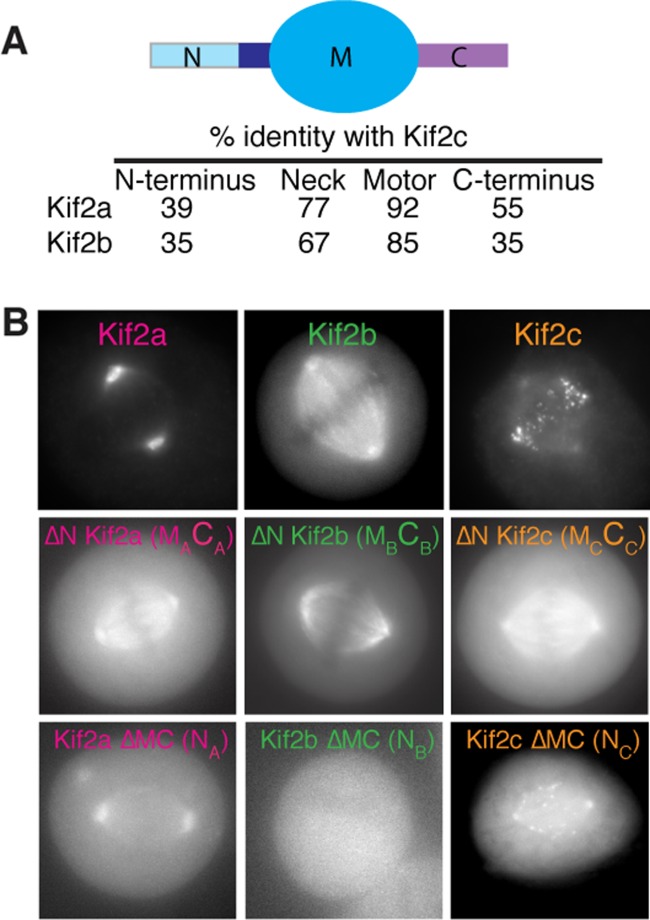
The N-terminus of Kinesin-13 is a primary determinant of kinesin localization. (A) Schematic diagram showing the Kinesin-13 domains and their percentage similarity with respect to Kif2c/MCAK. (B) Images of HeLa cells transiently expressing GFP fusions to full length and domains of Kif2a, Kif2b, and Kif2c. The ΔN and ΔMC domains represent GFP-fusions lacking the N-terminus or the motor and C-terminal domains of the kinesins, respectively. Adapted from Welburn and Cheeseman [2012].
Microtubule Interactions with Kinesin Non-motor Regions
Since the discovery of kinesins, multiple studies have identified a second microtubule binding site in kinesins in addition to the canonical motor domain that is essential for the correct function and targeting of the kinesin (Fig. 1D). The yeast Kar3 was the first motor proposed to have an N-terminus that can bind independently to microtubules, based on localization studies [Meluh and Rose, 1990]. The presence of a second nucleotide-insensitive microtubule domain has also been identified in Kinesin-1, -7, -8, -10, and -14 [Navone et al., 1992; Yen et al., 1992; Karabay and Walker, 1999; Shiroguchi et al., 2003; Wendt et al., 2003; Seeger and Rice, 2010; Moua et al., 2011]. The microtubule binding properties of the non-motor regions differ from those of the motor domains such that they are likely to provide a second low affinity binding site for the kinesin to increase its affinity for microtubules. I will focus here on the second microtubule binding domains of three families: (1) Kinesin-14, which are minus end directed motors and crosslink parallel microtubules, (2) Kinesin-5, which crosslinks anti-parallel microtubules, and (3) Kinesin-8, which depolymerizes and/or regulates microtubule plus ends.
Early biochemical work on the Kinesin-14 Ncd showed that this minus end directed motor had a second microtubule-binding site, independent of its C-terminal motor domain. The N-terminal domain can bind to and bundle microtubules in vitro [Chandra et al., 1993]. In fact, the N-terminal 200 amino acid tail contains two separate microtubule binding sites that are ATP-insensitive [Karabay and Walker, 1999; Wendt et al., 2003]. The mammalian, S. cerevisiae, and S. pombe Kinesin-14 counterparts HSET, Kar3, and Klp2 also have N-terminal microtubule binding domains in addition to the C-terminal motor domain [Meluh and Rose, 1990; Ando et al., 1994; Kuriyama et al., 1995; Braun et al., 2009]. Kinesin-14 family members possess two microtubule-binding sites at the N- and C-terminus of the polypeptide, which function in focusing microtubules at the spindle poles. The current working model for Kinesin-14 function is that the two microtubule binding sites enable parallel crosslinking of microtubules and pole focusing [Mountain et al., 1999; Goshima and Vale, 2003; Goshima et al., 2005a]. Indeed, recent work in vitro demonstrated that S. pombe Klp2 and Drosophila Ncd are sufficient to crosslink parallel microtubules [Braun et al., 2009; Fink et al., 2009]. It remains to be determined how Kinesin-14 functions in reconstituted assays in presence of Eg5 and other microtubule crosslinkers, to examine the contribution of Kinesin-14 to spindle formation, and define the mechanistic basis for their ability to preferentially crosslink parallel microtubules rather than microtubules of mixed polarity.
The function of a second microtubule-binding region in Xenopus Kinesin-5 (the functional homologue of Eg5 in humans) has also been well characterized. Kinesin-5 possesses a second microtubule-binding region downstream of the motor domain that is essential for Kinesin-5 microtubule crosslinking function and kinesin processivity. This activity is required even though Kinesin-5 forms tetrameric assemblies that already contain multiple microtubule binding sites [Weinger et al., 2011]. The second binding site does not alter the motility of the Kinesin-5 assembly, but instead increases its processivity. Collectively, the non-motor regions of Kinesin-5 associate with microtubules to maintain kinesin association with both antiparallel microtubules, despite stochastic dissociation of the Kinesin-5 motor heads from microtubules. This ensures robust sliding of antiparallel microtubules with respect to each other.
