Abstract
An association between low serum vitamin D levels and poorer melanoma survival has been reported. We have studied inheritance of a polymorphism of the GC gene, rs2282679, coding for the vitamin D-binding protein, which is associated with lower serum levels of vitamin D, in a meta-analysis of 3137 melanoma patients. The aim was to investigate evidence for a causal relationship between vitamin D and outcome (Mendelian randomization). The variant was not associated with reduced overall survival (OS) in the UK cohort, per-allele hazard ratio (HR) for death 1.23 (95% confidence interval (CI) 0.93, 1.64). In the smaller cohorts, HR in OS analysis was 1.07 (95% CI 0.88, 1.3) and for all cohorts combined, HR for OS was 1.09 (95% CI 0.93, 1.29). There was evidence of increased melanoma-specific deaths in the seven cohorts for which these data were available. The lack of unequivocal findings despite the large sample size illustrates the difficulties of implementing Mendelian randomization.
Keywords: vitamin D, melanoma, survival analysis, mendelian randomization, GC
Significance
Mendelian randomization utilizes inheritance of a genetic variant associated with the exposure of interest (here the gene coding for vitamin D-binding protein) to investigate causality: the concept being that a relationship between rs2282679 and survival would establish that low vitamin D levels impacted directly on outcome rather than the association being due to confounding e.g. by healthier people spending more time outdoors. We show no unequivocal evidence for causality even in a large multicentre study, but we cannot exclude a variable (J-shaped) effect in the presence of different population vitamin D levels. The value of Mendelian randomization is in practice limited by the small effect of genes and conceivably by non-linear relationships.
Introduction
25-Hydroxyvitamin D (henceforth referred to as Vitamin D) levels shortly after diagnosis were shown to be inversely associated with Breslow thickness and positively associated with survival within thickness groups in cases recruited to the Leeds Melanoma cohort (Newton-Bishop et al., 2009). A subsequent small study in stage IV melanoma patients reported that lower vitamin D levels were associated with poorer survival (Nurnberg et al., 2009), but in the absence of further published data, the Leeds study remains unvalidated. The role of vitamin D in bone health is clear, and there is much evidence in the literature that it plays an important role in many other aspects of health, including cancer survival (Goodwin et al., 2009) (Autier and Gandini, 2007). A meta-analysis of all eligible randomized controlled trials (RCTs) for vitamin D given for a variety of reasons showed an overall survival advantage for supplementation (Autier and Gandini, 2007), but as vitamin D levels tend to be higher in leaner fitter people, vitamin D levels, in observational studies at least, may simply be a marker of better health, rather than causally related to survival.
There are also data suggestive of increased mortality associated with high vitamin D levels in a large Danish study of patients tested in primary care (Durup et al., 2012) and particular concerns about supplementation in melanoma patients because of a theoretical risk that high levels of vitamin D could be harmful due to its reported immunosuppressive effects. We have investigated the viability of Mendelian randomization (Katan, 1986) as a method of using inherited variation in genes of known function (in this case variation known to affect vitamin D-binding protein levels and therefore vitamin D levels in the blood) to examine the evidence for a causal effect of vitamin D level on melanoma survival, given the difficulties of establishing causation in the absence of a randomized clinical trial (RCT). Genome-wide association (GWA) studies have shown that the strongest genetic determinant of serum vitamin D levels is the single nucleotide polymorphism (SNP) rs2282679, located in intron 12 of GC, the gene coding for the vitamin D-binding protein (VDBP) (Ahn et al., 2010; Wang et al., 2010). We postulated that if inheritance of the minor allele of rs2282679, which is associated with lower levels of vitamin D (Davies et al., 2011), was also associated with poorer outcome from melanoma, then, using the principle of Mendelian randomization, the observation would support the hypothesis that vitamin D has a causal relationship with melanoma survival. SNPs in other genes in the vitamin D metabolism pathway have been reported to be associated with serum vitamin D levels (Ahn et al., 2010). In a Leeds study previously reported however, these SNPs explained each <1% of the variance in serum levels and were therefore not tested.
Results
The association of the rs2282679 SNP with OS in the Leeds melanoma cohort reported in 2009
The minor allele of the SNP was not significantly associated with overall survival in the cohort reported in 2009 (n = 800, 119 deaths, Hazard ratio (HR) for death 1.18, 95% confidence interval (CI) 0.89, 1.56, P = 0.2 adjusted for age, sex, site of primary; and HR 1.23, 95% CI 0.93, 1.64, P = 0.1 when also adjusted for Breslow thickness) or in the larger Leeds melanoma cohort of 1390 cases, (HR for death 1.10, 95% CI 0.90, 1.35, P = 0.3 adjusted for age, sex and site; and HR 1.11, 95% CI 0.91, 1.36, P = 0.3 additionally adjusting for Breslow thickness). When follow-up was truncated at the end of 2009 (in view of adoption of advice to avoid vitamin D depletion in Leeds after 2009) the HR was similar to that seen in the original cohort (n = 1358, HR 1.17, 95% CI 0.94, 1.48, P = 0.2 adjusted for age, sex and site; HR 1.20, 95% CI 0.95, 1.51, P = 0.1 when also adjusted for Breslow thickness, data not shown).
Meta-analysis of the association of the rs2282679 SNP with survival in the additional BioGenoMEL cohorts and the Leeds cohort
Figure 1 and Table 1 show the association of rs2282679 with overall survival (OS) in each of the nine additional BioGenoMEL cohorts. For six of the nine cohorts, the direction of effect was to see a nonsignificantly poorer outcome in association with the rarer allele. We saw evidence of heterogeneity between the cohorts (Cochran's Q, P = 0.1, I2 = 41% for all smaller cohorts, Cochran's Q, P = 0.1, I2 = 37% when combined with the larger Leeds cohort). Contrary to the other cohorts, in the Lund cohort, the variant was observed to have a protective effect (HR 0.49, 95% CI 0.27, 0.89, P = 0.02). When this cohort was dropped, there was no evidence of heterogeneity (Cochran's Q, P = 0.6, I2 = 0% for the remaining eight BioGenoMEL cohorts and Leeds).
