Abstract
The molecular diagnosis of Y-chromosomal microdeletions is a common routine genetic test which is part of the diagnostic workup of azoospermic and severe oligozoospermic men. Since 1999, the European Academy of Andrology (EAA) and the European Molecular Genetics Quality Network (EMQN) have been actively involved in supporting the improvement of the quality of the diagnostic assays by publication of the laboratory guidelines for molecular diagnosis of Y-chromosomal microdeletions and by offering external quality assessment trials. The present revision of the 2004 laboratory guidelines summarizes all the clinical novelties related to the Y chromosome (classic, partial and gene-specific deletions, genotype–phenotype correlations, methodological issues) and provides an update on the results of the quality control programme. These aspects also reflect the consensus of a large group of specialists present at a round table session during the recent Florence-Utah-Symposium on ‘Genetics of male infertility’ (Florence, 19–21 September, 2013). During the last 10 years the gr/gr deletion has been demonstrated as a significant risk factor for impaired sperm production. However, the screening for this deletion type in the routine diagnostic setting is still a debated issue among experts. The original basic protocol based on two multiplex polymerase chain reactions remains fully valid and appropriate for accurate diagnosis of complete AZF deletions and it requires only a minor modification in populations with a specific Y chromosome background. However, in light of novel data on genotype–phenotype correlations, the extension analysis for the AZFa and AZFb deletions is now routinely recommended. Novel methods and kits with excessively high number of markers do not improve the sensitivity of the test, may even complicate the interpretation of the results and are not recommended. Annual participation in an external quality control programme is strongly encouraged. The 12-year experience with the EMQN/EAA scheme has shown a steep decline in diagnostic (genotyping) error rate and a simultaneous improvement on reporting practice.
Keywords: AZF, azoospermia, genetics, gr/gr deletion, male infertility, oligozoospermia, quality control, spermatogenesis, Y chromosome microdeletion
Introduction
After the Klinefelter syndrome, Y-chromosomal microdeletions are the second most frequent genetic cause of male infertility. In the last decade, many investigators have described the occurrence of microdeletions in infertile patients around the world and the molecular diagnosis of deletions has become an important test in the diagnostic workup of male infertility (Vogt et al., 1996; Krausz & Degl'Innocenti, 2006; Simoni et al., 2008).
Microdeletions occur in about one in 4000 men in the general population but its frequency is significantly increased among infertile men. Azoospermic men have a higher incidence of microdeletions than oligozoospermic men and consequently deletion frequency found in different laboratories may vary from 2 to 10% (or even higher, Fig. 1) reflecting the composition of the study population (Krausz et al., 2001; Simoni et al., 2008; Lo Giacco et al., 2013). Typically, routine laboratories receiving referrals from outside institutions, without controlled patient selection, have a much lower incidence, <2%.
Figure 1.
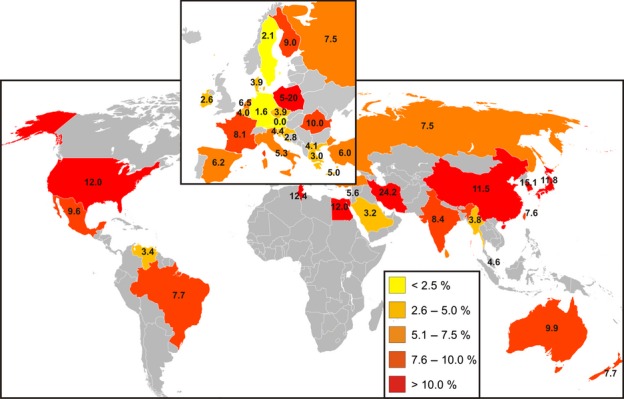
Worldwide frequencies of AZF deletions in infertile men (reprinted from Simoni et al., 2008 with publisher's permission). Percentages are coded in colours according to the legend. Note: Sweden, Germany and Austria show the lowest incidence. However, the composition of the study populations differed in terms of the proportion of azoospermic vs. oligozoospermic patients which may also contribute to the observed differences.
The published data and the quality control programme experience showed that diagnostic protocols can be quite different and that inaccurate or wrong diagnoses occur as well, suggesting the necessity of both standardization and quality control (Simoni, 2001). Therefore, the European Academy of Andrology (EAA) and the European Molecular Genetics Quality Network (EMQN) jointly supported the publication of two ‘Laboratory guidelines for molecular diagnosis of Y-chromosomal microdeletions’ (Simoni et al., 1999, 2004) and started offering external quality assessment (EQA).
During the last 9 years, novel data concerning gene-specific deletions, partial AZFc deletions/duplications and genotype–phenotype correlations have been accumulated. All these issues together with some methodological aspects which urged to be clarified, and an update on the EAA/EMQN AZF quality control scheme's activity is summarized in this study.
Structure of the male-specific region of the Y chromosome (MSY)
The complete physical map and sequence of MSY have been available since 2003 (Skaletsky et al., 2003). This information was obtained by sequencing and mapping 220 BAC clones containing portions of the MSY from one man. The use of only one individual was necessary because, owing to the presence of repetitive sequences with only minute differences characterizing the individual copies of each sequence (sequence family variants, SFV), interindividual allelic variation or polymorphisms would have prevented the accurate mapping of SFV necessary to allocate the BAC clones. Three classes of sequences were found in MSY: X-transposed (with 99% identity to the X chromosome), X-degenerate (single-copy genes or pseudogene homologues of X-linked genes) and ampliconic. Ampliconic sequences are characterized by sequence pairs showing nearly complete (>99.9%) identity, organized in massive palindromes. According to current knowledge, the reference MSY contains 156 transcription units including 78 protein-coding genes encoding 27 proteins. Ampliconic sequences comprise 60 coding genes and 74 non-coding transcription units mostly grouped in families and expressed mainly or only in the testis. Ampliconic sequences recombine through gene conversion, that is, non-reciprocal transfer of sequence information occurring between duplicated sequences within the chromosome, a process which maintains the >99.9% identity between repeated sequences organized in pairs in inverted orientation within palindromes.
Besides maintaining the gene content, this peculiar sequence organization provides the structural basis for deletions and rearrangements. It is widely accepted that complete AZF deletions arise invariably through Non-Allelic Homologous Recombination (NAHR) which takes place between highly homologous repeated sequences with the same orientation leading to loss of the genetic material between them. Considering the architecture of the MSY, several different deletions are hypothetically possible (Kuroda-Kawaguchi et al., 2001; Yen, 2001; Repping et al., 2003) and those which are clinically relevant for male infertility, based on current knowledge, are briefly described below.
Mechanism and type of deletions
Three discrete AZFa, AZFb and AZFc regions were originally characterized by careful mapping of the MSY of a large number of men with microdeletions when the sequence of the Y chromosome was not completely known (Vogt et al., 1996). Subsequently, thanks to the fine molecular characterization of the deletions, a new model of deletions, in which the AZFb and AZFc regions are overlapping, have been proposed (Fig. 2). In addition, the AZFb and AZFbc deletions have been suggested to be the consequence of at least three different deletions patterns (Repping et al., 2002) (Fig. 2). While the new nomenclature is more appropriate in biological terms, from the practical, clinical point of view either nomenclature can be adopted for the complete AZFb (P5/proximal P1) and AZFc (b2/b4) deletions. On the other hand, the distinction between the two AZFbc subtypes (P5/distal P1 and P4/distal P1) does have clinical relevance (see below). We additionally provide information on the genomic localization of the sY-loci used for the AZF analyses (Appendix C), which should be used to describe deletions following the HGVS nomenclature as part of a standard practice. The description of the AZFc (b2/b4) deletion is given in ‘report examples’.
Figure 2.
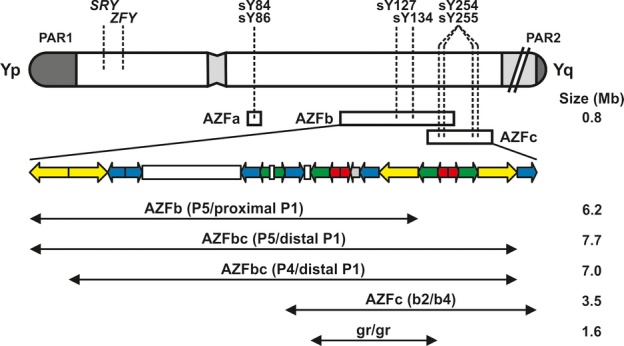
Schematic representation of the Y chromosome and the current microdeletion model (Repping et al., 2002). Repetitive sequences (colour coded palindromes) explain the origin of deletions in the AZFbc region by homologous recombination between identical sequences. The location of the STS primers suggested by the present guidelines is indicated by dashed lines. As four copies of the DAZ gene are normally present on the Y chromosome, the STS primers sY254, sY255 amplify four loci in AZFc. The AZFc (b2/b4) deletion is by far the most frequent type (∼80%) of Y-chromosomal microdeletions found in men with severe oligo/azoospermia.
The AZFa region is about 1100 kb long and contains the single-copy genes USP9Y (former DFFRY) and DDX3Y (former DBY). Recent data obtained simultaneously by different groups identify the origin of complete AZFa deletions in the homologous recombination between identical sequence blocks within the retroviral sequences in the same orientation HERVyq1 and HERVyq2 (Blanco et al., 2000; Kamp et al., 2000; Sun et al., 2000). Within these retroviruses, recombination can occur in either one of two identical sequence blocks (ID1 and ID2), giving rise to two major pattern of deletions slightly different in their precise breakpoints (Kamp et al., 2000; Sun et al., 2000; Kamp et al., 2001). In any case, the complete deletion of the AZFa region removes about 792 kb including both USP9Y and DDX3Y genes, the only two genes in the AZFa region.
The type and mechanism of deletions of the AZFb and AZFc region have been clarified by Kuroda-Kawaguchi et al. (2001). Both regions together comprise 24 genes, most of which are present in multiple copies for a total of 46 copies. The complete deletion of AZFb removes 6.2 Mb (including 32 copies of genes and transcription units) and results from homologous recombination between the palindromes P5/proximal P1 (Repping et al., 2002). The AZFc region includes 12 genes and transcription units, each present in a variable number of copies making a total of 32 copies (Repping et al., 2003). The classical complete deletion of AZFc, the most frequent pattern among men with deletions of the Y chromosome, removes 3.5 Mb, originates from the homologous recombination between amplicons b2 and b4 in palindromes P3 and P1, respectively, and removes 21 copies of genes and transcription units (Kuroda-Kawaguchi et al., 2001). Deletions of both AZFb and AZFc together occur by two major mechanisms involving homologous recombination between P5/distal P1 (7.7 Mb and 42 copies removed) or between P4/distal P1 (7.0 Mb, 38 copies removed) (Repping et al., 2002).
