Abstract
Loss-of-heterozygosity (LOH) analysis of archival tumor tissue can aid in determining the clinical significance of BRCA variants. Here we describe an approach for assessing LOH in formalin fixed paraffin embedded (FFPE) tissues using variant specific probes and droplet digital PCR (ddPCR). We evaluated LOH in two related breast cancer patients harboring a rare missense BRCA2 variant of unknown clinical significance (c.6966G>T; M2322I). Conventional PCR followed by Sanger sequencing suggested a change in allelic abundance in the FFPE specimens. However, we found no evidence of LOH as determined by the allelic ratio (wild type:variant) for BRCA2 in both patients' archival tumor specimens and adjacent normal control tissues using ddPCR. In summary, these experiments demonstrate the utility of ddPCR to quickly and accurately assess LOH in archival FFPE tumor tissue.
Keywords: droplet digital PCR, LOH, familial breast cancer, BRCA2, VUS
Introduction
The breast and ovarian cancer susceptibility genes, BRCA1 and BRCA2, lead to a significantly increased risk of developing breast and ovarian cancer for carriers [1]. Consequently, at risk individuals often pursue intensive screening and/or prophylactic treatments, such as oophorectomy and/or mastectomy, as these procedures have been shown to significantly reduce cancer incidence [2,3]. Unfortunately, many studies over the past two decades have demonstrated that both BRCA1 and BRCA2 are highly polymorphic genes, with many variants of unclear clinical significance (VUS) existing within the human population [4]. As a result, BRCA variant classification studies remain ongoing. Loss-of-heterozygosity (LOH) analysis of resected tumor tissue has been tremendously helpful for classifying particular BRCA alleles not clearly linked to disease [5–8]. Yet, there exist several technical hurdles that hinder archival tissue-based studies: namely, poor tumor DNA quality, normal DNA contamination within tumor DNA preparations and inherent shortcomings to the conventional methods used for studying LOH.
In particular, formalin fixation leads to a high degree of tissue damage, yielding a limited amount of usable DNA molecules for downstream studies. Moreover, contaminating normal DNA present within formalin fixed paraffin embedded (FFPE) tumor DNA preparations can hinder many genetic analyses. Additionally, fluorescence in situ hybridization (FISH) has intrinsic shortcomings that make analysis of archival tissue challenging. Specifically, FISH is subjective, prone to inter-observer variability, has limited sensitivity, can be technically difficult, time consuming, and cannot identify certain genomic alterations, such as loss with duplication [9]. To circumvent some of these challenges and facilitate future LOH studies, we used droplet digital PCR (ddPCR) to study genomic DNA derived from archival tumor tissues from two members of a cancer prone family harboring a BRCA2 VUS.
Case Study
Combined Clinical History
A 46-year-old woman of non-Ashkenazi ancestry and a family history of cancer, presented to the clinic for genetic counseling and testing for hereditary breast and ovarian cancer syndromes. Three years prior to her presentation, the proband was diagnosed with a grade II invasive ductal carcinoma (IDC) of the right breast with ductal carcinoma in situ (DCIS). The proband's tumor was found to co-express estrogen and progesterone receptors (ER+/PR+), and did not over-express the human epidermal growth factor receptor 2 (HER2-). The proband's mother was also diagnosed with a grade I IDC of the left breast with DCIS at the age of 76. Following a consultation with the proband, she and her mother were encouraged to pursue germline testing for mutations/alterations of the BRCA1 and BRCA2 genes. Genetic testing for the proband and her mother revealed the presence of a BRCA2 variant of uncertain clinical significance (c.6966G>T; M2322I). Patients were consented and enrolled in an IRB approved protocol at Johns Hopkins that allows for obtaining tissues and bodily fluids in a prospective fashion from breast cancer patients.
