Abstract
While primordial life is thought to have been RNA-based (Cech, Cold Spring Harbor Perspect. Biol. 4 (2012) a006742), all living organisms store genetic information in DNA, which is chemically more stable. Distinctions between the RNA and DNA worlds and our views of “DNA” synthesis continue to evolve as new details emerge on the incorporation, repair and biological effects of ribonucleotides in DNA genomes of organisms from bacteria through humans.
Keywords: DNA polymerase, DNA replication, Ribonucleotides, Genome instability, DNA repair
1. Introduction
The central dogma of molecular biology states that genetic information flows from DNA to RNA to protein. In reviewing this dogma, Crick [1] pointed out the well-known essential loop (highlighted in Fig. 1A) involving the transfer of genetic information from one DNA molecule to another via DNA synthesis. To the extent that this loop avoids use of ribonucleotides, DNA-based organisms have an advantage over RNA-based organisms [2] because DNA is a more stable storage medium for genetic information. This is because ribonucleotides contain a reactive 2′-hydroxyl on the ribose ring that greatly sensitizes the sugar-phosphate backbone to hydrolysis [3] (Fig. 1B). Moreover, ribonucleotides in DNA alter nucleic acid geometry (e.g., Fig. 1C, ([4–7] and references therein). Thus ribonucleotides in DNA could potentially influence cellular DNA transactions and alter the information stored in DNA. This possibility leads to the subject of this review, which considers the following questions. Are ribonucleotides introduced into DNA, and if so, how and how many? Are they removed, and if so, how? If not, what are the consequences?
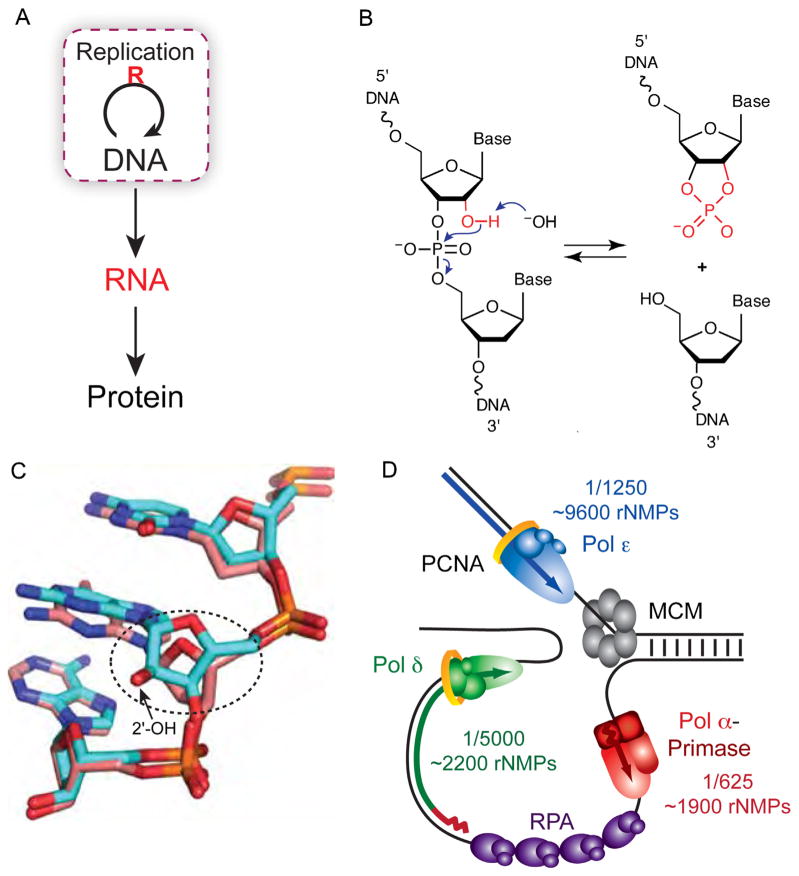
Fig. 1.
Ribonucleotides in DNA. (A) The central dogma of molecular biology describes how genetic information is transferred [1]. The essential loop has been modified to highlight the discovery that ribonucleotides are incorporated into DNA during replication. (B) Hydrolysis of the DNA backbone occurs at the site of a ribonucleotide in the presence of alkali when the deprotonated 2′ hydroxyl of a ribonucleotide cleaves the DNA strand via nucleophilic attack of the phosphate backbone. The resulting 2′–3′ cyclic phosphate is opened in the presence of water to yield equimolar amounts of 2′ and 3′ phosphate (not shown). (C) The Dickerson dodecamer NMR solution structure from [7] containing all DNA (dd-DNA, coral) or a single riboG (rG4-DNA, cyan). Shown is the view looking into the minor groove. Encircled is the alteration in sugar pucker from C2′-endo in the dd-DNA structure to C3′-endo in the rG4-DNA structure (Fig. 3 in [7]). (D) Model of the yeast replication fork with the frequency and estimated number of ribonucleotides incorporated by each DNA polymerase (adapted from Fig. 2D in [31]).
2. Origins of ribonucleotides in DNA
Hypothetically, ribonucleotides could be incorporated into DNA during any DNA synthesis reaction in a cell. Here we consider several possibilities, in order of potential abundance.
2.1. Ribonucleotide incorporation by RNA primase
In their initial description of DNA polymerization [8], Kornberg and colleagues pointed out that DNA synthesis does not begin de novo, but rather requires a “primer”. For replication in many DNA-based organisms (highlighted loop in Fig. 1A), this primer is a short RNA chain synthesized by an RNA primase (reviewed in [9]). For example, during replication in eukaryotes (Fig. 1D), a primer of approximately 10 ribonucleotides is used to initiate DNA synthesis of Okazaki fragments (OFs) of about 200 nucleotides (reviewed in [10–12]). Thus, lagging strand replication of the three billion base pair human nuclear DNA genome requires synthesis of about 150 million ribonucleotides. This is undoubtedly the major source of ribonucleotides initially introduced during DNA replication. Additional but less abundant sources of consecutive ribonucleotides in DNA include RNA chains used to restart stalled replication forks (reviewed in [13], e.g., mRNA [14] or RNA primers synthesized by PrimPol [15–18]. Because RNA primers are efficiently removed during OF maturation (see below), their presence in DNA is normally transient.
2.2. Ribonucleotide incorporation by DNA polymerases in vitro
Early work demonstrated that DNA polymerases incorporate ribonucleotides into DNA, to the point that when tricked by substituting Mn2+ for Mg2+ as the activating metal or when engineered, DNA polymerases can even behave as very weak RNA polymerases (e.g., see [19–22] and references therein). Numerous studies (reviewed in [23] and also see [24–26] and references therein) have quantified discrimination against insertion of rNTPs. These studies revealed variations among DNA polymerases ranging from as little as 10-fold to greater than a million-fold. Such wide variation in inserting a noncanonical sugar into DNA is similar to the million-fold variation among DNA polymerases for misinserting incorrect bases [27]. Structure–function analyses of sugar discrimination (reviewed in [20,23,28] and also see [24,26]) have provided insights into the mechanisms that prevent rNTP insertion. This prevention partly results from steric clashes between the 2′ oxygen of the ribonucleotide and a polymerase, often involving functionally conserved “steric gate” residues. In addition to studies of prevention, a recent study has also defined the catalytic cycle for stable incorporation of a ribonucleotide into DNA by Pol λ [22], which fills short DNA gaps during repair [29].