The Kinesin-8 family also uses a non ATP-sensitive microtubule-binding site for its function. In humans, Kif18a has been implicated in chromosome alignment [Mayr et al., 2007; Stumpff et al., 2008; Savoian and Glover, 2010], whereas Kif18b regulates the dynamics of astral microtubules, particularly during prometaphase [Stout et al., 2011; Tanenbaum et al., 2011]. The motor domains of Kif18b and Kif18a are highly conserved, whereas their C-terminal regions are more divergent. Kinesin-8 function is conserved across eukaryotes including yeast (Kip3, Klp5/6) and Drosophila (Klp67), despite some species-specific differences. The C-terminus of Kif18a homologues is essential for kinesin targeting across species [Savoian and Glover, 2010]. Recent studies have shown that the C-terminal region of Kif18a and Kip3 contains a microtubule-binding domain that is distinct from the motor domain [Mayr et al., 2011; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011]. This microtubule-binding site diffuses along the microtubule lattice and increases the dwell time of the motor on microtubules, which thereby increases the processivity of the kinesin. This allows Kinesin-8 to target to the end of microtubules. In addition, the Kinesin-8 C-terminal tail modulates its catalytic activity. In yeast, Kip3 can act as both a microtubule polymerase and a depolymerase, while the human Kif18a motor domain acts as a capping factor in vitro at the microtubule plus ends to stabilize microtubules [Du et al., 2010; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011]. Interestingly, in yeast this non-motor microtubule-binding domain interacts both with both tubulin dimers and the microtubule lattice. Thus, to stabilize the plus ends of microtubules, Kip3 may act on tubulin conformations found at plus ends of microtubules to straighten the protofilaments by cross-linking adjacent tubulin dimers [Su et al., 2011]. Kip3 may also locally increase the free tubulin concentration, thereby promoting microtubule polymerization, similarly to plus end binding proteins such as TOG and CLASP [Al-Bassam and Chang, 2011]. This second microtubule-binding site modulates the functional properties of Kinesin-8 motors, which results in differential localization and function. In total, it appears that a second microtubule binding region that is distinct from the motor domain of a kinesin contributes a pivotal role to the functional properties of a motor without interfering with its processivity and is widely used by kinesins.
Kinesin-Associated Proteins
In addition to the intrinsic properties and interactions of kinesin proteins, extrinsic associations with interacting proteins are critical for kinesin function. Recent targeted and broad-scale proteomic studies have analyzed the interactions of kinesin proteins to identify associated factors [Hutchins et al., 2010; Maliga et al., 2013]. Although many of these interactions remain to be fully characterized, it appears that such kinesin interactors play diverse roles in modulating kinesin function and localization. Here, I will discuss how kinesin associated proteins have defined properties to generate kinesin protein complexes with unique functions (Supporting Information Table S1).
Kinesins and Microtubule Associated Proteins
Although kinesin motors have been extensively studied in isolation, multiple kinesins associate with additional proteins for correct function (Fig. 1E). In some cases, these extrinsic associated factors display their own microtubule binding activity, which then modify the function of the kinesin.
A subset of microtubule-associated proteins enhances the targeting of multiple kinesins to microtubules. The spindle assembly factor Tpx2 associates with microtubules as the spindle assembles [Wittmann et al., 2000]. Tpx2 was first identified as a Targeting protein for Xklp2 in Xenopus [Wittmann et al., 1998]. Kif15, the human counterpart of Xklp2 essential for spindle bipolar maintenance, also localizes to the spindle in a Tpx2 manner [Tanenbaum et al., 2009; Vanneste et al., 2009]. In addition, the C-terminus of Tpx2 interacts with Eg5 to promote spindle bipolarity and K-fiber assembly [Eckerdt et al., 2008; Ma et al., 2010]. Interestingly, despite the microtubule cross-linking properties of the Eg5 tetramer, Tpx2 is required to localize Eg5 to the spindle microtubules and this interaction is essential for correct spindle formation [Ma et al., 2011]. Tpx2 is a large protein that acts as a platform to recruit Eg5 and Kif15 through distinct binding sites. This potentially allows Tpx2 to recruit them simultaneously to ensure robust spindle elongation and maintenance.
In the Kinesin-10 family, the chromokinesin Kid localizes to chromosomes and contributes to chromosome alignment. Kid shows microtubule plus end directed motility and contributes to polar ejection forces [Yajima et al., 2003; Stumpff et al., 2012; Wandke et al., 2012]. However, to localize to the spindle, Kid requires its interacting partner CHICA [Santamaria et al., 2008]. CHICA displays microtubule binding activity in vitro and localizes to spindles independently of Kid. In absence of CHICA, Kid does not target to spindles, but to date, this does not impair spindle dynamics and mitotic progression.
Other kinesins also associate with proteins that possess their own microtubule-binding domain to target to microtubules in different ways. For example, within the Kinesin-13 family of microtubule depolymerases, Kif2b and Kif2c/MCAK both associate with distinct microtubule-associated proteins. MCAK/Kif2c associates with the “end binding” (EB) family of proteins, which bind specifically to the plus ends of growing microtubules [Mennella et al., 2005; Lee et al., 2008]. MCAK binds to EB1 through a SXIP binding motif [Honnappa et al., 2009], and plus end binding is further enhanced through interactions with Tip150 and Kif18b [Jiang et al., 2009; Stout et al., 2011; Tanenbaum et al., 2011]. Both the motor domain and the MCAK-interacting domains of Kif18b are required for robust depolymerization, suggesting that Kif18b enhances MCAK activity at microtubule plus ends, rather than have intrinsic depolymerase activity [Tanenbaum et al., 2011]. In vitro MCAK enrichment at the plus ends does not prevent microtubule growth under these conditions, but increases the number of catastrophes, making microtubules more dynamic [Montenegro Gouveia et al., 2010]. It is possible that the in vitro system is lacking an MCAK activation partner such as Kif18b to promote depolymerization. In total, this plus end targeting mechanism provides MCAK with unique functional properties to control microtubule dynamics.
Despite the presence of a conserved catalytic core, the Kinesin-13 family member Kif2b, is regulated differently. Kif2b associates with the microtubule binding protein Cep170 and a related protein Cep170R. The Cep170 microtubule binding activity provides a second high-affinity microtubule-binding site for Kif2b and facilitates Kif2b targeting to the mitotic spindle [Welburn and Cheeseman, 2012]. Interestingly, Cep170 also interacts with Kifc3 and facilitates Kifc3 targeting to microtubules. Cep170 may also interact with Kif2a and Kif2c [Maliga et al., 2013], but the nature of these interactions is unclear. Overall, Cep170 may act as a general kinesin interactor, similarly to Tpx2, to recruit and coordinate several motor activities on the spindle [Hutchins et al., 2010; Welburn and Cheeseman, 2012; Maliga et al., 2013]. Kif2c/MCAK and Kif2a also associate with the microtubule-associated protein ICIS in Xenopus extracts, which stimulates their activity at the inner centromere [Ohi et al., 2003; Knowlton et al., 2009]. Thus, extrinsic factors that control Kinesin-13 specificity are unique to each kinesin, thereby creating functional diversity within a highly conserved kinesin family.