Tissue Processing, Nucleic Acid Preparation & Sanger Sequencing
Buccal and FFPE derived genomic DNA (gDNA) was isolated using QIAamp® DNA Blood Mini and QIAamp® FFPE tissue kits, respectively (Qiagen, Valencia, CA). Genomic DNA was isolated from FFPE tissue stored from 8 to 24 months duration using standard protocols. Briefly, H&E stained histology slides were examined by the study pathologist (P.A.) to identify areas of at least 70% breast carcinoma cells and adjacent benign lobular epithelial cells with 0% tumor cells greater than 10mm from any invasive component, hereafter termed FFPE tumor and FFPE normal, respectively. Five-micron thick unstained slides were deparaffinized and identified regions of interest were macrodissected using the Zymo pen and Pinpoint solution (Zymo Research, Irvine, CA), per the manufacturer's protocol. Sanger sequencing of gDNA was carried out following PCR amplification of respective loci using Phusion® High-Fidelity DNA Polymerase (New England BioLabs, Ipswich MA). PCR and nested sequencing primers for each locus described herein are listed in supplemental Table S1.
Droplet Digital PCR Experiments
All droplet digital PCR experiments were carried out using the QX100™ Droplet Digital PCR System according to the manufacturer's protocols (Bio-Rad, Hercules, CA), as previously described [10]. Droplet Digital PCR primers were purchased from Integrated DNA Technologies (Coralville, IA) and fluorescently labeled TaqMan® probes were purchased through Life Technologies (Carlsbad, CA). Primer and probe sequences are shown in supplemental Table S2. Prior to analysis of patient DNA samples, optimized thermo-cycling conditions were determined for allele specific binding of the fluorescent probes by initially cloning 381 base pairs of genomic sequence encompassing BRCA2 exon 13 (both the wild-type and VUS alleles) with flanking intronic sequence into a high-copy bacterial plasmid. The temperature range of 56-58°C provided strong fluorescent signals and yielded no probe cross-reactivity. Therefore, we carried out the manufacturers' recommended thermo-cycling protocol with a 58°C annealing/extension step. Supplemental Figure S1 shows experimental data from a representative ddPCR titration experiment demonstrating 100% probe specificity using linear plasmid DNA as PCR template. All data analysis was performed using the accompanying platform software, QuantaSoft (Bio-Rad), with all reported values calculated using Poisson statistics.
Targeted Next-Generation DNA Sequencing
The proband's archival tissue derived genomic DNA (both tumor and surrounding normal) was subjected to targeted next-generation DNA sequencing using a custom 484 cancer gene panel. Targeted coding exon enrichment was carried out using Agilent's SureSelect technology and DNA sequencing was performed on an Illumina HiSeq 2000 platform. Resultant data was aligned to human genome 19 using BWA 0.6.1 [11] and variants were called using GATK 1.4 [12].
Results
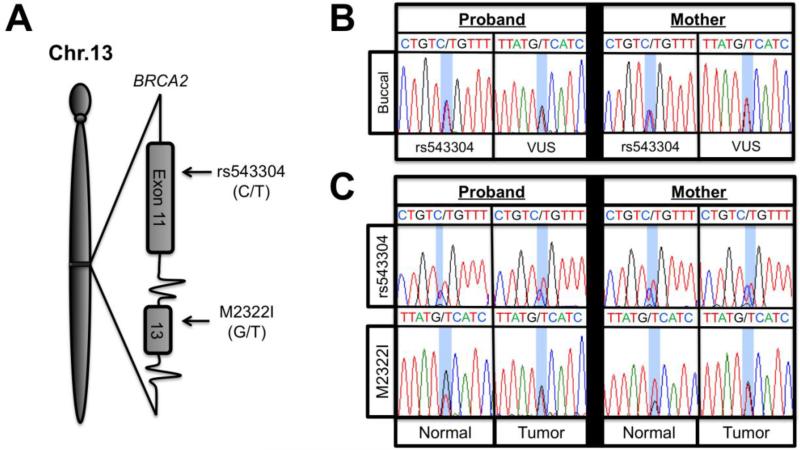