Figure 1.
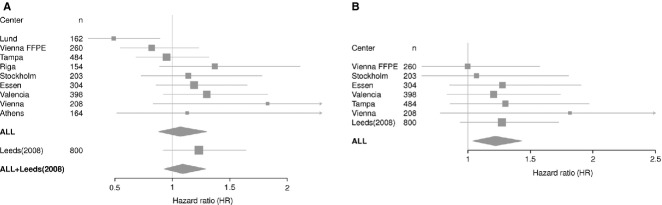
Forest plot showing the estimates per-allele HR for SNP rs2282679 in each of the ten cohorts with and without the Leeds cohort for (A) Overall survival (truncated at 8 yr), (B) Melanoma specific survival. Estimates were generated from Cox proportional hazard models and adjusted for age, sex, site of the primary tumour and Breslow thickness.
Table 1.
Association of the rs2282679 SNP with overall survival (truncated at 8 yr) in ten melanoma cohorts. Cox proportional hazard models were fitted assuming an additive effect. In the majority of cohorts, inheritance of the minor allele was associated with poorer outcome
| Centre | Cases | Minor allele frequencya | No. of deaths | HR (95% CI)b per minor allele | P-value | HR (95% CI)c per minor allele | P-value |
|---|---|---|---|---|---|---|---|
| Leeds (2008) | 800 | 0.29 | 119 | 1.18 (0.89, 1.56) | 0.2 | 1.23 (0.93, 1.64) | 0.1 |
| Leeds (2012) | 1390 | 0.29 | 233 | 1.10 (0.90, 1.35) | 0.3 | 1.11 (0.91, 1.36) | 0.3 |
| Valencia | 398 | 0.33 | 78 | 1.26 (0.90, 1.77) | 0.2 | 1.30 (0.92, 1.83) | 0.1 |
| Essen | 304 | 0.28 | 97 | 1.23 (0.89, 1.70) | 0.2 | 1.19 (0.86, 1.65) | 0.3 |
| Stockholm | 203 | 0.27 | 53 | 1.15 (0.75, 1.77) | 0.5 | 1.14 (0.74, 1.78) | 0.5 |
| Vienna | 208 | 0.27 | 18 | 1.40 (0.69, 2.83) | 0.4 | 1.83 (0.84, 4.01) | 0.1 |
| Riga | 154 | 0.31 | 53 | 1.42 (0.93, 2.16) | 0.1 | 1.37 (0.89, 2.11) | 0.2 |
| Tampa | 484 | 0.29 | 97 | 0.85 (0.62, 1.18) | 0.3 | 0.95 (0.68, 1.32) | 0.7 |
| Lund | 162 | 0.26 | 45 | 0.47 (0.26, 0.86) | 0.01 | 0.49 (0.27, 0.89) | 0.02 |
| Vienna FFPE | 260 | 0.28 | 75 | 0.88 (0.60, 1.30) | 0.5 | 0.82 (0.55, 1.23) | 0.3 |
| Athens | 164 | 0.25 | 18 | 1.15 (0.54, 2.45) | 0.7 | 1.13 (0.51, 2.47) | 0.8 |
| BioGenoMEL without Leeds (2008)d | 2337 | 534 | 1.06 (0.87, 1.28) | 0.6 | 1.07 (0.88, 1.30) | 0.5 | |
| BioGenoMEL with Leeds (2008)d | 3137 | 653 | 1.08 (0.91, 1.27) | 0.4 | 1.09 (0.93, 1.29) | 0.3 |
Frequency in CEU population of G allele in HapMap = 0.26.
Cases adjusted for age, sex, site of primary and a single primary melanoma recruited no more than 2 yr after diagnosis.
Cases additionally adjusted for Breslow thickness data > 0.75 mm.
Meta-analysis results assume a random effects model.
In a meta-analysis of the nine additional BioGenoMEL cohorts combined, the minor allele was not significantly associated with poorer survival under a random effects model (HR 1.06, 95% CI 0.87, 1.28, P = 0.6, adjusted for age, sex and site; HR 1.07, 95% CI 0.88, 1.30, P = 0.5, when also adjusted for Breslow thickness).
When the additional BioGenoMEL cohorts were also combined with the Leeds cohort in a random effects model, the estimated effect did not support a significant association between the rare allele and poorer outcome (HR 1.07, 95% CI 0.88, 1.30, P = 0.5 adjusted for age, sex, site; HR 1.09, 95% CI 0.93, 1.29, P = 0.3 when additionally adjusted for Breslow thickness). There was no meaningful difference in the results if the larger Leeds cohort was used instead.
Melanoma-specific Survival (MSS)
We compared MSS and OS where possible (Figure 1, Supporting information, Table S2). For most cohorts, the results were similar, but for the Tampa and Essen cohorts the association with the minor allele and MSS was stronger than for OS. Overall, in the cohorts for which MSS data were available, there was a statistically significant increased risk for melanoma deaths associated with the SNP associated with lower vitamin D levels, HR 1.22 95% CI 1.04–1.43, P = 0.01, assuming a random effects model.
Haplotype analysis
The rs2282679 SNP was in almost perfect linkage disequillibrium (LD) with rs4588 in the Leeds data (D’ = 1, R2 = 0.99), as has been reported, and was in strong LD with rs7041 (D’ = 1, R2 = 0.5). We subsequently typed both SNPs in the rest of the BioGenoMEL cohort and fitted a joint Cox regression of haplotypes Gc1s and Gc2 for overall survival taking the ancestral Gc1f haplotype as baseline (Table 2, Figure 2).
Table 2.
