Abstract
High-affinity binding of testosterone or dihydrotestosterone to the androgen receptor (AR) triggers the androgen-dependent AR NH2- and carboxyl-terminal (N/C) interaction between the AR NH2-terminal FXXLF motif and the activation function 2 (AF2) hydrophobic binding surface in the ligand-binding domain. The functional importance of the AR N/C interaction is supported by naturally occurring loss-of-function AR AF2 mutations where AR retains high-affinity androgen binding but is defective in AR FXXLF motif binding. Ligands with agonist activity in vivo such as testosterone, dihydrotestosterone, and the synthetic anabolic steroids induce the AR N/C interaction and increase AR transcriptional activity in part by slowing the dissociation rate of bound ligand and stabilizing AR against degradation. AR ligand-binding domain competitive antagonists inhibit the agonist-dependent AR N/C interaction. Although the human AR N/C interaction is important for transcriptional activity, it has an inhibitory effect on transcriptional activity from AF2 by competing for p160 coactivator LXXLL motif binding. The primate-specific AR coregulatory protein, melanoma antigen gene protein-A11 (MAGE-A11), modulates the AR N/C interaction through a direct interaction with the AR FXXLF motif. Inhibition of AF2 transcriptional activity by the AR N/C interaction is relieved by AR FXXLF motif binding to the F-box region of MAGE-11. Described here are methods to measure the androgen-dependent AR N/C inter-domain interaction and the influence of transcriptional coregulators.
Keywords: Androgen receptor, steroid receptor, N/C interaction, mammalian two-hybrid assay, MAGE-11, MAGE-A11
1. Introduction
The androgen receptor (AR) is a ligand-dependent transcription factor essential for male sex development and a critical factor in prostate cancer. AR binds the two biologically active androgens, testosterone and dihydrotestosterone (DHT), with similar high affinity. Androgen binding causes AR to translocate to the nucleus, bind to DNA-response elements, and interact with coregulatory proteins to promote the transcription of androgen-dependent genes required for male sex development and reproductive function. Early studies investigating the kinetics of androgen binding demonstrated that DHT dissociates from AR more slowly than does testosterone, a property that contributes to the greater physiological potency of DHT (1, 2). The greater effectiveness of DHT during development is evident from the incomplete masculinization of genetic males deficient in DHT due to naturally occurring gene mutations in the 5α-reductase enzyme that converts testosterone to DHT (3).
Both testosterone and DHT dissociate with slower kinetics from full-length AR than from an AR carboxyl-terminal, ligand-binding domain (LBD) fragment that lacks the NH2-termimal region (4). In the presence of androgen, an AR NH2-terminal fragment interacts with an AR DNA and ligand-binding domain fragment, and the complex binds DNA and activates the prostate-specific antigen (PSA) enhancer/promoter (5). These observations, together with the results of mammalian two-hybrid interaction assays (6, 7), provided the first evidence for an androgen-dependent AR NH2- and carboxyl-terminal (N/C) interaction.
The importance of the AR N/C interaction in male reproductive physiology is suggested by naturally occurring AR gene mutations in the LBD that disrupt the AR N/C interaction without altering equilibrium androgen-binding affinity (7–13). These LBD mutations cause the androgen insensitivity syndrome that results in partial or complete failure of masculinization in 46XY genetic males by disrupting the AR N/C interaction and p160 coactivator binding (6–8). Ligands that induce the AR N/C interaction display complete agonist activity in vivo, which indicates that the mammalian two-hybrid N/C interaction assay is a useful method to identify ligands that function as active androgens (14) (see Note 1). The N/C interaction contributes to agonist potency in part by slowing the ligand dissociation rate and stabilizing AR (6). The AR N/C interaction assay is also useful in conjunction with transcription assays to identify AR antagonists (14).
The androgen-dependent AR N/C interaction is mediated by the AR NH2-terminal FXXLF motif 23FQNLF27 binding to a hydrophobic cleft in the LBD surface known as activation function 2 (AF2) (9, 15, 16). Both the AR FXXLF motif and AF2 are flanked by complementary charged amino acid residues that facilitate their interaction (17). Assays of the AR N/C interaction require the expression of an AR NH2-terminal fragment that contains the AR-20–30 NH2-terminal region with 23FQNLF27 sequence (17). The coexpressed interacting LBD fragment must contain AR LBD residues 658–919, an AR fragment that retains high-affinity androgen binding but displays rapid androgen dissociation kinetics when expressed alone (2). The two-hybrid AR N/C interaction assay is performed in mammalian cells (see Note 2) using GAL4 DNA-binding domain and VP16 activation domain fusion proteins, and a GAL4–luciferase reporter gene. Deletion of the hinge region from the carboxyl-terminal LBD fragment minimizes an inhibitory effect (see Note 3). The assay can be performed using AR NH2-terminal and carboxyl-terminal fragments with an androgen-responsive luciferase reporter vector such as PSA-Enh-Luc (see Note 4).