The fact that many DNA polymerases discriminate well against ribonucleotide insertion implies that incorporation into DNA might be so rare as to be of little consequence. This implication may partly explain why the subjects of this review have not been studied nearly as extensively as has misincorporation of incorrect bases. However, just as the probabilities of base misincorporation and misalignment depend on the absolute and relative concentrations of the four dNTPs (e.g., see [30] and references therein), so too does the probability of ribonucleotide insertion and stable incorporation depend on the cellular concentrations of rNTPs and dNTPs. Measurements in yeast [31] and mammalian cells [32,33] indicate that rNTP concentrations are much higher than dNTP concentrations. This greatly increases the probability that ribonucleotides will be incorporated into DNA despite strong polymerase discrimination against this outcome. This point was nicely illustrated in a study [34] of human DNA polymerase μ, an enzyme that fills 1–2 nucleotide gaps during non-homologous end joining of double strand breaks (DSBs). Increasing the rNTP:dNTP ratio to that estimated in mammalian cells resulted in a several-fold increase in ribonucleotide incorporation in vitro. This led to the suggestion [22,34] that rNTPs may be frequently incorporated during gap-filling synthesis associated with DNA repair, especially in non-proliferating cells where dNTP concentrations are low and rNTP:dNTP ratios are high. This idea may be relevant to diseases, e.g., AOA1 (see below).
Following the study of Pol μ, it was shown [31] that in reactions containing rNTP and dNTP concentrations estimated to be present in Saccharomyces cerevisiae, yeast replicative polymerasesα, δ and ε stably incorporate one ribonucleotide (rNMP) for every 625, 5000 or 1250 deoxyribonucleotides (dNMPs), respectively. Given the current model for the division of labor among these three polymerases at the replication fork [35], these results suggest that more than 13,000 ribonucleotides might be incorporated in a single round of nuclear DNA replication in yeast (Fig. 1D). This extrapolates to the possible incorporation of more than three million ribonucleotides into much larger mammalian nuclear genomes. This prediction is supported by the fact that human Pol δ [36] and Pol ε [37] have ribonucleotide incorporation propensities similar to those of the yeast replicases. Further evidence in vitro for the evolutionary conservation of ribonucleotide incorporation during replication comes from recent studies of the replicase for the mitochondrial genome, DNA polymerase γ (see [25] and references therein] and the replicase for the Escherichia coli genome, DNA polymerase III [38].
The amount of ribonucleotides incorporated into DNA by the yeast replicases varies over a several-fold range depending on the polymerase, the identity of the base, and the local sequence context [31]. Moreover, the cellular concentrations of the four dNTPs are not equal, they change in the cell cycle (e.g., see [39]), and they are much lower in quiescent cells [33,40], thereby altering rNTP to dNTP ratios and the probability of ribonucleotide incorporation. Thus the probability of ribonucleotide incorporation into DNA in vivo may vary by location and over time, which has implications for the repair and biological consequences of ribonucleotides in DNA (see below).
Ribonucleotide incorporation into DNA is not limited to the major replicases. In addition to the studies mentioned above, ribonucleotides are readily incorporated by Tetrahymena telomerase [41], by terminal transferase [42], a template-independent Family X sibling of Pol μ with an important role in V(D)J recombination, and by E. coli Pol V, which participates in translesion synthesis [43].
2.3. Ribonucleotide incorporation by DNA polymerases in vivo
To determine if ribonucleotide incorporation during nuclear DNA replication occurs in vivo, a study was performed that took advantage of prior work suggesting that if a ribonucleotide was incorporated by a DNA polymerase, it would be removed through the combined action of RNase H2 and FEN1 ([44,45] and references therein). In budding yeast strains lacking the gene encoding the catalytic subunit of RNase H2 (see [46] for review of RNases H), genomic DNA was found to contain a large number of alkali-sensitive sites [47]. Alkali-sensitive sites were much less abundant in an RNase H2-proficient strain, implying that they are largely due to ribonucleotides in the genome. Importantly, the number of such sites was increased in a strain encoding a variant of the leading strand replicase Pol ε that incorporates a larger number of ribonucleotides in vitro, and conversely, was decreased by a variant that incorporates fewer ribonucleotides [47]. These results indicate that ribonucleotides are incorporated into the nuclear genome by Pol ε, and that as predicted [45], they are removed in an RNase H2-dependent manner. Both conclusions hold in a similar study of fission yeast [48]. That study further demonstrated that the ribonucleotides in the genome of the Pol ε variant strain were preferentially located in the nascent leading strand. This strand preference was later confirmed in budding yeast and extended to preferential incorporation of ribonucleotides into the nascent lagging strand by a homologous variant of Pol δ [49]. Such strand-specific ribonucleotide incorporation strongly supports the model for the division of labor among polymerases at the replication fork (Fig. 1B) that was inferred from studies of polymerase error signatures [35]. The fact that ribonucleotides serve as “biomarkers” for polymerase-specific DNA synthesis in vivo may be useful for future studies of replication, repair and recombination. Ribonucleotides have also been detected in mitochondrial DNA [50], consistent with their incorporation during replication by Pol γ [25]. A recent study demonstrated that ribonucleotides are present in the nuclear genome of RNase H2-deficient mouse cells [51], and in the genome of a Bacillus subtilis strain deleted for RnhB [38], supporting the idea that ribonucleotide incorporation is an evolutionarily conserved property of DNA replication.
How many ribonucleotides are incorporated during replication in vivo? The results in yeast suggest an average of one ribonucleotide is incorporated per ~6500 deoxyribonucleotdes [49], a value remarkably similar to the 1:7600 ratio observed in the nuclear genome of RNase H2-defective mouse cells [51]. Biochemical studies of E. coli Pol III suggest that one ribonucleotide will be incorporated per 2300 base pairs [38].
3. Removal of ribonucleotides from DNA
The total number of ribonucleotides introduced into DNA genomes during DNA replication exceeds the total of all other DNA lesions studied in the DNA repair field (see Table 1 in [52]). This predicts that, as for other common non-canonical constituents in the genome (e.g., 8-oxo-G and abasic sites), ribonucleotides in DNA are processed in multiple ways. Rapidly emerging evidence indicates that is indeed the case.