To confirm the presence of the BRCA2M2322I (c.6966G>T) variant in the two affected family members, we sequenced exon 13 of BRCA2 from buccal cell derived gDNA. Consistent with previous genetic testing, both individuals diagnosed with cancer harbored the VUS, as shown in Figure 1B. In addition, we identified the presence of a heterozygous single nucleotide polymorphism (SNP) located within exon 11 of BRCA2, rs543304, for the proband and her mother (Figure 1B). The rs543304 SNP is located in exon 11 approximately 8.7 kilobases upstream on gDNA from the VUS, as illustrated in Figure 1A. Next, we purified gDNA from FFPE tissue blocks for the proband and her mother to perform LOH and sequencing analysis on macrodissected tumor and adjacent normal breast tissue. Using Sanger sequencing we found that both the proband and her mother were heterozygous for the rs543304 SNP (Figure 1C). However, as seen in the sequencing traces in Figure 1C, there was a high degree of variability in Sanger sequencing results for the VUS locus. Both tumor samples appeared to have a 1:1 ratio of wild type to VUS alleles. In contrast, the proband's normal adjacent tissue, appeared to have a 2:1 wild type to VUS allelic ratio, while the mother's normal tissue seemingly contained a 1:2 wild type to VUS ratio. In our hands, this type of variability in PCR sequencing is common with the use of FFPE derived DNA, particularly when using minute quantities of DNA. In order to confirm that our tumor and normal samples were not inadvertently switched, since allelic ratios are more often changed in tumors, we verified that the DNA used for these analyses were indeed from cancerous tissues by identifying somatic mutations. First, we sequenced exons 9 and 20 of PIK3CA and all coding exons of TP53, as these genes are frequently mutated in breast cancers [13]. Initial sequencing results revealed a heterozygous somatic mutation in exon 20 of PIK3CA (c.3140A>T; H1047L) in the mother's tumor gDNA, as shown in supplemental Figure S2. The proband's DNA preparations exhibited no PIK3CA mutations and neither patient demonstrated a TP53 mutation by Sanger sequencing. Since no somatic mutations were identified for the proband for PIK3CA or TP53, we used the tumor and adjacent normal genomic DNA for targeted next generation sequencing with a custom 484 cancer gene panel. Somatic alterations in the proband's tumor were identified and are shown in the Supplemental Information.
Figure 1.
A) Diagrammatic representation of BRCA2 exons 11 and 13 showing the rs543304 SNP and c.6966G>T VUS, respectively. B) Sanger sequencing traces for both patient' buccal cell derived gDNA. C) BRCA2 Sanger sequencing traces for both patients' FFPE derived gDNA.
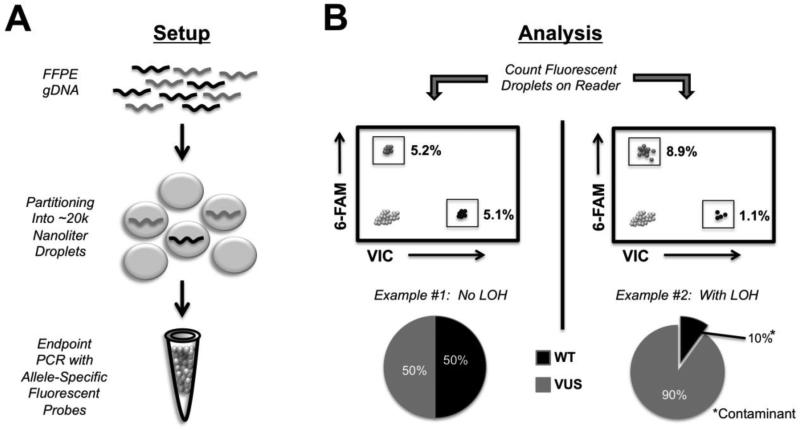
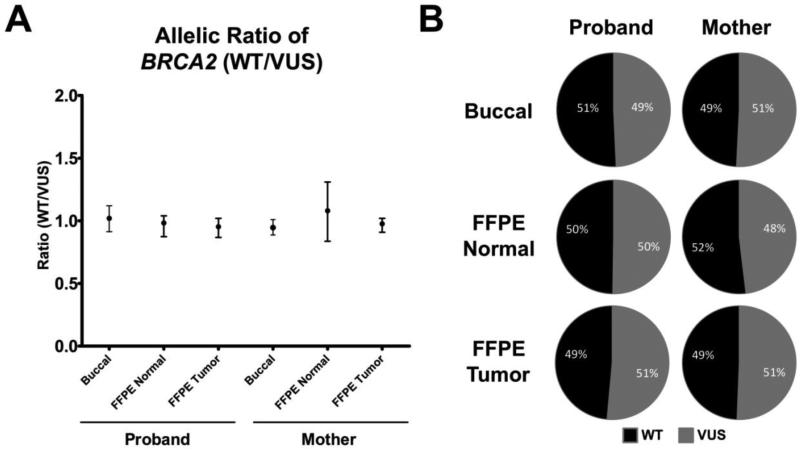
Our results with PCR and Sanger sequencing underscored the difficulty of determining allelic ratios and LOH using FFPE derived DNA. Therefore, to more accurately measure the BRCA2 allelic ratios, we then tested the relatively new technology of ddPCR to perform LOH analyses. Briefly, ddPCR is carried out through the creation of nanoliter droplets acting as parallel single molecule PCR reactions (Figure 2A) [10]. The identity of the DNA molecules within each droplet can then be resolved using conventional dual labeled fluorescent oligonucleotide probes. Following end-point PCR on a standard thermo-cycler, all droplets are then counted using a fluorescent droplet reader (Figure 2B). We reasoned that ddPCR would be well suited for LOH analysis of FFPE derived DNA for a number of reasons. First, it requires low amounts of input DNA with very short PCR amplicon sizes (<100 bp). Second, ddPCR uses dual labeled probes that are capable of reliably discriminating