Association of the per haplotype effect of Gc1s and Gc2 with overall survival (truncated at 8 yr) in nine melanoma cohorts. Cox proportional hazard models were fitted assuming an additive effect. In the majority of cohorts, inheritance of the minor allele was associated with poorer outcome. Both Gc1s and Gc2 were jointly fitted in a multivariable model to look at the effect of each haplotype adjusted for the other two
| Gc1s (baseline Gc1f) | Gc2 (baseline Gc1f) | ||||
|---|---|---|---|---|---|
| Centre | Number of cases (deaths) | HR (95% CI)a per minor allele | P-value | HR (95% CI)a per minor allele | P-value |
| Leeds (2008) | 783 (120) | 1.10 (0.76, 1.59) | 0.6 | 1.31 (0.87, 1.96) | 0.2 |
| Valencia | 373 (76) | 1.13 (0.67, 1.90) | 0.7 | 1.41 (0.81, 2.47) | 0.2 |
| Essen | 244 (76) | 1.38 (0.83, 2.31) | 0.2 | 1.45 (0.81, 2.61) | 0.2 |
| Vienna | 189 (18) | 3.58 (0.92, 13.91) | 0.07 | 4.11 (1.01, 16.79) | 0.05 |
| Riga | 151 (51) | 1.72 (0.71, 4.19) | 0.2 | 2.32 (0.89, 6.04) | 0.09 |
| Lund | 157 (43) | 0.59 (0.30, 1.15) | 0.1 | 0.29 (0.12, 0.70) | 0.006 |
| Vienna FFPE | 241 (68) | 1.15 (0.69, 1.94) | 0.6 | 0.85 (0.47, 1.53) | 0.6 |
| Barcelona | 259 (39) | 1.42 (0.59, 3.40) | 0.4 | 2.12 (0.89, 5.06) | 0.09 |
| Athens | 168 (15) | 1.32 (0.39, 4.48) | 0.7 | 1.34 (0.31, 5.83) | 0.7 |
| Overallb | 2565 (506) | 1.17 (0.95, 1.43) | 0.1 | 1.28 (0.88, 1.86) | 0.2 |
Cases adjusted for age, sex, site of primary, the other haplotype (Gc1s or Gc2) and Breslow thickness data. Cases have a single primary melanoma recruited no more than 2 yr after diagnosis.
Meta-analysis results assume a random effects model.
Figure 2.
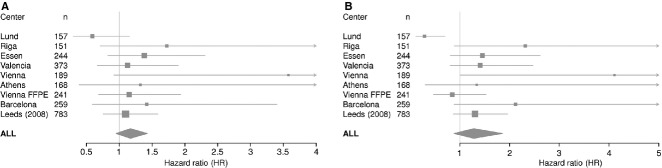
Forest plot showing the per-haplotype HR estimates for Gc1s and Gc2 in nine cohorts for overall survival (truncated at 8 yr). Estimates were generated from Cox proportional hazard models adjusted by age, sex, site, Breslow thickness and the other two haplotypes.
Neither analysis showed a statistically significant association with outcome, but the observed effect sizes were larger than for the single SNP analysis and the direction of effect was consistent with our hypothesis regarding the effect of vitamin D. We saw evidence that possessing more copies of the Gc1s variant is associated with a poor outcome (HR 1.17 per haplotype adjusted for age, sex, site, Breslow, Gc2, 95% CI 0.95, 1.43, P = 0.1). Possessing more copies of the Gc2 haplotype was associated with an even poorer outcome (HR 1.28 per haplotype adjusted for age, sex, site, Breslow, Gc1s, 95% CI 0.88, 1.86, P = 0.2).
As with the rs2282679 SNP, a strong effect is seen for the Lund cohort in the opposite direction. (HR = 0.59 per haplotype of Gc1s, 95% CI 0.30, 1.15, P = 0.1, adjusted for age, sex, site, Breslow, Gc2 and HR = 0.2 per haplotype of Gc2, 95% CI 0.12, 0.70, P = 0.006, adjusted for age, sex, site, Breslow, Gc1s). No significant heterogeneity was seen for the Gc1s estimates (Cochrane's Q, P = 0.4, I2 = 1%). However, substantial heterogeneity was seen for the Gc2 estimates (Cochrane's Q, P = 0.02, I2 = 56%), which was removed if the Lund cohort was omitted (Cochrane's Q = 0.4, I2 = 1%).
Testing the assumption that the GC SNP is independent of survival, conditional on vitamin D levels
A key assumption of Mendelian randomization is that the genetic variant only influences survival through its effect on the exposure variable (see Data S1, Figure S2). In the original Leeds cohort for cases with measured serum levels (n = 761), adjusting for vitamin D did reduce the observed effect size (HR 1.19, 95% CI 0.89, 1.58, P = 0.2 adjusted for age, sex and site; HR 1.09, 95% CI 0.82, 1.45, P = 0.6 when additionally adjusted for serum vitamin D as a continuous measure and season of blood draw, data not shown), suggesting that GC may have effects independent of its effect on vitamin D which would invalidate the Mendelian randomization approach.
Analysis of cases with primaries in sun-protected body sites
The reported association between rs2282679 and survival was stronger for melanomas arising in sun-protected sites than overall (Table S3) in the original Leeds cohort (HR 2.83, 95% CI 1.03, 7.73, P = 0.04). The Essen, Vienna FFPE and Valencia cohort data were similar (HR 3.48, 95% CI 0.65, 18.54, P = 0.1, HR 4.13, 95% CI 1.07, 15.89, P = 0.04 and HR 1.63, 95% CI 0.76, 3.47, P = 0.2 respectively). None of the other cohorts have enough rare cases to produce reliable estimates, though there was some evidence of a deleterious effect of the variant allele in the limited data from the Tampa cohort.