3.1. Ribonucleotide removal during OF maturation
The maturation of OFs into a continuous nascent lagging strand involves removal of RNA primers. Two main ideas have been proposed for how this is done (Fig. 2A). The biochemical properties of RNase H2 are consistent with multiple incisions to digest most of the RNA primer, with the exception of the final rNMP at the 5′-RNA–DNA-3′ junction, which is removed by FEN1 in a reaction like that used for ribonucleotide excision repair (RER, see below). Nonetheless, genetic studies (e.g., [53]) show that RNase H2 is not essential for removing RNA primers. This fact is consistent with the demonstration that ribonucleotides can be removed by nick-translation involving strand displacement synthesis by Pol δ, coordinated with consecutive cleavage of the resulting flaps by FEN1 ([54] and Fig. 2A). OF maturation is likely the most frequent of all DNA transactions in a cell, so functional redundancy and tight regulation is to be expected and has been observed (reviewed in [10–12]). For example, maturation can occur in the absence of FEN1, via a Dna2-dependent “long-flap” pathway suggested to not only remove the RNA primer, but also incorrect bases incorporated into the adjacent DNA by Pol α, which cannot proofread any mismatches it may generate. Theoretically, this long-flap pathway could also remove ribonucleotides incorporated by Pol α, which is the most promiscuous of the three replicases for ribonucleotide incorporation.
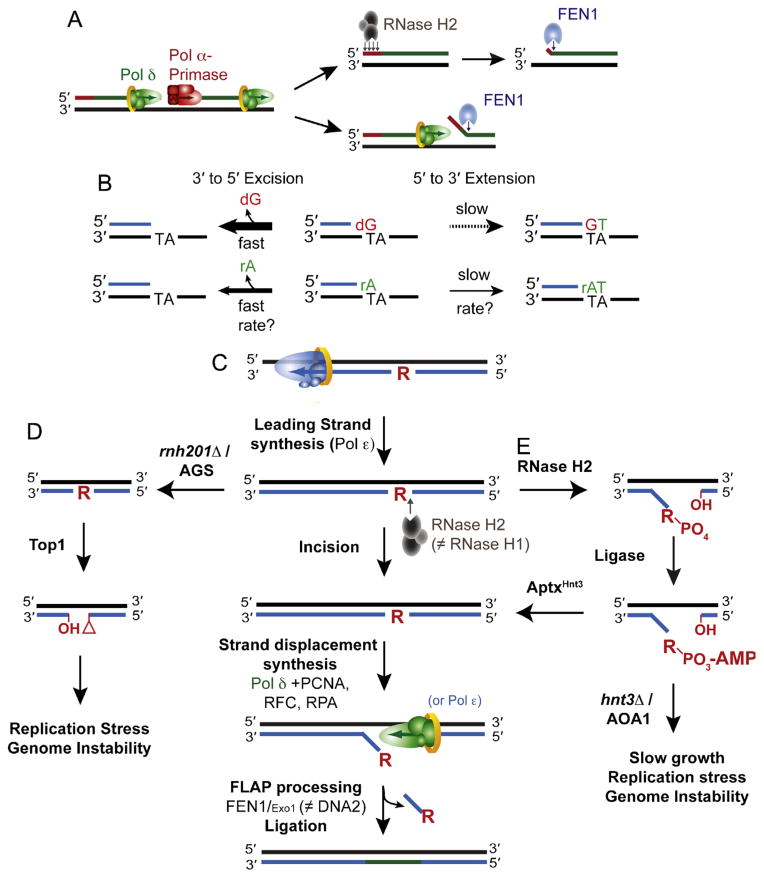
Fig. 2.
Origins and removal of ribonucleotides in DNA. (A) Incorporation and removal of ribonucleotides incorporated by RNA primase during OF synthesis and maturation. Two of the pathways by which the RNA primer synthesized by Pol α-primase is removed are displayed. RNase H2 is able to digest and remove the RNA primer up until the terminal 3′ ribonucleotide that is removed by FEN1. Alternatively, Pol δ can perform strand displacement synthesis to generate a flap that is degraded by FEN1. (B) Proofreading efficiency is determined by the balance between excision and extension. The polymerase exonuclease activity can excise ribonucleotides from the 3′ terminus in primer-template DNA, an event that is more efficient on a mismatched 3′ terminal dNMP primer than it is on an rNMP. The kinetics of proofreading (both excision and extension) by a replicative DNA polymerase in the presence of a terminal rNMP remain to be biochemically determined. (C) The ribonucleotide excision repair (RER) pathway is initiated by incision 5′ to the ribonucleotide by RNase H2. Strand displacement synthesis by the polymerase is followed by nucleolytic flap processing and ligation. There are redundant functions of the polymerases (Pol δ and Polε) as well as the nucleases (FEN1 and Exo1; Adapted from Fig. 4 in [56]). (D) Loss of RNase H2 activity due to deletion of RNH201 or RNase H2-deficiency in AGS leaves unrepaired genomic ribonucleotides that may become targets for Top1-cleavage and removal. The resulting unligatable DNA ends possess a 2′–3′-cyclic phosphate and a 5′-hydroxyl that may promote the formation of DNA breaks and/or recombination and cause genome instability. (Adapted from Fig. 4 in [60]). (E) The pathway by which RNase H2 cleavage at ribonucleotides can generate abortive ligation intermediates (adenylated 5′-RNA–DNA junctions) that require APTXHNT3 for resolution. Failure of this pathway due to deletion of HNT3 or APTX deficiency in AOA1 causes phenotypes that may reflect the persistence of adenylated RNA–DNA junctions (Adapted from Fig. 2E in [57].
3.2. Proofreading of ribonucleotides
Some replicases have 3′-exonuclease activity that can efficiently excise an incorrect base inserted during DNA replication. Recent studies [37,55] demonstrated that the 3′-exonuclease of human and yeast Pol ε can proofread newly inserted ribonucleotides (Fig. 2B), but does so much less efficiently than for editing base-base mismatches. A subsequent study showed that human and yeast Pol δ are even less efficient at proofreading ribonucleotides [36]. Together, these studies suggest that proofreading provides little protection against ribonucleotide incorporation during DNA replication in vivo. Failure to efficiently edit newly incorporated ribonucleotides during replication is consistent with the idea that their transient presence in the genome has positive consequences (see below). Decades of work (reviewed in [27]) have demonstrated that proofreading of primer terminal mismatches varies more than 1000-fold depending on a delicate balance between excision versus extension of the mismatched primer terminus (Fig. 2B). For example, fraying of the mismatched terminus during proofreading to allow partitioning of the single-strand DNA to the polymerase exonuclease site for excision may be more efficient on a mismatched 3′ terminal dNMP primer than it is on a correctly base-paired rNMP. However, it is possible that future studies will reveal more efficient proofreading of ribonucleotides under some circumstances.
3.3. Ribonucleotide excision repair (RER)
The demonstration that ribonucleotides are removed from the yeast nuclear genome by a repair process requiring RNase H2 [47] prompted a study [56] that has reconstituted Ribonucleotide Excision Repair (RER) reactions in vitro using proteins purified from budding yeast and DNA substrates containing ribonucleotides incorporated by Pol δ or Pol ε. The most efficient RER pathway (Fig. 2C) begins with RNase H2-dependent incision of the backbone 5′ to a ribonucleotide, generating a nick whose ends have a 3′-OH and a 5′-RNA–DNA junction. RNase H1 does not substitute for RNase H2 in this RER reaction. Incision is followed by strand displacement synthesis by Pol δ, assisted by PCNA and the RFC clamp loader. Pol ε is able to substitute for Pol δ in a reaction that is slightly less efficient. FEN1 then excises the resulting flap containing the RNA–DNA junction. Exo1 can substitute for FEN1, albeit less efficiently, but Dna2, which only acts on longer flaps, cannot substitute for FEN1. The resulting nick is sealed by DNA ligase 1 to complete RER. The enzymes involved in RER are conserved in organisms in all three kingdoms of life.
