Abstract
YB-1 is considered a negative prognostic marker for different types of cancer. Increased YB-1 protein levels in tumor cells indicate a worse prognosis. In a preceding study comparing the transcripts of CRPV-induced benign papillomas to mRNA levels of malignant epithelial tumors, we identified YB-1 as a gene that is up-regulated in papillomavirus-associated carcinomas and which causes an invasive phenotype in CRPV-positive cells in vitro. Here we demonstrate that YB-1 is a previously unknown factor required for papillomavirus-induced tumor development in the rabbit animal model system. By infecting the animals with a novel recombinant shRNA-expressing CRPV genome, we show that knock-down of YB-1 dramatically reduces papillomavirus-dependent tumor formation in vivo. Consistent with previous reports showing a nuclear distribution of YB-1 proteins as a hallmark of malignancy, we demonstrate a predominantly nuclear localization of YB-1 in CRPV-immortalized cells. Furthermore we give evidence of YB-1 regulating the CRPV URR and thereby viral gene expression and we identified YB-1 as a novel interactor of the CRPV regulatory protein E2. Taken together we hypothesize that YB-1 is essential for papillomavirus-induced tumor formation probably by regulating viral gene expression including expression of the oncogenes E6 and E7.
Keywords: Papillomavirus, CRPV, rabbit, in vivo, YB-1, E2, viral transcription, protein-protein interaction
Introduction
Cervical cancer is the fourth most common cancer among women worldwide [1]. The causative agents for cervical cancer are high-risk human papillomaviruses such as HPV16 or HPV18, which are recognized as class I carcinogens by the WHO [2]. Apart from cervical cancer, human papillomaviruses have also been implicated in other anogenital cancers and a subset of head and neck carcinomas [2-4].
In its natural host (Sylvilagus sp.), infection with Cottontail Rabbit Papillomavirus (CRPV) leads to benign tumors that spontaneously regress in the majority of the cases. However, CRPV infection of Oryctolagus sp. (New Zealand White rabbit) causes epithelial tumors which in 80% of the cases progress to carcinomas within 6 to 14 months post-infection (p.i.) without additional co-carcinogens. This model is the only in-vivo infection system to study high-risk papillomavirus-induced tumor progression [5-7].
In our previous gene profiling studies we identified YB-1 as a gene, which is up-regulated in CPRV-induced carcinomas as compared to benign papillomas in rabbits [8]. In humans, YB-1 has been found to be dramatically induced in a variety of cancers including cervical cancer as well as lung, colorectal, breast and ovary cancers where its expression significantly correlates with tumor stage and patient prognosis constituting a prognostic biomarker for tumor progression [9-14]. YB-1 is a member of the nucleic acid-binding protein family of Y-box proteins, which are highly conserved in prokaryotes and eukaryotes [15]. The common structural characteristic of these proteins is a conserved central nucleic acid binding domain known as the cold shock domain. This domain is thought to bind to inverted CCAT boxes within promoter regions, called Y-boxes [16]. YB-1 is involved in a wide variety of biological functions including DNA repair, transcription, RNA splicing and translation [17-19]. It translocates to the nucleus for its nuclear activities while the majority of YB-1 localizes to the cytoplasm [17]. Once in the nucleus, YB-1 regulates the transcription of numerous genes via direct interaction with Y-boxes, by associating with various other transcription factors or by binding to single stranded DNA, which is present in transcriptionally active promoters [20-22]. More recent reports argue against the association of YB-1 to promoter regions in vivo by showing that it preferably interacts with GC-rich regions in introns containing sites resembling Kozak sequences [23,24]. The majority of the YB-1-regulated genes are involved in cell proliferation stimulating both cell cycle progression and DNA replication. These include EGFR, cyclin A, cyclin B1 and DNA polymerase α [25-27]. Other tumor promoting genes regulated by YB-1 include MMP-2 and collagen α-1, which are involved in cell adhesion, motility and tumor invasion [28,29]. YB-1 is also involved in the development of chemoresistance in certain cancers by up-regulating the multi-drug resistance-1 gene (MDR1) [20]. Alteration of YB-1-dependent transcription influences a wide range of cellular functions. Not surprisingly these diverse functions of YB-1 are also exploited by a number of different viruses including adenovirus, dengue virus, hepatitis C virus, human immunodeficiency virus 1 (HIV-1), human T-cell lymphotropic virus type 1 (HTLV-1), influenza virus, JC polyomavirus (JCV) and Murine leukemia virus [30-41]. In HIV-1 replication, YB-1 enhances transcription of the long terminal repeat region after being recruited to the trans-activation response (TAR) element via direct association with the viral transcription factor Tat [42]. Also JCV transcription is regulated by the interaction of YB-1 with the viral proteins large T-antigen and agnoprotein [32,37]. Similar to JCV large T antigen, the papillomavirus E2-protein is involved in DNA replication and transcription [43,44]. CRPV E2 is a sequence specific DNA-binding protein, which associates with its palindromic recognition sequence ACCN6GGT present in multiple copies within the viral genome. E2 consists of a conserved N-terminal transactivation domain and a C-terminal DNA-binding/dimerization domain separated by a variable hinge region [43]. Mutations within the N-terminal domain of the E2-protein lead to a loss of tumor induction in vivo concluding that CRPV E2 is required for tumor formation in rabbits [5].
By employing a novel recombinant CRPV genome constructed to express shRNA targeting the host protein YB-1, we demonstrate that it is essential for CRPV-induced tumor development in vivo. As YB-1 has previously been shown to regulate transcription of viruses by binding to viral transcription factors, we investigated its role in CRPV gene expression and demonstrated that YB-1 is required for the activation of the viral regulatory region and showed that it binds to the viral transcription regulator CRPV E2.
Materials and methods
Cell culture
All rabbit keratinocyte cell lines (AVS, RK1-16E7/ras and RK-CRPVE6/E7) were cultured in supplemented keratinocyte serum-free medium (K-SFM; Invitrogen, Karlsruhe, Germany) without antibiotics as previously described [8]. HeLa cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) (life technologies, Darmstadt, Germany) containing 10% fetal bovine serum.