Discussion
We have previously reported an association between low serum levels of vitamin D soon after diagnosis and increased risk of death for melanoma patients (Newton-Bishop et al., 2009). The effects of vitamin D are pleiotropic, but vitamin D is clearly demonstrated to be antiproliferative for melanoma in vitro (Essa et al., 2010) and to promote cellular differentiation. We have looked at the association between inheritance of a variant in the GC gene encoding vitamin D-binding protein (VDBP) (associated with lower levels of vitamin D) and survival, to test for a causal relationship between vitamin D and survival. Although this gene is the strongest known genomic predictor of serum vitamin D levels in GWA studies, it does explain only 4% of the variance in blood levels of vitamin D (Davies et al., 2011).
The strength of the study is the size of the Leeds cohort and validation in multiple melanoma cohorts from Europe and North America, although some of those cohorts are individually small. A weakness of the study is that we could only use OS as the end point for outcome overall as melanoma specific survival data were not available for all cohorts, though in most cases the estimates under MSS and OS were similar. We did see a statistically significant association between the SNP and increased death from melanoma in the MSS analysis. However, we did not have MSS data from the Lund cohort, the cohort that contributed the majority of the heterogeneity observed in the OS analysis, and we are currently therefore unable to assess whether the Lund cohort would have a similar effect for MSS. A second weakness is that, in comparison to phenotypic factors such as Breslow thickness, genetic variants are likely to have only weak effects on outcome so that power is always an issue for studies of this type.
The haplotype analysis (for which functional data are published) was supportive of a relationship between the gene and survival, as there was a trend towards increased survival in those individuals who express one or more copies of the Gc1f protein isoform though no significant associations were found. We looked at this isoform as it has been shown, in vivo, to be more abundant in the plasma, to have a higher binding affinity for vitamin D metabolites and to be associated with higher serum vitamin D levels. Thus, this is consistent with the previous finding of this group that higher vitamin D levels, measured at diagnosis, are associated with increased survival times, but the result is not conclusive. In our secondary analysis of the larger Leeds cohort followed up after advice was given to avoid vitamin D depletion was recommended in 2009, the reported association of the variant was weaker.
There was some variation in the association between rs2282679 and survival between cohorts, which might be due to differences between populations, to differences in study design or to chance. The only significant outlier was the Lund cohort. When the Lund cohort is excluded, a significant association is seen between rs2282679 and outcome in the remaining cohorts (HR 1.15, 95% CI 1.01, 1.31, P = 0.03). Similarly, if the Lund cohort is dropped, a significant association is seen between the number of copies of Gc2 present and outcome (HR = 1.39, 95% CI 1.1, 1.76, P = 0.006, adjusted as above) and the number of copies of Gc1s and outcome (HR = 1.25, 95% CI 1.01, 1.54, P = 0.04, adjusted as above). Here, we consider possible explanations as to why the Lund cohort produces results so different from the other cohorts. The most likely, given the small size of the Lund cohort (162 cases), was that the inconsistent result is due to random variation. The Lund cohort was also recruited much earlier than other cohorts so that it may be that there is some unknown effect of study entry date on the association of the SNP with outcome that we have been unable to control for. However, it is also known that serum vitamin D levels in Scandinavia are the highest in Europe due to a diet rich in fish and mandatory fortification of foods (McKenna, 1992). In a population-based prospective cohort study of men living in Malmo (Southern Sweden near to Lund) born between 1923–1945, recruited in 1991–1996, designed to look at diet and cancer risk (Brandstedt et al., 2012), reported levels of vitamin D in the serum were extremely high compared to other populations. The average vitamin D level in the Swedish prostate cancer cases was 87.4 nmol/l and 86.4 in controls. In the Leeds melanoma cohort, the average level around diagnosis was 57 nmol/l in population controls and 60 in sibling controls (Davies et al., 2011). If high vitamin D levels were deleterious in melanoma as a result of immunosuppression, in a population with high dietary vitamin D, then a biological explanation for the difference between the Lund data and the others could conceivably be that in those circumstances inheritance of a genetic variant, further increasing levels might be associated with higher mortality. This biological hypothesis seems less likely than the possibility that the result in the Lund samples was a random event, but as vitamin D supplementation is common in the absence of measurement of levels, it is important to consider the possibility that high as well as low levels of vitamin D might be deleterious.
We report evidence of an effect on survival for melanoma patients whose primary was in sun-protected sites, for example, acral lentiginous tumours. This comparison was carried out because, although in case-control comparisons previously reported for serum vitamin D levels we showed no overall evidence that melanoma patients had lower levels than controls (Newton-Bishop et al., 2011), this sub-group in the Leeds cohort did have lower levels (paper in preparation).
Even taking into account the size of our large dataset by melanoma cohort standards, insufficient power remains problematic. Using results from our previous analyses, we are able to quantify the magnitude of this problem; we have previously shown in the Leeds melanoma cohort that there is a mean decrease of 5.7 nmol/l serum vitamin D levels per allele of rs2282679 (Davies et al., 2011) and that the hazard ratio for death is 0.83 for an increase of 20 nM (for OS) (Newton-Bishop et al., 2009). From these estimates, we would expect the per-allele effect of the SNP on survival to have a HR of 1.05, assuming the SNP had no effect on survival independent of its influence on serum vitamin D. Assuming that 20% of cases will die and a minor allele frequency (MAF) of 0.26, we would require over 42 000 cases to have 80% power to investigate an association of this magnitude and over 20 000 cases to have 50% power. This demonstrates that while Mendelian Randomization is in theory an excellent tool for investigating causality, since genetic effect sizes are usually modest, very large cohort sizes would be required to reach definitive conclusions. Our findings corroborate those of a recent analysis, where a similar approach was used to determine a causal role for vitamin B12 in the risk of developing prostate cancer (Collin et al., 2011). The authors determined that to have 90% power to detect a genotype-cancer association they would have needed 65 000 cases and 65 000 controls assuming the B12-prostate cancer association they had previously reported was not due to confounding. A Mendelian Randomization study has recently successfully been conducted where a variant at the FTO locus was shown to be associated with multiple cardiovascular disease related outcomes, containing data from 198 502 individuals (Fall et al., 2013). For diseases such as melanoma, however, recruiting so many cases is not currently feasible. An alternative method for boosting power is to combine a set of genetic variants that captures more of the variation in the phenotype of interest. We have presented an analysis where we looked at haplotypes of GC, the strongest known determinant of vitamin D levels. Even though the environment largely determines vitamin D levels, greater understanding of variation in genes involved in vitamin D metabolism may eventually lead to the development of a set of variants that has enough power to overcome the limitations of Mendelian Randomization in this setting.