Therefore, according to the present knowledge, the following recurrent microdeletions of the Y chromosome are clinically relevant and are found in men with severe oligo- or azoospermia (Fig. 2):
AZFa,
AZFb (P5/proximal P1),
AZFbc (P5/distal P1 or P4/distal P1),
AZFc (b2/b4).
The most frequent deletion type is the AZFc region deletion (∼80%) followed by AZFa (0.5–4%), AZFb (1–5%) and AZFbc (1–3%) deletion. Deletions which are detected as AZFabc are most likely related to abnormal karyotype such as 46,XX male or iso(Y) (Lange et al., 2009).
Gr/gr deletion
The AZFc region is particularly susceptible to NAHR events which may cause the formation of both partial deletions and duplications leading to gene dosage variations (Kuroda-Kawaguchi et al., 2001; Yen, 2001; Krausz et al., 2011). Although a number of different partial AZFc deletions have been described, only one of them is of potential clinical interest. This is the ‘gr/gr’ deletion, named after the fluorescent probes (‘green’ and ‘red’) used when first described (Repping et al., 2003). Although it removes half of the AZFc gene content (genes with exclusive or predominant expression in the germ cells), its clinical significance is still a matter of debate, because carriers may exhibit highly variable spermatogenic phenotypes ranging from azoo- to normozoospermia. Clearly the effect of the deletion is largely dependent on the ethnic and geographic origin of the study population. In fact, the frequency and phenotypic effect may vary among different ethnic groups, on the basis of the Y chromosome background; for example, in specific Y haplogroups, such as D2b, Q3 and Q1, common in Japan and certain areas of China, the deletion is fixed and apparently does not have negative effects on spermatogenesis (Sin et al., 2010; Yang et al., 2010). Controversies are also related to selection biases (lack of ethnic/geographic matching of cases and controls; inappropriate selection of infertile and control men) and methodological issues (lack of confirmation of gene loss). Many efforts have been done to clarify the molecular basis for the highly variable phenotypic presentation of this deletion type. It has been previously described that the loss of DAZ1/DAZ2 and CDY1 is prevalent (or even specific) in carriers with impaired sperm production (Fernandes et al., 2002; Ferlin et al., 2005; Giachini et al., 2005) while it was hypothesized that the restoration of normal AZFc gene dosage in case of gr/gr deletion followed by b2/b4 duplication may explain the lack of effect on sperm count (Repping et al., 2003). In this regard, a large multicentre study, based on a combined method (gene dosage, definition of the lost DAZ and CDY1 genes, Y hgr definition), was performed on Caucasians (Krausz et al., 2009). Notwithstanding the detailed characterization of subtypes of gr/gr deletions based on the type of missing gene copies and the detection of secondary rearrangements (deletion followed by b2/b4 duplication) together with the definition of Y haplogroups, it was impossible to define a specific pattern which would be associated with either a ‘neutral’ or a ‘pathogenic’ effect. On the contrary, studies dealing with Asian populations seem to support the hypothesis about a deletion subtype-dependent phenotypic effect and about the importance of the Y background on which the deletion arises (Yang et al., 2010; Choi et al., 2012). In addition to classic case/control studies aiming to define whether the gr/gr deletion confers a risk for spermatogenic disturbances, the analysis of consecutive patients through cross-sectional cohort analysis indicates that the gr/gr deletion has an effect even within the normal range of sperm count. It was observed, indeed, that normozoospermic carriers have a significantly lower sperm count, compared to men with intact Y chromosome (Visser et al., 2009). In addition, Yang et al. (2006) reported that, in the Chinese population, the deletion frequency drastically decreases in subgroups with sperm concentrations >50 × 106/mL.
The screening for gr/gr deletion is based on a PCR plus/minus method of two markers (sY1291 and sY1191) (Repping et al., 2003) and the diagnosis is based on the absence of marker sY1291 and presence of sY1191. It is worth noticing that a 5% false deletion rate has been detected in the multicenter study (Krausz et al., 2009), underlining the importance of the optimization of the PCR conditions and of additional confirmatory steps such as simplex PCR and eventually gene dosage analysis (Giachini et al., 2005; Choi et al., 2012). The definition of the Y haplogroup is indicated in Asian patients to exclude constitutive deletions which are unlikely to affect spermatogenesis (see above).
Clinical implications
As stated above, the heterogeneity of the study populations available in the literature complicates a reliable meta-analysis. However, four meta-analyses have been attempted on this topic all achieving significant odds ratios reporting on average 2- to 2.5-fold increased risks of reduced sperm output/infertility (Tüttelmann et al., 2007; Visser et al., 2009; Navarro-Costa et al., 2010; Stouffs et al., 2011). Therefore, the gr/gr deletion represents a unique example in andrology of a confirmed significant genetic risk factor for impaired sperm production. For instance, in the Italian population, gr/gr deletions confers a 7.9-fold increased risk for spermatogenic impairment (OR = 7.9, 95% CI 1.8–33.8) (Ferlin et al., 2005; Giachini et al., 2008).
A gr/gr deletion (i.e. a genetic risk factor for impaired sperm production) will be obligatorily transmitted to the male offspring. The partial deletion may expand to a complete AZFc deletion (i.e. a clear-cut causative factor for spermatogenic impairment) in the next generations (Zhang et al., 2007), but data are currently sparse to draw final conclusions on this specific risk. Gr/gr deletions have also been proposed as genetic risk factor for testis cancer (Nathanson et al., 2005; Linger et al., 2007) but this association still awaits further confirmation on large independent study populations.
The low cost of the test may justify its routine testing in those populations for which robust and consistent data with risk estimate are available (at present Italian, Spanish, Dutch and Chinese). Currently, however, no general agreement to advise routine testing has been reached (Tüttelmann et al., 2007; Krausz et al., 2011; Stouffs et al., 2011).
Isolated AZF gene-specific deletions
Although some authors found an extraordinary high frequency of single AZF gene deletions (Ferlin et al., 1999; Foresta et al., 2000), these data are in stark contrast with the general experience accumulated in >2000 patients tested elsewhere (Silber et al., 1998; Sun et al., 1999; Krausz et al., 1999a,b1999b; Krausz et al., 2001; Simoni et al., 2008). Gene-specific deletions are extremely rare, and all the five confirmed deletions (with the definition of the breakpoints) removed totally or partially the USP9Y gene belonging to the AZFa region (Tyler-Smith & Krausz, 2009). None of the deletions was because of NAHR and thus are likely to be unique, supporting the extreme rarity of the occurrence of these events. The associated semen/testis phenotype is largely variable among USP9Y deletion carriers (from azoospermia caused by hypospermatogenesis to normozoospermia) indicating that this gene rather acts as a fine tuner than an essential factor for spermatogenesis. Based on the absence of other than USP9Y gene deletions in the literature, screening for isolated gene-specific deletions is not advised in the routine diagnostic setting. Given that some of the commercially available kits contain gene-specific markers, much care has to be taken both of the validation of suspected single-gene deletions as well as of the interpretation of the results.
Genotype/phenotype correlation of complete AZF deletions
AZF deletions are specific for spermatogenic failure as no deletions have been reported in a large number of normozoospermic men (Krausz et al., 2003; Simoni et al., 2008). Although ‘fertility’ can be compatible with these deletions, it simply reflects the fact that natural fertilization may occur even with low sperm counts depending on the female partner's fertility status. For this reason, it is more appropriate to consider Y deletions as a cause of oligo/azoospermia rather than a cause of ‘infertility’.
Deletions of the entire AZFa region invariably result in sertoli cell only syndrome (SCOS) and azoospermia (Vogt et al., 1996; Krausz et al., 2000; Kamp et al., 2001; Hopps et al., 2003; Kleiman et al., 2012). The diagnosis of a complete deletion of the AZFa region implies the virtual impossibility to retrieve testicular spermatozoa for intracytoplasmic sperm injection (ICSI).
Complete deletions of AZFb and AZFbc (P5/proximal P1, P5/distal P1, P4/distal P1) are characterized by a histological picture of SCOS or spermatogenetic arrest resulting in azoospermia. Several reports have shown that similar to the complete deletions of the AZFa region, no spermatozoa are found upon attempts of testicular sperm extraction (TESE) in these patients (Krausz et al., 2000; Hopps et al., 2003; Kleiman et al., 2011). However, in three cases, spermatid arrest and even crypto/oligozoospermia has been reported in association with complete AZFb or AZbc deletions (Longepied et al., 2010; Soares et al., 2012). The biological explanation of the unusual phenotypes remains unclear, both Y background effect and differences in the exact extent of the deletions may account for it. In fact, a smaller deletion, that is, a proximal breakpoint at P4 may be associated with the retention of AZFb gene copies such as XKRY, CDY2 and HSFY. It has been therefore proposed that the associated phenotype is more severe in case of complete removal of the AZFb region. With very few exceptions reported in the literature, the diagnosis of complete deletions of AZFb or AZFbc (P5/proximal P1, P5/distal P1, P4/distal P1) implies that the chance for testicular sperm retrieval is virtually zero even with micro-TESE (Brandell et al., 1998).
Deletions of the AZFc region (b2/b4) are associated with a variable clinical and histological phenotype (Reijo et al., 1996; Luetjens et al., 2002; Oates et al., 2002). In general, AZFc deletions are compatible with residual spermatogenesis and thus can be found also in men with severe oligozoospermia and, in rare cases, may even be transmitted naturally to the male offspring (Kühnert et al., 2004 and references therein). In men with azoospermia and AZFc deletion there is approximately 50% chance of retrieving spermatozoa from TESE and children can be conceived by ICSI (Kent-First et al., 1996; Mulhall et al., 1997; Kamischke et al., 1999; Jiang et al., 1999; Kleiman et al., 1999; Page et al., 1999; Cram et al., 2000; van Golde et al., 2001; Oates et al., 2002; Peterlin et al., 2002; Ferlin et al., 2007; Simoni et al., 2008; Lo Giacco et al., 2013). The TESE success rate largely depends on the technique used and can be as low as 9% (Lo Giacco et al., 2013) and as high as 70–80% following micro-TESE (Hopps et al., 2003). According to one report, the presence of 45,X cell lines in blood may be a negative predictive factor for spermatogenesis (Jaruzelska et al., 2001).