The androgen-dependent AR N/C interaction promotes the expression of AR target genes (18, 19). However, not all androgen-responsive enhancer/promoter regions require the AR N/C interaction. Most notably, the mouse mammary tumor virus (MMTV) and sex-limited gene enhancer/promoters do not require the AR N/C interaction that was required for maximal induction of the PSA and other androgen-responsive genes (18).
The human AR N/C interaction inhibits p160 coactivator LXXLL motif binding to AF2, which decreases overall AR transcriptional activity derived from AF2 (20, 21). Competitive inhibition at the AF2 site in the LBD occurs between the AR FXXLF motif and the p160 coactivator LXXLL motifs that bind the same AF2 hydrophobic cleft on the LBD surface (22). The ~10 fold higher binding affinity for the AR FXXLF motif relative to p160 coactivator LXXLL motifs imposes an inhibitory effect on AF2 activity. Through this mechanism, the AR N/C interaction shifts the dominant activation region from AF2 in the LBD to activation function 1 (AF1) in the human AR NH2-terminal region. In normal physiology, AF1 is androgen-dependent because of the inhibitory effect of the unliganded LBD. An AR NH2-terminal and DNA-binding domain fragment that lacks the LBD is constitutively active (23).
The AR N/C interaction is modulated by AR coregulatory proteins (see Note 5). One recently characterized AR coregulator that influences the AR N/C interaction is melanoma antigen gene protein-A11 (MAGE-11). MAGE-11 is a member of the MAGE gene family of cancer-testis antigens that evolved in a species-specific manner. MAGE-11 is expressed only in humans and other primates and is not expressed in rodents. MAGE-11 binds the AR FXXLF motif and directly recruits p160 coactivators to the AR NH2-terminal region (24, 25). MAGE-11 binds the AR FXXLF motif through a MAGE-11 F-box that is post-translationally modified. MAGE-11 is phosphorylated at Thr-360 within the F-box (amino acid residues 329–369) by cell cycle checkpoint kinase Chk1 in response to epidermal growth factor (EGF). MAGE-11 is phosphorylated at Ser-174 outside the F-box by MAP kinase in response to serum stimulation (25). In contrast to the minimal AR-20–30 amino acid region required to bind AF2 in the AR N/C interaction, a longer AR-16–36 region that contains the FXXLF motif is required to bind MAGE-11. Thus, overlapping AR FXXLF motif regions mediate interactions with AR AF2 in the AR N/C interaction and with MAGE-11. This suggests different flanking sequence requirements for AR FXXLF motif interactions. MAGE-11 interaction with AR also depends on the EGF-stimulated monoubiquitinylation of MAGE-11 outside the F-box that is triggered by phosphorylation at Thr-360 within the F-box (26). The recent evolutionary appearance of MAGE-11 as an AR coregulator in primates provides important and novel regulatory control on AR function.
Thus, the AR N/C interaction regulates AR function in response to agonists, is inhibited by AR antagonists, and modulated by coactivators. Two-hybrid assays of the AR N/C interaction in mammalian cells provide a measure of ligand potency for agonists or antagonists, and the functional effects of AR coregulators and naturally occurring and targeted AR mutations.
2. Materials
2.1. AR N/C Interaction Assay Reagents
HeLa epithelial cells derived from a human cervix adenocarcinoma (CCL-2; American Type Culture Collection, Rockville, MD) (see Note 2).
HeLa cell medium: Minimum essential medium with or without phenol red contains 10% fetal bovine serum (FBS); 2 mM l-glutamine (5.5 ml of 200 mM 100 × L-glutamine, added to 500 ml media); penicillin; and streptomycin (5.5 ml of 10,000 IU/ml 100× penicillin and streptomycin, added to 500 ml media).
-
Eukaryotic expression and reporter vectors:
GAL-AR-658–919 is a fusion protein of the GAL4 DNA-binding domain amino acid residues 1–147 and human AR carboxyl-terminal amino acid residues 658–919 that constitutes the AR LBD (2, 27).
VP-AR-1–660 is a fusion protein of VP16 transactivation domain amino acid residues 411–456 and human AR NH2-terminal amino acid residues 1–660 that includes the DNA-binding domain (7).
5XGAL4Luc3 is a luciferase reporter vector with five copies of the GAL4 upstream enhancer element (17, 28).
Fugene 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN) is stored at 4°C.
Phosphate buffered saline (PBS).