Although the proteins described above direct the localization of kinesins to microtubule structures throughout mitosis, some microtubule-associated proteins are important in recruiting kinesins to substructures, such as kinetochores, centrosomes, or the spindle mid-zone. At kinetochores, Clasp1 recruits multiple kinesins such as CENP-E and Kif2b during prometaphase [Hannak and Heald, 2006; Maffini et al., 2009; Manning et al., 2010]. The microtubule binding protein PRC1 recruits Kif14, MKLP1/Kif23/Centralspindlin, MKLP2/Kif20 and Kif14 at the central spindle and the mid-zone during the late stages of mitosis, as described previously [Kurasawa et al., 2004; Gruneberg et al., 2006; Bassi et al., 2013]. Such kinesin-interacting proteins therefore create a temporal and spatial switch for kinesin recruitment and activity to a particular structure.
Interestingly, the yeast minus-end directed motor Kar3 complex can form a heterodimer with two alternative partners Cik1 or Vik1 that have a motor-like domain, but not nucleotide-binding properties [Manning et al., 1999; Allingham et al., 2007]. The presence of the second non-catalytic microtubule-binding site increases the affinity of Kar3 for microtubules and binding becomes cooperative. Association of Kar3 with either Cik1 or Vik1 creates distinct kinesin complexes with different biochemical properties, thereby increasing functional diversity of the Kar3 motor complex [Page et al., 1994; Manning et al., 1999; Barrett et al., 2000]. Overall, in vitro and in vivo approaches have provided a molecular understanding for how extrinsic kinesin associated proteins can facilitate timely kinesin targeting to specific locations and modify the properties of the kinesin motor to enhance its affinity for microtubules.
Kinesin Interactions with Chromatin and Regulation by Nuclear Import
Chromokinesin Interactions with DNA
Chromokinesins associate with both chromosomal DNA and spindle microtubules during mitosis and are implicated in various functions during mitosis (Fig. 1F). In metazoans, there are three families of chromokinesins (Kinesin-4, -10, and -12) that are important for bipolar spindle formation, chromosome condensation, chromosome alignment, and chromosome segregation [Mazumdar and Misteli, 2005; Vanneste et al., 2011]. The Kinesin-12 family chromokinesin Kif15 is important for maintaining bipolar spindle assembly, particularly in absence of Eg5 [Tanenbaum et al., 2009; Vanneste et al., 2009]. The Xenopus homologue Xklp2 is also important for spindle maintenance, but it localizes to centrosomes, rather than chromosomes [Boleti et al., 1996]. Within the Kinesin-10 family, the Drosophila NOD plays a role in meiosis [Afshar et al., 1995; Matthies et al., 2001; Cochran et al., 2009]. Kid (Kinesin-like DNA binding protein) is present in mitotic cells, where it contributes to chromosome alignment [Tokai et al., 1996; Levesque and Compton 2001; Levesque et al., 2003]. Its Xenopus counterpart xKid (also termed Kif22) plays a role in chromosome alignment during mitosis [Antonio et al., 2000; Funabiki and Murray, 2000]. Both Kinesin-4 and the Kinesin-10 have been proposed to cooperate positively to align chromosomes [Brouhard and Hunt, 2005; Bieling et al., 2010a]. In Drosophila, Klp3 and NOD also synergize their activities to align chromosomes [Goshima and Vale, 2003]. Kinesin-10/Kid is the major contributor to polar ejection force for chromosomes [Brouhard and Hunt, 2005; Bieling et al., 2010a; Wandke et al., 2012; Cane et al., 2013]. Kinesin-4 inhibits microtubule dynamics and promotes pausing of the plus ends by altering the structure of the microtubule lattice [Bringmann et al., 2004; Stumpff et al., 2012]. However, recent studies in cells have challenged the view that the chromokinesins Kid and Kif4a work cooperatively, and have instead suggested that Kif4a and Kid play antagonistic roles for chromosome oscillations in human cells [Stumpff et al., 2012]. Kif4a has also been implicated in chromosome condensation and cytokinesis [Kurasawa et al., 2004; Mazumdar et al., 2004]. Its role in mitotic chromosome structure is dependent on the presence of the Kif4a motor domain [Samejima et al., 2012], although this function also likely depends on its chromatin-binding properties. Kid is also important for chromosome compaction during the later stages of mitosis [Ohsugi et al., 2008]. Taken together, each of the chromokinesin families performs unique functions in mitosis.
Despite the fact that the loading of each chromokinesin family onto chromosomes occurs at the onset of mitosis, each chromokinesin family has distinct DNA-targeting domains that create molecular specificity. Based on sequence analysis, NOD and Kid have a DNA binding helix–hairpin–helix (HhH) domain that binds to chromatin [Antonio et al., 2000; Yajima et al., 2003]. The Kinesin-4 family member Kif4 was first proposed to regulate vesicular trafficking in mice [Sekine et al., 1994]. Human Kif4a was then observed on chromosomes and proposed to be a chromokinesin [Lee et al., 2001]. Kif4a requires a basic zipper motif and a Cysteine-rich motif at its C-terminus to target to chromatin [Wu and Chen, 2008]. In addition, Kif4a associates with the chromosome condensation factors Condensin I and II to target to chromosomes [Samejima et al., 2012]. The Kif4a chromosome targeting mechanism therefore relies on additional chromatin-associated proteins and is tightly regulated to allow Kif4a relocalization to the mid-zone during the later stages of mitosis [Zhu and Jiang, 2005; Zhu et al., 2006]. Overall, chromokinesins utilize distinct DNA binding properties and unique protein associations that enhance their timely targeting to chromosomes.
Kinesin Interactions with Nuclear Import Machinery
The loading of chromokinesins onto chromosomes is also dependent upon chromosome-derived Ran-GTP signals, highlighting that the association of chromokinesins with DNA is controlled by both intrinsic and extrinsic factors [Levesque and Compton, 2001; Mazumdar et al., 2004; Vanneste et al., 2009]. Ran-GTP present in the vicinity of chromosomes disrupts the Kid-Importin α/β interaction, allowing Kid loading onto chromosomes [Tahara et al., 2008]. The Kinesin-12 member Kif15 associates with the chromatin factor KI-67 to load onto chromatin [Vanneste et al., 2009], and relies on Tpx2 (Targeting Protein for XKpl2) for its spindle targeting [Tanenbaum et al., 2009]. Interestingly, Tpx2 is itself tightly regulated by the Ran-GTP gradient and is critical for spindle assembly [Wittmann et al., 1998, 2000; Gruss et al., 2001]. Other kinesins may also be influenced by the chromosome-derived Ran-GTP gradient. The Kinesin-8 Kif18a has been reported recently to interact with HURP, a K-fiber associated protein important for spindle assembly [Ye et al., 2011]. HURP localization to the spindle is Ran-dependent [Koffa et al., 2006; Sillje et al., 2006]. It remains to be determined whether disruption of the Ran-GTP gradient would alter Kif18a localization through HURP mislocalization. Although the Kinesin-8 family has been reported to walk along microtubules and accumulates at the plus end, Kif18a selectively targets to K-fiber microtubules within the spindle microtubules. The Ran-dependent HURP association with Kif18a could explain this targeting specificity. However, in taxol-treated cells, Kif18a targets non-specifically to all microtubules. Thus, Kif18a's specific localization to K-fibers is at least partly due to their stability relative to non-kinetochore microtubules [Masuda et al., 2011; Stumpff et al., 2011]. In conclusion, the Ran gradient plays a role supporting spindle formation and chromosome alignment. The Ran pathway regulates a number of mitotic kinesins to achieve correct chromosome biorientation and spindle dynamics.