Indications for molecular screening of the Y chromosome
Diagnosis of a microdeletion of the Y chromosome permits the cause of the patient's azoospermia/oligozoospermia to be established and to formulate a prognosis. In which patients should molecular screening of the Y chromosome be performed? The world literature, now based on several thousands of patients screened, indicates that, as a rule, clinically relevant deletions are found in patients with azoospermia or severe oligozoospermia with sperm concentrations <2 × 106/mL. Very rarely, deletions can be found in infertile patients with sperm concentration between 2 and 5 × 106/mL (Maurer & Simoni, 2000; Lo Giacco et al., 2013). We provide a flow chart with these indications and including the recommended analytic steps in Fig. 3. The usual clinical parameters such as hormone levels, testicular volume, varicocoele, maldescended testis, infections, etc. do not have any predictive value (Maurer et al., 2001; Oates et al., 2002; Tomasi et al., 2003; Simoni et al., 2008). In general, molecular analysis of the Y chromosome is not indicated in patients with chromosomal abnormalities (except 46,XY/45,X karyotype), obstructive azoospermia (unless FSH is above the normal limit) or hypogonadotropic hypogonadism. However, in the literature there are a number of examples of deletion carriers among non-idiopathic infertile men, for example, with a testicular tumour or after chemo-/radiotherapy, which would be considered to explain the spermatogenic failure. Therefore, the presence of any diagnosis accompanied by azoo- or severe oligozoospermia should be an indication for AZF testing. For instance, in men belonging to the above semen categories, AZF screening is important before varicocoelectomy because deletion carriers will most likely not benefit from the surgical procedure.
Figure 3.
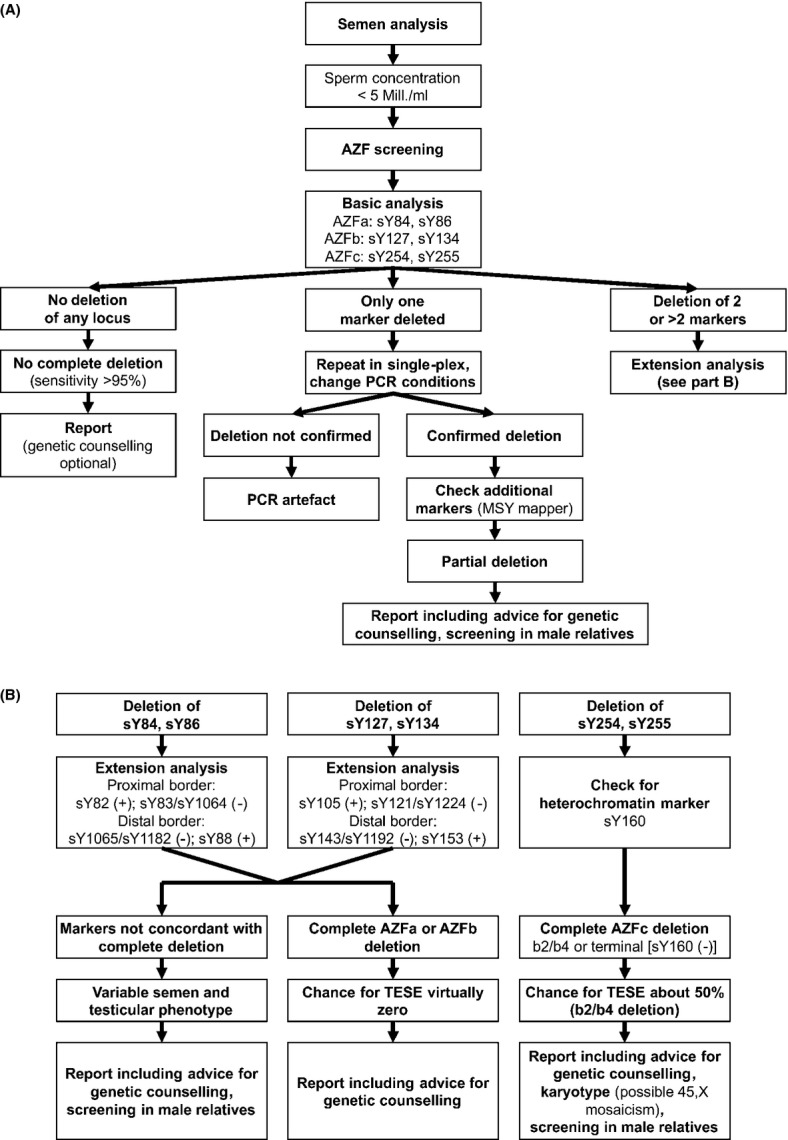
Flow chart with indication for AZF screening, common analytical steps and consequences: (A) basic analyses, (B) extension analyses.
After a high incidence of AZF deletions in Klinefelter patients had been reported in two small studies (Mitra et al., 2006; Hadjkacem-Loukil et al., 2009), the question arose whether deletion screening should be routinely be performed in these men. However, the described deletions were mainly diagnosed by only isolated markers of the AZFa and/or the AZFb region, not confirmed by additional analyses, and should probably be regarded as methodological artefacts. In contrast, three much larger studies did not find any AZF deletions in Klinefelter patients (Choe et al., 2007; Simoni et al., 2008; Rajpert-De Meyts et al., 2011).
Patients with azoospermia who may be candidate for TESE/ICSI should be offered deletion screening because TESE should not be recommended in cases of complete deletion of the AZFa region. Micro-TESE in azoospermic carriers of deletions of the AZFb or AZFbc regions with proximal breakpoint in the P4 palindrome may be eventually attempted. However, the patient should be fully informed about the very low/virtually zero chance to retrieve spermatozoa. A standard biopsy (without microsurgical equipment) should never be attempted in these cases. Therefore, the diagnosis of a deletion has prognostic value and can influence therapeutic options.
Genetic counselling
Genetic counselling is mandatory to provide information about the risk of conceiving a son with impaired spermatogenesis. In case of partial AZFa or AZFb and AZFc deletion, the counselling (with AZF testing) is relevant also for other male members of the family as transmission of these type of deletions has been reported in the literature (Krausz et al., 2006; Luddi et al., 2009; Plotton et al., 2010). Complete AZFa, AZFb, AZFbc or AZFabc deletion are generally incompatible with sperm production thus family screening is not indicated.
Several studies have been published about ICSI performed in couples with male partners carrying AZFc deletions (Kent-First et al., 1996; Mulhall et al., 1997; Jiang et al., 1999; Kamischke et al., 1999; Kleiman et al., 1999; Page et al., 1999; Cram et al., 2000; van Golde et al., 2001; Oates et al., 2002; Peterlin et al., 2002; Stouffs et al., 2005; Mau Kai et al., 2008; Simoni et al., 2008; Mateu et al., 2010; Lo Giacco et al., 2013). Although a lower fertilization rate and embryo quality (van Golde et al., 2001), a significantly impaired blastocyst rate (Mateu et al., 2010) and lower overall success of ICSI (Simoni et al., 2008) have been reported, the majority of studies report no significant differences in fertilization and pregnancy rates between men with or without Y deletion. While the deletion of the father will be obligatory transmitted to the son, who will have impaired sperm production, the exact testicular phenotype cannot be predicted because of the different genetic background and the impact of environmental factors on reproductive functions and on the fertility potential of father and son.
Concerns have been raised about the potential risk for Turner's syndrome (45,X) in the offspring and other phenotypic anomalies associated with sex chromosome mosaicism, including ambiguous genitalia. Data on men with Y microdeletions (Siffroi et al., 2000; Rajpert-De Meyts et al. 2011) and in patients bearing a mosaic 46,XY/45,X karyotype with sexual ambiguity and/or Turner stigmata (Patsalis et al., 2002) suggest that some Yq microdeletions are associated with an overall Y-chromosomal instability which might result in the formation of 45,X cell lines. The number of reported ICSI babies born from fathers affected by Yq microdeletions is still relatively low being close to 50 (Kent-First et al., 1996; Mulhall et al., 1997; Jiang et al., 1999; Kamischke et al., 1999; Kleiman et al., 1999; Cram et al., 2000; van Golde et al., 2001; Oates et al., 2002; Peterlin et al., 2002; Choi et al., 2004; Kihaile et al., 2004; Stouffs et al., 2005; Mau Kai et al., 2008; Simoni et al., 2008; Mateu et al., 2010; Lo Giacco et al., 2013). It appears that the children are phenotypically normal, except for one son born with pulmonary atresia and a hypoplastic right ventricle (Page et al., 1999) and no ambiguous genitalia or Turner syndrome have been observed among them. Considering that embryos bearing a 45,X karyotype have a very high risk of spontaneous abortion, it would be important to know whether there is a higher incidence of spontaneous abortion among the partners of Y deleted men. Two studies provide data on the aneuploidy rate in embryos by performing preimplantation genetic diagnosis in embryos derived from Y chromosome deletion carriers. In the first study, no sex chromosome anomalies have been found (Stouffs et al., 2005), whereas in the other a high percentage of abnormal embryos was observed, with a significant increase in the percentage of embryos with monosomy X in respect to oligozoospermic patients without Y deletion (Mateu et al., 2010). This implies that caution has to be taken when this risk is discussed with the patients and before pre-implantation or pre-natal diagnosis is proposed.
Recently, Jorgez et al. (2011) reported the detection of haploinsufficiency of the SHOX (Short-stature HOmeoboX-containing) gene located in the pseudoautosomal region 1 (PAR1) on the short arm of the Y chromosome in 5.4% of men with AZF deletions and a normal karyotype. They raised the question about the importance of screening for SHOX-linked copy number variations in men carrying Yq microdeletions. However, a subsequent much larger multicentre study did not find an association between Y-chromosomal microdeletions and SHOX haploinsufficiency, implying that deletion carriers have no augmented risk of SHOX-related pathologies (short stature and skeletal anomalies) (Chianese et al., 2013).