Testosterone, DHT (Steraloids, Inc., Newport, RI) and the synthetic androgen, methyltrienolone (R1881) (PerkinElmer, Waltham, MA) 10 mg/ml stocks, are prepared fresh each month in 100% ethanol and stored at −20°C. Dilutions of steroid stocks are prepared fresh each week in 100% ethanol.
Luciferase lysis buffer: 1% Triton X-100, 2 mM EDTA, and 25 mM Tris-phosphate, pH 7.8.
d-Luciferin, monopotassium salt (Thermo Scientific, Rockford, IL); 0.1 ml of 0.1 M d-luciferin is added automatically per well in 96-well assay plates in a luminometer.
Luciferase reading buffer: 25 mM Glycylglycine, 15 mM MgCl2, 5 mM ATP, and 0.5 mg/ml bovine serum albumin, pH 7.8; 0.1 ml reading buffer is added automatically per well in 96-well assay plates in a luminometer.
Twelve-well treated nonpyrogenic polystyrene tissue culture plates (Corning, Inc., Corning, NY).
96-Well, nontreated, flat-bottomed white polystyrene microtiter plates (Costar; Corning, Inc., Corning, NY).
15-ml Sterile RNase/DNase-free nonpyrogenic polypropylene centrifuge tubes (Corning, Inc., Corning, NY).
Automated Lumistar Galaxy multi-well plate luminometer (BMG Labtech, Germany).
3. Methods
3.1. HeLa Cell Culture
HeLa cells (see Note 2) are propagated in MEM supplemented with 10% FBS, l-glutamine, penicillin, and streptomycin. Cells are passaged twice each week at 1:7 dilution. Cells are harvested by washing T150 flasks with 10 ml PBS, adding 2 ml of a 0.05% trypsin and 0.53 mM EDTA solution/flask, and incubating at 37°C for 5 min to release cell adhesion. MEM containing 10% FBS is added to inactivate trypsin. Cells are counted using a hemocytometer, plated at 5 × 104 cells/well in 12-well plates, and transfected using FuGene 6 Transfection Reagent.
3.2. Experimental Design
The AR N/C interaction is performed as a mammalian two-hybrid assay. Cotransfection of an AR NH2-terminal FXXLF motif-containing fragment with an AR carboxyl-terminal fragment that contains the LBD and AF2 binding surface increases reporter gene activity in the presence of an AR agonist. The AR N/C interaction requires the addition of an active androgen such as testosterone, DHT, or synthetic androgen R1881 (Fig. 8.1), mibolerone, or anabolic steroid (see Note 1). The concentration of steroid that increases luciferase light units by greater than fivefold is indicative of ligand potency and a ligand-dependent AR intramolecular and/or intermolecular interaction (see Note 6). The AR N/C interaction is not induced by antagonists such as hydroxyflutamide or Casodex (bicalutamide). These antagonists inhibit the agonist-induced AR N/C interaction in a dose-dependent manner (Fig. 8.2). The effects of naturally occurring AR mutations identified in patients with the androgen insensitivity syndrome, somatic mutations in prostate cancer tissue (21), or targeted mutations designed to establish the AR sequence requirements for the N/C interaction can be tested in the two-hybrid assay when mutations are introduced into the AR NH2-terminal or LBD fragments. Assays are set in duplicate or triplicate with an agonist dose response range between 0.01 and 10 nM and an antagonist dose response range between 50 nM and 1 μM. The optimal dose response concentration for the AR N/C interaction in the two-hybrid assay is 10 nM testosterone, DHT, or synthetic androgen (Fig. 8.1). The N/C interaction assay has also been demonstrated for other steroid receptors using NH2- and carboxyl-terminal fragments (see Note 7).
Fig. 8.1.
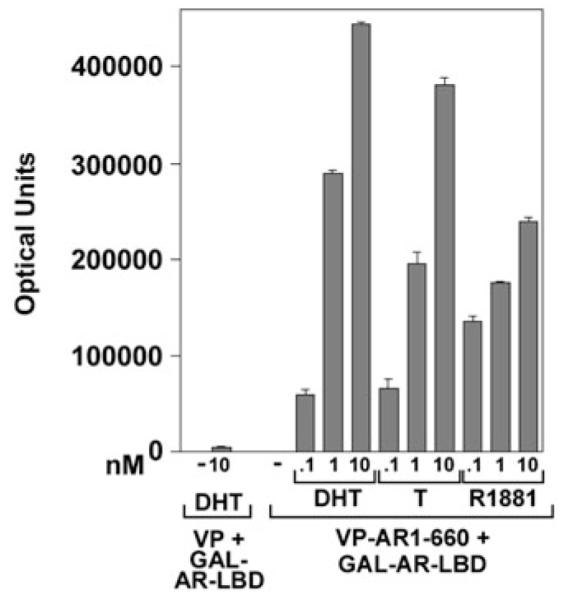
Androgen-dependent AR N/C interaction. HeLa cells (5 × 104/well of 12-well plates) were transfected using Fugene 6 with (per well) 0.1 μg 5XGAL4Luc3 reporter and 50 ng GAL-AR-658–919 (GAL-AR-ligand-binding domain (−LBD)) in the presence of 50 ng pVP16 empty vector (VP) or 50 ng VP-AR-1–660 that codes for the AR NH2-terminal and DNA-binding domains. The day after transfection, the medium was exchanged with phenol red-free, serum-free medium in the absence and the presence of 0.1, 1, and 10 nM dihydrotestosterone (DHT), testosterone (T), or the synthetic androgen methyltrienolone (R1881), respectively. Cells were incubated overnight at 37°C and luciferase activity was determined. The data are representative of the androgen-dependent mammalian two-hybrid AR N/C interaction assay.