between single base pair changes, such as SNPs and mutations. Third, ddPCR has the capacity to easily and accurately determine differences in copy number below 2 fold, which is the lower limit of reliable detection using conventional quantitative real time PCR (qPCR). Therefore, we used this approach to effectively count the number of BRCA2 alleles present within our patients' DNA samples using allele specific probes. By comparing the number of BRCA2 alleles present within our patients' samples we reasoned we could simultaneously assess LOH and quantify contaminating normal DNA, as illustrated in Figure 2B. Using conventional allele-specific fluorescent probes, differing by only one nucleotide at BRCA2 c.6966G/T, we quantified the BRCA2 allelic ratios within the normal and tumor FFPE gDNA for both individuals. As shown in Figure 3A, we determined the allelic ratios (wild type:VUS) to be approximately one in all tissue samples assayed. Absolute percentages over multiple experiments, corresponding to the percentage wild type DNA containing and VUS DNA containing droplets, are shown in Figure 3B. These data clearly show no evidence of LOH in either individual's tumor or normal sample, and importantly underscore the inability of conventional PCR sequencing to accurately determine allelic ratios using FFPE gDNA.
Figure 2.
Schematic of ddPCR workflow for assessing LOH. A) The ddPCR reaction involves combining fragmented DNA to be studied with droplet oil and PCR reagents (PCR master mix, primers and probes). The droplet generator machine partitions the required reagents and DNA fragments into approximately 2 × 104 individual droplets per well. Following droplet generation, end-point PCR is carried out on a standard thermo-cycler. B) After PCR amplification and generation of unquenched fluorophores, the droplets are read on a droplet reader. Example #1 illustrates the expected distribution of fluorescent droplets for no LOH, where 50% of the gated droplets are positive for the wild-type allele and 50% are positive for the VUS. Example #2 illustrates the approximate distribution of fluorescent droplets for LOH (with loss of the wild-type allele) with contaminating wild type from surrounding normal tissue.
Figure 3.
LOH analysis for the proband and her mother. A) Plot showing the allelic ratio (Wild type:VUS) measured for the buccal and FFPE DNA patient samples with standard error shown using Poisson statistics. B) Average percentage of fluorescent droplets corresponding to the VUS and wild type alleles for all patient samples.
Discussion
LOH information, in combination with other methods, is useful in classifying uncertain BRCA variants [5,8]. Although loss of the wildtype allele in tumor tissues provides strong evidence for a deleterious germline mutation, it should be noted that absence of LOH cannot be used as a definitive criterion, since other mechanisms of inactivation of the wildtype allele exist. This would include epigenetic silencing as well as a de novo inactivating mutation within the wildtype allele. Nonetheless, given that loss of the second allele is an accepted common mechanism of carcinogenesis mediated by tumor suppressor genes in familial cancer syndromes, the studies described here have great potential for definitively assessing LOH and helping to ascribe a functional significance to germline VUS and deleterious mutations.