In conclusion, the indication for molecular diagnosis of Y-chromosomal microdeletions is based on sperm concentration and it is strongly advised in patients affected by azoospermia and severe oligozoospermia (<5 × 106/mL). AZF testing has prognostic value for sperm retrieval and in case spermatozoa can be found in the ejaculate or by testicular biopsy (micro-TESE instead of conventional TESE is strongly advised) the deletion will be obligatory transmitted to the male offspring. The fertilization rate and pregnancy rate seem to be similar to that obtained in men without Y microdeletion, but a lower embryo quality and blastocyst rate have also been described. We still do not have conclusive information about the real risk for Turner syndrome, ambiguous genitalia or other chromosomal anomalies because data are scarce and discordant. Analysis of the male members of the family is advised in case of AZFc or partial AZFb or AZFa deletions. Moreover, karyotype is indicated in the presence of AZFc or Yq terminal deletions to rule out 46,XY/45,X mosaicism.
Guidelines for diagnostic testing
Diagnostic testing for deletions is performed by PCR amplification of selected regions of the Y chromosome. MSY-specific STS primers amplify both anonymous sequences of the chromosome or genes and can be now mapped precisely (Skaletsky et al., 2003). Although the map of the MSY is now known, still virtually nothing is known about the role of the individual genes and transcription units in spermatogenesis and their causal role for infertility. It has been shown that using STS primers specific for discrete genes does not increase the detection rate of clinically relevant microdeletions in DNA samples from ICSI candidates (Silber et al., 1998; Krausz et al., 1999a, 2001; Peterlin et al., 2002). Therefore, it remains basically unimportant whether the STS primers used amplify anonymous regions or specific MSY genes. What is important for the diagnosis is that the panel of STS primers is derived from regions of the Y chromosome which are not polymorphic and are well-known to be deleted specifically in men affected by oligo-/azoospermia according to the known, clinically relevant microdeletion pattern. The sequence of the MSY and the mechanism underlying the microdeletions have shown definitely that a putative fourth AZFd region postulated by Kent-First et al. (1999) (and considered in a popular commercial kit) does not exist.
PCR format and internal quality control
The PCR amplification of genomic DNA for clinical diagnosis requires strict compliance with good laboratory practice and basic principles of quality control. Guidelines for internal quality control should be carefully followed when implementing the diagnostics of Y-chromosomal microdeletions.
In parallel to the patient's DNA sample, a female sample has to be processed as a control for DNA contamination during the whole procedure. Each set of PCR reactions should be carried out at least in duplex or, even better, multiplex PCR. The multiplex format is helpful to distinguish a negative result from a technical failure through the use of an internal control. An appropriate internal PCR control in AZF diagnostics is the ZFX/ZFY gene because the primers amplify a unique fragment both in male and female DNA respectively. Positive and negative controls must be run in parallel with each multiplex, that is, with each set of primers. Appropriate positive and negative controls are a DNA sample from a man with normal spermatogenesis and from a woman respectively. In addition, a water sample, which contains all reaction components but water instead of DNA, must be run with each set of primers as control for contamination.
In summary, the diagnostics of Y-chromosomal microdeletions should be performed by multiplex (at least duplex) PCR amplification of genomic DNA, using the ZFX/ZFY as internal PCR control. A DNA sample from a fertile male and from a women and a blank (water) control should be run in parallel with each multiplex.
Basic set of STS primers
In principle, the analysis of only one non-polymorphic STS locus in each AZF region is sufficient to determine whether any STS deletion is present in AZFa, AZFb or AZFc. However, analysing two STS loci in each region reinforces diagnostic accuracy, as deletions involve well-defined regions including many STS loci. Therefore, the concept that at least two STS loci in each AZF region should be analysed remains valid. Based on the experience of many laboratories, the results of external quality control and considering the multiplex PCR format, the first choice of STS primers recommended in the previous versions of the guidelines remains valid (Fig. 3A). These primers include:
For AZFa: sY84, sY86
For AZFb: sY127, sY134
For AZFc: sY254, sY255 (both in the DAZ gene)
These STS primers have been shown to give robust and reproducible results in multiplex PCR reactions by several laboratories and in external quality control trials. However, it must be noted that according to the latest high-quality sequencing, there is a mismatch in the middle of the sequences of the primer sY84-F (which does not preclude the efficacy of amplification) and thus the sequence has been changed in the table accordingly. Concerning the sY84-R a SNP (rs72609647) is present in the 5th nucleotide of the primer sequence. In case of amplification failure, an alternative STS nearby sY84 should be tested; for neighbouring markers see the ‘MSY breakpoint mapper’, http://breakpointmapper.wi.mit.edu/ (Lange et al., 2008). The SRY gene should be included in the analysis as a control for the testis-determining factor on the short arm of the Y chromosome and for the presence of Y-specific sequences when the ZFY gene is absent (e.g. in XX males). Testing for ZFX is relevant not only for the female control DNA but also in SRY negative 46,XX males as it will be the only positive marker.
In summary, the set of PCR primers which should be used in multiplex PCR reactions as best choice for the diagnosis of microdeletion of the AZFa, AZFb and AZFc region includes: sY14 (SRY), ZFX/ZFY, sY84, sY86, sY127, sY134, sY254, sY255. The location of these primers on the Y chromosome is indicated in Fig. 2. The sequence of the primers and an example of a PCR protocol are reported in the Appendix. The use of this primer set will enable the detection of almost all clinically relevant deletions and of over 95% of the deletions reported in the literature in the three AZF regions and is sufficient for routine diagnostics. Adoption of this favourite set of primers by all laboratories is strongly recommended as it allows a minimal standardization and good comparison of laboratory performance and interlaboratory variability.
Significance of the basic primer set and extension analysis
AZFa
The molecular analysis of the AZFa region involves the use of the two STS markers sY84 and sY86. Both markers are located upstream of the USPY9 and DDX3Y genes and are anonymous. According to the pathogenic mechanism of the deletion and current experience, once a deletion of both sY84 and sY86 is detected, the probability of dealing with a complete deletion is very high. However, as partial AZFa deletions have been described in the literature and their phenotypic expression is milder than the complete ones (Krausz et al., 2006), the definition of the extension of the deletion is now compulsory (in contrast to previous guidelines).
The determination of the extension of the deletion (complete/not complete) should be performed by using the STS primers sY82 (present), sY83 (absent or present depending on the type of breakpoint) or sY1064 (absent) for the proximal border and sY1065 or sY1182 (absent), sY88 (present) for the distal border (Fig. 3B). The marker sY87 is not recommended anymore because it is located between the two AZFa genes. A more sophisticated determination of the breakpoints can be obtained with the protocol suggested by Kamp et al. (2001). If only one of the two AZFa STS loci (only sY84 or only sY86) is deleted and amplification failures can be excluded, the AZFa region should be studied in more detail testing for the presence/absence of the two AZFa genes (DDX3Y and USP9Y) and the borders according to the map provided by Kamp et al. (2001) or the definition of the breakpoints can be performed by consulting the previously mentioned publicly available database ‘MSY breakpoint mapper’. This event, however, is presently considered to be extraordinarily rare.
AZFb (P5/proximal P1)
The two anonymous markers sY127 and sY134 are located in the median and distal part of the AZFb region. According to the present knowledge, in the vast majority of cases the deletion of both markers indicates a complete deletion of the AZFb region. However, as mentioned before, for predictive purposes prior to TESE it is now – and in contrast to the previous guidelines – mandatory to perform additional analyses with the following second choice markers: sY105 (present) and sY121 or sY1224 (absent) for the proximal border and sY143 or sY1192 (absent) and sY153 (present) for the distal border (Fig. 3B). The markers sY114 and sY152 are not recommended anymore because these are mapping to more than one genomic region. A more accurate definition of the breakpoints can be defined by consulting the above-mentioned ‘MSY breakpoint mapper’.
AZFc (b2/b4)
The two markers sY254 and sY255 are specific for the DAZ gene, which is present in the reference Y chromosome sequence in four copies arranged in two complexes of two genes each in head-to-head orientation located in the palindromes P2 and P1, respectively, in the reference MSY sequence (Saxena et al., 2000). The absence of both markers indicates deletion of the entire AZFc region, which removes all copies of DAZ. According to current knowledge, the deletion of only one of these two markers is impossible and should be always regarded as a methodological error.
The vast experience accumulated until now has shown that when both markers sY254 and sY255 are deleted, a diagnosis of complete deletion of the AZFc region can be made. Some studies have shown that the AZFc deletion pattern is rather constant, although not always identical (Kuroda-Kawaguchi et al., 2001; Luetjens et al., 2002). The primers indicated by Kuroda-Kawaguchi et al. (2001) and the analysis of sY160 (heterochromatin marker) permit the laboratory to determine if the deletion corresponds to the b2/b4 pattern (Fig. 3B). Terminal deletions (absence of sY160) are more often associated with mosaic karyotype (46,XY/45,X) and thus karyotype analysis should be requested. The presence of 45,X cell lines has been considered a negative prognostic factor for the presence of testicular spermatozoa (Jaruzelska et al., 2001).
AZFbc (P5/distal P1 or P4/distal P1)
The complete deletion of both AZFb and AZFc regions is indicated by the lack of amplification of all four markers sY127, sY134, sY254 and sY255. The use of more specific markers as indicated by Repping et al. (2002) determine whether the deletion corresponds to the P5/distal P1 or P4/distal P1 pattern (sY116 is positive in case of P4/distal P1 and absent in case of P5/distal P1). This definition has clinical prognostic value as stated above (Kleiman et al. 2011). Also, in these patients it is worthwhile to test for sY160 (heterochromatine marker) to define whether it is a terminal deletion.
Interpretation of the results, control and repetition of the test
The protocol suggested by these guidelines (see appendix) has been conceived and optimized so that each of the two multiplex reactions contains a marker for each AZF region. Thus, when a complete deletion occurs in a sample both PCR reactions should show the lack of amplification for the marker specific for that region. While partial deletions of the AZFa and AZFb region, as indicated by the lack of amplification of only one marker for the relevant region, are possible, the elective deletion of only sY254 or sY255 should always be regarded as a methodological error. If only one marker for AZFa or AZFb is deleted, the deletion must first be carefully confirmed (see below) and then the entire region should be studied in more detail. This event, however, is presently regarded as exceptional. In case of AZFabc deletion (all the eight Yq markers are absent), the interpretation of the control markers is of outstanding importance (SRY and ZFX/Y) to rule out technical problems.