Methods
Data collection
The Leeds melanoma cohort
Population-ascertained incident melanoma cases were recruited to an ethically approved case-control study in a geographically defined area of the UK (Yorkshire and the Northern region south of the River Tyne, 67% participation rate); 960 cases (aged 18 to 76 yr) were recruited from September 2000 to December 2005 (Falchi et al., 2009; Randerson-Moor et al., 2009). Additional cases were recruited from 2005 to 2012 to build the Leeds melanoma cohort. Since January 2007, cases have also been recruited from outside Yorkshire to enhance for participants having had a sentinel node biopsy and for melanomas arising in sun-protected sites. As of March 2012, there were 2112 cases recruited, the majority of whom are still undergoing sample and data collection.
In this study, we present analyses in 800 of the cases in which we reported an association of serum vitamin D previously. Cases chosen for analysis had tumours thicker than 0.75 mm, a single primary melanoma and complete follow-up data available at the time of analysis. We report a secondary analysis in the larger cohort of 1390 with complete data in March 2012; interpretation of the results in the full dataset is complicated by the fact that vitamin D supplementation was recommended to many cases at the end of 2009, and it is unknown how many individuals followed this advice and to what degree. In previous studies, supplementation was an even more potent predictor of vitamin D levels than the inheritance of the rs2282679 SNP in the GC gene, which is strongly associated with serum levels (Davies et al., 2011).
Recruitment (and therefore blood sampling) took place, wherever possible, within a period of 3–6 months after diagnosis. Season of serum vitamin D sampling was calculated from the date of blood draw and grouped as a categorical variable by month: Jan–Mar, Apr–Jun, Jul–Sept and Oct–Dec. Relapse/survival data are collected by annual participant questionnaire, contact (bi-annual survey) with family doctors, extraction of data from clinical notes and tagging with the cancer registry, which produces regular updates of death registration and new cancer diagnoses. OS and melanoma-specific survival (MSS) data are available. The Leeds cohort was analysed at two specific time points: when the association with vitamin D was first reported (15) and the most recent update in March 2012. The original dataset consisted of cases with tumours thicker than 0.75 mm that were recruited before late 2006 and followed up until November 2008. In this analysis, but not in the original report, we also excluded from all analyses cases with multiple melanomas and those who were recruited 2 yr or more after diagnosis to produce consistency across all cohorts.
BioGenoMEL cohorts
Eleven cohorts, the Leeds cohort plus ten additional cohorts from Europe and the USA, contributed data to this analysis (Table 3). Additional details can be found in Data S1.
Table 3.
Cases eligible for analysis in the cohorts that comprise the meta-analysis of the association of the GC SNPs with outcome. Cases in each column also meet the criteria of all conditions to the left of it
| Cases with complete data on adjusting covariates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Centre | Whole cohort size | Incident case (recruited <2 yr after diagnosis) | Cases genotyped for the SNP | Number of cases with a single melanoma | Age | Sex | Site | Breslow | Number with Breslow > 0.75 mm | Cases with complete follow-up | Cases not in sun protected sites |
| Leedsa | 1157 | 1085 | 1031 | 1006 | 1006 | 1006 | 1003 | 991 | 819 | 800 | 751 |
| Valencia | 1440 | 1248 | 654 | 654 | 654 | 654 | 640 | 585 | 399 | 398 | 342 |
| Essen | 941 | 643 | 630 | 615 | 574 | 574 | 490 | 392 | 304 | 304 | 281 |
| Stockholm | 870 | 605 | 222 | 222 | 222 | 222 | 222 | 222 | 203 | 203 | 203 |
| Vienna | 1085 | 389 | 251 | 251 | 242 | 242 | 242 | 225 | 212 | 208 | 195 |
| Riga | 243 | 242 | 200 | 199 | 199 | 199 | 199 | 171 | 155 | 154 | 149 |
| Tampa | 585 | 585 | 505 | 505 | 505 | 505 | 503 | 500 | 484 | 484 | 463 |
| Lund | 355 | 355 | 338 | 338 | 338 | 338 | 327 | 327 | 162 | 162 | 160 |
| Vienna FFPE | 302 | 302 | 286 | 279 | 276 | 276 | 276 | 273 | 260 | 260 | 244 |
| Athens | 200 | 200 | 170 | 170 | 170 | 170 | 170 | 170 | 165 | 164 | 164 |
| Barcelona | 398 | 358 | 330c | 294 | 280 | 279 | 277 | 268 | 259 | 259 | 250 |
| Validation set total | 6419 | 4927 | 3586 | 3527 | 3460 | 3459 | 3346 | 3133 | 2603 | 2596 | 2451 |
| Leeds (2012) | 2112 | 1996 | 1699 | 1661 | 1661 | 1661 | 1661 | 1619 | 1430 | 1390 | 1289 |
This dataset was the initial Leeds cohort recruited up until 2006 and followed up until Nov 2008.
One case dropped because it had an improbably large recorded Breslow thickness that greatly affected the linear relationship of Breslow thickness with outcome in the multivariable model.
Cases genotyped for both rs7041 and rs4588.