Vectors and plasmids
The prokaryotic expression vector pMal-c2x was purchased from New England Biolabs. The constructs pMal-CRPVE2 and pMal-CRPVE7 were generated by PCR using the primers 5’-ATGGAGGCTCTCAGCCAG-3’ pMAL/XmnICRPVE2 and 5’-GGCCGCCAAGCTTCTAAAGCCCATAAA-3’ pMAL/HindIICRPVE2 for CRPV E2, 5’-ATGATAGGCAGAACTCCTAAG-3’ pMal/XmnICRPVE7 and 5’-TCCATTCTGCAGTCAGTTACAACACTCCGGG-3’ pMal-PstICRPVE7 for CRPVE7 and the plasmid pLA2-CRPV [5] as a template. The plasmids pMal-CRPVSE6 and pMal-CRPVLE6 were described previously [45]. The luciferase reporter plasmid pL1-pE7(CRPV)-pGL3 (pGL3-PL-P1-P2-P3-luc) and the luciferase reporter constructs MPM/2xSP1-BS and MPM/2xSP1-BS/4xE2BS have been described previously [46,47].
Generation of shRNA containing recombinant CRPV plasmids
We used the recombinant CRPV genomes CRPV-pLAIIdelXba1 or CRPV-Xba1-mcs in which nucleotides 4665 to 5775 (Xba1 fragment) were deleted by Xba1 digestion (Figure 1). CRPV-Xba1-mcs contains an additional multiple cloning site consisting of SnaB1, SacII, Not1 and Xho1 [48]. To generate the shRNA cassette, which was inserted into the recombinant CRPV genome, shYB-1-1, shYB-1-2 and shLuciferase were inserted into the shRNA expression vector pSuper [49], which was linearized by the restriction enzymes BglII and SalI and subsequently annealed by synthetic double stranded oligonucleotides coding for the respective shRNAs. The oligonucleotide sequences were as followed: 5’-GATCCCCgaaggtcatcgcaacgaagttcaagagacttcgttgcgatgaccttcTTTTTG-3’ for shYB-1-1; 5’-GATCCCCagaagaaatgaatatgaaattcaagagatttcatattcatttcttctTTTTTG-3’ for shYB-1-2 and 5’-GATCCCCcttacgctgagtacttcgattcaagagatcgaagtactcagcgtaagTTTTTG-3’ for shLuciferase.
Figure 1.
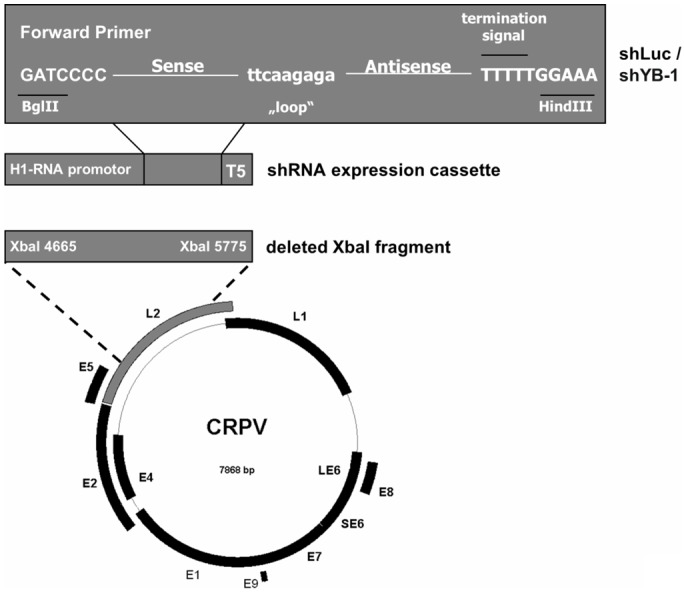
Illustration of the recombinant CRPV-based shRNA expression vector. A novel shRNA-containing recombinant CRPV genome was constructed to express shRNAs directed against YB-1 or firefly luciferase. A large portion of the L2 gene was replaced by the shRNA expression cassettes originating from the plasmid pSuper.
To generate the vectors CRPVshLuc-pLAII and CRPVshYB-1-1-pLAII the recombinant CRPV genome CRPV-pLAIIdelXba1 and the pSuper vectors containing shLuciferase and shYB-1-1 were digested using Ecl136II/HincII or NotI/XhoI. Each obtained shRNA-containing fragment was ligated with the linearized plasmid. To generate the vector CRPVshYB-1-2-pLAII, the plasmid CRPV-Xba1-mcs and the shYB-1-2 containing pSuper vector were digested with XbaI/XhoI and ligated with each other. The constructs were validated by DNA sequencing.
Western blot analysis
To obtain whole-cell extracts, cells were lysed in 2x SB buffer (4% (w/v) SDS, 12% (v/v) glycerin, 0.2 M DTT, 0.1% bromophenol blue, 125 mM Tris/HCl (pH 6.8)). Proteins were targeted using primary antibodies directed against YB-1 (Santa Cruz, sc101198) or α-Tubulin (Calbiochem, CP06). Secondary antibodies conjugated to horseradish peroxidase (Dako) were detected using SuperSignal West substrates (Thermo Scientific) and Fluor-S Max MultiImager (Bio-Rad).
For the subcellular fractionation assay blots were also probed with a TRIM 28 (BD, 610680) primary antibody and the MBP protein conjugates were detected using anti-MBP antibodies conjugated to horseradish peroxidase (HRP) (NEB, E8038).
Animal infection and monitoring
All experimental procedures were reviewed and approved (Permit Number: H1/08) by the responsible authority (Regierungspräsidium Tübingen, Baden-Württemberg, Germany) according to the German Animal Welfare Act (TierSchG §8 Abs. 1) and were performed according to institutional guidelines. Rabbits were maintained under specific-pathogen-free conditions in the animal facility of the University of Tübingen, Germany. Infection and monitoring of rabbits was conducted as described previously. The backs of New Zealand White rabbits, purchased from Charles River, Kisslegg, Germany, were infected as previously described [48,50,51]. 14 weeks later, the number and size of tumors was documented. Statistical analysis (Fisher’s exact test) was performed by using the http://graphpad.com/QuickCalcs/website.
Subcellular fractionation assay
Cytosolic extracts and nuclear extracts were obtained as follows. AVS cells were washed with PBS and incubated in hypotonic buffer (10 mM HEPES/KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, complete protease inhibitor) on ice for 10 min. Subsequently, cells were vortexed for 10 sec and the nuclei were pelleted at maximum speed for 10 sec. The supernatant constitutes the cytosol fraction. The nuclear pellet was lysed in high salt buffer (20 mM HEPES/KOH (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 25% (v/v) glycerin, 0.2 mM EDTA, 0.5 mM DTT, complete protease inhibitor). Lysates were centrifuged for 2 min at full speed. Cytoplasmic and nuclear extracts were subjected to western blot analysis.