PCR conditions should be carefully optimized in each laboratory according to the equipment available (e.g. type of Thermocycler) and DNA quality. If the result is ambiguous and/or a technical failure is suspected, the multiplex reaction should be repeated. If the multiplex does not work for a specific DNA sample, the primer set may be run in simplex reactions. If the results of both multiplex PCR consistently speak in favour of a deletion, the deletion is confirmed. If the results of the two multiplexes are not in agreement, the whole set of primers should be repeated in simplex PCR, as there is no reason to repeat the test in the same manner. It is known that simplex PCR is less subject to amplification failure and it is strongly advised to repeat the amplification at a lower annealing temperature. There is no general advice as to the number of repetitions. The test should be repeated until the results are clear and reproducible (good laboratory practice).
Reporting
Reports should be written in a standardized format and should be clear to the non-specialist. Guidelines on how to write reports on the outcome of molecular genetics investigations on a patient can be found at the EMQN web site. In general, reports must be clear, concise, accurate, fully interpretative, credible and authoritative. Hand-written reports are not acceptable. Reports must include the following information:
clear identification of the laboratory
date of referral and reporting
patient identification: full name, date of birth and unique laboratory accession/identification number
restatement in some form of the clinical question being asked (e.g. diagnosis of microdeletion of the Y chromosome), and the indication (e.g. azoospermia, ICSI, etc.)
tissue studied (e.g. blood, buccal smear, etc.)
method used (e.g. multiplex PCR amplification)
outcome of the analysis: a tabular form of the various STS loci analysed is preferred. Avoid the use of + and −, which can be misinterpreted. Use words instead (e.g. present/absent, or similar)
a written interpretation understandable by the non-specialist
signatures of two independent assessors.
Examples of reports concerning the most frequent deletions are provided in the supplementary materials.
Alternative methods for Y microdeletion testing
Since the publications of the first guidelines in 1999, several alternative methods have been published to assess Y-chromosomal deletions. In addition, there are commercial kits on the market, which, however, almost all contain an unnecessary high number of markers (see Appendix B). This may lead to detection of ‘false’ deletions, especially if the DNA quality and PCR conditions are suboptimal (Aknin-Seifer et al., 2003, 2005). Moreover, the large majority of kits do not allow the validation of suspected deletions by single PCR (see supplementary material). Recently, Vogt & Bender (2013) proposed a multiplex PCR based on gene-specific markers. Although this approach allows the detection of isolated gene-specific deletions, the extreme rarity and unclear clinical significance of these deletions (Tyler-Smith & Krausz, 2009) preclude its use in the routine diagnostics.
Alternative methods partially based on the guidelines protocols have been developed using capillary electrophoresis, real-time PCR, MLPA and array-CGH (Osborne et al., 2007; Kozina et al., 2011; Guo et al., 2012; Jiang et al., 2012; Segat et al., 2012) Using the EAA/EMQN multiplexes, but adding a fluorescent label to the primers, allows detection with capillary electrophoresis. Some laboratories have adapted the proposed protocol accordingly, and validated this in house (LH, personal communication). Real-time PCR has the advantage of being relatively fast, because the protocol does not involve running an agarose gel, but the equipment needed is not available in every laboratory. There is one publication comparing MLPA-based Y deletion detection with other methods, but unfortunately only the abstract is available in English (Jiang et al., 2012). Finally, microarray technology has also been proposed as an alternative assay, but it does not seem to be very cost-effective and includes many more markers than necessary (Osborne et al., 2007).
In conclusion, from the literature there is no evidence that the addition of more than the advised STSs is advantageous for clinical routine diagnostics. Any time a laboratory establishes a specific method, this needs to be validated on a large enough number of samples including positive and negative controls to estimate the specificity and sensitivity. For reporting, it is important to specifically mention the method(s) used and not just refer to them as ‘according to guidelines’, which only applies to the two multiplexes with all markers described herein.
Twelve-year experience of the EAA/EMQN EQA
The laboratories performing AZF diagnostics should annually join an EQA scheme. A respective scheme is available at the EMQN that is carried out in collaboration with the EAA; online registration to the scheme is available at http://www.emqn.org. During each EAA/EMQN AZF scheme, three validated DNA samples with mock clinical case descriptions are distributed to participating laboratories per year. It is fundamental that the DNA samples received from the organizers of the EQA programme are processed exactly in the same way as patients’ samples are handled, including reporting. The results are assessed by at least two independent reviewers. Both a general report summarizing overall performance and common problems as well as individual reports to each participant including specific recommendations are issued. Laboratories receive a certificate in which their performance is evaluated.
Between 2000 and 2012 the number of participating laboratories almost tripled from 57 to 148 (Fig. 4A). The diagnostic error rate (an incorrect genotype that would lead to a misdiagnosis) decreased steeply during the first 5 years from almost 8% and now fluctuates at around 1–2% (Fig. 4B). While more variable, an assessment of the quality of diagnostic report content also showed an increase and around 50% of analyses in the last 4 years have scored full marks which is a clear improvement on the previous earlier time of the scheme. Recurrent interpretation problems still arise owing to laboratories using an unnecessary high number of markers, which are specifically included in commercially available kits (see above). The two dips in interpretation scores in 2006 and 2012 (Fig. 4B) can be explained by an 46,XX male included as an extraordinary case, which lead to recurring problems in reporting positive markers, conclusions for further testing (karyotyping needs to be recommended) and prognostic value (no chance for TESE success).
Figure 4.
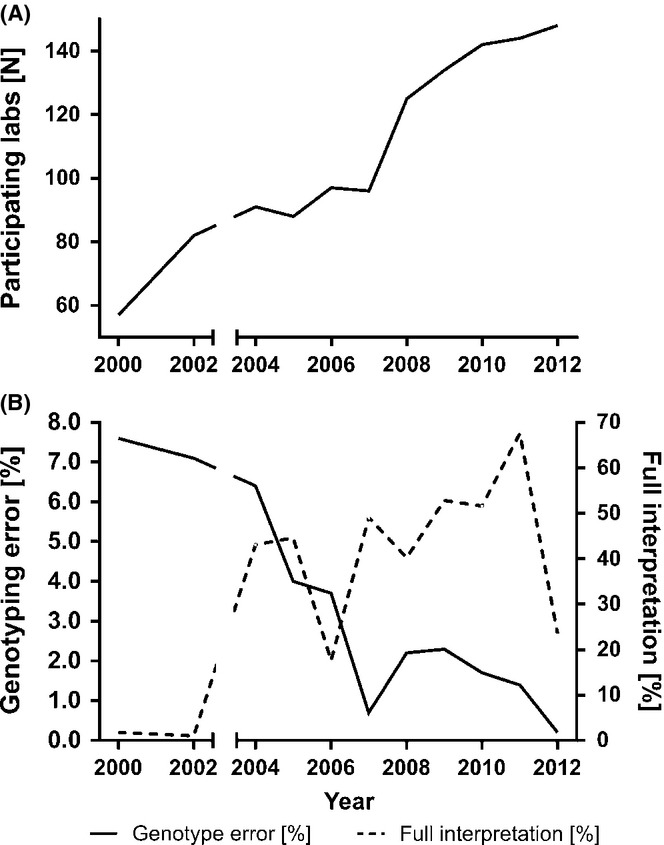
Twelve-year experience with the EAA/EMQN external quality control scheme. The number of participating labs has steadily increased (A). Genotyping error rates have steeply declined, while interpretation scores gradually increased (B).
Overall, the established EQA scheme is a successful tool to improve the performance of participating laboratories and has demonstrated an improvement on reporting practice and decreasing diagnostic error rates.
Authority statement
These guidelines have been worked out based on the long-lasting experience of the authors in the frame of the EAA/EMQN quality control AZF scheme and reflects the consensus of a large group of experts in genetics of male infertility present at the Florence-Utah-Symposium on ‘Genetics of male infertility’ (Florence, 19–21 September 2013). The document has also been approved by the EMQN board.
Acknowledgments
We are thankful to Dr Sandra Kleiman and Dr Helen Skaletsky for their precious contribution to the round table session of the Florence-Utah Symposium. We also gratefully acknowledge all the experts attending this session for their active participation and the continuous support of Dr. Simon Patton and Outi Kamarainen at EMQN.
Appendix A
Example of a PCR protocol
Two multiplex reactions were designed for the analysis of the three AZF deletion regions on the Y chromosome. Both multiplexes contain five fragments, that is, the three AZF loci and the two control fragments SRY and ZFX/Y. Each laboratory should set up and validate its own protocol. Here, we give an example of the protocol validated and currently in use at the Institute of Human Genetics in Münster.
PCR kit: Quiagen Multiplex PCR Kit (Cat.No. 206143, Quiagen, Hilden, Germany).
Preparation of 10× primer mix A and B (containing 2 μM each primer). Primer mixes are prepared in batches sufficient for about 100 reactions, and packaged in smaller size aliquots (sufficient for 10 or 20 reactions) for storage at −20 °C.
The 50-μL PCR reaction mix contains: 25 μL 2× Quiagen Multiplex PCR MasterMix [containing HotStarTaq DNA Polymerase, Qiagen Multiplex PCR Buffer (containing 6 mM MgCl2) and dNTP Mix], 5 μL 10× Primer mix (2 μM each primer), ∼1 μg template DNA, sterile distilled water to 50 μL. Amplification conditions (as established using a Hybaid Touch Down Thermocycler) start with an initial activation step of 15 min at 95 °C, followed by 35 cycles of 30-sec denaturation (94 °C), 90-sec annealing (57 °C) and 60-sec elongation (72 °C), ended by a last elongation step of 10 min and cooling to 4 °C. Reaction products (30 μL) are separated on a 2% Agarose (Peqbold Universal Agarose, Peqlab, Erlangen, Germany) plus 0.5% DNA Agar (Serva, Heidelberg, Germany) gels in 1 × TBE for 25V overnight. An example of both multiplexes is given in Fig. A1.
Fig A1.