Follow-up data are collected in the additional cohorts in a similar manner to the Leeds melanoma cohort with some local variations (Table S1). Only seven groups had MSS data and genotyping data for the rs2282679 SNP, so OS was used in the meta-analysis of these data sets.
There was concern that cases with primary melanomas arising in sun-protected body sites may have a different biology to those in sun-exposed sites. Where possible, these cases were also analysed in a separate subanalysis.
There was some variation between the cohorts, as has been reported before (Davies et al., 2011). Figure 3 shows variation in the Breslow thickness distribution for each of the cohorts, showing that thicker tumours were removed in the Riga cohort and (to a lesser degree) in the Essen cohort, which recruited from a clinic serving as a referral centre within the Ruhr region of Germany. Figure 4 shows Kaplan–Meier estimates for survival in each of the ten cohorts. The Kaplan–Meier curves are relatively similar in most centres, with the exception of the Riga and Essen cohorts where the survival curve is much steeper, as was expected given the thicker tumours at presentation. Only cases with invasive tumours thicker than 0.75 mm were included in the analysis.
Figure 3.
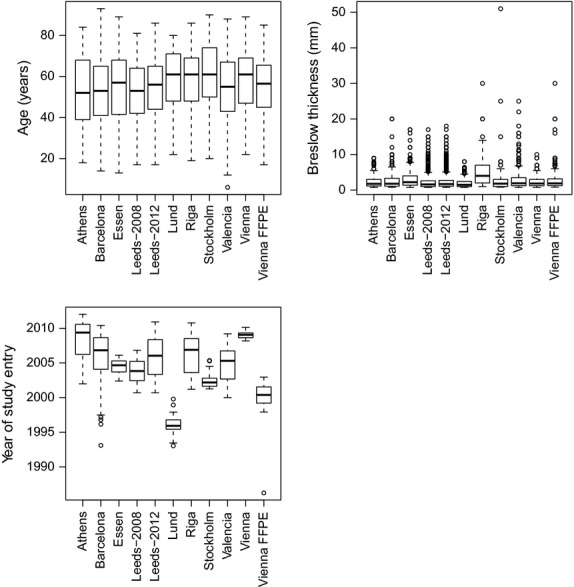
Box plots showing variation of Breslow thickness, age of diagnosis and date of study entry in each of the ten cohorts. Individual patient data was not available for the Tampa cohort. For the Lund cohort date of study entry was not available so date of diagnosis is presented; date of study entry was within 2 yr of diagnosis.
Figure 4.
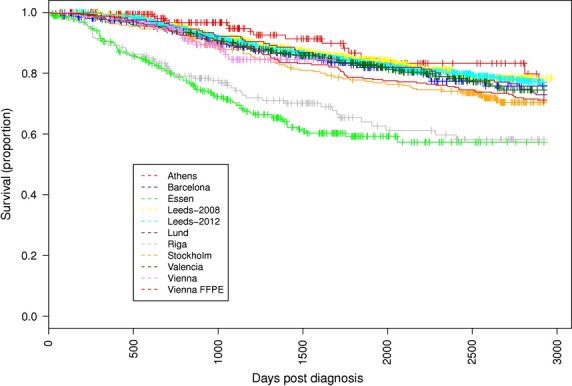
Kaplan–Meier estimator of overall survival (truncated at 8 yr) for each of the ten cohorts. Follow-up is truncated at 8 yr post-diagnosis.
SNP genotyping
The intronic GC SNP rs2282679 was genotyped in ten BioGenoMEL cohorts; residual DNA was insufficient to type this SNP in the Barcelona cohort. We subsequently typed two functional SNPs rs4588 and rs7041 in the Leeds cohort and nine of the BioGenoMEL cohorts (not including Tampa), the rs4588 SNP was poorly typed in the Stockholm cohort (call rate = 71%) so these data were discarded. These SNPs are both exonic and in strong linkage disequilibrium (LD) with the rs2282679 SNP and each other. The GC gene is highly polymorphic, leading to three common VDBP isoforms, termed Gc1f, Gc1s and Gc2, as well as over 120 rarer variants (Cleve and Constans, 1988). Variation in rs4588 and rs7041 is responsible for generating the three common isoforms of the GC protein (see Supporting information for further details). Serum levels of vitamin D metabolites are highest for Gc1f and lowest for Gc2 (Lauridsen et al., 2005). Similarly, binding affinity for vitamin D metabolites is also highest for Gc1f and lowest for Gc2 (Arnaud and Constans, 1993). The minor allele of rs2282679 is found exclusively in the Gc2 isoform (Figure S1). Different haplotypes are therefore associated with higher or lower levels of vitamin D, and we investigated the association of these two additional SNPs and the haplotypes in the Leeds cohort with survival.
Statistical analysis
Link-anonymized data were centralized in Leeds from the ten European BioGenoMEL cohorts. Analyses were carried out in a similar manner separately by the Tampa group to comply with their institutional review board approval. Initial analyses were carried out in the Leeds data alone, and validation was subsequently sought in the other data sets.
Survival time was defined as the period between the date of surgical excision of the primary and date of death or last date of follow-up (at which point records were censored). Multivariate survival analyses were performed using Cox's proportional hazards model. Models were fitted using the ‘coxph’ routine in the ‘survival’ package in R 2.10.1. Hazard ratio estimates were calculated for the effect of the rs2282679 SNP (and subsequently two closely related SNPs, rs7041 and rs4588) on OS adjusted for sex, site of melanoma (head/neck, trunk, limbs or other) and age at diagnosis. Since vitamin D levels were reported to be associated with Breslow thickness, we also ran parallel analyses additionally adjusting for thickness. MSS data were not available from all groups; therefore, to reduce the number of non-melanoma-related deaths reported, we truncated OS time after 8 yrs of follow-up. A log-additive genetic model was assumed and the ‘per-allele’ hazard ratios were estimated.