Immunofluorescence
AVS cells were grown on cover slips. Fixed cells were stained with YB-1 antibody (sc-101198; Santa Cruz Biotechnology) which was detected by Alexa fluor 488-conjugated anti-mouse IgG antibody. Nuclei were DAPI-stained. Fluorescence signals were visualized using the Zeiss Axiovert 200M microscope (Carl Zeiss MicroImaging GmbH).
WST-1 viability assay
The cell viability was assessed using the cell proliferation reagent WST-1 assay (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s instructions. Briefly, 5×103 rabbit keratinocytes were seeded per well of a 96-well microtitre plate; 24 h, 48 h and 72 h later WST-1 reagent was added to the cells and incubated for 2 hours at 37°C. Subsequently, the optical density (OD) was measured at 450 nm and 650 nm as the reference wavelength using a plate reader (DiaSorin/ETI-System Fast Reader S800).
Quantitative real-time PCR
RNA was isolated using the RNeasy kit (Qiagen), and cDNA was synthesized from 1 μg of total RNA by the QuantiTect Reverse Transcription Kit (Qiagen). PCR reactions were carried out using 20 μl reaction mixtures consisting of 10 μl of LightCycler 480 SYBR Green I Master (Roche), 50 ng of cDNA, and 3 μmol/l forward and reverse primers. Relative transcript levels were calculated using GAPDH transcripts as a reference [52]. Sequences for the primers used are GAPDH for 5’-TGCACCACCAACTGCTTAGC-3’, GAPDH rev 5’-GGCATGGACTGTGGTCATGAG-3’, CRPVE2 for 5’-CGCCTTAAAAGCAAGCACTC-3’ and CRPVE2 rev 5’-AGAAACTTATCGCGCTGCTC-3’.
Transient luciferase expression assay
3.5×104 AVS cells were seeded into 24-well dishes the day before transfection. Cells were transfected with 50 ng of the respective luciferase-reporter plasmid and FugeneHD transfection reagent (Promega, Madison, WI), 48 h later luciferase activity was measured as previously described [53].
When cells were transfected with a luciferase-reporter plasmid and siRNA, cells were first transfected with 75 ng of YB-1 siRNA [8] or a scrambled control siRNA (Allstars negative control Alexa488 siRNA; Qiagen no.1027284) in Hiperfect transfection reagent (Qiagen, Hilden, Germany) and OptiMEM (Invitrogen). 24 hours later cells were additionally transfected with 50 ng of the luciferase reporter and FugeneHD transfection reagent. Luciferase assays were carried out 48 hours post-transfection of the reporter plasmid. Statistical analysis (t-test) was performed using GraphPad Prism 5.
Maltose-binding protein pull-down assay
Maltose-binding protein (MBP) pull-down assays of MBP, CRPV E2, SE6, LE6 and E7 were performed as described previously [54]. Briefly, affinity purified MBP, MBP-CRPVE2, MBP-CRPVSE6, MBP-CRPVLE6 and MBP-CRPVE7 proteins were incubated with whole cell extracts derived from 12×106 HeLa cells each and lysed in 50 mM NaCl, 50 mM Tris (pH 8.0), 0.1% (v/v) β-mercaptoethanol, 0.1% (v/v) Igepal® CA-630 and complete protease inhibitor (Roche). After extensive washing, the retained proteins were eluted with 20 mM Tris (pH 7.5), 12.5 mM MgCl2, 0.1% (v/v) Igepal CA-630, 0.1% (v/v) β-mercaptoethanol, 20 mM maltose, and protease inhibitors and analyzed by immunoblotting.
Results
Construction of a recombinant CRPV genome expressing shRNA targeting YB-1
We have previously observed, that malignant progression of CRPV-induced tumors is accompanied by an up-regulation of YB-1 [8]. We therefore investigated whether down regulation of YB-1 has an effect on tumor formation in vivo by constructing a recombinant CRPV genome containing an shRNA cassette (Figure 1). The CRPV backbone is based on the vector CRPV-Xba1-mcs which was previously described [48]. This vector lacks a functional copy of L2, but still efficiently induces papillomas [48]. A fragment containing the H1-promoter and the shRNA cassette originating from the shRNA expression vector pSuper were cloned into this vector. Two different shRNAs based on functional siRNA sequences were used to create the plasmids CRPVshYB-1-1-pLAII and CRPVshYB-1-2-pLAII, respectively. Preparatory, we designed siRNAs targeting YB-1, which were analyzed for their knock-down capacity via transient transfection of AVS cells. A strong decrease of YB-1 protein levels was observed for both siYB1-1 and siYB-1-2 (Figure 2A). Specificity of shYB-1-1 was determined in previous work [55]. As a control a vector containing shRNA sequences targeting the non-rabbit control gene firefly luciferase was included (Figure 1).
Figure 2.
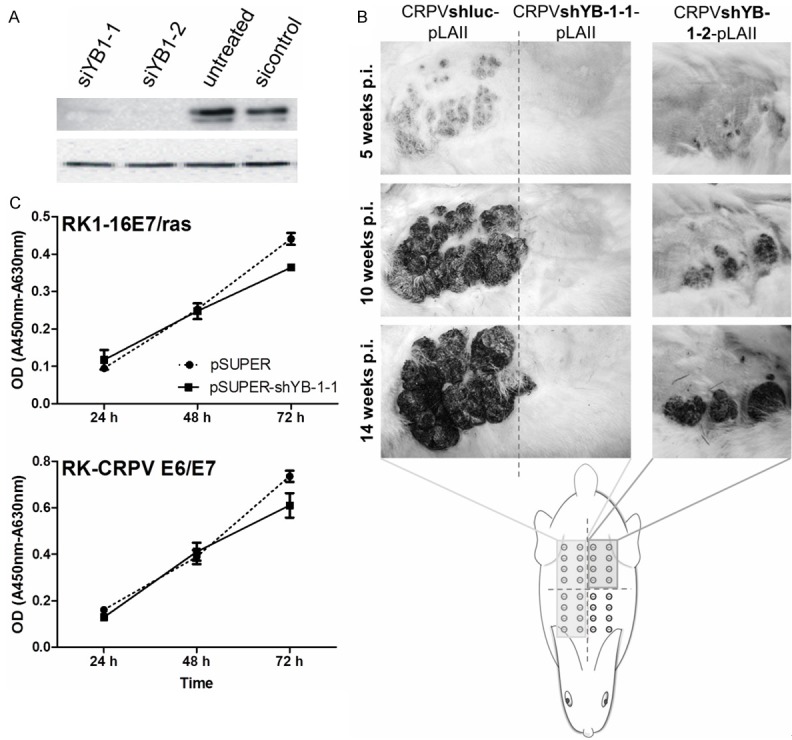
Knock-down of YB-1 leads to reduced tumor formation in vivo. A. Western blot analysis of YB-1 protein levels in AVS cells transiently transfected with siRNAs targeting YB-1 or a scrambled control siRNA. B. Photographs of tumor development on the back of one representative rabbit infected with CRPV either containing shRNA targeting YB-1 (CRPVshYB-1-1-pLAII or CRPVshYB1-2-pLAII) or expressing a non-specific control shRNA (CRPVshluc-pLAII); 5 to 14 weeks post infection. C. WST cell viability assay of immortalized rabbit keratinocytes stably transfected with pSuper-shYB-1-1 versus the empty vector control cells.