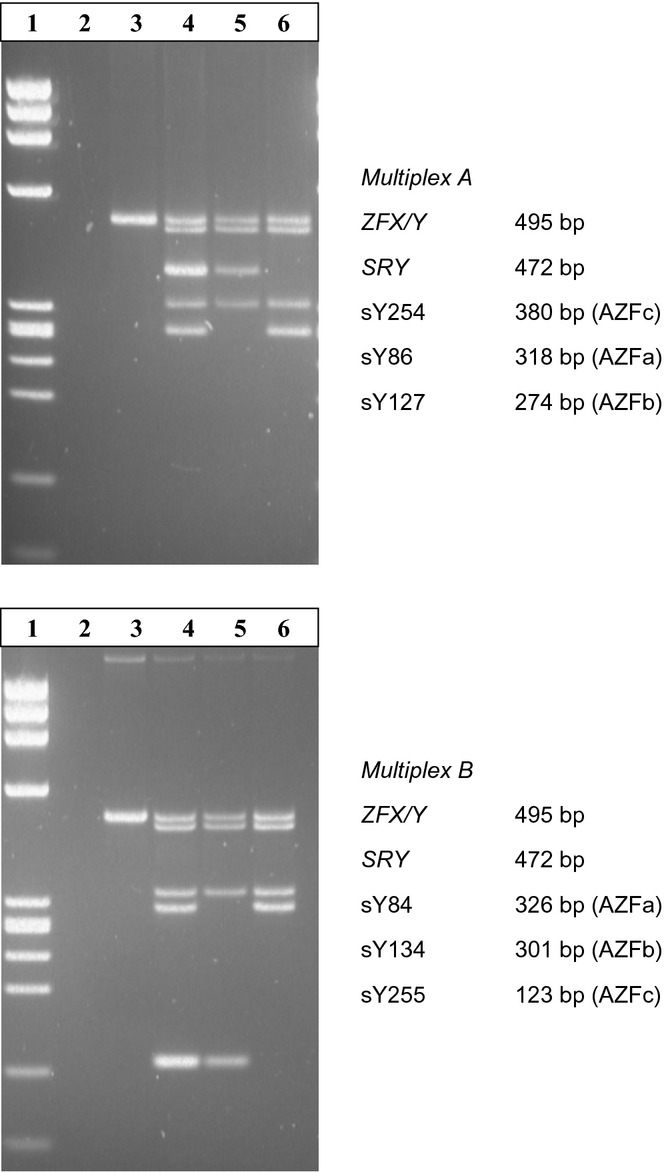
Examples of both Multiplex PCRs. Multiplex A: lane 1 phi X-HeaIII size marker, lane 2 water, lane 3 female DNA, lane 4 DNA of normal male, lane 5 DNA of AZFb (P5/proximal P1)-deleted patient, lane 6 DNA of AZFc (b2/b4)-deleted patient.
Appendix B Locus and sequence of the PCR primers (for further info see also ‘MSY Breakpoint Mapper’)
| Locus | Primer | Sequence | Product size [bp] | Genomic locus UCSC ChrY.hg19a | Status in classic, complete deletion |
|---|---|---|---|---|---|
| Format A and B | |||||
| ZFX/Y | ZFX/Y-F | 5′-ACC RCT GTA CTG ACT GTG ATT ACA C-3′ | 495 | Present | |
| ZFX/Y-R | 5′-GCA CYT CTT TGG TAT CYG AGA AAG T-3′ | ||||
| SRY | sY14-F | 5′-GAA TAT TCC CGC TCT CCG GA-3′ | 472 | Present | |
| sY14-R | 5′-GCT GGT GCT CCA TTC TTG AG-3′ | ||||
| Format A | |||||
| AZFa | sY86-F | 5′-GTG ACA CAC AGA CTA TGC TTC-3′ | 318 | Absent | |
| sY86-R | 5′ - ACA CAC AGA GGG ACA ACC CT - 3′ | ||||
| AZFb | sY127-F | 5′-GGC TCA CAA ACG AAA AGA AA-3′ | 274 | 22570359–22570742 | Absent |
| sY127-R | 5′-CTG CAG GCA GTA ATA AGG GA-3′ | ||||
| AZFc | sY254-F | 5′-GGG TGT TAC CAG AAG GCA AA-3′ | 380 | 25316193–25316572b | Absent |
| sY254-R | 5′-GAA CCG TAT CTA CCA AAG CAG C-3′ | ||||
| Format B | |||||
| AZFa | sY84-F | 5′-AGA AGG GTC CTG AAA GCA GGT-3′ | 326 | Absent | |
| sY84-R | 5′-GCC TAC TAC CTG GAG GCT TC-3′ | ||||
| AZFb | sY134-F | 5′-GTC TGC CTC ACC ATA AAA CG-3′ | 301 | 23555947–23556406 | Absent |
| sY134-R | 5′-ACC ACT GCC AAA ACT TTC AA-3′ | ||||
| AZFc | sY255-F | 5′-GTT ACA GGA TTC GGC GTG AT-3′ | 123 | 26999443–26999566b | Absent |
| sY255-R | 5′-CTC GTC ATG TGC AGC CAC-3′ | ||||
| Extension analysis | |||||
| AZFa | |||||
| AZFa | sY82-F | 5′-ATC CTG CCC TTC TGA ATC TC-3′ | 264 | Present | |
| sY82-R | 5′-CAG TGT CCA CTG ATG GAT GA-3′ | ||||
| AZFa1 | sY83-F | 5′-CTT GAA TCA AAG AAG GCC CT-3′ | 275–277 | Absent | |
| sY83-R | 5′-CAA TTT GGT TTG GCT GAC AT-3′ | ||||
| AZFa1 | sY1064-F | 5′-GGG TCG GTG CAC CTA AAT AA-3′ | 110 | Absent | |
| sY1064-R | 5′-TGC ACT AAA GAG TGA TAA TAA ATT CTG-3′ | ||||
| AZFa2 | sY1065-F | 5′-TCA GGT ACT GTG ATG CCG TT-3′ | 239 | Absent | |
| sY1065-R | 5′-TGA AGA GGA CAC AAA GGG AAA-3′ | ||||
| AZFa2 | sY1182-F | 5′-ATG GCT TCA TCC CAA CTG AG-3′ | 247 | Absent | |
| sY1182-R | 5′-CAT TGG CCT CTC CTG AGA CT-3′ | ||||
| AZFa | sY88-F | 5′-TTG TAA TCC AAA TAC ATG GGC-3′ | 123 | Present | |
| sY88-R | 5′-CAC CCA GCC ATT TGT TTT AC-3′ | ||||
| AZFb | |||||
| AZFb | sY105-F | 5′-AAG GGC TTC TTC TCT TGC TT-3′ | 301 | 19357220–19357589 | Present |
| sY105-R | 5′-AGG GAG CTT AAA CTC ACC GT-3′ | ||||
| AZFb3 | sY121-F | 5′-AGT TCA CAG AAT GGA GCC TG-3′ | 190 | 21052033–21052360 | Absent |
| sY121-R | 5′-CCT GTG ACT CCA GTT TGG TC-3′ | ||||
| AZFb3 | sY1224-F | 5′-GGC TTA AAC TTG GGA GGG TG-3′ | 640 | 20611625–20612264 | Absent |
| sY1224-R | 5′-CAA AGA GCC TCC CAG ACC A-3′ | ||||
| AZFb4 | sY143-F | 5′-GCA GGA TGA GAA GCA GGT AG-3′ | 311 | 23977880–23978312 | Absent |
| sY143-R | 5′-CCG TGT GCT GGA GAC TAA TC-3′ | ||||
| AZFb4 | sY1192-F | 5′-ACT ACC ATT TCT GGA AGC CG-3′ | 255 | 24872541–24873141 | Absent |
| sY1192-R | 5′-CTC CCT TGG TTC ATG CCA TT-3′ | ||||
| AZFb | sY153-F | 5′-GCA TCC TCA TTT TAT GTC CA-3′ | 139 | 24912639–25112794b | Present |
| sY153-R | 5′-CAA CCC AAA AGC ACT GAG TA-3′ | ||||
| gr/gr | sY1291-F | 5′-TAA AAG GCA GAA CTG CCA GG-3′ | 527 | Absent | |
| sY1291-R | 5′-GGG AGA AAA GTT CTG CAA CG-3′ | ||||
| gr/gr | sY1191-F | 5′-CCA GAC GTT CTA CCC TTT CG-3′ | 385 | Present | |
| sY1191-R | 5′-GAG CCG AGA TCC AGT TAC CA-3′ | ||||
| Hetero-chromatin | sY160-F | 5′-TAC GGG TCT CGA ATG GAA TA-3′ | 236 | 58911807–58912042b | |
| sY160-R | 5′-TCA TTG CAT TCC TTT CCA TT-3′ | ||||
1,2,3,4 markers with same numbers are interchangeable.
There are some differences in the genomic position of STSs between UCSC (hg19) and MSY breakpoint mapper, which is currently still based on hg18.
Multicopy STSs. Only the most proximal/distal position needed to describe deletions are given.
Appendix C Commercially available kits and their characteristics with respect to the EAA/EMQN Guidelines
| Name of the kit (producer) | Fully respects the Guidelines (STSs) | Confirmation step by simplex or duplex PCR |
|---|---|---|
| AB Analitica | Noa | No |
| Devyser | Noa | No |
| Diachem/Bird | Yes | Yes |
| Euroclone Strip test | Yes | No |
| Euroclone | Yes | No |
| Experteam | Yes | No |
| Promega 2.0 | Noa | No |
| Qiagen | Nob | No |
Excessive number of markers.
Different STS panel, only one marker for AZFa.
cKits which are based on the standard gel electrophoresis method are shaded.
Appendix D
Useful web sites:
European Academy of Andrology (EAA): http://www.andrologyacademy.net/
European Molecular Genetics Quality Network (EMQN): http://www.emqn.org/
MSY breakpoint mapper: http://breakpointmapper.wi.mit.edu/
E-mail addresses of the authors:
C. Krausz: csillagabriella.krausz@unifi.it
L. Hoefsloot: L.Hoefsloot@antrg.umcn.nl
M. Simoni: manuela.simoni@unimore.it
F. Tüttelmann: frank.tuettelmann@ukmuenster.de
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Data S1. Examples of reports concerning the most frequent deletions (English/German).