Fig. 8.2.
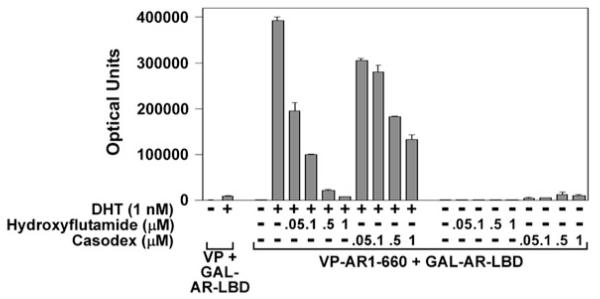
Inhibition of the DHT-induced AR N/C interaction by AR antagonists. HeLa cells were transfected with 5XGAL4Luc3 reporter vector and GAL-AR-658–919 (GAL-AR-LBD) with pVP16 empty vector (VP) or VP-AR-1–660 as described in Fig. 8.1. The next day, cells were incubated in phenol red-free, serum-free media in the absence and the presence of 1 nM dihydrotestosterone (DHT) with and without 0.05, 0.1, 0.5, and 1 μM hydroxyflutamide or Casodex (bicalutamide), and the same increasing concentrations of hydroxyflutamide or Casodex alone. The data show that the DHT-induced AR N/C interaction measured in a mammalian two-hybrid assay is inhibited in a dose-dependent manner by increasing concentrations of AR antagonists, hydroxyflutamide and Casodex, and that these antagonists alone do not induce the AR N/C interaction.
When the AR N/C interaction is performed using GAL4 and VP16 fusion vectors, a 5XGAL4Luc reporter vector is used. The AR N/C interaction may also be performed using the androgen-responsive luciferase reporter vector PSA-Enh-Luc, or the less active MMTV-Luc, transfected into HeLa cells with the AR DNA and ligand-binding domain fragment AR-507–919 and AR NH2-terminal fragment AR-1–503 that lacks the DNA-binding domain (see Note 4). Experimental procedures for both assays are otherwise identical.
3.3. Transfection of HeLa Cells Using FuGene 6
DNAs are aliquoted into microfuge tubes and stored for not more than 1 week at −20°C using per well:
50 ng Gal-AR-658–919 (see Note 3)
50 ng VP-AR-1–660
0.1 μg 5XGAL4Luc3
Coregulator expression plasmid DNA (25–50 ng/well of 12-well plates) can be added to test for effects on the AR N/C interaction by including equivalent amounts of empty vector DNA added to controls (see Note 5).
Day 1: Plate 5 × 104 HeLa cells/well in 12-well plates with 1 ml HeLa cell medium containing phenol red and incubate overnight in a 5% CO2 cell culture incubator.
Day 2:
Aspirate the medium and add 0.75 ml/well fresh HeLa cell medium containing phenol red using a sterile repeat pipette and return the plates to the cell culture incubator.
Calculate 43 μl × number of wells (include four extra wells in the calculation) of serum-free, phenol red-free medium and add to a 15-ml centrifuge tube.
Briefly warm the Fugene reagent and vortex for 1 s.
Add 0.6 μl Fugene reagent × total number of wells directly to the aliquoted serum-free media avoiding contact with the plastic tube.
Vortex for 1 s and incubate for 5 min at room temperature.
Thaw the previously aliquoted expression plasmid and luciferase reporter DNA.
Add sufficient Fugene cell medium solution for 43 μl/well to the aliquoted DNA.
Vortex each tube for 1 s and incubate for 15 min at room temperature.
Vortex briefly and add 40 μl of DNA–Fugene cell medium solution to each well.
Return the plates to the 37°C cell culture incubator and incubate overnight.