YB-1 is necessary for tumor formation in vivo
In order to determine whether the knock-down of YB-1 has an impact on tumor formation in vivo, New Zealand White rabbits were treated with the recombinant shRNA cassette-containing CRPV genome constructs. Four rabbits received injections of CRPVshYB-1-1-pLAII DNA into a total of 96 sites, two rabbits were injected with CRPVshYB-1-2-pLAII DNA at a total of 12 sites and another four rabbits were treated with CRPVshluc-pLAII DNA at 36 sites. After 5-14 weeks a strongly diminished tumor growth at sites treated with shYB-1 compared to shLuc treated sites was observed (p<0.0001 for shYB-1-1 and p=0.0003 for shYB1-2; Figure 2B). In detail, only 4% of the sites injected with CRPVshYB-1-1-pLAII and 41.6% of sites injected with CRPVshYB-1-2-pLAII showed tumor growth. In contrast, the control CRPV genome encoding a firefly luciferase-specific shRNA, which induces tumor formation as potently as the wildtype CRPV genome (data not shown), showed tumor development in 94% of injection sites (Table 1).
Table 1.
Summary of tumor induction in all rabbits 14 weeks post infection. The number of rabbits, infected sites and the percentage of papilloma induction (sites with visible papillomas compared to the total number of infected sites) are shown
| Recombinant CRPV genome | Number of rabbits | Number of injected sites | % papilloma induction |
|---|---|---|---|
| CRPVshluc-pLAII | 4 | 36 | 94 |
| CRPVshYB-1-1-pLAII | 4 | 96 | 4 |
| CRPVshYB-1-2-pLAII | 2 | 12 | 42 |
In order to evaluate, whether the reduced tumor growth is due to cytotoxic effects of the shYB-1, cell proliferation assays where conducted using rabbit keratinocytes immortalized by HPV16 E6/ras (RK1-16E7/ras) or by CRPV E6 and E7 (RK-CRPVE6/E7) stably transduced with either pSuper-YB-1-1 or the empty vector control. No differences in cell viability between shYB-1- and the empty vector control-containing cells were observed (Figure 2C) thereby excluding a cytotoxic effect of the YB-1-specific shRNA.
YB-1 is localized within the nucleus in AVS cells
Because the majority of YB-1 localizes to the cytoplasm, it is required to translocate to the nucleus for its transcriptional activities [56]. AVS cells are rabbit primary keratinocytes, which have been immortalized by the integration of full length CRPV into the genome [8]. To examine YB-1’s subcellular localization within the AVS cell line, cell fractionation assays were performed comparing cytosolic to nuclear extracts. Immunoblot assays were then conducted detecting α-tubulin as a control for cytosolic localization, TRIM28, which localizes to the nuclear fraction [57] and the candidate protein YB-1. The majority of YB-1 was detected within the nuclear extract evidencing a predominantly nuclear localization of YB-1 in AVS cells (Figure 3A) allowing it to act as a transcriptional regulator. Immunofluorescence studies confirmed its nuclear distribution (Figure 3B).
Figure 3.
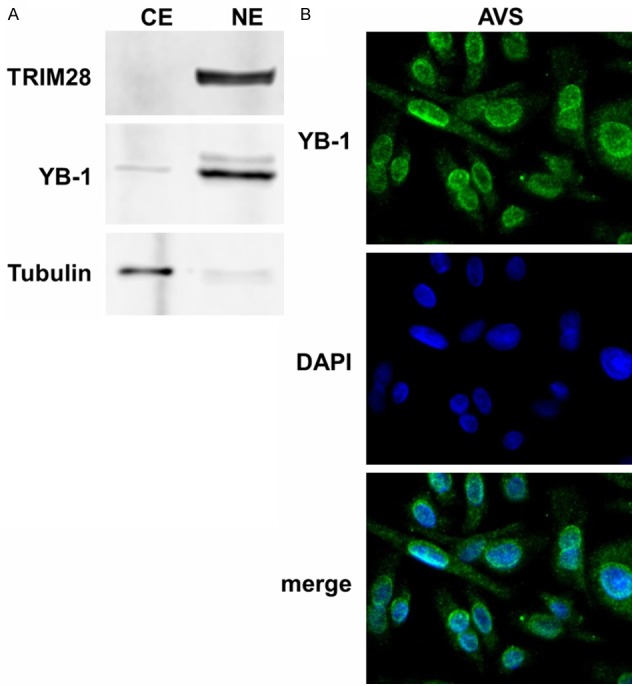
YB-1 localizes to the nucleus of AVS cells. A. Cell fractionation assay of AVS cells. Immunoblotting was conducted detecting the nuclear marker TRIM28, the cytosolic marker α-Tubulin and YB-1. B. Immunofluorescence of AVS cells at 630x magnification. Cells were fixed and stained with DAPI and anti-YB-1 antibody.
YB-1 is necessary for transcription of CRPV genes
Previous studies showed, that YB-1 has an impact on viral transcription [30,31,58]. Accordingly, we investigated a possible role of YB-1 in the regulation of CRPV transcription. Therefore, the influence of an YB-1 knock-down on the activity of the luciferase reporter plasmid pL1-pE7 (CRPV)-pGL3 containing the CRPV URR was analyzed. For the reporter assays we used AVS cells, which are rabbit keratinocytes immortalized by CRPV, thus, expressing viral transcripts including E6 and E7 [54] as well as CRPV E2 which is essential for viral transcription.