3.4. Repair of adenylated RNA–DNA junctions by aprataxin (APTX)
The large number of ribonucleotides incorporated during replication provides an enormous potential to generate toxic RER intermediates. For example, premature attempts to ligate the nick generated by RNase H2 before the ribonucleotide is excised can generate an adenylated 5′-RNA–DNA junction (Fig. 2D). A recent structure–function study [57] shows that adenylated 5′-RNA–DNA junctions are indeed generated during attempted ligation. APTX efficiently deadenylates these junctions by a mechanism involving A-form RNA-binding and conformational changes. This ligation “proofreading” restores normal RER and avoids the toxic consequences (see below) of this compound lesion, i.e., an adenylated 5′-RNA–DNA junction.
3.5. Ribonucleotide removal via Topoisomerase 1 cleavage
In the absence of RNase H2-dependent RER (Fig. 2D), budding yeast removes ribonucleotides from DNA by a process initiated by Topoisomerase 1 (Top1) incision at ribonucleotides [58–60]. This incision generates a nick containing 5′-OH and cyclic 2′–3′ phosphate ends that cannot be ligated, thus requiring other, currently unknown, proteins to complete repair. Curiously, while Top1 can initiate removal of thousands of ribonucleotides from the yeast nuclear genome [60], this is only apparent when RNase H2-dependent RER is defective. It may be that, as it thought to be the case for mismatch repair (MMR; see below), RNase H2 dependent RER is coupled to replication. In this case, RNase H2 would be the first repair enzyme to encounter newly incorporated ribonucleotides, obviating the need for repair by Top1. It is also curious that the genomes of RNase H2-deficient yeast and mouse cells contain many ribonucleotides even when Top1 is proficient [47–49,51,60]. This indicates that Top1 only participates in removing a subset of ribonucleotides from the genome. This subset could possibly differ by nucleotide identity, configuration, the surrounding sequence context, genomic location or chromatin status. It is possible that Top1-initiated removal is related to transcription, because Top1 interacts with RNA polymerase II to relieve super coils generated during transcription [61,62] and RNase H2 resolves R-loops generated during transcription [63].
3.6. Evidence that other pathways process ribonucleotides
RNase H2-deficient yeast and mouse cells have phenotypes characteristic of DSB formation possibly resulting from Top1 incision and/or replication fork stalling (see below). Because DSBs can by repaired by recombination, it may be that in the absence of RNase H2-dependent RER, ribonucleotides embedded in DNA can be processed by recombination. Consistent with this possibility, an allele of the Rnh202 subunit of RNase H2 (hpr4-1) was isolated in a genetic screen for mutations conferring a hyper-recombination phenotype for intrachromosomal gene conversion [64], and the rnh202Δ mutant displays increased recombination and spontaneous mutagenesis (Potenski and Klein, unpublished). Further support comes from the discovery of increased loss-of-heterozygosity in diploid, RNase H2-deficient yeast (Williams, Lujan and Kunkel, unpublished), and by changes in expression of yeast genes involved in Rad52 focus formation and recombination [65]. Additionally, the fact that RAD52 is essential in rnh201Δ rnh1Δ cells [66] supports the importance of homologous recombination for survival when ribonucleotide-removal is impaired.
Several recent studies indicate that ribonucleotides embedded in DNA may also be processed by nucleotide excision repair (NER). Loss of yeast RNase H2 is associated with changes in expression of NER genes [65], and loss of NER in RNase H2-defective yeast elevates transcription-associated mutagenesis ([67] and see below). A hybrid bacterial UvrABC complex has recently been shown to incise DNA duplexes containing ribonucleotides [68], a fact that may be relevant to the observation that a defect in NER elevates base substitution mutagenesis in RNase H-defective E. coli strains encoding a Y11A allele the UmuC gene, the catalytic subunit of the translesion synthesis (TLS) enzyme, DNA polymerase V. Tyrosine 11 is the steric gate residue in the active site of Pol V that limits ribonucleotide incorporation, such that the Y11A variant is highly promiscuous for ribonucleotide incorporation in vitro, so much so that it can incorporate multiple consecutive ribonucleotides [69]. It has been suggested that ribonucleotides incorporated by Y11A Pol V are removed by RNase B-dependent repair, providing the opportunity to concomitantly remove mismatches generated by this relatively inaccurate TLS enzyme. The observation that a defect in NER elevates base substitution mutagenesis in a umuC Y11A ΔrnhB strain suggests that NER may serve as a backup pathway to remove ribonucleotides when RNase HII is defective [68]. Additional work suggests the involvement of the strand displacement and nuclease activities of DNA polymerase I (pol I) in RER in E. coli [70].
4. Consequences of ribonucleotides in DNA
Incorporation of ribonucleotides into DNA has several consequences, some resulting from their processing and others resulting from failure to process them (recently reviewed in [71]). For each circumstance, we consider both negative and positive consequences.
4.1. Negative consequences—RNA–DNA damage responses (RDDR)
The term “DNA damage response” (DDR) abounds in the extensive literature on DNA transactions. Therefore, it seems reasonable to use the term “RNA–DNA damage response” (RDDR) for emerging literature on the negative consequences of ribonucleotides in DNA genomes, as recently described for adenylated RNA–DNA repair by aprataxin [57]. Two major negative consequences are replication stress and genome instability.
4.1.1. Replication stress
Five phenotypes of replicative stress are exhibited by yeast strains that are defective in RNase H2-dependent RER and encode a Pol ε allele that increases ribonucleotide incorporation into the nascent leading strand during replication [47]. These strains grow slowly, accumulate in S-phase, have slightly elevated dNTP pools, have an activated S phase checkpoint and are sensitive to the replication inhibitor hydroxyurea (HU). These consequences are not seen in RER-defective strains encoding a Pol ε allele (pol2-M644L) that decreases ribonucleotide incorporation, indicating that they result from ribonucleotides incorporated during replication. Importantly, deleting Top1 largely alleviates these effects [60], suggesting that they result from the “dirty” (i.e., unligatable) DNA ends created by Top1 incision (Fig. 2D). A second example [57] of replication stress involves failure of aprataxin to resolve adenylated 5′-RNA–DNA junctions arising during attempted ligation too early in the RER pathway (Fig. 2E). Growth is severely impaired for a pol2-M644G strain (but not a pol2-M644L strain) from which the HNT3APTX gene is deleted. This strain is also HU-sensitive and has an activated S phase checkpoint. Importantly, these effects are eliminated by loss of RNase H2, implying that they result from failure to deadenylate 5′-RNA–DNA junctions created by RNase H2 incision at ribonucleotides incorporated during leading strand replication.