Day 3
Aspirate the medium and replace with 1 ml serum-free, phenol red-free HeLa cell medium with and without ligand.
Return the plates to the 37°C cell culture incubator and incubate overnight.
Day 4
Aspirate the media and wash each well with 1 ml PBS.
Aspirate PBS twice to dryness and add 0.25 ml luciferase lysis buffer using a repipetter.
Gently rock the plates on a platform shaker for 30 min at room temperature.
Aliquot 0.1 ml from each well into a 96-well microtiter assay plates and measure luciferase activity using an automated luminometer.
4. Notes
N/C interaction screen for AR agonists and antagonists: Androgens that induce the AR N/C interaction have agonist activity in vivo. The AR N/C interaction is inhibited by classical AR antagonists such as hydroxyflutamide and Casodex that bind the AR LBD with moderate affinity (14). Examples of AR agonists that induce the AR N/C interaction are testosterone, DHT, and the anabolic steroids oxandrolone and fluoxymesterone which bind with lower affinity than testosterone or DHT but are potent agonists in vivo (14). These findings suggest that the AR N/C two-hybrid interaction assay can be used to identify agonists that increase AR transcriptional activity in vivo when performed in cells with suitable ligand uptake and lack of metabolism (see Note 2). The AR N/C interaction has been used to screen AR agonists and antagonists, establish ligand dependence and motif-binding specificity, and investigate AR gene mutations that cause the androgen insensitivity syndrome (10, 29). While AR antagonists competitively inhibit the agonist-induced N/C interaction (Fig. 8.2), high concentrations of an AR antagonist, e.g., 10 μM hydroxyflutamide, may have agonist activity with wild-type AR in transient transfection experiments, but only weakly induce the AR N/C interaction (6, 14, 30). Somatic AR mutations in prostate cancer can enhance the ability of antagonists to induce the AR N/C interaction and increase AR transcriptional activity.
AR N/C interaction assay in other cell lines: The AR N/C interaction has been performed in Chinese hamster ovary (CHO) (6, 7, 9, 15, 31), human hepatocellular carcinoma HepG2 (17, 32), and HeLa cells (2, 18, 21). HeLa cells are advantageous because they contain low levels of steroid-metabolizing enzymes. This is evident by the similar activities of the naturally occurring androgens testosterone and DHT, and the synthetic androgen methyltrienolone (R1881), which is less susceptible to metabolism than are naturally occurring androgens. HepG2 cells derive from a human hepatocellular carcinoma and thus may express liver-derived, steroid-metabolizing enzymes. This is supported by the weaker activities of testosterone and DHT compared to synthetic androgens when assayed in HepG2 cells. The AR N/C interaction has also been performed in yeast (33). In this case, consideration should be given to ligand uptake and metabolism. While a variety of cell lines can be used to perform the AR N/C interaction assay, possible complications to be considered are ligand uptake and metabolism, and the influence of endogenous transcriptional coregulators that can differ between cell lines.
-
Inhibition of the AR N/C interaction by the hinge region: Human AR has 919 amino acids that include the NH2-terminal residues 1–558, DNA-binding domain residues 559–624, hinge region residues 625–676, and LBD residues 677–919 (34). However, the number of amino acid residues in human AR varies between individuals depending on the length of the polymorphic NH2-terminal glutamine repeat that begins at amino acid residue 58. Length of the AR CAG-encoded glutamine repeat influences AR transcriptional capacity through mechanisms that are not completely understood (35–37).
Initial experiments that identified and characterized the androgen-dependent AR N/C interaction made use of the AR LBD fragment AR-624–919, which contains the LBD and the entire hinge region (6, 7). It was shown subsequently that human AR hinge region residues 628–646, 624–658, or 629–636 have an inhibitory effect on the AR N/C interaction and AR transcriptional activity (2, 38, 39). The inhibitory region contains part of the bipartite AR nuclear-targeting signal at residues 617–633 (39, 40). However, inhibition may be independent of a detrimental effect on AR nuclear transport, since inhibition was also observed using a GAL4 DNA-binding domain–AR LBD fusion protein that has an independent nuclear-targeting signal (2). The inhibitory effect of the AR hinge region appears to be mediated through the AR AF2 site (2). A naturally occurring AR-R629W hinge mutation that disrupts the AR N/C interaction caused severe androgen insensitivity without altering androgen-binding affinity and nuclear localization (12). These findings support a negative influence of the hinge region on the AR N/C interaction and AR transcriptional activity in vivo. The phenotypic expression of decreased AR transcriptional activity in the androgen insensitivity syndrome resulting from mutations that disrupt the AR N/C interaction supports the functional importance of the AR N/C interaction in vivo.