To confirm that E2 is present in AVS cells, quantitative real-time PCR analysis of these cells was performed revealing high levels of CRPV E2 transcripts in AVS cells as compared to primary rabbit keratinocytes (data not shown). To test the functionality of CRPV E2, luciferase reporter assays using constructs with (MPM/2xSP1-BS/4xE2BS) or without E2 binding sites (E2BS; MPM/2xSP1-BS) were conducted. A significantly higher luciferase activity (p=0.033) in cells containing the E2BS reporter construct was observed confirming that CRPV E2 is indeed active and functional in AVS cells (Figure 4A).
Figure 4.
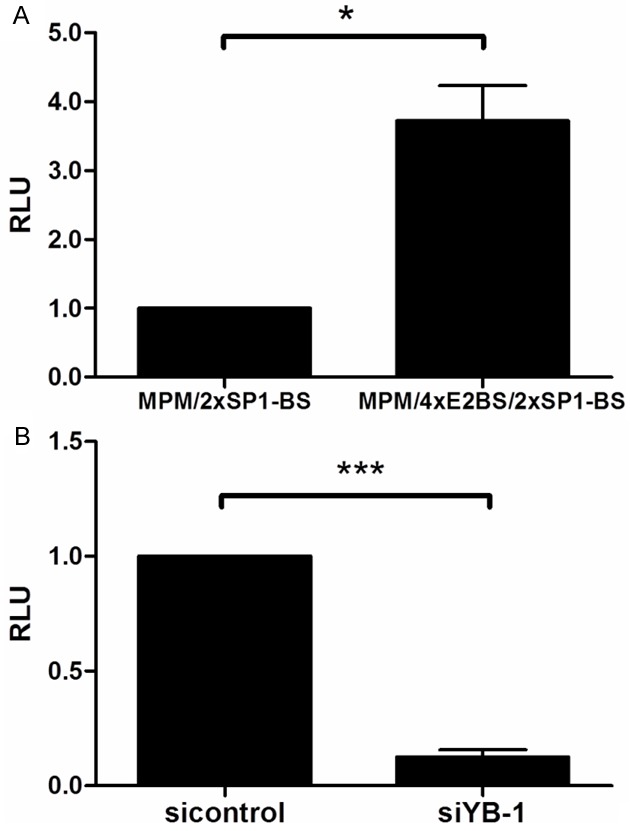
Transient knock down of YB-1 reduces activity of the CRPV URR in E2-expressing AVS cells. A. Luciferase assays were performed using AVS cells transfected with the luciferase-reporter constructs MPM/2xSP1-BS or MPM/4xE2BS/2xSP1-BS. Luciferase activity is shown in relative light units (RLU) relative to the control (MPM/2xSp-1-BS). B. Luciferase assays were performed using AVS cells transfected with siYB-1-1 or an siRNA control. The transfection with the luciferase-reporter construct pL1-pE7(CRPV)-pGL3 was performed 24 h later. Luciferase activities were measured in relative light units relative to the siRNA control. Results shown are from three and four independent experiments, respectively, each run in triplicates. The standard deviations are indicated by error bars and asterisks illustrate significance: *p<0.05, ***p<0.001.
To analyze the effect a reduction of the YB-1 protein levels has on CRPV transcription, we compared the activity of the CRPV URR linked to a luciferase gene in the presence of siYB-1-1 or a scrambled siRNA control. We observed a strong decrease (87.5%) in luciferase activity in the siYB-1-1-containing cells as compared to the control (p=0.0001), demonstrating that YB-1 is essential for CRPV transcription (Figure 4B).
YB-1 binds the viral transcription factor CRPV E2
It has previously been shown that YB-1 interacts with JCV and HIV-1 proteins in order to regulate viral transcription [32,42]. Since YB-1 is necessary for CRPV transcription and E2 is the essential viral transcription factor for papillomaviruses, we analyzed the possibility of a physical interaction between YB-1 and CRPV E2. We used other CRPV proteins in addition to E2 to control for the specificity of the in vitro binding assay. The viral genes E2, SE6, LE6 and E7 were expressed as maltose binding protein fusion proteins (MBP) in E. coli, affinity-purified and subsequently incubated with whole cell extracts derived from HeLa cells, which express the YB-1 protein. Immunoblotting revealed that CRPV E2 specifically pulled-down YB-1. No other viral protein associated with YB-1 confirming the specificity of the E2-YB-1 interaction (Figure 5).
Figure 5.
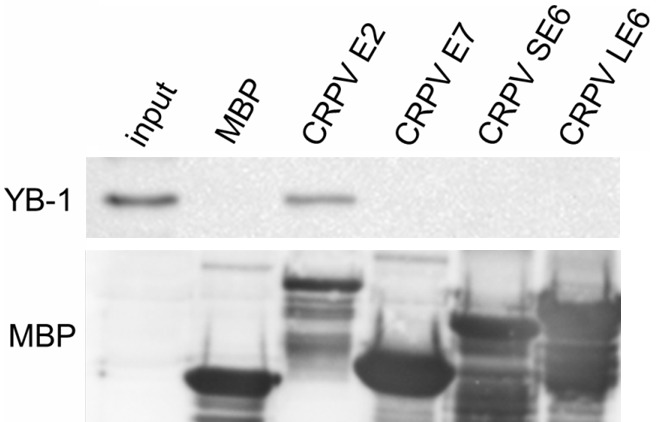
YB-1 interacts with the viral transcriptional regulator CRPV E2. MBP fusion proteins of CRPV E2, E7, SE6, LE6 and MBP were tested for YB-1 binding by MBP pull-down assay. Prokaryotic-expressed recombinant fusion proteins were incubated with HeLa cell extracts and eluted using maltose. YB-1 and MBP were analyzed by immunoblotting.
In summary our data demonstrate that YB-1 is required for CRPV-associated tumor formation. In addition we showed that YB-1 regulates the viral URR and that it physically associates with the viral transcription regulator E2.
