Abstract
Background & Aims
A genome wide association study and multiple pharmacogenetic studies have implicated the hepatic uptake transporter organic anion transporting polypeptide-1B1 (OATP1B1) in the pharmacokinetics and musculoskeletal toxicity of statin drugs. Other OATP uptake transporters can participate in the transport of pravastatin, partially compensating for the loss of OATP1B1 in patients carrying the polymorphism. Nonalcoholic steatohepatitis (NASH) in humans and in a diet-induced rodent model alter the expression of multiple OATP transporters.
Methods
To determine how genetic alteration in one Oatp transporter can interact with NASH-associated changes in Oatp expression we measured the disposition of intravenously administered pravastatin in Slco1b2 knockout (Slco1b2−/−) and wild-type (WT) mice fed either a control or a methionine and choline deficient (MCD) diet to induce NASH.
Results
Genetic loss of Oatp1b2, the rodent ortholog of human OATP1B transporters, caused a modest increase in pravastatin plasma concentrations in mice with healthy livers. Although a diet-induced model of NASH decreased the expression of multiple hepatic Oatp transporters, it did not alter the disposition of pravastatin compared to WT control mice. In contrast, the combination of NASH-associated decrease in compensatory Oatp transporters and Oatp1b2 genetic loss caused a synergistic increase in plasma area under the curve (AUC) and tissue concentrations in kidney and muscle.
Conclusions
Our data show that NASH alters the expression of multiple hepatic uptake transporters which, due to overlapping substrate specificity among the OATP transporters, may combine with the pharmacogenetic loss of OATP1B1 to increase the risk of statin-induced adverse drug reactions.
Keywords: Non-alcoholic steatohepatitis, statin, myopathy, adverse drug reaction
Introduction
Multiple biological factors can impact drug disposition and the occurrence of adverse drug reactions (ADRs), including pharmacogenetic variation in transporters and liver diseases [1-3]. Single nucleotide polymorphisms (SNPs) in SLCO1B1, the gene encoding human hepatic organic anion transporting polypeptide-1B1 (OATP1B1), have been shown to cause altered transporter function and altered disposition of multiple statin drugs, including pravastatin [2,4,5]. Statin drugs are among the most widely prescribed drugs worldwide and are primarily used to reduce hyperlipidemia and the risk of heart disease and stroke [6,7]. Although statins are relatively safe with few side effects, it is believed that these drugs are underutilized, partially due to the occurrence of statin-induced muscle toxicities [6,7]. The Prediction of Muscular Risk in Observational conditions (PRIMO) study showed that muscular symptoms were reported in 5.1%-18.2% of patients taking high-dosage statins, with 10.9% of patients taking pravastatin reporting symptoms of muscle toxicity [6]. Importantly, statin-induced muscle toxicities are dose-dependent and are related to plasma drug concentrations [4,7-10]. Pharmacogenetic variation in statin disposition has gained considerable attention as a potential risk factor for statin-induced muscle toxicities and recent studies have linked SNPs in SLCO1B1 to increased statin plasma concentrations and statin-induced muscle toxicities [9-11]. In addition to pharmacogenetic variation there are other variables, such as disease-specific alterations in drug transporters that can influence drug disposition and occurrence of ADRs.
It is well recognized that liver diseases can alter drug disposition and require dose adjustment to maintain drug concentrations within the therapeutic window [1,12]. Our group has shown that nonalcoholic steatohepatitis (NASH), a progressive form of nonalcoholic fatty liver disease (NAFLD), causes altered drug transporter function and contributes to altered drug disposition [3,13-16]. We have also shown that diet-induced NASH in rodents decreases the expression of multiple hepatic Oatp uptake transporters and increases the plasma concentrations of sulfobromophthalein [15]. There is a high degree of overlap in substrate specificity between the various OATP isoforms, and the coordinated down-regulation of numerous uptake transporters in NASH may reduce the potential for compensatory transport. We hypothesized that the combination of genetic- and disease-specific changes in drug transporters will synergistically alter the disposition of the OATP substrate pravastatin.
We utilized a genetic knockout model of Oatp1b2, the primary rodent uptake transporter for pravastatin, and a methionine and choline deficient (MCD) diet-induced model of NASH to test the impact of genetics (Oatp1b2 knockout), disease (NASH), and the combined gene-by-environment effect on pravastatin disposition.
Materials and Methods
Animals
Male C57Bl6 mice from Jackson Laboratory (Bar Harbor, ME) and Slco1b2−/− mice from Dr. Curtis Klaassen (University of Kansas Medical Center) at 5 months of age were housed on a 12 h light and 12 h dark cycle in the University of Arizona animal care facility. Mice were provided either a methionine and choline deficient (MCD) diet or a control diet replete with methionine and choline (control) from Dyets Inc. (Bethlehem, PA) ad libitum for six weeks. Animals were anesthetized and surgery performed to place cannulas into the carotid artery and jugular vein for pravastatin disposition studies as previously described [17]. Pravastatin (Sigma Aldrich, St. Louis, MO) was solubilized in sterile normal saline, and administered into the jugular vein (10 mg/kg, 5 ml/kg). Blood was collected from the carotid artery 5, 10, 25, and 40 min after pravastatin administration into heparinized tubes and plasma was isolated by centrifugation. After the 40 min blood collection the mice were euthanized and liver, kidney, gastrocnemius, and soleus tissues were collected. A slice of liver and kidney tissues were fixed in formalin and embedded in paraffin for histological analysis. Tissues were stained with hematoxylin and eosin and scored by a trained veterinary pathologist for lipid accumulation, necrosis, inflammation, fibrosis, and biliary hyperplasia. Pathology scores were as follows: 0, no significant lesions (0%); 1, minimal (<10%); 2, mild (10-25%); 3, moderate (25-40%); 4, marked (40-50%); 5, severe (>50%). The remaining tissues were snap frozen for mRNA or protein isolation or pravastatin quantification. The animal studies were approved by the University of Arizona Animal Care and Use Committee.