The arrival of commercially available digital PCR platforms has the potential to dramatically alter the speed and sensitivity of many molecular biology assays commonly used in the clinic. Others have shown that ddPCR can accurately determine both copy number changes and expression of critical oncogenes using patient samples [14,15]. This approach may prove to be a more objective and accurate measure for determining oncogene amplification and overexpression in cancer patients. Droplet digital PCR is especially useful for analysis of FFPE derived gDNA, since it relies on short DNA fragments and FFPE gDNA is generally highly fragmented. Moreover, since ddPCR queries individual DNA molecules, this approach should prove useful in several assays including determining the fractional abundance of somatic mutations within heterogeneous tumor tissue specimens, validation of somatic mutations identified using whole genome sequencing that are below the limit of detection by Sanger sequencing, and as demonstrated here, determining the allelic fraction of a given gene for LOH analysis. Although in principle, next generation sequencing using a targeted gene capture approach could also be used for LOH and copy number analysis, ddPCR offers significant advantages including a much faster and cost efficient platform, need for less input DNA, superior sensitivity, no artifactual errors inherent with next generation sequencing, and no requisite for a bioinformatics pipeline for variant calling. To our knowledge, this is the first report demonstrating the utility of using ddPCR for accurately assessing allelic ratios via SNPs in FFPE tumor samples. This approach should greatly facilitate not only BRCA VUS LOH studies, but studies for other variants of genes implicated in hereditary cancer susceptibility.
Supplementary Material
Acknowledgements
We would like to thank the patients described in this study for their graceful participation. We would also like to thank the Sandy Garcia Charitable Foundation. R.L.C. and S.C. are supported by National Institutes of Health Pre-Doctoral Service Awards (CA168180 and CA167939, respectively). B.H.P. acknowledges support from the Breast Cancer Research Foundation, the Commonwealth Foundation, the Santa Fe Foundation, the NIH/National Cancer Institute, and the Avon Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–76. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 2.Rebbeck TR, Friebel T, Lynch HT, Neuhausen SL, van 't Veer L, Garber JE, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22:1055–62. doi: 10.1200/JCO.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 3.Rebbeck T, Lynch H, Neuhausen S, Narod S, Veer L, Garber J, et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N Engl J Med. 2002;346:1616–22. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 4.Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–83. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, et al. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–27. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 6.Osorio A, de la Hoya M, Rodríguez-López R, Martínez-Ramírez A, Cazorla A, Granizo JJ, et al. Loss of heterozygosity analysis at the BRCA loci in tumor samples from patients with familial breast cancer. Int J Cancer. 2002;99:305–9. doi: 10.1002/ijc.10337. [DOI] [PubMed] [Google Scholar]
- 7.Osorio A, Milne R, Honrado E, Barroso A, Diez O, Salazar R, et al. Classification of missense variants of unknown significance in BRCA1 based on clinical and tumor information. Hum Mutat. 2007;28:477–85. doi: 10.1002/humu.20470. [DOI] [PubMed] [Google Scholar]
- 8.Spearman AD, Sweet K, Zhou X-P, McLennan J, Couch FJ, Toland AE. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J Clin Oncol. 2008;26:5393–400. doi: 10.1200/JCO.2008.17.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnan S, Mohseni M, Konishi Y, Tamaki A, Hosokawa Y, Park BH, et al. Controversial BRCA1 allelotypes in commonly used breast cancer cell lines. Breast Cancer Res Treat. 2010;119:249–51. doi: 10.1007/s10549-009-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson B, Ness K, Masquelier D, Belgrader P, Heredia N, Makarewicz A, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachman K, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA Gene is Mutated with High Frequency in Human Breast Cancers. Cancer Biol Ther. 2004;3:772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 14.Belgrader P, Tanner SC, Regan JF, Koehler R, Hindson BJ, Brown AS. Droplet Digital PCR Measurement of HER2 Copy Number Alteration in Formalin-Fixed Paraffin-Embedded Breast Carcinoma Tissue. Clin Chem. 2013;000:1–4. doi: 10.1373/clinchem.2012.197855. [DOI] [PubMed] [Google Scholar]
- 15.Heredia NJ, Belgrader P, Wang S, Koehler R, Regan J, Cosman AM, et al. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59:S20–3. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.