A third example of the RDDR is seen in mouse models, where knocking out genes encoding subunits of RNase H2 causes embryonic lethality [51,72]. RNase H2 null embryos grow slowly due to reduced cell proliferation. RNase H2-deficient mouse cells contain greater than 1,000,000 ribonucleotides in the genome and exhibit a p53-dependent DNA damage response characterized by reduced cellular proliferation and elevated levels of p21, cyclin G1, strand breaks, histone H2AX foci and genome instability.
These consequences are consistent with the encounter of a replication fork with unresolved nicks that ultimately result in DSBs. An additional, non-exclusive possibility for replication stress is replication fork stalling during attempts to bypass ribonucleotides in the template strand. This idea is consistent with the fact that replicases require normal DNA helix geometry to achieve efficient and accurate DNA synthesis, and with crystallographic and NMR studies (e.g., [4–7] and references therein) showing that ribonucleotides in protein-free DNA duplexes alter helix geometry. Moreover, recent studies in vitro show that ribonucleotides in DNA templates impede DNA synthesis by the E. coli replisome [38] and by four B Family replicases, bacteriophage RB69 Pol [73] and yeast Pols α, δ, and ε [31,36,47,66,73–75]. All these replicases can completely bypass a single ribonucleotide, with relative bypass efficiencies varying from ~10–90% of normal. However, compared to bypass by RB69 Pol and by Pol δ, Pol ε is progressively less efficient as the number of consecutive ribonucleotides in the template is increased from one to four (Fig. 3A). A recent structural study of a RB69 Pol variant [73] reveals that ribonucleotides in the template alter sugar pucker and the positions of two amino acids that are conserved in B Family polymerases and are important for interaction with the template strand (Fig. 3B). An interesting exception among B family polymerases is yeast Pol ζ. Renowned for its participation in TLS, Pol ζ efficiently bypasses even four consecutive ribonucleotides in a DNA template [66]. This ability appears to be biologically relevant in a yeast strain defective in both RNase H2 and RNase H1, which progresses slowly through S phase, accumulates ubiquitylated PCNA that is used for TLS, and is HU-sensitive [66]. In the presence of HU, survival of this strain depends on either of two post-replication repair pathways, MMS2-dependent template switching or Pol ζ-dependent TLS (see model in Fig. 6 [66]). Dependence on these two pathways is not observed for strains defective in either RNase H1 or RNase H2 alone. Because only RNase H2 can repair single ribonucleotides in DNA, but both RNases H1 and H2 can digest substrates containing multiple consecutive ribonucleotides in DNA [46], the results suggest that in the RNase H1/H2 double mutant strain, the replication fork may stall due to multiple consecutive unrepaired ribonucleotides in DNA, and that this stalling can be alleviated by either template switching or TLS by Pol ζ.
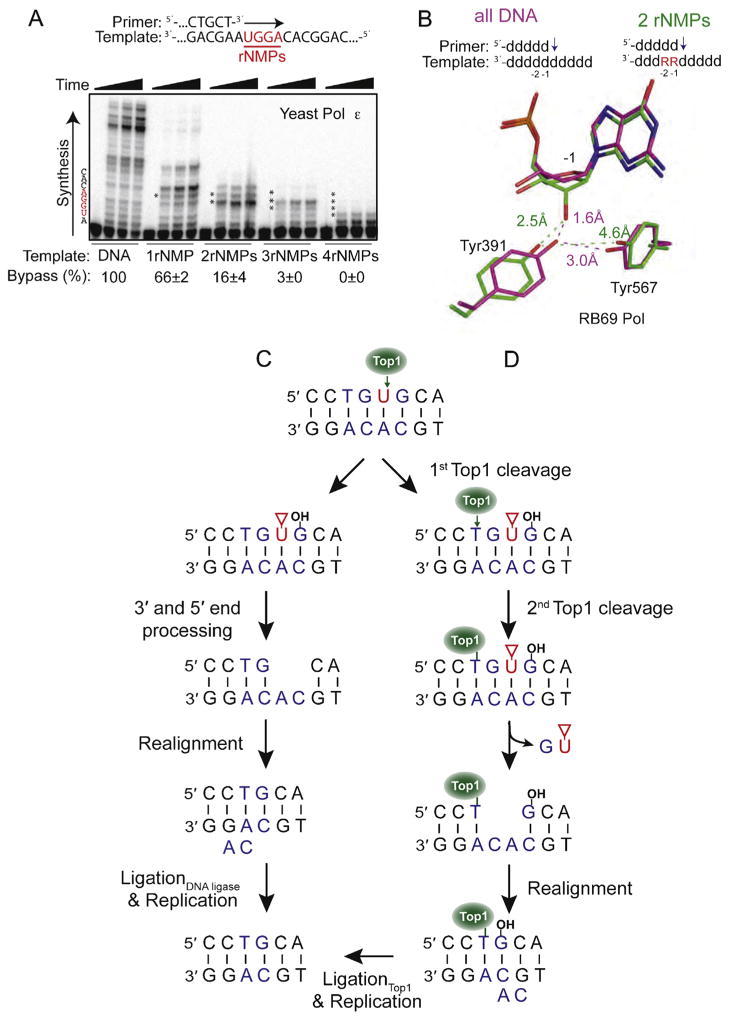
Fig. 3.
Consequences of ribonucleotides in DNA. (A) Bypass efficiency of ribonucleotides by yeast Pol ε. Primer extension was performed using five different primer-templates that varied in the number of template ribonucleotides (shown in red). Bypass efficiency (%) was calculated relative to the all-DNA control (Adapted from Fig. 1B in [73]). (B) The presence of two consecutive ribonucleotides in a template affects sugar pucker and positioning of two conserved active site tyrosine residues in RB69 Pol. The positioning of Tyr391 and Tyr567 in the presence of all-DNA template (magenta) is compared to the positions of these residues in the presence of a ribonucleotide at the −1 position (green). (Adapted from Fig. 4 in [73]). (C) The first of two models for generation of a two base deletion following Top1 cleavage at a ribonucleotide. In this pathway, end processing of the damaged 3′ and 5′ ends generates a gap within the repeat unit. Following misalignment of the complementary strands, ligation and replication cause loss of one repeat unit (Adapted from Fig. 4 in [59]). The red triangle denotes a 2′–3′-cyclic phosphate. (D) The second model involves sequential Top1-cleavages for generation of an rNMP-associated deletion. Following the first incision, there is a second Top1-cleavage reaction on the 5′ side of the nick that causes release of the DNA between the two cleavage sites and trapping of Top1. Realignment of the complementary DNA strands moves the free 5′-OH in close proximity to Top1 so that it can catalyze religation (Adapted from Fig. 6 in [83]).