Discussion
We previously demonstrated that the Y-box protein YB-1 is up-regulated in CRPV-associated tumors in rabbits [8]. In the present study we investigated the role of YB-1 in papillomavirus-induced tumor formation in vivo. We infected rabbits with a specifically designed novel shRNA-containing CRPV genome construct encoding shRNA directed against YB-1 or the non-specific control gene firefly luciferase. A strong reduction of tumor formation was observed at sites infected with the CRPV genome encoding shYB-1-2 and no tumor formation occurred at sites injected with the shYB-1-1-expressing CRPV construct. In contrast, virtually 100% of the sites infected with the control shRNA genome CRPV-shLuc developed tumors similar to wild type CRPV. We confirmed that the reduced tumor induction is not due to cytotoxic effects as proliferation assays in cells stably transfected with shYB-1-1 showed no differences in viability as compared to control cells. These results therefore implicate a specific role of YB-1 in CRPV-induced tumor development and carcinogenesis. In addition, our results demonstrate the applicability of a novel and unique CRPV vector system which allows the analysis of the role of cellular genes in tumor development within the same cells that contain the tumor inducing virus.
Our results are also supported by data showing a primarily nuclear expression of YB-1 in cells infected with CRPV as previous reports linked a nuclear expression pattern of YB-1 to a poor prognosis of various cancer types [12,59-61]. In addition to its prognostic biomarker potential for carcinogenesis, YB-1 is reportedly involved in the transcription of different viruses such as HIV-1, HTLV-1, adenovirus and JCV [30,32,35,42]. We therefore hypothesized a potential participation of YB-1 in CRPV transcription as the possible mechanism underlying the requirement of YB-1 for papillomavirus-based tumor development. To investigate the role of YB-1 in viral gene expression, we examined the effect YB-1 silencing has on the activity of a construct containing the luciferase expression cassette under the control of the CRPV URR. We observed a strong decrease in luciferase activity upon reduced YB-1 protein levels, suggesting that YB-1 might be necessary for viral gene expression.
Direct interactions of YB-1 with viral proteins of HIV-1 and JCV have previously been reported [32,37,42]. These interactions involve viral proteins known to regulate viral transcription. It is thought that YB-1 is recruited by these proteins in order to usurp its functions as a transcription enhancer. We here demonstrate that YB-1 physically associates with the CRPV transcription factor E2 substantiating our hypothesis that YB-1 contributes to the transcriptional regulation of the CRPV early promoter. However, further investigation is necessary in order to map the CRPV E2–YB-1 binding domains and to establish the functional consequences of their interaction. Furthermore, it is currently not known whether the association of YB-1 and CRPV E2 is DNA or RNA-dependent. CRPV E2 binds sites within the viral promoter [43], whereas YB-1 recognizes sites within DNA and RNA sequences [16,23,24]. Importantly, YB-1 has previously been found to bind to a specific site within the HPV18 URR enhancer sequence [62]. We therefore speculate that the concerted binding of YB-1 and E2 to the long control region could result in the activation of viral oncogene (E6/E7) expression. In addition to YB-1, CRPV E2 interacts with the transcription co-activator Brd4 [63], which is an enhancer of the general RNA polymerase II-dependent transcription machinery through binding to core factors of the positive transcription elongation factor b (P-TEFb) [64,65]. Whether YB-1, CRPVE2 and Brd4 act as a transactivation complex for early viral gene expression is currently unknown. Brd4 has been shown to be involved in a number of E2 functions, including transcriptional gene expression regulation as well as viral genome maintenance and partitioning (summarized by [66]). Regarding other viruses, e.g. HIV-1, Brd4 has been shown to compete with Tat for the P-TEFb binding site [67] and therefore appears to function as a dose-dependent negative regulator of HIV-1 transcription. In contrast, YB-1 positively regulates HIV-1 gene expression via direct association with Tat and TAR [41], which contradicts the hypothesis of a common transactivation complex and rather points towards virus-specific mechanisms.
YB-1 has been shown to interact with p300 - a facilitator of chromatin remodeling and assembly of transcription complexes [68]. P300 is known to weakly associate with E2 [69,70]. This interaction is enhanced by direct binding of Gps2 to p300 causing a higher histone acetylation activity which leads to an enhanced transactivation of viral transcription [69]. A mechanism by which YB-1 associates with p300 and E2 with Gps2 stabilizing this transactivation complex may be another possibility to explain our findings of YB-1 playing an essential role in CRPV oncogene transcription. However, this hypothesis awaits full elucidation. In addition, literature links YB-1 to a wealth of different cellular processes, which complicates the examination of the underlying mechanism leading to the phenotype demonstrated in this study. Among others YB-1 has been shown to be involved in transcription, pre-mRNA splicing, mRNA translation and DNA repair [71]; and YB-1 deficiency might disrupt any of these processes leading to the observed lack of tumor development following CRPV infection.
In summary, we have demonstrated for the first time that a shRNA-expressing CRPV genome is functional in vivo and that it is a valuable tool for confirming the involvement of selected genes in papillomavirus-associated tumorigenesis. Finally, we demonstrate that the multifunctional cellular protein YB-1 is required for CRPV-induced tumor formation in rabbits in vivo. As YB-1 interacts with the viral transcription factor CRPV E2 and activates the URR, it appears to be a novel co-factor recruited by E2 in order to enhance viral gene expression. Therefore interference with YB-1 expression might constitute a novel therapeutic approach against HPV-associated cancers.