Human liver samples
Human liver tissue was acquired from the National Institutes of Health-funded Liver Tissue Cell Distribution System (LTCDS) which was funded by NIH Contract #N01-DK-7-0004/HHSN267200700004C. Clinical and demographic information of these human liver samples has been described previously [18]. Tissues were collected postmortem and preserved as either frozen or paraffin embedded tissue. The samples were diagnosed as normal, steatotic, and NASH by a Liver Tissue Cell Distribution System medical pathologist according to the NAFLD activity scoring categorization [19].
mRNA analysis
Total RNA was isolated from mouse livers using RNAzol B reagent from Tel-Test Inc. (Friendswood, TX) according to the manufacturer’s protocol. The branched DNA (bDNA) assay was used to quantify mRNA transcripts using gene specific oligonucleotide probes for Oatp1a1, Oatp1a4, Oatp1b2, Mrp2, Mrp3, Mrp4, OATP1B1, and OATP1B3. A QuantiGene 1.0 Discovery Assay Kit from Affymetrix Inc. (Santa Clara, CA) was used according to the manufacturer’s protocol and as previously described [20]. Luminescence was measured using a Quantiplex 320 bDNA luminometer interfaced with Quantiplex Data Management Software (version 5.02). OATP2B1 mRNA expression was quantified from a previously validated microarray dataset [13].
Western blot analysis
Whole cell protein lysates were prepared from mouse livers as previously described [16]. Portions of the whole cell lysates were subjected to ultracentrifugation at 100,000 × g for one h to collect a membrane enriched fraction. Sixty micrograms of whole cell lysate or 30 μg of membrane preparation were separated on 7.5% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes. The following antibodies were used for each protein: Oatp1a1, Santa Cruz Biotechnology (Santa Cruz, CA) sc-47265; Oatp1a4, sc-18436; Oatp1b2, sc-47270; Mrp2, M2III-5 clone Kamiya Biomedical Company (Seattle, WA) MC-267; Mrp3, sc-5775; Mrp4, M4I-10 clone generated by George L. Scheffer (Amsterdam, The Netherlands); OATP1B1, Progen Biotech (Heidelberg, Germany); OATP1B3, gift from Dr. Bruno Hagenbuch (University of Kansas Medical Center); Oatp2b1/OATP2B1 sc-135099. Relative protein levels were measured using Image J software from the National Institutes of Health (Bethesda, MD) and each protein was normalized to either Erk-2 (sc-154) or pan-cadherin (Abcam, Ab16505).
Pravastatin quantification
The methods for pravastatin quantification in mouse plasma, liver, kidney, gastrocnemius, soleus, and urine were adapted from previously published methods [21]. Pravastatin (catalog #P4498) and 3-α-isopravastatin (catalog #H952310) were purchased from Sigma Aldrich and Toronto Research Chemicals (Ontario, Canada), respectively. A deuterium labeled internal standard pravastatin-d3 (catalog #P702002) was purchased from Toronto Research Chemicals. A Waters (Milford, MA) Micromass Quattro Premier XE tandem mass spectrometer coupled to an Acquity UPLC was used in the Arizona Laboratory for Emerging Contaminants at the University of Arizona. The mobile phase consisted of 0.1% formic acid (A) and acetonitrile (B) and was pumped at a flow rate of 0.3mL/min through a Waters Acquity UPLC BEH C19 column (1.7 μm, 2.1 × 50mm). The UPLC gradient started at 20% B and increased to 36% B over four min, then was equilibrated to 20% B for one min before the next injection. Multiple reaction monitoring in negative mode was used to detect pravastatin at m/z 423.3 > 303.3, 3-α-isopravastatin at m/z 423.3 > 303.3, and pravastatin-d3 at m/z 426.3 > 303.3. Liver, kidney, gastrocnemius, and soleus tissues were homogenized by grinding the tissue in liquid nitrogen (~100mg for liver, kidney, and gastrocnemius, and ~10mg for soleus), followed by addition of 4% bovine serum albumin. The samples were vortexed vigorously, frozen, thawed, and then centrifuged at 13,000 rpm for 10 min. Twenty-microliters of either plasma or tissue homogenate or 30 μL of urine was mixed with 30 μL of acetonitrile that contained pravastatin-d3, vortexed, and centrifuged at 13,000 rpm for 10 min. Forty-microliters of the supernatant was mixed with 80 μL of water and 10 μL was injected onto the BEH column.
Statistical analyses
All results are represented as mean ± standard error of the mean (SEM). For all comparisons within the rodent studies Two-way ANOVA statistical analyses were employed with a Bonferroni multiple comparisons post-test to compare between control and NASH animals within each genotype. For the human OATP expression data, One-way ANOVA was employed with Dunnett’s multiple comparisons post-test to compare the Steatosis and NASH samples to control samples.