Some of the negative consequences of inactivating RNase H2, either alone or in combination with RNase H1, could reflect loss of the ability to degrade RNA–DNA hybrids. Such hybrids form during transcription and can stall replication forks, activate the S phase checkpoint and destabilize the genome (see [76,77] and references therein). RNase H2 has also been suggested to degrade endogenous retroelements [78]. RNases H1 and H2 process noncoding telomeric repeat-containing RNA (TERRA) that form RNA–DNA hybrids at telomeres to regulate telomere-length [79] and may repair ribonucleotides incorporated into telomeres during replication [41]. Supporting these possibilities, RNase H2 was detected in a screen for genes involved in telomere maintenance [80], and RNase H2-defective yeast have increased expression of several genes associated with telomere maintenance [65]. The HU sensitivity of the yeast strain defective in RNase H2 and RNase H1 may not be due to unresolved R-loops, because concomitant deletion of the HPR1 gene results in accumulation of R-loops [81] but does not further increase the HU sensitivity of RNase H2/RNase H1 double mutant yeast strains [66]. Nevertheless, it remains important for future studies to distinguish between negative consequences of RNase H defects due to ribonucleotides incorporated by DNA polymerases versus unresolved RNA–DNA hybrids. As discussed below, this distinction has recently been enabled by the availability of a separation-of-function mutant of yeast RNase H2 [82]. Some of the negative consequences of inactivating RNase H2 could also reflect increased DNA damage due to the presence of ribonucleotides in DNA. This possibility is consistent with studies showing altered helix parameters for DNA containing ribonucleotides (see [4–7] and references therein). Indeed, gene expression studies show that a defect in RNase H2 results in changes in expression of hundreds of yeast genes associated with response to several types of stress, including oxidative stress, osmotic and heat shock, changes in pH, diauxic shift and exposure to heavy metals and xenobiotics [65].
4.1.2. Genome instability
Loss of RNase H2 results in several types of genome instability, generated by mechanisms that are partly understood. Here these are considered in order of the number of base pairs involved. The smallest mutations, single base events, are discussed under positive consequences because they are suppressed by ribonucleotides in the DNA.
4.1.3. 2–5 bp deletions
Loss of RNase H2-dependent RER strongly increases the rate at which 2–5 base pairs are deleted from short, directly repeated sequences [47]. Deletion rates are increased by the pol2-M644G allele that increases ribonucleotide incorporation, and decreased by the pol2-M644L allele that decreases ribonucleotide incorporation, indicating that they result from ribonucleotides incorporated during replication [47,59,83]. That newly incorporated ribonucleotides rather than RNA–DNA hybrids underlie the deletions is supported by the high rate of 2–5 base pair deletions seen in a yeast strain encoding an allele of the catalytic subunit of RNase H2 that cannot repair single ribonucleotides in DNA but can still resolve RNA–DNA hybrids [82]. The rate of short deletions is increased by transcription [84,85]. Some of these deletions are ribonucleotide-dependent and some are not [59,83], but all strongly depend on Top1. As mentioned above, Top1 relieves supercoils arising during transcription and it can cleave the backbone at an RNA–DNA junction to create ends that must be processed before ligation can occur (Fig. 2D). The events following Top1 cleavage at ribonucleotides that result in short deletions are uncertain. One model (Fig. 3C) posits that a single cleavage is followed by end processing to create a gap in which strand misalignment occurs, followed by ligation [59]. A more recent study [83] proposes that the initial Top1 cleavage at the RNA–DNA junction is followed by a second cleavage adjacent to the first that traps Top1 in a covalent complex with the DNA. Misalignment then allows Top1 to catalyze re-ligation, leading to the deletion (Fig. 3D).
4.1.4. Complex mutations templated by quasipalindromes
Loss of yeast RNase H2 also results in complex mutations that are most easily explained by DNA synthesis templated by one arm of an imperfect quasipalindrome, thus dubbed “QP” mutations [67]. These are only observed under conditions of high transcription. They are independent of Top1 and homologous recombination, but depend on the direction of DNA replication, on RNase H1 activity and on TLS, and as mentioned above, they are limited by NER. QP mutations are suggested to reflect either a transcription-associated perturbation of OF processing, or the use of a nascent transcript to resume replication following a transcription–replication conflict [67].
4.1.5. Large-scale genome instability
RNase H2-defective mouse embryonic fibroblasts contain increased levels of micronuclei, interchromosomal translocations and cytogenetic anomalies and chromosomal rearrangements in metaphase chromosomes [51]. The mechanisms responsible for these types of large-scale genome instability remain to be established, but likely involve DNA strand breaks that result from the presence of unrepaired ribonucleotides in DNA, from replication conflicts with unresolved R-loops, or both. Consistent with this, gross chromosomal rearrangements (GCRs) are observed in an RNase H2-defective yeast strain (rnh203Δ), and the GCR rate is increased when combined with mutations that affect DNA damage response and repair [86].
4.1.6. Disease relevance
Partial-loss-of-function mutations in the genes encoding any of the three RNase H2 subunits cause Aicardi–Goutières syndrome (AGS), an autosomal-recessive, early-onset neuroinflammatory disorder that mimics congenital viral infection [87]. Because innate immune-mediated inflammation is thought to result from the accumulation of endogenous nucleic acid species, the association between AGS and defective RNase H2 could reflect partial loss of RER, partial loss of the ability to degrade RNA–DNA hybrids (e.g., endogenous retroelements), or both.
The neurological disorder Ataxia with Oculomotor Apraxia 1 (AOA1) is linked to mutations in the Aptx gene encoding aprataxin [88], which resolves adenylated RNA–DNA junctions [57]. AOA1-associated Aptx mutations map to locations on the human aprataxin structure that are critical for function, including one mutation that has been shown to distort the substrate binding pocket and impair deadenylation of RNA–DNA junctions. These data suggest that a failure to resolve this toxic intermediate of RER may promote AOA1.
Theoretically, ribonucleotide incorporation into DNA could also be relevant to sickle cell anemia. Patients suffering from this disease are treated with HU [89], which inhibits ribonucleotide reductase. This inhibition decreases dNTP pools and increases rNTP:dNTP ratios. In turn, this increases the probability of ribonucleotide incorporation into DNA, as nicely demonstrated by the increased number of ribonucleotides in the genome of RNase H2-defective mouse cells treated with HU [51]. Theoretically, these newly incorporated ribonucleotides could be related to the negative side effects observed in HU-treated sickle cell anemia patients.
4.2. Positive consequences—Signaling roles for ribonucleotides in DNA
Because ribonucleotides in DNA alter helix parameters, they have the potential to act as positive signals for cellular DNA transactions.
4.2.1. Mating type switching
Schizosaccharomyces pombe employs two mating types for cellular differentiation, designated M and P. Switching between M and P occurs through recombination that replaces a cassette at the mat1 locus with one of the opposite mating-type, using either mat2P or mat3M. Current evidence (reviewed in [90,91]) supports a model wherein a two-ribonucleotide imprint made during lagging strand replication survives OF maturation (see Fig. 3 in [91]) and persists until the next round of replication, where it stalls leading strand replication by Pol ε [73], thereby initiating the recombination process used to switch the mating type (see Fig. 4 in [91]).