Additional checks were made to ensure that the formalin-fixed paraffin-embedded (FFPE) Vienna dataset was viable as an independent cohort. Normal DNA for cases in the FFPE Vienna cohort was extracted from archived material independent of patient survival. Kaplan–Meier estimates of survival were similar to those of the other participating cohorts (Figure 4). Violations of Hardy–Weinberg equilibrium were tested for the genotyped SNP, and no evidence of deviation was detected (P = 0.8). The cohort was therefore judged acceptable.
Relevant point estimates and standard errors for each study were taken from the fitted Cox's proportional hazards models. These data were then used to carry out a random effects meta-analysis using the ‘forestplot’ function in the ‘rmeta’ package in R. Models were reported adjusted only for age of diagnosis, sex, site and with/without Breslow thickness. We also compared the association of the SNP for OS and MSS in the seven cohorts where cause of death was available.
As reported previously (Davies et al., 2012), we did not adjust for tumour ulceration or American Joint Committee on Cancer (AJCC) stage because of incomplete data for ulceration across all the cohorts, pending central review of all pathology. More details of approaches taken to analysis of multiple cohorts can be found in Davies et al.(Davies et al., 2012).
For the haplotype analysis of the two closely related variants in the GC gene in the Leeds cohort, LD was calculated using the ‘pwld’ routine in Stata version 10. Since rs7041 and rs4588 are in very strong LD (D’ = 1) and are known to code specifically for three isoforms, we expect that the phase for double heterozygotes is known. To test this formally, we calculated haplotype frequencies in the Leeds Melanoma Cohort data using an expectation–maximization (EM) algorithm implemented in the ‘hapipf’ routine in Stata. This confirmed that only the three haplotypes Gc1f, Gc1s and Gc2 were seen in these data.
We fitted a Cox regression model for the per-haplotype association of Gc1s and Gc2 with overall survival adjusted for the other two haplotypes for each of the BioGenoMEL cohorts for which haplotype data were available. We constructed forest plots to test for a combined association in all cohorts as above.
The proportional hazards assumption was tested using the ‘phtest’ function in Stata, looking for an association of the Schoenfeld residuals with time in the global model and for each covariate. Heterogeneity measures were calculated using the ‘metaan’ function in Stata. Power analysis was conducted using the ‘stpower’ function in Stata.
Acknowledgments
Additional support for the analyses conducted at the H. Lee Moffitt Cancer Center & Research Institute, Tampa was provided by Hyun Park (technical laboratory support) from the Department of Cancer Epidemiology; Jane L. Messina (dermatopathologist) and Vernon K. Sondak (program leader) from the department of Cutaneous Oncology. Dr. Katerina Kypreou (biochemist) collected and prepared the biological samples of the Athens cohort (A. Sygros Hospital) and Dr. Dimitrios Bafaloukos (medical oncologist, Department of Medical Oncology, Metropolitan Hospital, Athens) assisted with patient recruitment.
Author contributions
Jennifer H. Barrett, Remco van Doorn, D Timothy Bishop and Julia Newton-Bishop: Designed research. Mark Harland, Juliette Randerson-Moor, Rajiv Kumar, Sinead Field, Eduardo Nagore, Susana Puig, Johan Hansson, Veronica Höiom, Göran Jönsson, Guan Jian, P. Sivaramakrishna Rachakonda, Jong Y. Park, Judith Wendt, Ichiro Okamoto, Dirk Schadendorf, Dace Pjanova, Håkan Olsson, Zaida García-Casado, Simona Donina, Antje Sucker, Kathleen M. Egan, Julia Newton-Bishop: Conducted research. Eduardo Nagore, Johan Hansson, Veronica Höiom, Göran Jönsson, Guan Jian, P. Sivaramakrishna Rachakonda, Jong Y. Park, Judith Wendt, Ichiro Okamoto, Joan Anton Puig-Butille, Dirk Schadendorf, Dace Pjanova, Håkan Olsson, Zaida García-Casado, Simona Donina, Antje Sucker, Kathleen M. Egan, Alexander J. Stratigos, Elizabeth Kodela, Zighereda Ogbah, Ismail Hosen: Provided essential reagents/materials. John R. Davies, Gabriella M. Anic: Analysed data or performed statistical analysis. John R. Davies, Julia Newton-Bishop, Jennifer H. Barrett, Paul Affleck, Remco van Doorn, Nelleke A. Gruis: Wrote paper. John R. Davies: Primary responsibility for final content.
Funding
The BioGenoMEL consortium is supported by a Skin Cancer Research Fund (SCaRF, http://www.skin-cancer-research-fund.org.uk) grant, a pump priming award by Yorkshire Cancer Research (YCR, http://www.yorkshirecancerresearch.org.uk) and a World Universities Net- work/University of Leeds International Matched Partnership Fund award. Through membership of the GenoMEL consortium, several groups have received support from the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and from the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115). The Leeds team received funding for the melanoma cohort study from Cancer Research UK (project grant C8216/A6129, programme awards C588/A4994 and C588/A10589, and Centre Award C37059/A11941). The Stockholm team received funding from the Swedish Cancer Society, the Radiumhemmet Research Funds and the Karolinska Institutet Research Funds. The Riga team received funding from the Latvian Council of Science: ESF Project No.1DP/1.1.1.2.0/09/APIA/VIAA/150, project No.10.0010.8. The Leiden team received a grant provided by European Biobanking and Biomolecular Resources Research Infrastructure (BBMRI)−Netherlands hub (CO18). The Heidelberg and Essen teams received funding from DFG (Scha 422/11-1). The Vienna team were supported by the Österreichische Nationalbank (project number 12161 and 13036) and the Hans und Blanca Moser Stiftung. The Barcelona team were supported by Fondo de Investigaciones Sanitarias (grants 05/0302, 06/0265, 09/1393 and 12/00840). Zighereda Ogbah received financial support provided by Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). The Lund team received additional funding from grants from the Swedish Cancer Society, the Swedish Research Council, the Gunnar Nilsson Foundation, The Berta Kamprad Foundation and European Research Council Advanced Grant ERC-2011-294576. No funders had any influence on the findings of this research.