References
- Aknin-Seifer IE, Touraine RL, Lejeune H, Laurent JL, Lauras B, Levy R. A simple, low cost and non-invasive method for screening Y-chromosome microdeletions in infertile men. Hum Reprod. 2003;18:257–261. doi: 10.1093/humrep/deg067. [DOI] [PubMed] [Google Scholar]
- Aknin-Seifer IE, Touraine RL, Faure AK, Fellmann F, Chouteau J, Levy R. Two fast methods for detection of Y-microdeletions. Fertil Steril. 2005;84:740–742. doi: 10.1016/j.fertnstert.2005.03.050. [DOI] [PubMed] [Google Scholar]
- Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J Med Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandell RA, Mielnik A, Liotta D, Ye Z, Veeck LL, Palermo GD, et al. AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod. 1998;13:2812–2815. doi: 10.1093/humrep/13.10.2812. [DOI] [PubMed] [Google Scholar]
- Chianese C, Lo Giacco D, Tüttelmann F, Ferlin A, Ntostis P, Vinci S, et al. Y-chromosome microdeletions are not associated with SHOX haploinsufficiency. Hum Reprod. 2013;28:3155–3160. doi: 10.1093/humrep/det322. [DOI] [PubMed] [Google Scholar]
- Choe JH, Kim JW, Lee JS, Seo JT. Routine screening for classical azoospermia factor deletions of the Y chromosome in azoospermic patients with Klinefelter syndrome. Asian J Androl. 2007;9:815–820. doi: 10.1111/j.1745-7262.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- Choi JM, Chung P, Veeck L, Mielnik A, Palermo GD, Schlegel PN. AZF microdeletions of the Y chromosome and in vitro fertilization outcome. Fertil Steril. 2004;81:337–341. doi: 10.1016/j.fertnstert.2003.06.030. [DOI] [PubMed] [Google Scholar]
- Choi J, Song SH, Bak CW, Sung SR, Yoon TK, Lee DR, et al. Impaired spermatogenesis and gr/gr deletions related to Y chromosome haplogroups in Korean men. PLoS One. 2012;7:e43550. doi: 10.1371/journal.pone.0043550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram DS, Ma K, Bhasin S, Arias J, Pandjaitan M, Chu B, et al. Y chromosome analysis of infertile men and their sons conceived through intracytoplasmic sperm injection: vertical transmission of deletions and rarity of de novo deletions. Fertil Steril. 2000;74:909–915. doi: 10.1016/s0015-0282(00)01568-5. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Moro E, Garolla A, Foresta C. Human male infertility and Y chromosome deletions: role of the AZF-candidate genes DAZ, RBM and DFFRY. Hum Reprod. 1999;14:1710–1716. doi: 10.1093/humrep/14.7.1710. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, et al. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42:209–213. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, et al. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92:762–770. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, Rajpert De Meyts E, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8:286–298. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- Giachini C, Guarducci E, Longepied G, Degl'Innocenti S, Becherini L, Forti G, et al. The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet. 2005;42:497–502. doi: 10.1136/jmg.2004.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124:399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- van Golde RJ, Wetzels AM, de Graaf R, Tuerlings JH, Braat DD, Kremer JA. Decreased fertilization rate and embryo quality after ICSI in oligozoospermic men with microdeletions in the azoospermia factor c region of the Y chromosome. Hum Reprod. 2001;16:289–292. doi: 10.1093/humrep/16.2.289. [DOI] [PubMed] [Google Scholar]
- Guo Q, Lan F, Xu L, Jiang Y, Xiao L, Huang H, et al. Quadruplex real-time polymerase chain reaction assay for molecular diagnosis of Y-chromosomal microdeletions. Fertil Steril. 2012;97:864–869. doi: 10.1016/j.fertnstert.2012.01.088. [DOI] [PubMed] [Google Scholar]
- Hadjkacem-Loukil L, Ghorbel M, Bahloul A, Ayadi H, Ammar-Keskes L. Genetic association between AZF region polymorphism and Klinefelter syndrome. Reprod Biomed Online. 2009;19:547–551. doi: 10.1016/j.rbmo.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18:1660–1665. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- Jaruzelska J, Korcz A, Wojda A, Jedrzejczak P, Bierla J, Surmacz T, et al. Mosaicism for 45, X cell line may accentuate the severity of spermatogenic defects in men with AZFc deletion. J Med Genet. 2001;38:798–802. doi: 10.1136/jmg.38.11.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang MC, Lien YR, Chen SU, Ko TM, Ho HN, Yang YS. Transmission of de novo mutations of the deleted in azoospermia genes from a severely oligozoospermic male to a son via intracytoplasmic sperm injection. Fertil Steril. 1999;71:1029–1032. doi: 10.1016/s0015-0282(99)00150-8. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang WB, Guo QW, Sha YW, Ouyang HG, Zhou YL. Multiplex ligation-dependent probe amplification for detecting AZF microdeletions on the Y chromosome in infertile men with azoospermia or severe oligozoospermia. Zhonghua Nan Ke Xue. 2012;18:115–121. [PubMed] [Google Scholar]
- Jorgez CJ, Weedin JW, Sahin A, Tannour-Louet M, Han S, Bournat JC, et al. Aberrations in pseudoautosomal regions (PARs) found in infertile men with Y-chromosome microdeletions. J Clin Endocrinol Metab. 2011;96:E674–E679. doi: 10.1210/jc.2010-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamischke A, Gromoll J, Simoni M, Behre HM, Nieschlag E. Transmission of a Y chromosomal deletion involving the deleted in azoospermia (DAZ) and chromodomain (CDY1) genes from father to son through intracytoplasmic sperm injection: case report. Hum Reprod. 1999;14:2320–2322. doi: 10.1093/humrep/14.9.2320. [DOI] [PubMed] [Google Scholar]
- Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol Hum Reprod. 2001;7:987–994. doi: 10.1093/molehr/7.10.987. [DOI] [PubMed] [Google Scholar]
- Kent-First MG, Kol S, Muallem A, Ofir R, Manor D, Blazer S, et al. The incidence and possible relevance of Y-linked microdeletions in babies born after intracytoplasmic sperm injection and their infertile fathers. Mol Hum Reprod. 1996;2:943–950. doi: 10.1093/molehr/2.12.943. [DOI] [PubMed] [Google Scholar]
- Kent-First M, Muallem A, Shultz J, Pryor J, Roberts K, Nolten W, et al. Defining regions of the Y-chromosome responsible for male infertility and identification of a fourth AZF region (AZFd) by Y-chromosome microdeletion detection. Mol Reprod Dev. 1999;53:27–41. doi: 10.1002/(SICI)1098-2795(199905)53:1<27::AID-MRD4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kihaile PE, Kisanga RE, Aoki K, Kumasako Y, Misumi J, Utsunomiya T. Embryo outcome in Y-chromosome microdeleted infertile males after ICSI. Mol Reprod Dev. 2004;68:176–181. doi: 10.1002/mrd.20074. [DOI] [PubMed] [Google Scholar]
- Kleiman SE, Yogev L, Gamzu R, Hauser R, Botchan A, Paz G, et al. Three-generation evaluation of Y-chromosome microdeletion. J Androl. 1999;20:394–398. [PubMed] [Google Scholar]
- Kleiman SE, Yogev L, Lehavi O, Hauser R, Botchan A, Paz G, et al. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994–2010) Fertil Steril. 2011;95:2005–2012. doi: 10.1016/j.fertnstert.2011.01.162. 2012.e1-4. [DOI] [PubMed] [Google Scholar]
- Kleiman SE, Almog R, Yogev L, Hauser R, Lehavi O, Paz G, et al. Screening for partial AZFa microdeletions in the Y chromosome of infertile men: is it of clinical relevance? Fertil Steril. 2012;98:43–47. doi: 10.1016/j.fertnstert.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Kozina V, Cappallo-Obermann H, Gromoll J, Spiess AN. A one-step real-time multiplex PCR for screening Y-chromosomal microdeletions without downstream amplicon size analysis. PLoS One. 2011;6:e23174. doi: 10.1371/journal.pone.0023174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Degl'Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci. 2006;11:3049–3061. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- Krausz C, Bussani-Mastellone C, Granchi S, McElreavey K, Scarselli G, Forti G. Screening for microdeletions of Y chromosome genes in patients undergoing intracytoplasmic sperm injection. Hum Reprod. 1999a;14:1717–1721. doi: 10.1093/humrep/14.7.1717. [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, Barbaux S, Siffroi JP, Rouba H, Delafontaine D, et al. A high frequency of Y chromosome deletions in males with nonidiopathic infertility. J Clin Endocrinol Metab. 1999b;84:3606–3612. doi: 10.1210/jcem.84.10.6040. [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, McElreavey K. Prognostic value of Y deletion analysis: what is the clinical prognostic value of Y chromosome microdeletion analysis? Hum Reprod. 2000;15:1431–1434. doi: 10.1093/humrep/15.7.1431. [DOI] [PubMed] [Google Scholar]
- Krausz C, Rajpert-De Meyts E, Frydelund-Larsen L, Quintana-Murci L, McElreavey K, Skakkebaek NE. Double-blind Y chromosome microdeletion analysis in men with known sperm parameters and reproductive hormone profiles: microdeletions are specific for spermatogenic failure. J Clin Endocrinol Metab. 2001;86:2638–2642. doi: 10.1210/jcem.86.6.7527. [DOI] [PubMed] [Google Scholar]
- Krausz C, Forti G, McElreavey K. The Y chromosome and male fertility and infertility. Int J Androl. 2003;26:70–75. doi: 10.1046/j.1365-2605.2003.00402.x. [DOI] [PubMed] [Google Scholar]
- Krausz C, Degl'Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–2681. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
- Krausz C, Giachini C, Xue Y, O'Bryan MK, Gromoll J, Rajpert-de Meyts E, et al. Phenotypic variation within European carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. J Med Genet. 2009;46:21–31. doi: 10.1136/jmg.2008.059915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G. The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest. 2011;34:376–382. doi: 10.1007/BF03347463. [DOI] [PubMed] [Google Scholar]