4.2.2. Signaling for MMR
MMR must be targeted to the nascent strand by one or more strand-discrimination signals. In E. coli, the nascent strand is marked for repair when MutH nicks the backbone at transiently unmethylated adenines in GATC sequences. In bacteria that lack MutH, and in eukaryotes, the identity of the DNA ends used for strand discrimination is less certain. The transient 5′-DNA end of an OF may be one signal for lagging strand MMR [92], while signals for the more continuously replicated leading strand have remained uncertain. Four years ago, we suggested that ribonucleotides incorporated by replicases might mark the nascent strand for MMR [31]. This hypothesis is supported by two recent studies [49,93] leading to the model in Fig. 4, in which ribonucleotides incorporated into the nascent leading strand are incised by RNase H2, creating a signal that directs MMR to correct nearby replication errors. The genetic evidence in budding yeast accounts for MMR of a small but significant fraction of leading strand errors, but not lagging strand errors. Inactivation of RNase HII elevates the spontaneous mutation rate in B. subtilis [38], consistent with the possibility that nicking of newly incorporated ribonucleotides may provide a signal for MMR in those bacteria that lack MutH. The model in Fig. 4 further implies that replication, MMR and RER may all be tightly coupled, especially since all three processes use several of the same proteins.
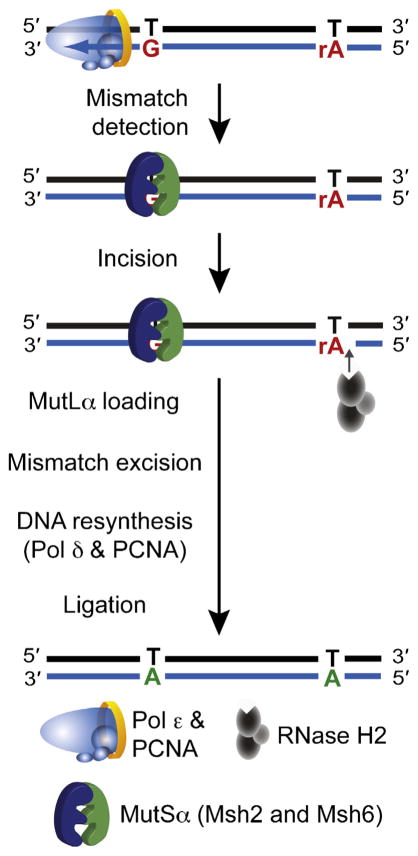
Fig. 4. Ribonucleotides may act as strand-discrimination signals for MMR of leading strand errors.
A model depicting how ribonucleotides incorporated by Pol ε act as a signal for MMR of errors generated in nascent leading strand DNA. Following recognition of the mismatch by the MutSα heterodimer, incision 5′ to a ribonucleotide by RNase H2 creates a nick that can be used as a loading site for MutLα. This nick can be distant from the site of the mismatch, because MutLα is able to translocate along the DNA and incise at a site close to the mismatch to allow excision, resynthesis and ligation.
5. Speculations and perspectives
Ribonucleotides embedded in DNA may have other consequences. They may influence telomere function, because they are incorporated by telomerase [41], their presence in DNA reduces DNA binding by the telomere binding protein Pot1 [94], loss of RNase H2 in yeast is associated with increased expression of genes involved in telomere maintenance [65], and the yeast RNH201 gene was recovered in two screens [80,95] for genes that affect telomere length. Ribonucleotides in DNA reduce nucleosome binding [96,97], possibly influencing histone re-loading behind the replication fork and/or nucleosome positioning or dynamics. This idea could possibly be related to the fact that Pol ε, which incorporates ribonucleotides into the nascent leading strand, is involved in gene silencing and one of its non-catalytic subunits is involved in chromatin remodeling (reviewed in [98]). Moreover, mutations in the S. pombe cdc22 and tds1 genes, encoding the large subunit of ribonucleotide reductase and a putative thymidylate synthase, respectively, cause spreading of silencing across heterochromatic barriers in the mating-type switching region [99]. These two factors regulate dNTP pools, such that defects in cdc22 and tds1 may alter dNTP:rNTP ratios to promote increased incorporation of ribonucleotides. Thus, in a conceptually similar manner to DNA methylation, ribonucleotides may modulate epigenetic change.
In considering the known and theoretical consequences of ribonucleotides in DNA, it will undoubtedly be useful to define their identity (rA, rC, rG or rU), their density (spacing and whether singlets or consecutive), their locations within genomes (e.g., in telomeres, in the highly transcribed rDNA locus, relative to nucleosomes, by DNA strand), and their persistence (e.g., transient for MMR, persistent for mating type switching). These parameters will likely depend on the properties of the polymerases responsible for their incorporation, whether a primase or replicase, or a DNA polymerase involved in TLS or DNA repair. Thus it will be of interest to determine the ribonucleotide incorporation capacity and the ribonucleotide bypass capacity of the many DNA polymerases not yet been examined, and to determine if these properties are biologically relevant.
The fact that that Top1 [58,59] and a bacterial UvrABC complex [68] incise DNA duplexes containing ribonucleotides, and the fact that ribonucleotides alter helix geometry (Fig. 1C) leads one to wonder how well other enzymes involved in nucleic acid transactions may process (or not) DNA containing ribonucleotides. This includes a long list of DNA repair enzymes already characterized for their ability to process lesions in “all-DNA” duplexes. As one example, repair of an abasic site in DNA requires the participation of several enzymes. It would be interesting to determine how well these enzymes repair an abasic site derived from a ribonucleotide in DNA, especially since an abasic lesion is more stable in RNA [100]. Another example involves DNA ligases, which vary widely in the ability to ligate RNA–DNA junctions (see [57,101] and references therein). Remarkably, it has also been reported that ligation of an RNA–DNA junction in vitro can generate a 2′–5′ linkage in the DNA backbone [102], a linkage that is well known in the literature on RNA transactions. Unpublished results (Watt and Kunkel) demonstrated that a 2′–5′ linkage in a DNA template completely blocks DNA synthesis in vitro by several DNA polymerases. Thus, if this linkage was generated in DNA in vivo, its consequences could be as severe as a DNA crosslink or a DSB, leading one to wonder if and how it might be repaired or tolerated. Also of potential interest are enzymes known to process RNA molecules, but have not yet been examined for possible roles in DNA transactions. As but one example, TRL1 contributes to tRNA maturation by processing 5′-OH and 2′–3′-cyclic phosphate RNA ends [103], the same end modifications generated by Top1 incision at a ribonucleotide in DNA (Fig. 2D).
One also wonders whether the helical perturbations caused by ribonucleotides in DNA (Fig. 1C) increase the probability of DNA damage. This possibility is consistent with the increased expression of a large number of stress response genes in RNase H2-defective yeast [65]. Curiously, these include OGG1, which encodes a DNA glycosylase that excises 8-oxo-guanine to initiate base excision repair (BER). Does the presence of a ribonucleotide in DNA increase the probability of oxidative (or other) damage to DNA? Conversely, it has been suggested [104,105] that ribonucleotides could arise in DNA through oxidation at the 2′-position of deoxyribonucleotides. A yet more speculative possibility for which there is no evidence, is that ribonucleotides embedded in DNA might be directly reduced to deoxyribonucleotides, conceptually akin to reduction of rNDPs to dNDPs by ribonucleotide reductase.