Acknowledgements
Financial support was provided by the German research Foundation (DFG), grant number SFB 773 B4.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. GLOBOCAN 2012 v1.0 2013. doi: 10.1002/ijc.29210. http://globocan.iarc.fr. [DOI] [PubMed] [Google Scholar]
- 2.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M, Clifford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer. 2009;4:8. doi: 10.1186/1750-9378-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray F. Chapter 2: The burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S3/S11–25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 5.Jeckel S, Huber E, Stubenrauch F, Iftner T. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. J Virol. 2002;76:11209–11215. doi: 10.1128/JVI.76.22.11209-11215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Syverton JT. The pathogenesis of the rabbit papilloma-to-carcinoma sequence. Ann N Y Acad Sci. 1952;54:1126–1140. doi: 10.1111/j.1749-6632.1952.tb39983.x. [DOI] [PubMed] [Google Scholar]
- 7.Zeltner R, Borenstein LA, Wettstein FO, Iftner T. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J Virol. 1994;68:3620–3630. doi: 10.1128/jvi.68.6.3620-3630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber E, Vlasny D, Jeckel S, Stubenrauch F, Iftner T. Gene profiling of cottontail rabbit papillomavirus-induced carcinomas identifies upregulated genes directly Involved in stroma invasion as shown by small interfering RNA-mediated gene silencing. J Virol. 2004;78:7478–7489. doi: 10.1128/JVI.78.14.7478-7489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto K, Bay BH. Significance of the Y-box proteins in human cancers. J Mol Genet Med. 2005;1:11–17. doi: 10.4172/1747-0862.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamura T, Yahata H, Amada S, Ogawa S, Sonoda T, Kobayashi H, Mitsumoto M, Kohno K, Kuwano M, Nakano H. Is nuclear expression of Y box-binding protein-1 a new prognostic factor in ovarian serous adenocarcinoma? Cancer. 1999;85:2450–2454. doi: 10.1002/(sici)1097-0142(19990601)85:11<2450::aid-cncr21>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Tan PH, Li KB, Matsumoto K, Tsujimoto M, Bay BH. Y-box binding protein, YB-1, as a marker of tumor aggressiveness and response to adjuvant chemotherapy in breast cancer. Int J Oncol. 2005;26:607–613. [PubMed] [Google Scholar]
- 12.Gessner C, Woischwill C, Schumacher A, Liebers U, Kuhn H, Stiehl P, Jurchott K, Royer HD, Witt C, Wolff G. Nuclear YB-1 expression as a negative prognostic marker in nonsmall cell lung cancer. Eur Respir J. 2004;23:14–19. doi: 10.1183/09031936.03.00033203. [DOI] [PubMed] [Google Scholar]
- 13.Shibao K, Takano H, Nakayama Y, Okazaki K, Nagata N, Izumi H, Uchiumi T, Kuwano M, Kohno K, Itoh H. Enhanced coexpression of YB-1 and DNA topoisomerase II alpha genes in human colorectal carcinomas. Int J Cancer. 1999;83:732–737. doi: 10.1002/(sici)1097-0215(19991210)83:6<732::aid-ijc6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Reng SR, Wang L, Lu L, Zhao ZH, Zhang ZK, Feng XD, Ding XD, Wang J, Feng G, Dai TZ, Pu J, Du XB. Overexpression of Y-box binding protein-1 in cervical cancer and its association with the pathological response rate to chemoradiotherapy. Med Oncol. 2012;29:1992–1997. doi: 10.1007/s12032-011-0062-2. [DOI] [PubMed] [Google Scholar]
- 15.Wolffe AP, Tafuri S, Ranjan M, Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992;4:290–298. [PubMed] [Google Scholar]
- 16.Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 17.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 18.Evdokimova V, Ovchinnikov LP, Sorensen PH. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle. 2006;5:1143–1147. doi: 10.4161/cc.5.11.2784. [DOI] [PubMed] [Google Scholar]
- 19.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohga T, Uchiumi T, Makino Y, Koike K, Wada M, Kuwano M, Kohno K. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto T, Izumi H, Imamura T, Takano H, Ise T, Uchiumi T, Kuwano M, Kohno K. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000;19:6194–6202. doi: 10.1038/sj.onc.1204029. [DOI] [PubMed] [Google Scholar]
- 22.Dhalla AK, Ririe SS, Swamynathan SK, Weber KT, Guntaka RV. chk-YB-1b, a Y-box binding protein activates transcription from rat alpha1(I) procollagen gene promoter. Biochem J. 1998;336:373–379. doi: 10.1042/bj3360373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolfini D, Mantovani R. YB-1 (YBX1) does not bind to Y/CCAAT boxes in vivo. Oncogene. 2013;32:4189–90. doi: 10.1038/onc.2012.521. [DOI] [PubMed] [Google Scholar]
- 24.Dolfini D, Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20:676–85. doi: 10.1038/cdd.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stratford AL, Habibi G, Astanehe A, Jiang H, Hu K, Park E, Shadeo A, Buys TP, Lam W, Pugh T, Marra M, Nielsen TO, Klinge U, Mertens PR, Aparicio S, Dunn SE. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007;9:R61. doi: 10.1186/bcr1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.En-Nia A, Yilmaz E, Klinge U, Lovett DH, Stefanidis I, Mertens PR. Transcription factor YB-1 mediates DNA polymerase alpha gene expression. J Biol Chem. 2005;280:7702–7711. doi: 10.1074/jbc.M413353200. [DOI] [PubMed] [Google Scholar]
- 27.Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278:27988–27996. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 28.Mertens PR, Alfonso-Jaume MA, Steinmann K, Lovett DH. YB-1 regulation of the human and rat gelatinase A genes via similar enhancer elements. J Am Soc Nephrol. 1999;10:2480–2487. doi: 10.1681/ASN.V10122480. [DOI] [PubMed] [Google Scholar]
- 29.Norman JT, Lindahl GE, Shakib K, En-Nia A, Yilmaz E, Mertens PR. The Y-box binding protein YB-1 suppresses collagen alpha 1(I) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J Biol Chem. 2001;276:29880–29890. doi: 10.1074/jbc.M103145200. [DOI] [PubMed] [Google Scholar]
- 30.Kashanchi F, Duvall JF, Dittmer J, Mireskandari A, Reid RL, Gitlin SD, Brady JN. Involvement of transcription factor YB-1 in human T-cell lymphotropic virus type I basal gene expression. J Virol. 1994;68:561–565. doi: 10.1128/jvi.68.1.561-565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawaya BE, Khalili K, Amini S. Transcription of the human immunodeficiency virus type 1 (HIV-1) promoter in central nervous system cells: effect of YB-1 on expression of the HIV-1 long terminal repeat. J Gen Virol. 1998;79:239–246. doi: 10.1099/0022-1317-79-2-239. [DOI] [PubMed] [Google Scholar]