Conflict of interest
The authors have no conflicts of interests to declare. All authors have read and approved the final manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Additional detail on the BioGenoMEL cohorts, functional variants in the GC gene, Mendelian randomization and Melanoma specific survival (MSS).
Figure S1. Cartoon representation of the three GC gene haploytypes that code for the major isoforms of the Vitamin D binding protein.
Figure S2. Directed acyclic graph showing the assumed dependencies between the rs2282679 SNP, vitamin D status, lifestyle and survival.
Table S1. Follow-up protocol in the BioGenoMEL cohorts.
Table S2. Overall and Melanoma Specific survival for the rs2282679 SNP in seven cohorts.
Table S3. Association of the rs2282679 SNP with overall survival for each of the rare cases in each participating cohort.
References
- Ahn J, Yu K, Stolzenberg-Solomon R, et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 2010;19:2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP) Hum. Genet. 1993;92:183–188. doi: 10.1007/BF00219689. [DOI] [PubMed] [Google Scholar]
- Autier P, Gandini S. Vitamin D supplementation and total mortality: a meta-analysis of randomized controlled trials. Arch. Intern. Med. 2007;167:1730–1737. doi: 10.1001/archinte.167.16.1730. [DOI] [PubMed] [Google Scholar]
- Brandstedt J, Almquist M, Manjer J, Malm J. Vitamin D, PTH, and calcium and the risk of prostate cancer: a prospective nested case-control study. Cancer Causes Control. 2012;23:1377–1385. doi: 10.1007/s10552-012-9948-3. [DOI] [PubMed] [Google Scholar]
- Cleve H, Constans J. The mutants of the vitamin-D-binding protein: more than 120 variants of the GC/DBP system. Vox Sang. 1988;54:215–225. doi: 10.1111/j.1423-0410.1988.tb03908.x. [DOI] [PubMed] [Google Scholar]
- Collin SM, Metcalfe C, Palmer TM, et al. The causal roles of vitamin B(12) and transcobalamin in prostate cancer: can Mendelian randomization analysis provide definitive answers? Int. J. Mol. Epidemiol. Genet. 2011;2:316–327. [PMC free article] [PubMed] [Google Scholar]
- Davies JR, Chang YM, Snowden H, et al. The determinants of serum vitamin D levels in participants in a melanoma case-control study living in a temperate climate. Cancer Causes Control. 2011;22:1471–1482. doi: 10.1007/s10552-011-9827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JR, Randerson-Moor J, Kukalizch K, et al. Inherited variants in the MC1R gene and survival from cutaneous melanoma: a BioGenoMEL study. Pigment Cell Melanoma Res. 2012;25:384–394. doi: 10.1111/j.1755-148X.2012.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durup D, Jorgensen HL, Christensen J, Schwarz P, Heegaard AM, Lind B. A reverse J-shaped association of all-cause mortality with serum 25-hydroxyvitamin D in general practice: the CopD study. J. Clin. Endocrinol. Metab. 2012;97:2644–2652. doi: 10.1210/jc.2012-1176. [DOI] [PubMed] [Google Scholar]
- Essa S, Denzer N, Mahlknecht U, Klein R, Collnot EM, Tilgen W, Reichrath J. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J. Steroid Biochem. Mol. Biol. 2010;121:110–113. doi: 10.1016/j.jsbmb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Falchi M, Bataille V, Hayward NK, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall T, Hägg S, Mägi R, et al. The role of adiposity in cardiometabolic traits: a mendelian randomization analysis. PLoS Med. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PJ, Ennis M, Pritchard KI, Koo J, Hood N. Prognostic effects of 25-hydroxyvitamin D levels in early breast cancer. J. Clin. Oncol. 2009;27:3757–3763. doi: 10.1200/JCO.2008.20.0725. [DOI] [PubMed] [Google Scholar]
- Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- Lauridsen AL, Vestergaard P, Hermann AP, Brot C, Heickendorff L, Mosekilde L, Nexo E. Plasma concentrations of 25-hydroxy-vitamin D and 1,25-dihydroxy-vitamin D are related to the phenotype of Gc (vitamin D-binding protein): a cross-sectional study on 595 early postmenopausal women. Calcif. Tissue Int. 2005;77:15–22. doi: 10.1007/s00223-004-0227-5. [DOI] [PubMed] [Google Scholar]
- McKenna MJ. Differences in vitamin D status between countries in young adults and the elderly. Am. J. Med. 1992;93:69–77. doi: 10.1016/0002-9343(92)90682-2. [DOI] [PubMed] [Google Scholar]
- Newton-Bishop JA, Beswick S, Randerson-Moor J, et al. Serum 25-hydroxyvitamin D3 levels are associated with breslow thickness at presentation and survival from melanoma. J. Clin. Oncol. 2009;27:5439–5444. doi: 10.1200/JCO.2009.22.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Bishop JA, Chang YM, Elliott F, et al. Relationship between sun exposure and melanoma risk for tumours in different body sites in a large case-control study in a temperate climate. Eur. J. Cancer. 2011;47:732–741. doi: 10.1016/j.ejca.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg B, Graber S, Gartner B, Geisel J, Pfohler C, Schadendorf D, Tilgen W, Reichrath J. Reduced serum 25-hydroxyvitamin D levels in stage IV melanoma patients. Anticancer Res. 2009;29:3669–3674. [PubMed] [Google Scholar]
- Randerson-Moor JA, Taylor JC, Elliott F, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur. J. Cancer. 2009;45:3271–3281. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Zhang F, Richards JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