Results
Methionine and choline deficient diet induces NASH in wild-type and Slco1b2−/− mice
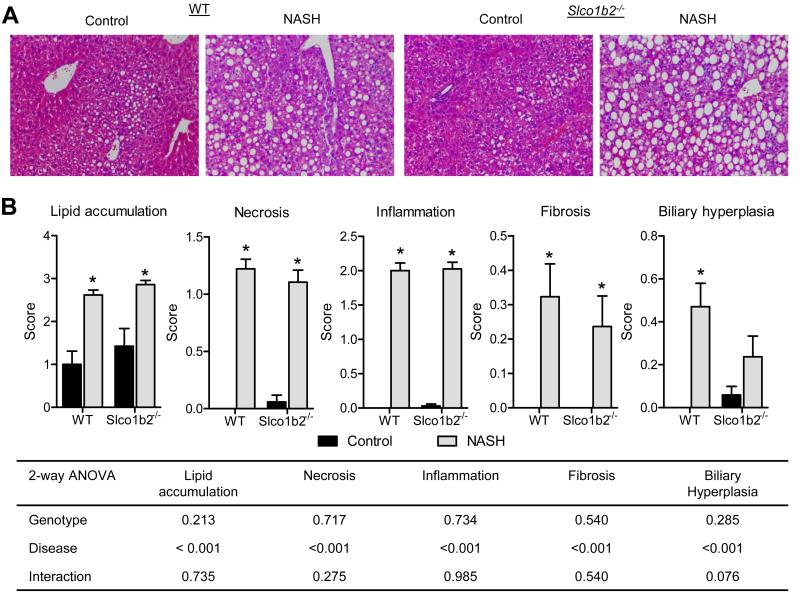
The MCD diet is a common model used to study NASH because it recapitulates many of the characteristic liver pathology features and expression patterns in drug metabolizing enzymes and transporters observed in the human condition [22]. Although this model does not reproduce the physiological steps that are observed in the human NASH progression, a recent publication from our lab shows that the MCD diet in rats and mice demonstrates similarity in transporter regulation and may be the most useful NASH model for predicting the disposition of drugs in humans, thus increasing the translatability of our data to the human condition [23]. After six weeks of MCD diet treatment, the WT and Slco1b2−/− mice exhibited the same liver pathology scores for characteristic features of NASH (Fig. 1), including increased lipid accumulation, necrosis, inflammation, fibrosis, and biliary hyperplasia.
Fig. 1. The MCD diet induces NASH pathology in WT and Slco1b2−/− mice.
Hematoxylin and Eosin stained liver sections from WT or Slco1b2−/− mice fed either a control diet or MCD diet (NASH). (B) Liver sections were scored for lipid accumulation, necrosis, inflammation, fibrosis, and biliary hyperplasia. Data represent mean ± SEM. WT control n=14, WT NASH n=17, Slco1b2−/− control n=17, Slco1b2−/− NASH n=18. Two-way ANOVA p-values for each pathology score are shown in the table. *p-value <0.05 according to a Two-way ANOVA Bonferroni multiple comparison post-test.
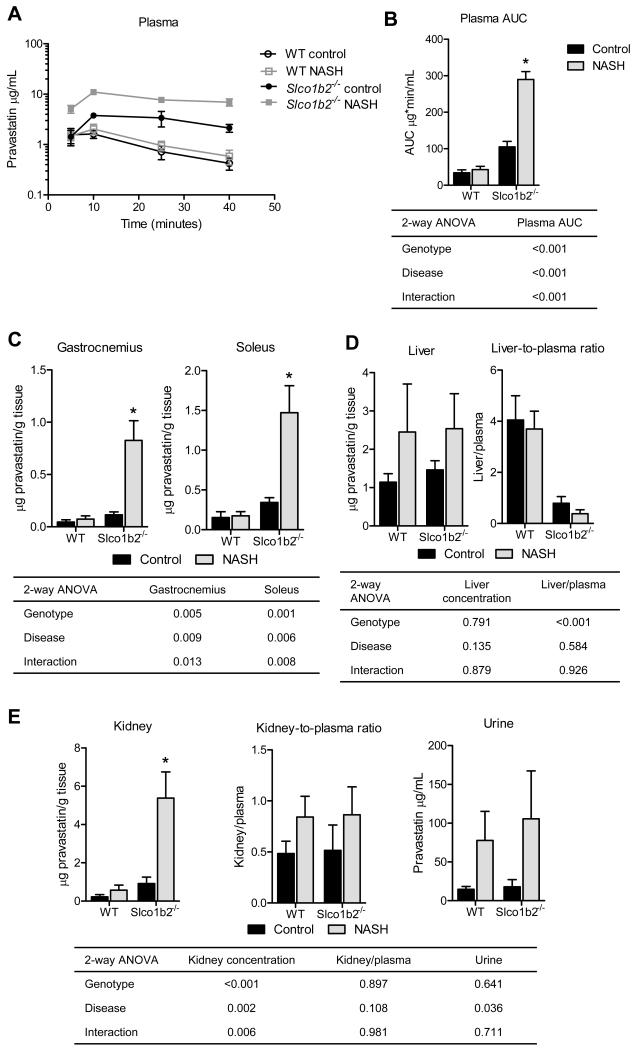
Slco1b2−/− and NASH synergistically increase the plasma concentrations of pravastatin
Diet-induced NASH in WT mice did not change the plasma concentration-time profile or the area under the curve (AUC) of pravastatin compared to the control WT mice (Fig. 2A,B). Genetic loss of Oatp1b2 in control mice resulted in a 3-fold increase in plasma AUC, confirming the central role of Oatp1b2 in pravastatin hepatic uptake and elimination (Fig. 2A,B). The combination of diet-induced Slco1b2 knockout and NASH altered the pravastatin plasma concentration-time profile and resulted in a synergistic increase (8.5-fold) in pravastatin AUC compared to WT control (Fig. 2A,B).
Fig. 2. The combination of Slco1b2−/− and diet-induced NASH dramatically increases pravastatin exposure.