- Kühnert B, Gromoll J, Kostova E, Tschanter P, Luetjens CM, Simoni M, et al. Case report: natural transmission of an AZFc Y-chromosomal microdeletion from father to his sons. Hum Reprod. 2004;19:886–888. doi: 10.1093/humrep/deh186. [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, Bell GW, Page DC. MSY Breakpoint Mapper, a database of sequence-tagged sites useful in defining naturally occurring deletions in the human Y chromosome. Nucleic Acids Res. 2008;36:D809–D814. doi: 10.1093/nar/gkm849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger R, Dudakia D, Huddart R, Easton D, Bishop DT, Stratton MR, et al. A physical analysis of the Y chromosome shows no additional deletions, other than Gr/Gr, associated with testicular germ cell tumour. Br J Cancer. 2007;96:357–361. doi: 10.1038/sj.bjc.6603557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giacco D, Chianese C, Sánchez-Curbelo J, Bassas L, Ruiz P, Rajmil O, et al. Clinical relevance of Y-linked CNV screening in male infertility: new insights based on the 8-year experience of a diagnostic genetic laboratory. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.253. doi: 10.1038/ejhg.2013.253. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longepied G, Saut N, Aknin-Seifer I, Levy R, Frances AM, Metzler-Guillemain C, et al. Complete deletion of the AZFb interval from the Y chromosome in an oligozoospermic man. Hum Reprod. 2010;25:2655–2663. doi: 10.1093/humrep/deq209. [DOI] [PubMed] [Google Scholar]
- Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, et al. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360:881–885. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- Luetjens CM, Gromoll J, Engelhardt M, Von Eckardstein S, Bergmann M, Nieschlag E, et al. Manifestation of Y-chromosomal deletions in the human testis: a morphometrical and immunohistochemical evaluation. Hum Reprod. 2002;17:2258–2266. doi: 10.1093/humrep/17.9.2258. [DOI] [PubMed] [Google Scholar]
- Mateu E, Rodrigo L, Martinez MC, Peinado V, Milan M, Gil-Salom M, et al. Aneuploidies in embryos and spermatozoa from patients with Y chromosome microdeletions. Fertil Steril. 2010;94:2874–2877. doi: 10.1016/j.fertnstert.2010.06.046. [DOI] [PubMed] [Google Scholar]
- Mau Kai C, Juul A, McElreavey K, Ottesen AM, Garn ID, Main KM, et al. Sons conceived by assisted reproduction techniques inherit deletions in the azoospermia factor (AZF) region of the Y chromosome and the DAZ gene copy number. Hum Reprod. 2008;23:1669–1678. doi: 10.1093/humrep/den124. [DOI] [PubMed] [Google Scholar]
- Maurer B, Simoni M. Y chromosome microdeletion screening in infertile men. J Endocrinol Invest. 2000;23:664–670. doi: 10.1007/BF03343791. [DOI] [PubMed] [Google Scholar]
- Maurer B, Gromoll J, Simoni M, Nieschlag E. Prevalence of Y chromosome microdeletions in infertile men who consulted a tertiary care medical centre: the Münster experience. Andrologia. 2001;33:27–33. doi: 10.1046/j.1439-0272.2001.00406.x. [DOI] [PubMed] [Google Scholar]
- Mitra A, Dada R, Kumar R, Gupta NP, Kucheria K, Gupta SK. Y chromosome microdeletions in azoospermic patients with Klinefelter's syndrome. Asian J Androl. 2006;8:81–88. doi: 10.1111/j.1745-7262.2006.00083.x. [DOI] [PubMed] [Google Scholar]
- Mulhall JP, Reijo R, Alagappan R, Brown L, Page D, Carson R, et al. Azoospermic men with deletion of the DAZ gene cluster are capable of completing spermatogenesis: fertilization, normal embryonic development and pregnancy occur when retrieved testicular spermatozoa are used for intracytoplasmic sperm injection. Hum Reprod. 1997;12:503–508. doi: 10.1093/humrep/12.3.503. [DOI] [PubMed] [Google Scholar]
- Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Costa P, Goncalves J, Plancha CE. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update. 2010;16:525–542. doi: 10.1093/humupd/dmq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates RD, Silber S, Brown LG, Page DC. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum Reprod. 2002;17:2813–2824. doi: 10.1093/humrep/17.11.2813. [DOI] [PubMed] [Google Scholar]
- Osborne EC, Lynch M, McLachlan R, Trounson AO, Cram DS. Microarray detection of Y chromosome deletions associated with male infertility. Reprod Biomed Online. 2007;15:673–680. doi: 10.1016/s1472-6483(10)60534-2. [DOI] [PubMed] [Google Scholar]
- Page DC, Silber S, Brown LG. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod. 1999;14:1722–1726. doi: 10.1093/humrep/14.7.1722. [DOI] [PubMed] [Google Scholar]
- Patsalis PC, Sismani C, Quintana-Murci L, Taleb-Bekkouche F, Krausz C, McElreavey K. Effects of transmission of Y chromosome AZFc deletions. Lancet. 2002;360:1222–1224. doi: 10.1016/s0140-6736(02)11248-7. [DOI] [PubMed] [Google Scholar]
- Peterlin B, Kunej T, Sinkovec J, Gligorievska N, Zorn B. Screening for Y chromosome microdeletions in 226 Slovenian subfertile men. Hum Reprod. 2002;17:17–24. doi: 10.1093/humrep/17.1.17. [DOI] [PubMed] [Google Scholar]
- Plotton I, Ducros C, Pugeat M, Morel Y, Lejeune H. Transmissible microdeletion of the Y-chromosome encompassing two DAZ copies, four RBMY1 copies, and both PRY copies. Fertil Steril. 2010;94:2770.e11–2770.e16. doi: 10.1016/j.fertnstert.2010.04.038. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Ottesen AM, Garn ID, Aksglaede L, Juul A. Deletions of the Y chromosome are associated with sex chromosome aneuploidy but not with Klinefelter syndrome. Acta Paed. 2011;100:900–902. doi: 10.1111/j.1651-2227.2011.02169.x. [DOI] [PubMed] [Google Scholar]
- Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- Saxena R, de Vries JW, Repping S, Alagappan RK, Skaletsky H, Brown LG, et al. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- Segat L, Padovan L, Doc D, Petix V, Morgutti M, Crovella S, et al. A real-time polymerase chain reaction-based protocol for low/medium-throughput Y-chromosome microdeletions analysis. Genet Test Mol Biomarkers. 2012;16:1349–1355. doi: 10.1089/gtmb.2012.0220. [DOI] [PubMed] [Google Scholar]
- Siffroi JP, Le Bourhis C, Krausz C, Barbaux S, Quintana-Murci L, Kanafani S, et al. Sex chromosome mosaicism in males carrying Y chromosome long arm deletions. Hum Reprod. 2000;15:2559–2562. doi: 10.1093/humrep/15.12.2559. [DOI] [PubMed] [Google Scholar]
- Silber SJ, Alagappan R, Brown LG, Page DC. Y chromosome deletions in azoospermic and severely oligozoospermic men undergoing intracytoplasmic sperm injection after testicular sperm extraction. Hum Reprod. 1998;13:3332–3337. doi: 10.1093/humrep/13.12.3332. [DOI] [PubMed] [Google Scholar]
- Simoni M. Molecular diagnosis of Y chromosome microdeletions in Europe: state-of-the-art and quality control. Hum Reprod. 2001;16:402–409. doi: 10.1093/humrep/16.3.402. [DOI] [PubMed] [Google Scholar]
- Simoni M, Bakker E, Eurlings MC, Matthijs G, Moro E, Muller CR, et al. Laboratory guidelines for molecular diagnosis of Y-chromosomal microdeletions. Int J Androl. 1999;22:292–299. doi: 10.1046/j.1365-2605.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl. 2004;27:240–249. doi: 10.1111/j.1365-2605.2004.00495.x. [DOI] [PubMed] [Google Scholar]
- Simoni M, Tüttelmann F, Gromoll J, Nieschlag E. Clinical consequences of microdeletions of the Y chromosome: the extended Münster experience. Reprod Biomed Online. 2008;16:289–303. doi: 10.1016/s1472-6483(10)60588-3. [DOI] [PubMed] [Google Scholar]
- Sin HS, Koh E, Shigehara K, Sugimoto K, Maeda Y, Yoshida A, et al. Features of constitutive gr/gr deletion in a Japanese population. Hum Reprod. 2010;25:2396–2403. doi: 10.1093/humrep/deq191. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Soares AR, Costa P, Silva J, Sousa M, Barros A, Fernandes S. AZFb microdeletions and oligozoospermia–which mechanisms? Fertil Steril. 2012;97:858–863. doi: 10.1016/j.fertnstert.2012.01.099. [DOI] [PubMed] [Google Scholar]
- Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. The choice and outcome of the fertility treatment of 38 couples in whom the male partner has a Yq microdeletion. Hum Reprod. 2005;20:1887–1896. doi: 10.1093/humrep/deh847. [DOI] [PubMed] [Google Scholar]
- Stouffs K, Lissens W, Tournaye H, Haentjens P. What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update. 2011;17:197–209. doi: 10.1093/humupd/dmq046. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, et al. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- Tomasi PA, Oates R, Brown L, Delitala G, Page DC. The pituitary-testicular axis in Klinefelter's syndrome and in oligo-azoospermic patients with and without deletions of the Y chromosome long arm. Clin Endocrinol (Oxf) 2003;59:214–222. doi: 10.1046/j.1365-2265.2003.01828.x. [DOI] [PubMed] [Google Scholar]
- Tüttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M. Gene polymorphisms and male infertility - a meta-analysis and literature review. Reprod Biomed Online. 2007;15:643–658. doi: 10.1016/s1472-6483(10)60531-7. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Krausz C. The will-o’-the-wisp of genetics–hunting for the azoospermia factor gene. N Engl J Med. 2009;360:925–927. doi: 10.1056/NEJMe0900301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser L, Westerveld GH, Korver CM, van Daalen SK, Hovingh SE, Rozen S, et al. Y chromosome gr/gr deletions are a risk factor for low semen quality. Hum Reprod. 2009;24:2667–2673. doi: 10.1093/humrep/dep243. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Bender U. Human Y chromosome microdeletion analysis by PCR multiplex protocols identifying only clinically relevant AZF microdeletions. Methods Mol Biol. 2013;927:187–204. doi: 10.1007/978-1-62703-038-0_17. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xiao CY, A ZC, Zhang SZ, Li X, Zhang SX. DAZ1/DAZ2 cluster deletion mediated by gr/gr recombination per se may not be sufficient for spermatogenesis impairment: a study of Chinese normozoospermic men. Asian J Androl. 2006;8:183–187. doi: 10.1111/j.1745-7262.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ma M, Li L, Su D, Chen P, Ma Y, et al. Differential effect of specific gr/gr deletion subtypes on spermatogenesis in the Chinese Han population. Int J Androl. 2010;33:745–754. doi: 10.1111/j.1365-2605.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- Yen P. The fragility of fertility. Nat Genet. 2001;29:243–244. doi: 10.1038/ng1101-243. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lu C, Li Z, Xie P, Xia Y, Zhu X, et al. Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet. 2007;44:437–444. doi: 10.1136/jmg.2007.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