- 32.Safak M, Gallia GL, Ansari SA, Khalili K. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J Virol. 1999;73:10146–10157. doi: 10.1128/jvi.73.12.10146-10157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3’-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 34.Chatel-Chaix L, Melancon P, Racine ME, Baril M, Lamarre D. Y-box-binding protein 1 interacts with hepatitis C virus NS3/4A and influences the equilibrium between viral RNA replication and infectious particle production. J Virol. 2011;85:11022–11037. doi: 10.1128/JVI.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm PS, Bergmann S, Jurchott K, Lage H, Brand K, Ladhoff A, Mantwill K, Curiel DT, Dobbelstein M, Dietel M, Gansbacher B, Royer HD. YB-1 relocates to the nucleus in adenovirus-infected cells and facilitates viral replication by inducing E2 gene expression through the E2 late promoter. J Biol Chem. 2002;277:10427–10434. doi: 10.1074/jbc.M106955200. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Wang X, Gao G. Expression of YB-1 enhances production of murine leukemia virus vectors by stabilizing genomic viral RNA. Protein Cell. 2012;3:943–949. doi: 10.1007/s13238-012-2090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J Virol. 2002;76:3828–3838. doi: 10.1128/JVI.76.8.3828-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen NN, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaguchi A, Matsumoto K, Nagata K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J Virol. 2012;86:11086–11095. doi: 10.1128/JVI.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerr D, Chang CF, Chen N, Gallia G, Raj G, Schwartz B, Khalili K. Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J Virol. 1994;68:7637–7643. doi: 10.1128/jvi.68.11.7637-7643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ansari SA, Safak M, Gallia GL, Sawaya BE, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J Gen Virol. 1999;80:2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 42.Ansari SA, Safak M, Gallia GL, Sawaya BE, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J Gen Virol. 1999;80:2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 43.McBride AA, Romanczuk H, Howley PM. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 44.Rapp B, Pawellek A, Kraetzer F, Schaefer M, May C, Purdie K, Grassmann K, Iftner T. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J Virol. 1997;71:6956–6966. doi: 10.1128/jvi.71.9.6956-6966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muench P, Probst S, Schuetz J, Leiprecht N, Busch M, Wesselborg S, Stubenrauch F, Iftner T. Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res. 2010;70:6913–6924. doi: 10.1158/0008-5472.CAN-10-1307. [DOI] [PubMed] [Google Scholar]
- 46.Behren A, Simon C, Schwab RM, Loetzsch E, Brodbeck S, Huber E, Stubenrauch F, Zenner HP, Iftner T. Papillomavirus E2 protein induces expression of the matrix metalloproteinase-9 via the extracellular signal-regulated kinase/activator protein-1 signaling pathway. Cancer Res. 2005;65:11613–11621. doi: 10.1158/0008-5472.CAN-05-2672. [DOI] [PubMed] [Google Scholar]
- 47.Zobel T, Iftner T, Stubenrauch F. The papillomavirus E8-E2C protein represses DNA replication from extrachromosomal origins. Mol Cell Biol. 2003;23:8352–8362. doi: 10.1128/MCB.23.22.8352-8362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Probst S, Notz E, Wolff M, Buehlmann J, Stubenrauch F, Iftner T. A recombinant cottontail rabbit papillomavirus genome for ectopic expression of genes in cells infected with virus in vivo. J Virol Methods. 2013;187:110–3. doi: 10.1016/j.jviromet.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 50.Xiao W, Brandsma JL. High efficiency, long-term clinical expression of cottontail rabbit papillomavirus (CRPV) DNA in rabbit skin following particle-mediated DNA transfer. Nucleic Acids Res. 1996;24:2620–2622. doi: 10.1093/nar/24.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeckel S, Loetzsch E, Huber E, Stubenrauch F, Iftner T. Identification of the E9/E2C cDNA and functional characterization of the gene product reveal a new repressor of transcription and replication in cottontail rabbit papillomavirus. J Virol. 2003;77:8736–8744. doi: 10.1128/JVI.77.16.8736-8744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stubenrauch F, Zobel T, Iftner T. The E8 domain confers a novel long-distance transcriptional repression activity on the E8E2C protein of high-risk human papillomavirus type 31. J Virol. 2001;75:4139–4149. doi: 10.1128/JVI.75.9.4139-4149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganzenmueller T, Matthaei M, Muench P, Scheible M, Iftner A, Hiller T, Leiprecht N, Probst S, Stubenrauch F, Iftner T. The E7 protein of the cottontail rabbit papillomavirus immortalizes normal rabbit keratinocytes and reduces pRb levels, while E6 cooperates in immortalization but neither degrades p53 nor binds E6AP. Virology. 2008;372:313–324. doi: 10.1016/j.virol.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Schittek B, Psenner K, Sauer B, Meier F, Iftner T, Garbe C. The increased expression of Y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. Int J Cancer. 2007;120:2110–2118. doi: 10.1002/ijc.22512. [DOI] [PubMed] [Google Scholar]
- 56.Braithwaite AW, Del Sal G, Lu X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006;13:984–993. doi: 10.1038/sj.cdd.4401924. [DOI] [PubMed] [Google Scholar]
- 57.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ 3rd. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen NN, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oda Y, Sakamoto A, Shinohara N, Ohga T, Uchiumi T, Kohno K, Tsuneyoshi M, Kuwano M, Iwamoto Y. Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human osteosarcoma. Clin Cancer Res. 1998;4:2273–2277. [PubMed] [Google Scholar]
- 60.Dahl E, En-Nia A, Wiesmann F, Krings R, Djudjaj S, Breuer E, Fuchs T, Wild PJ, Hartmann A, Dunn SE, Mertens PR. Nuclear detection of Y-box protein-1 (YB-1) closely associates with progesterone receptor negativity and is a strong adverse survival factor in human breast cancer. BMC Cancer. 2009;9:410. doi: 10.1186/1471-2407-9-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyogotani A, Ito K, Yoshida K, Izumi H, Kohno K, Amano J. Association of nuclear YB-1 localization with lung resistance-related protein and epidermal growth factor receptor expression in lung cancer. Clin Lung Cancer. 2012;13:375–384. doi: 10.1016/j.cllc.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Spitkovsky DD, Royer-Pokora B, Delius H, Kisseljov F, Jenkins NA, Gilbert DJ, Copeland NG, Royer HD. Tissue restricted expression and chromosomal localization of the YB-1 gene encoding a 42 kD nuclear CCAAT binding protein. Nucleic Acids Res. 1992;20:797–803. doi: 10.1093/nar/20.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 64.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 66.McBride AA, Jang MK. Current understanding of the role of the brd4 protein in the papillomavirus lifecycle. Viruses. 2013;5:1374–1394. doi: 10.3390/v5061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J Biol Chem. 2003;278:43470–43479. doi: 10.1074/jbc.M302339200. [DOI] [PubMed] [Google Scholar]
- 69.Peng YC, Breiding DE, Sverdrup F, Richard J, Androphy EJ. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J Virol. 2000;74:5872–5879. doi: 10.1128/jvi.74.13.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kruppel U, Muller-Schiffmann A, Baldus SE, Smola-Hess S, Steger G. E2 and the co-activator p300 can cooperate in activation of the human papillomavirus type 16 early promoter. Virology. 2008;377:151–159. doi: 10.1016/j.virol.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 71.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]