(A) Pravastatin plasma concentrations. (B) Plasma area under the curve (AUC) (0-40 min). (C) Pravastatin concentrations in gastrocnemius and soleus muscles. (D) Liver pravastatin concentrations and liver to plasma ratios. (E) Kidney pravastatin concentrations and kidney to plasma ratios. Data represent mean ± SEM. WT control n=6, WT NASH n=4, Slco1b2 KO control n=4, Slco1b2 KO NASH n=4. Two-way ANOVA p-values are shown in the table. *p-value <0.05 according to a Two-way ANOVA Bonferroni multiple comparison post-test.
Slco1b2−/− and NASH synergistically increase the muscle concentrations of pravastatin
Because statin-induced muscle toxicities are reported to occur in a concentration dependent manner [24] we determined the concentrations of pravastatin in gastrocnemius and soleus muscles of mice in this study. Similar to what was observed in the plasma, we found that Slco1b2−/− or NASH alone did not cause dramatic changes in pravastatin concentrations in muscle but the combination of Slco1b2−/− and NASH resulted in a synergistic increase in pravastatin concentrations in muscle (Fig. 2C).
Oatp1b2 is an important determinant of pravastatin plasma clearance in control and NASH mice
Neither Slco1b2−/− nor NASH altered the liver concentrations of pravastatin, while the liver-to-plasma ratio was significantly decreased in Slco1b2−/− mice in both control and diet-induced NASH treatment groups (Fig. 2D). This indicates that hepatic uptake, rather than efflux, is a major determinant of pravastatin elimination, which is consistent with several previously published reports [25-27]. In contrast, kidney concentrations of pravastatin mirror the plasma concentrations, with a dramatic increase in Slco1b2−/− mice with NASH, resulting in no significant differences in the kidney-to-plasma ratios for any groups (Fig. 2E). The presence of NASH significantly increased the amount of pravastatin in the urine of WT and Slco1b2−/− mice (Fig. 2E).
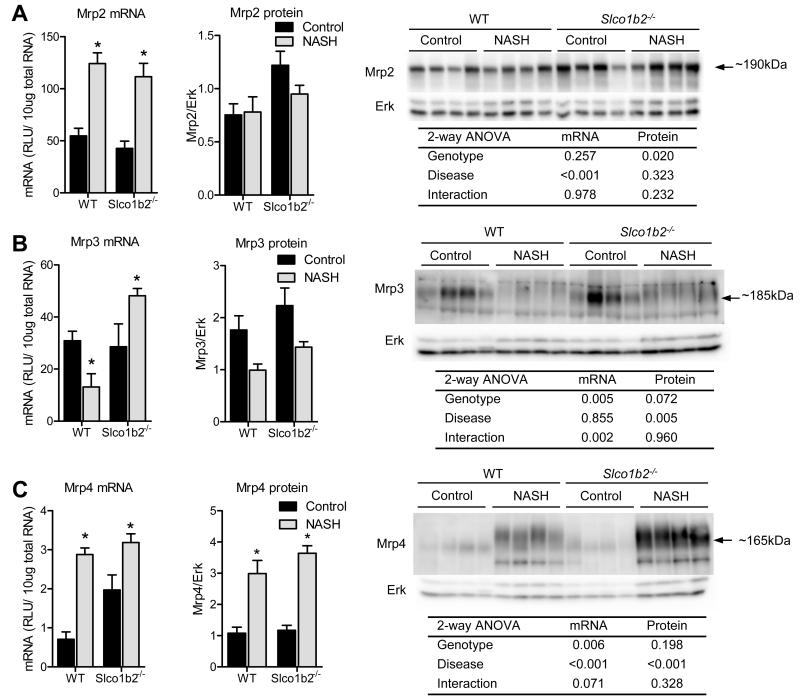
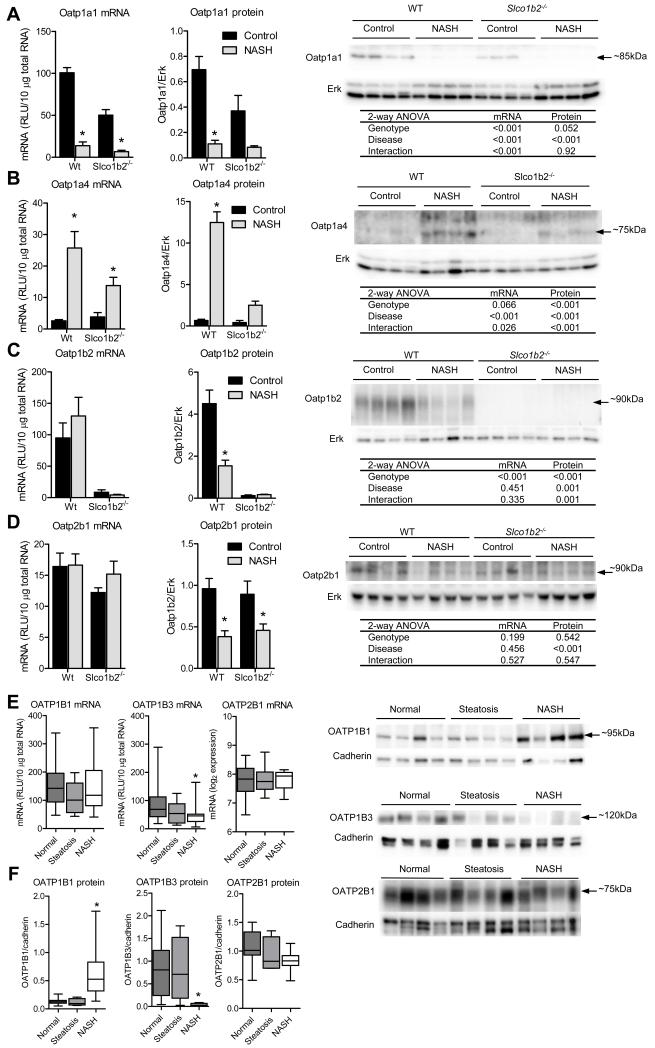
NASH causes alterations in expression of hepatic uptake and efflux transporters
To determine whether other hepatic transporters involved in pravastatin disposition contribute to the altered disposition observed in Slco1b2−/− mice with NASH, mRNA and protein expression analyses were performed. We found that diet-induced NASH caused similar alterations in Mrp efflux (Fig. 3) and Oatp uptake (Fig. 4) transporter expression in WT and Slco1b2−/− mice livers. The mRNA expression of the efflux transporters Mrp2 and Mrp4, which are localized to the canalicular and sinusoidal membranes, respectively, were increased by NASH in both genotypes (Fig. 3A,C). The protein expression of Mrp2 was slightly increased in the Slco1b2−/− mice, while Mrp4 protein expression was increased by NASH (Fig. 3A,C). mRNA for Mrp3, which is localized to the sinusoidal membrane, was decreased in WT mice with NASH but increased in Slco1b2−/− mice with NASH. At the protein level, Mrp3 was decreased in mice from both genotypes with NASH (Fig. 3B). NASH caused a significant decrease in Oatp1a1 (Fig. 4A) and an increase in Oatp1a4 (Fig. 4B) mRNA and protein expression. Interestingly, the Oatp1a4 induction observed in Slco1b2−/− NASH mice was smaller than the induction observed in WT NASH mice, potentially contributing to the altered pravastatin disposition. As expected, Oatp1b2 mRNA and protein were absent in the Slco1b2−/− mice. In WT mice with NASH, Oatp1b2 mRNA expression did not change but the protein expression decreased (Fig. 4C), suggesting a different regulation mechanism for decreased Oatp1b2 expression compared to the Oatp1a transporters studied herein. Similary, NASH did not alter the mRNA expression of Oatp2b1 but caused a significant decrease in Oatp2b1 protein expression (Fig. 4D).