Interactions of Kinesins with Other Proteins to Modulate their Activity
Kinesins can associate with microtubule associated proteins or DNA, as described above. In addition, there are examples of kinesins associating with proteins that have catalytic properties to provide additional and new functions to kinesin complex. A well-studied example is the centralspindlin complex, which has two key enzymatic activities. Centralspindlin is composed of the Kinesin-6 family member MKLP1 and a kinesin-associated protein that has a RhoGTPase activating protein domain (RacGAP1). The C. elegans centralspindlin counterpart ZEN-4-CYK4 is particularly well studied biochemically and structurally, revealing that the formation of a heterodimeric stable complex is essential for its function [Jantsch-Plunger et al., 2000; Mishima et al., 2002; Pavicic-Kaltenbrunner et al., 2007]. As a result, the centralspindlin complex has multiple activities conferred by its different properties. Centralspindlin positions the spindle division plane, clusters microtubules in the mid-zone, regulates the mid-zone, and contributes to abscission by controlling Rho family GTPases [White and Glotzer, 2012]. Thus, association of a kinesin with a protein bearing enzymatic activity provides the kinesin complex with new catalytic functions to perform accurate mitosis.
Mitotic Kinesins as Recruitment Hubs
Molecular motors can generate force to assemble and maintain the mitotic spindle and align the chromosomes. However, a subset of kinesins also plays a role in recruiting molecules to cellular structures such as kinetochores in mitosis. For example, kinesins have been reported to interact and recruit signaling and checkpoint proteins [Hutchins et al., 2010; Kim et al., 2010; Meadows et al., 2011]. Mitotic motors including the Kinesin-7 CENP-E and Kinesin-8 Klp5/6 have a PP1 phosphatase binding motifs. Current models propose that these motors act as platforms at the kinetochore to recruit the checkpoint silencing phosphatase PP1, which is essential for bi-orientation and stabilization of kinetochore-microtubule attachments [Kim et al., 2010; Meadows et al., 2011]. Both Kinesin-8 and CENP-E localize to kinetochores during prometaphase. CENP-E leaves the kinetochore as chromosomes become biorientated [Yen et al., 1991, 1992]. CENP-E and Kinesin-8 could therefore recruit PP1 to kinetochores as microtubule attachments are being stabilized. At stable end-on attachments, KNL-1, the primary PP1 receptor at kinetochores is spatially separated from Aurora B at the inner centromere and dephosphorylated, thereby creating additional PP1 binding sites [Liu et al., 2010]. In fission yeast, both Klp5/6 and Spc7/Knl-1 are necessary to silence the checkpoint [Meadows et al., 2011]. In addition, the recruitment of a phosphatase such as PP1 to kinetochore-targeted motors can fine-tune the activity of these motors by opposing Aurora B and Plk1 activity as tension is established [Liu et al., 2010]. At kinetochores, CENP-E has also been proposed to act as a “cyclin” for BubR1 kinase activity, [Mao et al., 2003; Weaver et al., 2003; Mao et al., 2005] although whether BubR1 has kinase activity that is required for mitosis is debated [Suijkerbuijk et al., 2012; Elowe, 2011]. In total, processive kinesins may use their unique non-motor domains to act as an interaction platform and recruit molecules to a particular structure to drive mitotic progression.
Regulation of Interactions by Post-translational Modifications
In addition to associating with other proteins, kinesin activity is controlled by post-translational modifications (Fig. 1G). The most extensively studied post-translational modification that affects mitotic motors is phosphorylation. A large subset of kinesins only associate with microtubules in mitosis. These microtubule-kinesin associations coincide with a dramatic increase in protein phosphorylation that occurs upon mitotic entry [Dephoure et al., 2008]. In some cases, mitotic kinesins are retained in the nucleus in interphase by their nuclear localization signal preventing their association with the cytoplasmic microtubule cytoskeleton, such as Kid and Kif18a [Tokai et al., 1996; Du et al., 2010]. However, other kinesins that play roles in mitosis are present in the cytoplasm during interphase and do not bind to microtubules [Houliston et al., 1994; Vanneste et al., 2009]. Often, phosphorylation directly controls the ability of motors to associate with microtubules [Syred et al., 2013]. Based on large-scale phospho-proteomic analyses, the motor domain of kinesins is rarely post-translationally modified. In contrast, the non-motor regions are often phosphorylated, which is key for differentially regulating the members of a kinesin family. Recent work has provided molecular insights into the role of phosphorylation on mitotic motor function and regulation for a small number of kinesins as a rapidly reversible way to fine-tune molecular motors.
Phosphorylation May Regulate Kinesin Affinity for Microtubules
Phosphorylation may dramatically modify the electrostatic surface properties of the kinesin to increase its overall negative charge. As a result, phosphorylation can reduce the affinity of a kinesin for microtubules through electrostatic interference with the acidic, negatively charged tubulin lattice. Both motile and depolymerizing kinesins use electrostatic complementarity to facilitate directional and processive movement on the lattice [Thorn et al., 2000]. Kinesins have a positively charged neck region that interacts with the microtubule to promote kinesin diffusion on the lattice through electrostatic interactions [Thorn et al., 2000; Niederstrasser et al., 2002; Ovechkina et al., 2002]. Addition of negative charges in the kinesin neck reduces the run length of conventional kinesin [Thorn et al., 2000]. The positively charged neck region of the Kinesin-13 MCAK/Kif2c is critical for depolymerase activity [Cooper et al., 2010]. Phosphorylation of the neck region (Ser192 MCAK in humans, S196 in Xenopus) by Aurora B reduces its depolymerase activity [Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004]. The negative charge may neutralize the positively charged neck region to ultimately reduce kinesin binding to the microtubule and reduce depolymerase activity. Thus, regulation by phosphorylation of the neck region modulates the activity of kinesins dramatically.