The inhibitory effect of the hinge region does not appear to result from structural artifacts associated with expression of the AR NH2-terminal and LBD fragments, since inhibition was also observed in full-length AR. While the precise mechanism for AR hinge region inhibition of AF2 is not known, the inhibitory effect decreases AF2 binding of the AR FXXLF and p160 coactivator LXXLL motifs (2). This suggests that AR AF2 may have greater inherent transcriptional capacity than previously recognized, especially when p160 coactivator levels are increased as in castration-recurrent prostate cancer (41). In HeLa cells in the absence of overexpressed p160 coactivators, AR AF2 transcriptional activity was almost undetectable when GAL-AR-LBD was expressed in the presence of 10 nM DHT in the absence of an interacting AR NH2-terminal fragment (Fig. 8.1). This reflects in part the genetic changes in the AR LBD AF2 site during evolution that have weakened the binding affinity for p160 coactivator LXXLL motifs and improved binding of the FXXLF motifs (22). However, in prostate cancer cells, such as CWR-R1 cells that have higher levels of coactivators, expression of GAL-AR-LBD alone can have significant activity (2).
Thus, the contribution of AF2 to AR transcriptional activity relative to AR NH2-terminal AF1 is limited by the inhibitory effect of the AR N/C interaction, by genetic changes in AF2 that weaken p160 coactivator LXXLL motif binding relative to the FXXLF motif, through inhibitory mechanisms involving the hinge region, and through the complement of cell-specific coregulatory proteins.
Promoter specificity of the AR N/C interaction: Androgen-responsive enhancer/promoters differ in the requirement for the AR N/C interaction. For example, transcriptional activation of the PSA enhancer is increased through mechanisms that are increased by the AR N/C interaction (18, 19), whereas some androgen-responsive enhancers/promoters do not require the AR N/C interaction. The most notable example is MMTV which is activated to a similar extent in transient transfection assays whether or not the AR N/C interaction is disrupted by mutations in the AR FXXLF motif (18). The extent of activation of PSA-Enh-Luc or MMTV-Luc by the coexpression of AR-507–919 and AR-1–503 is influenced by the concentration of endogenous coregulators which differs between cell lines. The full complement of transcriptional regulators that influence the AR N/C interaction and AR transcriptional activity remains to be defined.
-
AR FXXLF motif and influence of coregulators: The effect of coregulatory proteins on the AR N/C interaction can be assessed in the mammalian two-hybrid assay. However, nonspecific inhibitory effects associated with the addition of increasing amounts of coactivator DNA require appropriate controls. Expression vector DNA concentrations should be kept to a minimum (100 ng or less/well in 12-well plates) to avoid nonspecific inhibition. Transient transfections may be performed in monkey kidney CV1 cells using calcium phosphate precipitation of DNA when assessing the effects of coregulatory proteins on AR transcriptional activity (24).
The AR N/C interaction assay measures the androgen-dependent AR NH2-terminal FXXLF motif 23FQNLF27 interaction with the AF2 site in the LBD. The AR FXXLF motif also serves as the principal interaction site for MAGE-11, an AR coregulator that increases AR transcriptional activity by relieving inhibition of p160 coactivator LXXLL motif binding at AF2 and through direct interactions with transcriptional coregulators (20, 25). The AR FXXLF motif was also implicated as a site involved in AR degradation by the proteasome (42). An NH2-terminal WXXLF (433WHTLF437) contributes to ligand-independent AR transcriptional activity and to the AR N/C interaction through mechanisms that remain to be established (15, 18, 43).
FXXLF-like motifs that mediate androgen-dependent interactions with AR AF2 are present in the AR coregulatory proteins ARA54, ARA55, and ARA70 (32, 44–46). While the affinity of these FXXLF-related motifs has not been reported, their inability to slow the dissociation rate of bound androgen when inserted to replace the AR FXXLF motif suggests that AR AF2 affinity is weaker for these coregulator FXXLF motifs than for the AR FXXLF motif (32). Furthermore, MAGE-11 does not interact with FXXLF-related motifs present in these AR coregulators, indicating specificity for the AR FXXLF motif (24). MAGE-11 contains an FXXIF motif that serves as part of a recognition sequence for p160 coactivators. Interaction between the MAGE-11 FXXIF and an F-box-like region in transcriptional mediator protein 2 (TIF2), and AR FXXLF motif binding to the MAGE-11 F-box, suggests a novel protein–protein interaction paradigm of FXXLF motif binding to the F-box (25).