Fig. 3. Diet-induced NASH causes similar changes in efflux transporter protein expression between WT and Slco1b2−/− mice.
mRNA and protein expression of Mrp2 (A), Mrp3 (B), and Mrp4 (C). Representative western blots and the Two-way ANOVA p-values are shown. Data represent mean ± SEM. WT control n=4, WT NASH n=4, Slco1b2 KO control n=4, Slco1b2 KO NASH n=4. *p-value <0.05 according to a Two-way ANOVA Bonferroni multiple comparison post-test.
Fig. 4. Diet-induced NASH and human NASH causes altered expression of OATP transporters.
mRNA and protein expression of Oatp1a1 (A), Oatp1a4 (B), Oatp1b2 (C), and Oatp2b1 (D). Representative western blots and the Two-way ANOVA p-values are shown. *indicates a p-value <0.05 according to a Two-way ANOVA Bonferroni multiple comparison post-test. Data represent mean ± SEM. WT control n=4, WT NASH n=4, Slco1b2−/− control n=4, Slco1b2−/− NASH n=4. (E) OATP1B1, OATP1B3, and OATP2B1 mRNA expression levels in normal (n=20), steatosis (n=10), and NASH (n=19) livers. (F) OATP1B1, OATP1B3, and OATP2B1 protein expression levels in normal (n=10), steatosis (n=4), and NASH (n=12) livers. Box and whisker plots represent the median and the min and max. *indicates a p-value <0.05 according to a One-way ANOVA Dunnett’s multiple comparison post-test. Representative human OATP blots are shown on the right.
To gain insight into the potential clinical implications of our preclinical data for compensatory Oatp-mediated transport, we measured the mRNA and protein expression of OATP1B1, OATP1B3, and OATP2B1 in human livers categorized as normal, steatosis, or NASH. We found that OATP1B1 expression was increased, OATP1B3 was decreased, whereas OATP2B1 was unchanged in NASH compared to normal (Fig. 4E,F).
Discussion
With the widespread and growing use of statins for a variety of maladies, including hyperlipidemia, the health and economic impacts of the statin-induced muscle toxicities is significant. For example, it has been estimated that up to 60% of elderly patients will stop taking their statin within two years of starting therapy, partially due to muscle toxicities [7]. This cessation of therapy potentially places a large number of patients at greater risk of heart disease and stroke. Understanding the true occurrence of statin-associated muscle toxicities has been challenging and is still an ongoing debate because there is an apparent discrepancy in the occurrence of statin-induced ADRs between clinical trials and observational studies or clinical experience [6,7,28,29]. One potential explanation for this discrepancy is the exclusion of risk populations from these clinical trials, such as patients with renal or hepatic insufficiency. Despite this debate, statins are widely prescribed and statin-induced muscle toxicities are a concern in the clinic, providing the impetus to identify risk factors for these toxicities.