CENP-E phosphorylation of a region close to the neck region on T422 is critical for mitosis [Kim et al., 2010]. Both Aurora A and B phosphorylate this residue to reduce the affinity of the motor for microtubules and its processivity. Aurora A-phosphorylated CENP-E can rapidly search and capture of chromosomes close to the poles, due to its lower affinity for microtubules which allows for greater sampling. As CENP-E moves the chromosomes along existing K-fibers to the equator of the cell, CENP-E is dephosphorylated by its binding partner PP1, which increases the processivity of the motor and allows chromosome alignment. Thus, CENP-E phosphorylation and binding to a phosphatase allows the fine-tuning the activity of the motor during chromosome congression and alignment. Thus phosphorylation of regions that interact with microtubules may also directly affect the processive properties of the kinesin.
Surprisingly, phosphorylation of a kinesin in a non-motor region can also increase its affinity for microtubules, through a yet unknown mechanism. For example, phosphorylation of the Kinesin-5 Eg5 by Cdk1 in its C-terminal region (T927 in humans, T937 in Xenopus) increases its affinity for microtubules in vitro [Cahu et al., 2008]. This phosphorylation site is present in a highly conserved region known as the BimC box and is necessary for timely targeting of Eg5 to the spindle [Heck et al., 1993; Blangy et al., 1995; Sawin and Mitchison, 1995] to ensure that spindle bipolar assembly occurs robustly at the onset of mitosis. However, this regulatory site is not functionally conserved throughout species. Mutation of the BimC region and this phosphorylation site in the fission yeast Eg5 homologue Cut7 does not impair spindle formation and mitosis [Drummond and Hagan, 1998]. Interestingly, this region also contains the second microtubule binding site in Eg5 and is essential for its correct function [Weinger et al., 2011]. Thus phosphorylation provides a controlled way to increase the activity and targeting of Eg5 specifically at the onset of mitosis. How phosphorylation contributes mechanistically to increasing the affinity of Eg5 is still not well understood.
Phosphorylation May Disrupt Protein–Protein Interactions
The addition of a phosphate group to an acceptor site dramatically changes the properties of that amino acid. Phosphorylation can disrupt protein–protein interactions through electrostatic repulsion or steric hindrance following the addition of a phosphate group. In this way, the targeting of Kif2c/MCAK to microtubule plus ends can be selectively abolished by phosphorylation of its N-terminal targeting domain. An SXIP motif in the N-terminal region of MCAK supports its interaction with EBs [Honnappa et al., 2009], and targets MCAK to the plus ends of microtubules. The region close to this EB-binding motif is phosphorylated by Aurora B [Andrews et al., 2004; Lan et al., 2004], which disrupts the MCAK-EB interaction [Honnappa et al., 2009]. Consequently, phosphorylation prevents MCAK localization to microtubule plus ends in the vicinity of chromosome-bound Aurora B, spatially restricting MCAK depolymerase activity [Moore et al., 2005; Tanenbaum and Medema 2011]. Aurora B phosphorylation of MCAK also weakens the MCAK–Kif18b interaction, although it is not clear if Kif18b associates with MCAK or a MCAK–EB1 complex to target to the plus ends [Tanenbaum et al., 2011]. Overall, Aurora B phosphorylation of MCAK reduces the presence of both MCAK and Kif18b destabilizing enzymes at the plus ends of microtubules by abolishing protein–protein interactions.
Phosphorylation Can Inhibit Intramolecular Interactions
To control the activity of kinesins in a rapid and reversible manner, phosphorylation can act to control kinesin conformation. For example, the mitotic Kinesin-7 CENP-E adopts a self-inhibitory conformation with the C-terminal tail folding onto the motor region to inhibit its ATPase activity. The C-terminal tail binds to the motor region to abolish motility. This inhibition is alleviated by CDK and Mps1-dependent phosphorylation [Espeut et al., 2008]. Phosphorylation of the non-motor regions of MCAK might also regulate intramolecular interactions [Zhang et al., 2011].
Phosphorylation to Create Protein–Protein Interactions
Phosphorylation can create binding motifs for phospho-binding proteins such as FHA domains and 14-3-3 proteins. The phospho-binding domain can then promote kinesin targeting and dimerization or sequester the phospho-protein to alter its function. For example, Kif23/MKLP1 activity is controlled by its phosphorylation and association with 14-3-3 [Mishima et al., 2004; Douglas et al., 2010]. MKLP1/Kif23 (part of the Centralspindlin complex) plays a role at the spindle mid-zone to complete cytokinesis. Phosphorylation in the C-terminus of MKLP1/Kif23 creates a binding site for 14-3-3. 14-3-3 then sequesters Centralspindlin and prevents its aberrant clustering, but also reduces the affinity of Centralspindlin for microtubules. Importantly, this interaction is further regulated by the spatially restricted Aurora B kinase, which phosphorylates Kif23/MKLP1 on the neighboring serine at P-2 to abolish the Kif23/MKLP1-14-3-3 interaction and to allow centralspindlin clustering at the mid-zone where Aurora B activity is present [Douglas et al., 2010]. This phospho-dependent 14-3-3-Kif23/MKLP1 interaction is also antagonized by ARF6 to further control the activity of the Centralspindlin complex [Joseph et al., 2012]. 14-3-3 proteins have also been reported to associate with other motors, such as KLC and Kif1c, but not in the context of mitosis [Dorner et al., 1999; Johnson et al., 2011].
Phosphorylation to Regulate Kinesin Localization
Kinesin phosphorylation may affect its localization or targeting properties. ZEN-4/MKLP1 targeting to the spindle is inhibited by CDK1/cyclinB phosphorylation [Mishima et al., 2004]. Phosphorylation on T9 and T450 reduces the affinity of the motor for microtubules. The counteracting Cdc14 phosphatase, found at the central spindle dephosphorylates MKLP1/ZEN-4 to localize MKLP1/ZEN-4 to the mid-zone. Mutations of CDK consensus sites in Drosophila Pav (MKLP1/ZEN4 homologue) also regulate the timely targeting of Pav to the central spindle [Goshima and Vale, 2005]. MCAK localization is also regulated both by Aurora A and Aurora B through yet unknown mechanisms. Aurora A phosphorylates MCAK in the C-terminus (S719 in Xenopus, equivalent to S715 in humans) and Kif2a to regulate their targeting to the spindle pole, without affecting their depolymerase activity [De Luca et al., 2006; Zhang et al., 2008; Jang et al., 2009]. In Xenopus egg extracts, Aurora B phosphorylation of the N-terminus through a combinatorial phosphorylation controls its targeting. For example, phosphorylation of T95 downregulates targeting to chromatin targeting while S110 phosphorylation promotes MCAK recruitment to the chromosome arms [Zhang et al., 2007]. Xenopus MCAK requires Aurora B phosphorylation to target to the centromere [Lan et al. 2004; Ohi et al. 2004]. MCAK is particularly recruited to merotelically attached kinetochores in an Aurora B dependent fashion, with an increase in active MCAK at kinetochores [Knowlton et al., 2006]. This would enhance the destabilization of erroneous kinetochore-microtubule attachments.