The above speculations and this review of rapidly emerging literature on the origins, repair and consequences of ribonucleotides in DNA demonstrate that within the central dogma of molecular biology, there is still much to be learned about the transfer of genetic information from one DNA molecule to another via DNA synthesis, and about possible relationships between the DNA and RNA worlds [2]. What we know is satisfying. What we hope to discover is likely to be exhilarating, as we celebrate 50 years of research since the discovery of excision repair.
Acknowledgments
Funding
This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS).
For brevity, and with apologies to the many investigators who have contributed for over 50 years to the subjects in this review, we largely focus on recent publications. Many other relevant studies are cited within these publications, especially the reviews. We thank Scott Williams and Scott Lujan for helpful comments on this work.
Footnotes
Conflict of interest statement
The authors have no conflict of interest to declare.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. The RNA worlds in context. Cold Spring Harbor Perspect Biol. 2012;4:a006742. doi: 10.1101/cshperspect.a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc. 1999;121:5326–5372. [Google Scholar]
- 4.Egli M, Usman N, Rich A. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA–RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 5.Jaishree TN, van der Marel GA, van Boom JH, Wang AH. Structural influence of RNA incorporation in DNA: quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32:4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- 6.Ban C, Ramakrishnan B, Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA—crystal structure of the DNA–RNA chimeric decamer duplex d(CCGGC)R(CCGG) with a novel intermolecular G-center-dot-C base-paired quadruplet. J Mol Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 7.DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–198. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- 9.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harbor Perspect Biol. 2013;5:a010173. doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harbor Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerantz RT, O’Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, Sun Y, Lou H, Huang J. hPrimpol1/CCDC111 is a human DNA primase–polymerase required for the maintenance of genome integrity. EMBO Rep. 2013;14:1104–1112. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, Terrados G, Powell C, Salido E, Mendez J, Holt IJ, Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, Doherty AJ. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L, Mendez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 19.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Nat Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Nat Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinkai A, Patel PH, Loeb LA. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J Biol Chem. 2001;276:18836–18842. doi: 10.1074/jbc.M011472200. [DOI] [PubMed] [Google Scholar]
- 22.Gosavi RA, Moon AF, Kunkel TA, Pedersen LC, Bebenek K. The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda. Nucleic Acids Res. 2012;40:7518–7527. doi: 10.1093/nar/gks413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavanaugh NA, Beard WA, Batra VK, Perera L, Pedersen LG, Wilson SH. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J Biol Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286:31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Wu EY, Hellinga HW, Beese LS. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J Biol Chem. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harbor Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nick McElhinny SA, Watts B, Kumar D, Watt DL, Lundström E-B, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Nat Acad Sci USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 33.Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goksenin AY, Zahurancik W, LeCompte KG, Taggart DJ, Suo Z, Pursell ZF. Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J Biol Chem. 2012;287:42675–42684. doi: 10.1074/jbc.M112.422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O’Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Nat Acad Sci USA. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 40.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 41.Collins K, Greider CW. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 1995;14:5422–5432. doi: 10.1002/j.1460-2075.1995.tb00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boule JB, Rougeon F, Papanicolaou C. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J Biol Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 43.Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40:6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 45.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Nat Acad Sci USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc Nat Acad Sci USA. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkel TA. The high cost of living. Trends Genet; American Association for Cancer Research Special Conference: endogenous sources of mutations; Fort Myers, Florida, USA. 11–15 November, 1998; 1999. pp. 93–94. [DOI] [PubMed] [Google Scholar]
- 53.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams JS, Clausen AR, Nick McElhinny SA, Watts BE, Johansson E, Kunkel TA. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair (Amst) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA–DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 59.Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arana ME, Kerns RT, Wharey L, Gerrish KE, Bushel PR, Kunkel TA. Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae. DNA Repair (Amst) 2012;11:933–941. doi: 10.1016/j.dnarep.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim N, Cho JE, Li YC, Jinks-Robertson S. RNA:DNA hybrids initiate quasi-palindrome-associated mutations in highly transcribed yeast DNA. PLoS Genet. 2013;9:e1003924. doi: 10.1371/journal.pgen.1003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaisman A, McDonald JP, Huston D, Kuban W, Liu L, Van Houten B, Woodgate R. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 2013;9:e1003878. doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 2012;8:e1003030. doi: 10.1371/journal.pgen.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaisman A, McDonald JP, Noll S, Huston D, Loeb G, Goodman MF, Woodgate R. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res. 2014;761:21–33. doi: 10.1016/j.mrfmmm.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caldecott KW. Molecular biology. Ribose—an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 72.Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clausen AR, Murray MS, Passer AR, Pedersen LC, Kunkel TA. Structure–function analysis of ribonucleotide bypass by B family DNA replicases. Proc Nat Acad Sci USA. 2013;110:16802–16807. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA–DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 80.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Nat Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 82.Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst) 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O’Shea SH, Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Nat Acad Sci USA. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Nat Acad Sci USA. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allen-Soltero SL, Martinez CD, Putnam RD, Kolodner A. Saccharomyces cerevisiae Ribonuclease H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 88.Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brawley OW, Cornelius LJ, Edwards LR, Gamble VN, Green BL, Inturrisi C, James AH, Laraque D, Mendez M, Montoya CJ, Pollock BH, Robinson L, Scholnik AP, Schori M. National Institutes of Health Consensus Development Conference statement: hydroxyurea treatment for sickle cell disease. Ann Intern Med. 2008;148:932–938. doi: 10.7326/0003-4819-148-12-200806170-00220. [DOI] [PubMed] [Google Scholar]
- 90.Sayrac S, Vengrova S, Godfrey EL, Dalgaard JZ. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS Genet. 2011;7:e1001328. doi: 10.1371/journal.pgen.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc Nat Acad Sci USA. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci USA. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 96.Dunn K, Griffith JD. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980;8:555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hovatter KR, Martinson HG. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc Nat Acad Sci USA. 1987;84:1162–1166. doi: 10.1073/pnas.84.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pursell ZF, Kunkel TA. DNA polymerase epsilon: a polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Singh G, Klar AJ. Mutations in deoxyribonucleotide biosynthesis pathway cause spreading of silencing across heterochromatic barriers at the mating-type region of the fission yeast. Yeast. 2008;25:117–128. doi: 10.1002/yea.1569. [DOI] [PubMed] [Google Scholar]
- 100.Kupfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bartlett EJ, Brissett NC, Doherty AJ. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc Nat Acad Sci USA. 2013;110:E1984–E1991. doi: 10.1073/pnas.1302616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986;261:2978–2986. [PubMed] [Google Scholar]
- 104.Vengrova S, Dalgaard JZ. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 2004;18:794–804. doi: 10.1101/gad.289404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Randerath K, Reddy R, Danna TF, Watson WP, Crane AE, Randerath E. Formation of ribonucleotides in DNA modified by oxidative damage in vitro and in vivo. Characterization by 32P-postlabeling. Mutat Res. 1992;275:355–366. doi: 10.1016/0921-8734(92)90038-q. [DOI] [PubMed] [Google Scholar]