Pharmacogenetic variation in statin metabolism and disposition has gained considerable attention as a risk factor for statin-induced muscle toxicities. Recent pharmacogenetics studies have linked SLCO1B1 SNPs to statin-induced myopathy [9-11]. The rs4149056 SNP in SLCO1B1 causes an amino acid change (V174A) that is associated with increased plasma concentrations of pravastatin [4,30-33]. In vitro transport experiments showed that impaired membrane localization of OATP1B1 was the molecular mechanism by which rs4149056 increased pravastatin plasma AUC [5]. Interestingly, the STRENGTH (Statin Response Examined by Genetic Haplotype Markers) study showed that greater than or equal to one rs4149056 allele did not increase the risk of composite adverse events for pravastatin [10]. Although rs4149056 does not appear to increase the risk of muscle-induced toxicities, multiple reports have shown that this SNP reduced pravastatin’s cholesterol lowering capacity, which is also dose-dependent. It is notable that this attenuated cholesterol lowering capacity may be modest and some conflicting results have been reported [4,8,34,35]. The lack of strong data for pravastatin-induced muscle toxicities and attenuated cholesterol lowering capacity in the context of the rs4149056 SNP have raised a cautionary flag but have not led to clear clinical guidelines. These data indicate that genetic variation in one OATP transporter (OATP1B1) has an equivocal effect on pravastatin pharmacodynamics suggesting that other factors must be contributing to the occurrence of pravastatin-induced myopathy.
The data presented here clearly indicate a synergistic interaction between NASH and genetic loss of Oatp1b2 on pravastatin disposition in mice. The mechanism we have identified behind this interaction is the ability of other fully functional hepatic Oatp transporters that can compensate for the genetic loss of one Oatp transporter.This idea is supported by the fact that there is the high degree of overlap in substrate specificity between the various OATP isoforms. In fact, Pravastatin is known to be a substrate for the main hepatic OATPs, OATP1B1, OATP1B3, and OATP2B1 [36,37]. This overlap in substrate specificity could explain the modest effect of rs4149056 on pravastatin pharmacokinetics, pravastatin-induced muscle toxicity, and cholesterol lowering capacity, because OATP1B3 could compensate for the reduced transport capacity of the polymorphic OATP1B1. This overlap becomes salient when other factors, such as disease, modulate the expression of the compensatory transporters. Our group has shown previously that human NASH causes a coordinated down-regulation of hepatic uptake transporters [13], and here we show that OATP1B3 is dramatically reduced, while OATP2B1 is unchanged in NASH. Because the rs4149056 SNP impairs membrane localization of OATP1B1, it is possible that the NASH-induced increase in its expression may not actually increase the amount of transporter at the membrane and, consequently, the amount of pravastatin transported, although this needs to be investigated. Together these data indicate a potential for reduced compensatory uptake transport by OATP1B3 when patients carry the rs4149056 SNP in OATP1B1 who also have NASH, which may place these patients at greater risk of statin-induced myopathy. This assertion for compensatory uptake transport of pravastatin by other fully functional transporters is supported by the data shown here and from previous reports in Oatp knockout mice. Similar to human hepatic uptake transporters, pravastatin is known to be transported by multiple rodent Oatps including Oatp1b2, the single rodent ortholog of the human OATP1B isoforms, and Oatp1a1, Oatp1a4, and Oatp2b1 [38,39]. Herein we show that genetic knockout of Oatp1b2 causes only a modest 2- to 3-fold increase in pravastatin plasma AUC and muscle concentrations, which is consistent with a previously published study showing that steady state subcutaneous infusion of pravastatin in Slco1b2−/− mice exhibited only a 1.8-fold increase in plasma concentrations [25]. This modest, rather than dramatic increase in pravastatin concentration is likely due to compensatory transport from the other Oatp isoforms in the liver. As further evidence for compensatory transport of pravastatin, the WT mice with diet-induced NASH exhibited decreased Oatp1b2, Oatp1a1, and Oatp2b1 protein expression but no significant changes in pravastatin exposure, due to a strong increase in Oatp1a4 protein expression. Finally, Iusuf et. al. showed a more robust 7-fold increase in plasma AUC in Slco1a/1b−/− mice, which have the entire Oatp1a/1b cluster knocked out, thus eliminating Oatp-related compensatory transport [26]. Similarly, we observed that Slco1b2−/− mice which also had NASH-associated decreased expression of Oatp1a1 and Oatp2b1 with minimal increased expression of Oatp1a4, exhibited an 8.5-fold increase in plasma AUC. Although we observed changes in hepatic Mrp efflux transporters, these changes did not affect pravastatin disposition because the changes were not different between genotypes. Furthermore, our present results, along with data from others, have shown that hepatic uptake is the rate-limiting step in pravastatin elimination [25-27]. Collectively, these data clearly indicate that the combination of genetic loss of Oatp1b2 and NASH-associated changes in other compensatory uptake transporters causes a synergistic increase in pravastatin exposure, potentially increasing the risk of the dose-dependent statin-induced muscle toxicities. Translation of our data directly into human populations is complicated by the fact that the expression patterns and othrology of rodent versus human hepatic OATPs is not straight forward. Further studies are needed in the appropriate populations (i.e. patients with SLCO1B1 SNPs and NASH) before strong conclusions can be made regarding the impact of these transporter changes in humans.
The findings from this study are highly relevant to the clinic because they raise several important cautionary points. First, NASH is only beginning to be recognized as a liver pathology that can impact drug metabolism, disposition, and the occurrence of ADRs. Because proper diagnosis of NASH still requires an invasive liver biopsy, and clinical markers of hepatic function are not consistently altered in NASH, it may be difficult to identify patients at risk. Second, although the overlap in patients harboring the rs4149056 SNP (allele frequency 14% of European Americans [3]) and NASH (present in 5% to 17% of Adults in the U.S. [40]) does not represent a large patient population, our data provide the rationale for investigating whether severe statin-induced muscle toxicities previously believed to be idiosyncratic are, in fact, a result of underlying synergistic interaction between genetics and disease. Third, and perhaps most important, there have been several small clinical trials investigating the therapeutic value of statins, including pravastatin, in NAFLD and NASH [41]. The data from our study indicate that caution should be taken as those investigations continue, because patients harboring the rs4149056 SNP may be at increased risk of statin-induced muscle toxicity. These results strengthen the case for individualized medicine based not only on pharmacogenetics but also on diagnosis of underlying disease conditions, such as NASH.