This is an exciting time for dissecting how motors are regulated, given the number of post-translational modifications for kinesins that have been identified and are unique to each member, but have not yet been characterized. Although cell biological approaches provide insights to the physiological significance of these phosphorylation events, defining mechanistically the role of phosphorylation on kinesin function requires recombinant proteins as well as biochemical and biophysical tools.
Conclusions
Unlike cytoplasmic dynein, which uses multiple accessory proteins to create functional specificity and targeting, there are many functionally distinct kinesins that co-exist within each species and work non-redundantly during mitosis. Mitotic kinesins are specific to cell division and represent strong anti-cancer targets [Rath and Kozielski, 2012]. However, redundant pathways for spindle assembly and function that exist during mitosis may allow resistance to anti-mitotic drugs [Raaijmakers et al., 2012]. Defining how kinesins work together in parallel pathways and their unique functional properties would allow researchers to evaluate what combination of drug targets would be required to kill cancer cells. Unlike kinases, the ATP binding site of kinesins does not form a closed pocket, which makes designing inhibitors to target the conserved catalytic core of kinesins a challenge. Understanding how the non-motor regions regulate kinesin activity could uncover protein–protein interactions as well as protein–ATP interactions that could be evaluated as new anti-mitotic drug targets [Welburn and Endicott, 2005].
Acknowledgments
I thank Iain Cheeseman, Gohta Goshima, Jason Stumpff, Marvin Tanenbaum and the members of the Welburn lab for critical reading of the manuscript. J. W. is supported by a CRUK Career Development Fellowship (C40377/ A12840). The Wellcome Trust Centre for Cell Biology is supported by core funding from the Wellcome Trust (092076).
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- Afshar K, Barton NR, Hawley RS, Goldstein LS. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995;81(1):129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21(10):604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128(6):1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando A, Kikuti YY, Kawata H, Okamoto N, Imai T, Eki T, Yokoyama K, Soeda E, Ikemura T, Abe K, et al. Cloning of a new kinesin-related gene located at the centromeric end of the human MHC region. Immunogenetics. 1994;39(3):194–200. doi: 10.1007/BF00241260. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6(2):253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102(4):425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302(5653):2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JG, Manning BD, Snyder M. The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p. Mol Biol Cell. 2000;11(7):2373–2385. doi: 10.1091/mbc.11.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, D'Avino PP. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci USA. 2013;110(24):9782–9787. doi: 10.1073/pnas.1301328110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Kronja I, Surrey T. Microtubule motility on reconstituted meiotic chromatin. Curr Biol. 2010a;20(8):763–769. doi: 10.1016/j.cub.2010.02.067. [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010b;142(3):420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83(7):1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92(9):2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleti H, Karsenti E, Vernos I. Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84(1):49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317(6032):73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11(6):724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303(5663):1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Hunt AJ. Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc Natl Acad Sci USA. 2005;102(39):13903–13908. doi: 10.1073/pnas.0506017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE. 2008;3(12):e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176(1):51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane S, Ye AA, Luks-Morgan SJ, Maresca TJ. Elevated polar ejection forces stabilize kinetochore-microtubule attachments. J Cell Biol. 2013;200(2):203–218. doi: 10.1083/jcb.201211119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NJ, Cross RA. Kinesin's moonwalk. Curr Opin Cell Biol. 2006;18(1):61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90(5):959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Chandra R, Salmon ED, Erickson HP, Lockhart A, Endow SA. Structural and functional domains of the Drosophila ncd microtubule motor protein. J Biol Chem. 1993;268(12):9005–9013. [PubMed] [Google Scholar]
- Cochran JC, Sindelar CV, Mulko NK, Collins KA, Kong SE, Hawley RS, Kull FJ. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell. 2009;136(1):110–122. doi: 10.1016/j.cell.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A "slow" homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269(37):22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17(1):77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1(5):288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Cross RA. The kinetic mechanism of kinesin. Trends Biochem Sci. 2004;29(6):301–309. doi: 10.1016/j.tibs.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci. 2004;117(Pt 1):3–7. doi: 10.1242/jcs.00875. [DOI] [PubMed] [Google Scholar]
- De Luca M, Lavia P, Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 2006;5(3):296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159(4):549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner C, Ullrich A, Haring HU, Lammers R. The kinesin-like motor protein KIF1C occurs in intact cells as a dimer and associates with proteins of the 14-3-3 family. J Biol Chem. 1999;274(47):33654–33660. doi: 10.1074/jbc.274.47.33654. [DOI] [PubMed] [Google Scholar]
- Douglas ME, Davies T, Joseph N, Mishima M. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr Biol. 2010;20(10):927–933. doi: 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hagan IM. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J Cell Sci. 1998;111(Pt 7):853–865. doi: 10.1242/jcs.111.7.853. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20(4):374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18(7):519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S. Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Mol Cell Biol. 2011;31(15):3085–3093. doi: 10.1128/MCB.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA. Determinants of molecular motor directionality. Nat Cell Biol. 1999;1(6):E163–E167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13(11):2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Waligora KW. Determinants of kinesin motor polarity. Science. 1998;281(5380):1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60(6):1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Espeut J, Gaussen A, Bieling P, Morin V, Prieto S, Fesquet D, Surrey T, Abrieu A. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol Cell. 2008;29(5):637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11(6):717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1(5):293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Friel CT, Howard J. Coupling of kinesin ATP turnover to translocation and microtubule regulation: one engine, many machines. J Muscle Res Cell Motil. 2012;33(6):377–383. doi: 10.1007/s10974-012-9289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102(4):411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147(5):1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005a;171(2):229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162(6):1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16(8):3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005b;15(22):1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172(3):363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104(1):83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Hackney DD, Baek N, Snyder AC. Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry. 2009;48(15):3448–3456. doi: 10.1021/bi8022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD, Levitt JD, Suhan J. Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem. 1992;267(12):8696–8701. [PubMed] [Google Scholar]