A number of AR coregulatory proteins are reported to influence the AR N/C interaction, some of which involve interactions with FXXLF-related motifs. Cyclin D1 binds the AR NH2-terminal FXXLF motif and inhibits the AR N/C interaction and AR transcriptional activity independent of its ability to recruit histone deacetylases (47). p53 contains an α-helical FXXLF-like recognition motif in its activation domain and inhibits the AR N/C interaction (48, 49). hRAD9 contains a carboxyl-terminal FXXLF motif, inhibits the AR N/C interaction, and represses AR transcriptional activity (50). FoxO1 binding to the AR NH2-terminal region inhibits the AR N/C interaction (51). Inhibition of the AR N/C interaction is caused by corepressor SMRT that interacts with the AR NH2-terminal region (52, 53) and by N-CoR that interacts with the AR carboxyl-terminal region (54, 55).
MAGE-11 contains an FXXIF motif and increases AR transcriptional activity by competitively inhibiting AR FXXLF motif binding to AF2 in the AR N/C interaction, and by directly recruiting p160 coactivators (24). c-Jun, although not reported to contain an FXXLF motif, enhanced the AR N/C interaction, AR binding to DNA, and AR transcriptional activity (56). Effects of AR coregula-tory proteins can be assessed in two-hybrid interaction assays as described here. Endogenous levels of these and other coregulators yet to be defined influence the AR N/C interaction. Experiments using small inhibitory RNAs to knock down endogenous protein expression can therefore provide additional evidence for the effects of coregulatory proteins in the AR N/C interaction and AR function.
Intramolecular versus intermolecular AR N/C interaction: The AR N/C interaction may occur as an intramolecular and an intermolecular interaction. Early studies on androgen insensitivity syndrome AR mutations suggest that an intermolecular AR N/C interaction facilitates AR binding to DNA as an anti-parallel dimer (5–7). Structural analysis using fluorescence resonance energy transfer (FRET) suggests an intermolecular AR N/C interaction in the nucleus and an intramolecular interaction in the cytoplasm (57). Additional FRET analysis suggests that the AR N/C interaction occurs when AR is mobile, but not when transiently bound to DNA (58), and an intramolecular N/C interaction for transcriptionally active AR bound to DNA (59). Under some conditions, an intermolecular AR N/C interaction could contribute to runaway domain swapping (60), in which amyloid-like fibrils are associated with the degenerative phenotype that results from glutamine expansion diseases, such as spinal bulbar muscular atrophy which is caused by an expanded AR NH2-terminal glutamine repeat (61). However, the length of the polymorphic glutamine repeat in the human AR NH2-terminal region has not been shown to influence the AR N/C interaction (6).
-
N/C interaction in other steroid receptors: A ligand-dependent N/C interaction has been reported for the progesterone receptor (PR) (62), estrogen receptor-α (63), and mineralocorticoid receptor (MR) (64), although none has been as extensively characterized as the AR N/C interaction. The glucocorticoid receptor (GR) does not undergo an agonist-induced N/C interaction. However, introducing a p160 coactivator LXXLL motif into the NH2-terminal region to create a GR chimera slows the dissociation rate of dexamethasone (t1/2 168 min), a synthetic glucocorticoid, compared to wild-type GR (t1/2 31 min) (20). This demonstrates the ability of an N/C interaction to slow ligand dissociation and stabilize a receptor.
The longest PR-B contains an extended NH2-terminal LXXLL-like motif upstream region absent from the shorter PR-A. This unique PR-B upstream region imparts greater transcriptional activity through cooperative interactions involving a PR-B N/C interaction when bound to DNA (65).
The MR N/C interaction displays striking ligand specificity, with strong induction by aldosterone and the synthetic agonist 9α-fludrocortisol, and weak or no interaction with other physiologically relevant ligands such as deoxycorticosterone or cortisol (64, 66). While the precise nature of the MR NH2-terminal interaction motif has yet to be characterized, evidence suggests that like AR, the MR N/C interaction contributes to MR transcriptional activity by discriminating ligand-specific effects in vivo. An MR corepressor recruited by agonist-bound MR inhibited the MR N/C interaction (67).