Acknowledgments
Financial support
This work was supported by The National Institute of Environmental Health Science Toxicology Training Grant [ES007091] and National Institutes of Health Grants [HD062489], [AI083927], and [ES019487].
Abbreviations
- ADR
adverse drug reaction
- Oatp
organic anion transporting polypeptide
- SNP
single nucleotide polymorphism
- NASH
nonalcoholic steatohepatitis
- NAFLD
nonalcoholic fatty liver disease
- MCD
methionine and choline deficient
- AUC
area under the curve
- Mrp
multidrug resistance-associated protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- [1].Gandhi A, Moorthy B, Ghose R. Drug disposition in pathophysiological conditions. Curr Drug Metab. 2012;13:1327–44. doi: 10.2174/138920012803341302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63:157–81. doi: 10.1124/pr.110.002857. [DOI] [PubMed] [Google Scholar]
- [3].Clarke JD, Cherrington NJ. Genetics or environment in drug transport: the case of organic anion transporting polypeptides and adverse drug reactions. Expert Opin Drug Metab Toxicol. 2012;8:349–60. doi: 10.1517/17425255.2012.656087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Igel M, Arnold KA, Niemi M, Hofmann U, Schwab M, Lütjohann D, et al. Impact of the SLCO1B1 polymorphism on the pharmacokinetics and lipid-lowering efficacy of multiple-dose pravastatin. Clin Pharmacol Ther. 2006;79:419–26. doi: 10.1016/j.clpt.2006.01.010. [DOI] [PubMed] [Google Scholar]
- [5].Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15:513–22. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- [6].Bruckert E, Hayem G, Dejager S, Yau C, Bégaud B. Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients--the PRIMO study. Cardiovasc Drugs Ther. 2005;19:403–14. doi: 10.1007/s10557-005-5686-z. [DOI] [PubMed] [Google Scholar]
- [7].Fernandez G, Spatz ES, Jablecki C, Phillips PS. Statin myopathy: a common dilemma not reflected in clinical trials. Cleve Clin J Med. 2011;78:393–403. doi: 10.3949/ccjm.78a.10073. [DOI] [PubMed] [Google Scholar]
- [8].Zhang W, Chen B-L, Ozdemir V, He Y-J, Zhou G, Peng D-D, et al. SLCO1B1 521T-->C functional genetic polymorphism and lipid-lowering efficacy of multiple-dose pravastatin in Chinese coronary heart disease patients. Br J Clin Pharmacol. 2007;64:346–52. doi: 10.1111/j.1365-2125.2007.02892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92:112–7. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J AmCollCardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Niemi M. Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther. 2010;87:130–3. doi: 10.1038/clpt.2009.197. [DOI] [PubMed] [Google Scholar]
- [12].Verbeeck RK. Pharmacokinetics and dosage adjustment in patients with hepatic dysfunction. Eur J Clin Pharmacol. 2008;64:1147–61. doi: 10.1007/s00228-008-0553-z. [DOI] [PubMed] [Google Scholar]
- [13].Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, et al. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic Fatty liver disease. Drug Metab Dispos. 2011;39:1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Merrell MD, Cherrington NJ. Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev. 2011;43:317–334. doi: 10.3109/03602532.2011.577781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fisher CD, Lickteig AJ, Augustine LM, Oude Elferink RP, Besselsen DG, Erickson RP, et al. Experimental non-alcoholic fatty liver disease results in decreased hepatic uptake transporter expression and function in rats. Eur.J.Pharmacol. 2009;613:119–127. doi: 10.1016/j.ejphar.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hardwick RN, Fisher CD, Street SM, Canet MJ, Cherrington NJ. Molecular mechanism of altered ezetimibe disposition in nonalcoholic steatohepatitis. Drug Metab Dispos. 2012;40:450–460. doi: 10.1124/dmd.111.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hardwick RN, Cherrington NJ. Measuring altered disposition of xenobiotics in experimental models of liver disease. Curr Protoc Toxicol. 2012 doi: 10.1002/0471140856.tx2301s52. Chapter 23:Unit 23.1. [DOI] [PubMed] [Google Scholar]
- [18].Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, et al. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos. 2009;37:2087–94. doi: 10.1124/dmd.109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kleiner DE, Brunt EM, Van NM, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- [20].Hardwick RN, Fisher CD, Canet MJ, Lake AD, Cherrington NJ. Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2010;38:2293–2301. doi: 10.1124/dmd.110.035006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sparidans RW, Iusuf D, Schinkel AH, Schellens JH, Beijnen JH. Liquid chromatography-tandem mass spectrometric assay for pravastatin and two isomeric metabolites in mouse plasma and tissue homogenates. J ChromatogrB Anal Sci. 2010;878:2751–2759. doi: 10.1016/j.jchromb.2010.08.015. [DOI] [PubMed] [Google Scholar]