- Hackney DD, Levitt JD, Wagner DD. Characterization of alpha 2 beta 2 and alpha 2 forms of kinesin. Biochem Biophys Res Commun. 1991;174(2):810–815. doi: 10.1016/0006-291x(91)91490-4. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347(6293):563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189(6):1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7(3):e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172(1):19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123(3):665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen U, Schliwa M. Reversal in the direction of movement of a molecular motor. Nature. 1997;389(6646):93–96. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006;17(2):700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Nitta R, Okada Y. The mechanisms of kinesin motor motility: lessons from the monomeric motor KIF1A. Nat Rev Mol Cell Biol. 2009a;10(12):877–884. doi: 10.1038/nrm2807. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009b;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56(5):867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138(2):366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Houliston E, Le Guellec R, Kress M, Philippe M, Le Guellec K. The kinesin-related protein Eg5 associates with both interphase and spindle microtubules during Xenopus early development. Dev Biol. 1994;164(1):147–159. doi: 10.1006/dbio.1994.1187. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328(5978):593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CY, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci. 2009;122(Pt 9):1334–1341. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149(7):1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Wang J, Liu J, Ward T, Wordeman L, Davidson A, Wang F, Yao X. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009;10(8):857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2(6):877–885. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- Johnson C, Tinti M, Wood NT, Campbell DG, Toth R, Dubois F, Geraghty KM, Wong BH, Brown LJ, Tyler J, et al. Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol Cell Proteomics. 2011;10(10) doi: 10.1074/mcp.M110.005751. M110 005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N, Hutterer A, Poser I, Mishima M. ARF6 GTPase protects the post-mitotic midbody from 14-3-3-mediated disintegration. EMBO J. 2012;31(11):2604–2614. doi: 10.1038/emboj.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333(6044):883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Karabay A, Walker RA. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38(6):1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5(12):1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996a;379(6562):270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature. 1996b;384(6606):225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181(3):411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142(3):444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16(17):1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol. 2009;19(9):758–763. doi: 10.1016/j.cub.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145(6):914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16(8):743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kozielski F, De Bonis S, Burmeister WP, Cohen-Addad C, Wade RH. The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations. Structure. 1999;7(11):1407–1416. doi: 10.1016/s0969-2126(00)80030-1. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380(6574):550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23(16):3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Kofron M, Essner R, Kato T, Dragas-Granoic S, Omoto CK, Khodjakov A. Characterization of a minus end-directed kinesin-like motor protein from cultured mammalian cells. J Cell Biol. 1995;129(4):1049–1059. doi: 10.1083/jcb.129.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14(4):273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167(1):19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;2(5):362–363. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- Lee JR, Shin H, Choi J, Ko J, Kim S, Lee HW, Kim K, Rho SH, Lee JH, Song HE, et al. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 2004;23(7):1506–1515. doi: 10.1038/sj.emboj.7600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Langford KJ, Askham JM, Bruning-Richardson A, Morrison EE. MCAK associates with EB1. Oncogene. 2008;27(17):2494–2500. doi: 10.1038/sj.onc.1210867. [DOI] [PubMed] [Google Scholar]
- Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J. 2001;360(Pt 3):549–556. doi: 10.1042/0264-6021:3600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Liu B. Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiol. 2004;136(4):3877–3883. doi: 10.1104/pp.104.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154(6):1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Howard L, Gordon MB, Compton DA. A functional relationship between NuMA and kid is involved in both spindle organization and chromosome alignment in vertebrate cells. Mol Biol Cell. 2003;14(9):3541–3552. doi: 10.1091/mbc.E03-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188(6):809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol. 2011;195(1):87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol Biol Cell. 2010;21(6):979–988. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini S, Maia AR, Manning AL, Maliga Z, Pereira AL, Junqueira M, Shevchenko A, Hyman A, Yates JR, III, Galjart N, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19(18):1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermudez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15(3):325–334. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142(3):787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Wagenbach M, Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J Biol Chem. 2001;276(37):34753–34758. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29(20):3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144(6):1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114(1):87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170(6):873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Hoenger A, Mandelkow E. Structures of kinesin motor proteins. Cell Motil Cytoskeleton. 2009;66(11):958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Shimodaira T, Shiu SJ, Tokai-Nishizumi N, Yamamoto T, Ohsugi M. Microtubule stabilization triggers the plus-end accumulation of Kif18A/kinesin-8. Cell Struct Funct. 2011;36(2):261–267. doi: 10.1247/csf.11032. [DOI] [PubMed] [Google Scholar]
- Matthies HJ, Baskin RJ, Hawley RS. Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol Biol Cell. 2001;12(12):4000–4012. doi: 10.1091/mbc.12.12.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17(6):488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS One. 2011;6(11):e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M, Misteli T. Chromokinesins: multitalented players in mitosis. Trends Cell Biol. 2005;15(7):349–355. doi: 10.1016/j.tcb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166(5):613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Goldstein LS. Identification and characterization of a gene encoding a kinesin-like protein in Drosophila. Cell. 1990;61(6):991–1000. doi: 10.1016/0092-8674(90)90064-l. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Stewart RJ, Goldstein LS. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63(6):1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, Millar JB. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20(6):739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60(6):1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7(3):235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15(9):467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2(1):41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Mishima M, Pavicic V, Gruneberg U, Nigg EA, Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430(7002):908–913. doi: 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157(7):1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia S, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, Wordeman L, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20(19):1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Moore A, Wordeman L. C-terminus of mitotic centromere-associated kinesin (MCAK) inhibits its lattice-stimulated ATPase activity. Biochem J. 2004;383(Pt 2):227–235. doi: 10.1042/BJ20040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169(3):391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18(3):207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Moua P, Fullerton D, Serbus LR, Warrior R, Saxton WM. Kinesin-1 tail autoregulation and microtubule-binding regions function in saltatory transport but not ooplasmic streaming. Development. 2011;138(6):1087–1092. doi: 10.1242/dev.048645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147(2):351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79(7):1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein HD, McCaffrey G, Vale RD. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117(6):1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neighbors BW, Williams RC, Jr, McIntosh JR. Localization of kinesin in cultured cells. J Cell Biol. 1988;106(4):1193–1204. doi: 10.1083/jcb.106.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E, Garcia-Saez I, DeBonis S, Wade RH, Kozielski F, Conway JF. Human kinetochore-associated kinesin CENP-E visualized at 17 A resolution bound to microtubules. J Mol Biol. 2006;362(2):203–211. doi: 10.1016/j.jmb.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Niederstrasser H, Salehi-Had H, Gan EC, Walczak C, Nogales E. XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J Mol Biol. 2002;316(3):817–828. doi: 10.1006/jmbi.2001.5360. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116(4):591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Ohi R, Coughlin ML, Lane WS, Mitchison TJ. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell. 2003;5(2):309–321. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol Biol Cell. 2004;15(6):2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Adachi K, Horai R, Kakuta S, Sudo K, Kotaki H, Tokai-Nishizumi N, Sagara H, Iwakura Y, Yamamoto T. Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell. 2008;132(5):771–782. doi: 10.1016/j.cell.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81(5):769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a "neckless" MCAK mutant. J Cell Biol. 2002;159(4):557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124(4):507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell. 2007;18(12):4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29(20):3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31(21):4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12(8):527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- Reddy AS, Day IS. Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics. 2001;2:2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera T, Ghenoiu C, Rodriguez-Corsino M, Mochida S, Funabiki H, Losada A. Xenopus Shugoshin 2 regulates the spindle assembly pathway mediated by the chromosomal passenger complex. EMBO J. 2012;31(6):1467–1479. doi: 10.1038/emboj.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332(6025):94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Vale RD, Fletterick RJ. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395(6704):813–816. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol. 2012;199(5):755–770. doi: 10.1083/jcb.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr Biol. 2008;18(10):723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J Cell Sci. 2010;123(Pt 5):767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA. 1995;92(10):4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Porter ME, Cohn SA, Scholey JM, Raff EC, McIntosh JR. Drosophila kinesin: characterization of microtubule motility and ATPase. Proc Natl Acad Sci USA. 1988;85(4):1109–1113. doi: 10.1073/pnas.85.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Porter ME, Grissom PM, McIntosh JR. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318(6045):483–486. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160(4):517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger MA, Rice SE. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem. 2010;285(11):8155–8162. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol. 1994;127(1):187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1(1):51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Shiroguchi K, Ohsugi M, Edamatsu M, Yamamoto T, Toyoshima YY. The second microtubule-binding site of monomeric kid enhances the microtubule affinity. J Biol Chem. 2003;278(25):22460–22465. doi: 10.1074/jbc.M212274200. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16(8):731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, Mori H, Hauser-Hahn I, Jiang CJ, Sonobe S, Lloyd CW, Hussey PJ. A new class of microtubule-associated proteins in plants. Nat Cell Biol. 2000;2(10):750–753. doi: 10.1038/35036390. [DOI] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15(15):1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem. 1999;274(21):14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22(17):3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43(5):764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14(2):252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22(5):1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by kip3/kinesin-8. Mol Cell. 2011;43(5):751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142(3):433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and Kinesin-4. Cell. 2013;154(3):337–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, van Dam TJ, Karagoz GE, von Castelmur E, Hubner NC, Duarte AM, Vleugel M, Perrakis A, Rudiger SG, Snel B, et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev Cell. 2012;22(6):1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Syred HM, Welburn J, Rappsilber J, Ohkura H. Cell cycle regulation of microtubule interactomes: multi-layered regulation is critical for the interphase/mitosis transition. 2013. Jul 26. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Tahara K, Takagi M, Ohsugi M, Sone T, Nishiumi F, Maeshima K, Horiuchi Y, Tokai-Nishizumi N, Imamoto F, Yamamoto T, et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J Cell Biol. 2008;180(3):493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27(24):3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19(20):1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21(16):1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Localized Aurora B activity spatially controls non-kinetochore microtubules during spindle assembly. Chromosoma. 2011;120(6):599–607. doi: 10.1007/s00412-011-0334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24(19):2169–2179. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151(5):1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 1996;15(3):457–467. [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297(5590):2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184(3):365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Case R, Sablin E, Hart C, Fletterick R. Searching for kinesin's mechanical amplifier. Philos Trans R Soc Lond B Biol Sci. 2000;355(1396):449–457. doi: 10.1098/rstb.2000.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Goldstein LS. One motor, many tails: an expanding repertoire of force-generating enzymes. Cell. 1990;60(6):883–885. doi: 10.1016/0092-8674(90)90334-b. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42(1):39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18(23):1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D, Ferreira V, Vernos I. Chromokinesins: localization-dependent functions and regulation during cell division. Biochem Soc Trans. 2011;39(5):1154–1160. doi: 10.1042/BST0391154. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19(20):1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8(9):957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Verbrugghe KJ, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14(19):1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- Verni F, Somma MP, Gunsalus KC, Bonaccorsi S, Belloni G, Goldberg ML, Gatti M. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr Biol. 2004;14(17):1569–1575. doi: 10.1016/j.cub.2004.08.054. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gan EC, Desai A, Mitchison TJ, Kline-Smith SL. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr Biol. 2002;12(21):1885–1889. doi: 10.1016/s0960-9822(02)01227-7. [DOI] [PubMed] [Google Scholar]
- Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol. 2012;198(5):847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162(4):551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, Leblanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21(17):1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21(2):154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Cheeseman IM. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell. 2012;23(24):4786–4795. doi: 10.1091/mbc.E12-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Endicott JA. Inhibition of the cell cycle with chemical inhibitors: a targeted approach. Semin Cell Dev Biol. 2005;16(3):369–381. doi: 10.1016/j.semcdb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Wendt T, Karabay A, Krebs A, Gross H, Walker R, Hoenger A. A structural analysis of the interaction between ncd tail and tubulin protofilaments. J Mol Biol. 2003;333(3):541–552. doi: 10.1016/j.jmb.2003.08.051. [DOI] [PubMed] [Google Scholar]
- White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken. 2012;69(11):882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A "holistic" kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17(4):1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol. 1998;143(3):673–685. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149(7):1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Chen PL. Structural requirements of chromokinesin Kif4A for its proper function in mitosis. Biochem Biophys Res Commun. 2008;372(3):454–458. doi: 10.1016/j.bbrc.2008.05.065. [DOI] [PubMed] [Google Scholar]
- Yajima J, Edamatsu M, Watai-Nishii J, Tokai-Nishizumi N, Yamamoto T, Toyoshima YY. The human chromokinesin Kid is a plus end-directed microtubule-based motor. EMBO J. 2003;22(5):1067–1074. doi: 10.1093/emboj/cdg102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LS. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989;56(5):879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yang JT, Saxton WM, Goldstein LS. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc Natl Acad Sci USA. 1988;85(6):1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Tan L, Yang Q, Xia Y, Deng LW, Murata-Hori M, Liou YC. HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr Biol. 2011;21(18):1584–1591. doi: 10.1016/j.cub.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10(5):1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359(6395):536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Zhang L, Shao H, Huang Y, Yan F, Chu Y, Hou H, Zhu M, Fu C, Aikhionbare F, Fang G, et al. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J Biol Chem. 2011;286(4):3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19(7):2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18(9):3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA. 2005;102(2):343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci USA. 2006;103(16):6196–6201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