- [22].Larter CZ, Yeh MM. Animal models of NASH: getting both pathology and metabolic context right. J GastroenterolHepatol. 2008;23:1635–1648. doi: 10.1111/j.1440-1746.2008.05543.x. [DOI] [PubMed] [Google Scholar]
- [23].Canet MJ, Hardwick RN, Lake AD, Dzierlenga AL, Clarke JD, Cherrington NJ. Modeling Human Nonalcoholic Steatohepatitis-Associated Changes in Drug Transporter Expression Using Experimental Rodent Models. Drug Metab Dispos. 2014 doi: 10.1124/dmd.113.055996. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harper CR, Jacobson TA. Evidence-based management of statin myopathy. Curr Atheroscler Rep. 2010;12:322–30. doi: 10.1007/s11883-010-0120-9. [DOI] [PubMed] [Google Scholar]
- [25].Zaher H, Meyer zu, Schwabedissen HE, Tirona RG, Cox ML, Obert LA, Agrawal N, et al. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Mol Pharmacol. 2008;74:320–9. doi: 10.1124/mol.108.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Iusuf D, Sparidans RW, van Esch A, Hobbs M, Kenworthy KE, van de Steeg E, et al. Organic anion-transporting polypeptides 1a/1b control the hepatic uptake of pravastatin in mice. Mol Pharm. 2012;9:2497–504. doi: 10.1021/mp300108c. [DOI] [PubMed] [Google Scholar]
- [27].Varma MVS, Lai Y, Feng B, Litchfield J, Goosen TC, Bergman A. Physiologically based modeling of pravastatin transporter-mediated hepatobiliary disposition and drug-drug interactions. Pharm Res. 2012;29:2860–73. doi: 10.1007/s11095-012-0792-7. [DOI] [PubMed] [Google Scholar]
- [28].Di Stasi SL, MacLeod TD, Winters JD, Binder-Macleod SA. Effects of statins on skeletal muscle: a perspective for physical therapists. Phys Ther. 2010;90:1530–42. doi: 10.2522/ptj.20090251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McKenney JM, Davidson MH, Jacobson TA, Guyton JR. Final conclusions and recommendations of the National Lipid Association Statin Safety Assessment Task Force. Am J Cardiol. 2006;97:89C–94C. doi: 10.1016/j.amjcard.2006.02.030. [DOI] [PubMed] [Google Scholar]
- [30].Ho RH, Choi L, Lee W, Mayo G, Schwarz UI, Tirona RG, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics. 2007;17:647–56. doi: 10.1097/FPC.0b013e3280ef698f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mwinyi J, Johne A, Bauer S, Roots I, Gerloff T. Evidence for inverse effects of OATP-C (SLC21A6) 5 and 1b haplotypes on pravastatin kinetics. Clin Pharmacol Ther. 2004;75:415–21. doi: 10.1016/j.clpt.2003.12.016. [DOI] [PubMed] [Google Scholar]
- [32].Nishizato Y, Ieiri I, Suzuki H, Kimura M, Kawabata K, Hirota T, et al. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin Pharmacol Ther. 2003;73:554–65. doi: 10.1016/S0009-9236(03)00060-2. [DOI] [PubMed] [Google Scholar]
- [33].Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, et al. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1) Pharmacogenetics. 2004;14:429–440. doi: 10.1097/01.fpc.0000114750.08559.32. [DOI] [PubMed] [Google Scholar]
- [34].Tachibana-Iimori R, Tabara Y, Kusuhara H, Kohara K, Kawamoto R, Nakura J, et al. Effect of genetic polymorphism of OATP-C (SLCO1B1) on lipid-lowering response to HMG-CoA reductase inhibitors. Drug Metab Pharmacokinet. 2004;19:375–380. doi: 10.2133/dmpk.19.375. [DOI] [PubMed] [Google Scholar]
- [35].Takane H, Miyata M, Burioka N, Shigemasa C, Shimizu E, Otsubo K, et al. Pharmacogenetic determinants of variability in lipid-lowering response to pravastatin therapy. J Hum Genet. 2006;51:822–6. doi: 10.1007/s10038-006-0025-1. [DOI] [PubMed] [Google Scholar]
- [36].Seithel A, Eberl S, Singer K, Auge D, Heinkele G, Wolf NB, et al. The influence of macrolide antibiotics on the uptake of organic anions and drugs mediated by OATP1B1 and OATP1B3. Drug Metab Dispos. 2007;35:779–786. doi: 10.1124/dmd.106.014407. [DOI] [PubMed] [Google Scholar]
- [37].Nozawa T, Imai K, Nezu J-I, Tsuji A, Tamai I. Functional characterization of pH-sensitive organic anion transporting polypeptide OATP-B in human. J Pharmacol Exp Ther. 2004;308:438–45. doi: 10.1124/jpet.103.060194. [DOI] [PubMed] [Google Scholar]
- [38].Hsiang B, Zhu Y, Wang Z, Wu Y, Sasseville V, Yang WP, et al. A novel human hepatic organic anion transporting polypeptide (OATP2). Identification of a liver-specific human organic anion transporting polypeptide and identification of rat and human hydroxymethylglutaryl-CoA reductase inhibitor transporters. J Biol Chem. 1999;274:37161–8. doi: 10.1074/jbc.274.52.37161. [DOI] [PubMed] [Google Scholar]
- [39].Tokui T, Nakai D, Nakagomi R, Yawo H, Abe T, Sugiyama Y. Pravastatin, an HMG-CoA reductase inhibitor, is transported by rat organic anion transporting polypeptide, oatp2. Pharm Res. 1999;16:904–8. doi: 10.1023/a:1018838405987. [DOI] [PubMed] [Google Scholar]
- [40].Ali R, Cusi K. New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD) Ann Med. 2009;41:265–78. doi: 10.1080/07853890802552437. [DOI] [PubMed] [Google Scholar]
- [41].Nseir W, Mograbi J, Ghali M. Lipid-lowering agents in nonalcoholic fatty liver disease and steatohepatitis: human studies. Dig Dis Sci. 2012;57:1773–81. doi: 10.1007/s10620-012-2118-3. [DOI] [PubMed] [Google Scholar]