References
- 1.Wilson EM, French FS. Binding properties of androgen receptors: evidence for identical receptors in rat testis, epididymis, and prostate. J. Biol. Chem. 1976;251:5620–5629. [PubMed] [Google Scholar]
- 2.Askew EB, Gampe RT, Stanley TB, Faggart JL, Wilson EM. Modulation of androgen receptor activation function 2 by testosterone and dihydrotestosterone. J. Biol. Chem. 2007;282:25801–25816. doi: 10.1074/jbc.M703268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imperato-McGinley J, Guerrero L, Gautier T, Peterson RE. Steroid 5-alpha-reductase deficiency in man: an inherited form of male pseudo-hermaphroditism. Science. 1974;186:1213–1215. doi: 10.1126/science.186.4170.1213. [DOI] [PubMed] [Google Scholar]
- 4.Zhou ZX, Lane MV, Kemppainen JA, French FS, Wilson EM. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol. Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 5.Wong CI, Zhou ZX, Sar M, Wilson EM. Steroid requirement for androgen receptor dimerization and DNA binding. Modulation by intramolecular interactions between the NH2-terminal and steroid-binding domains. J. Biol. Chem. 1993;268:19004–19012. [PubMed] [Google Scholar]
- 6.Langley E, Zhou ZX, Wilson EM. Evidence for an antiparallel orientation of the ligand activated human androgen receptor dimer. J. Biol. Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 7.Langley E, Kemppainen JA, Wilson EM. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J. Biol. Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 8.Quigley CA, Tan JA, He B, Zhou ZX, Mebarki F, Morel Y, Forest M, Chatelain P, Ritzen EM, French FS, Wilson EM. Partial androgen insensitivity with phenotypic variation caused by androgen receptor mutations that disrupt activation function 2 and the NH2- and carboxyl-terminal interaction. Mech. Ageing Dev. 2004;125:683–695. doi: 10.1016/j.mad.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 9.He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 10.Ghali SA, Gottlieb B, Lumbroso R, Beitel LK, Elhaji Y, Wu J, Pinsky L, Trifiro MA. The use of androgen receptor amino/carboxyl-terminal interaction assays to investigate androgen receptor gene mutations in subjects with varying degrees of androgen insensitivity. J. Clin. Endocrinol. Metab. 2003;88:2185–2193. doi: 10.1210/jc.2002-021324. [DOI] [PubMed] [Google Scholar]
- 11.Thompson J, Saatcioglu F, Jänne OA, Palvimo JJ. Disrupted amino- and carboxyl-terminal interactions of the androgen receptor are linked to androgen insensitivity. Mol. Endocrinol. 2001;15:923–935. doi: 10.1210/mend.15.6.0647. [DOI] [PubMed] [Google Scholar]
- 12.Deeb A, Jääskeläinen J, Dattani M, Whitaker HC, Costigan C, Hughes IA. A novel mutation in the human androgen receptor suggests a regulatory role for the hinge region in amino-terminal and carboxy-terminal interactions. J. Clin. Endocrinol. Metab. 2008;93:3691–3696. doi: 10.1210/jc.2008-0737. [DOI] [PubMed] [Google Scholar]
- 13.Jääskeläinen J, Deeb A, Schwabe JW, Mongan NP, Martin H, Hughes IA. Human androgen receptor gene ligand-binding-domain mutations leading to disrupted interaction between the N- and C-terminal domains. J. Mol. Endocrinol. 2006;36:361–368. doi: 10.1677/jme.1.01885. [DOI] [PubMed] [Google Scholar]
- 14.Kemppainen JA, Langley E, Wong CI, Bobseine K, Kelce WR, Wilson EM. Distinguishing androgen receptor agonists and antagonists: distinct mechanisms of activation by medroxyprogesterone acetate and dihydrotestosterone. Mol. Endocrinol. 1999;13:440–454. doi: 10.1210/mend.13.3.0255. [DOI] [PubMed] [Google Scholar]
- 15.He B, Kemppainen JA, Wilson EM. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 16.He B, Wilson EM. The NH2- terminal and carboxyl-terminal interaction in the human androgen receptor. Mol. Gen. Metab. 2002;75:293–298. doi: 10.1016/S1096-7192(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 17.He B, Wilson EM. Electrostatic modulation of steroid receptor recruitment of the LXXLL and FXXLF motifs. Mol. Cell. Biol. 2003;23:2135–2150. doi: 10.1128/MCB.23.6.2135-2150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and carboxyl-terminal interaction. J. Biol. Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 19.Callewaert L, Verrijdt G, Christiaens V, Haelens A, Claessens F. Dual function of an amino-terminal amphipatic helix in androgen receptor-mediated trans-activation through specific and nonspecific response elements. J. Biol. Chem. 2003;278:8212–8218. doi: 10.1074/jbc.M210744200. [DOI] [PubMed] [Google Scholar]
- 20.He B, Bowen NT, Minges JT, Wilson EM. Androgen-induced NH2- and carboxyl-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 2001;276:42293–42301. doi: 10.1074/jbc.M107492200. [DOI] [PubMed] [Google Scholar]
- 21.He B, Gampe RT, Hnat AT, Faggart JL, Minges JT, French FS, Wilson EM. Probing the functional link between androgen receptor coactivator and ligand binding sites in prostate cancer and androgen insensitivity. J. Biol. Chem. 2006;281:6648–6663. doi: 10.1074/jbc.M511738200. [DOI] [PubMed] [Google Scholar]
- 22.He B, Gampe RT, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell. 2004;16:425–438. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 23.Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 24.Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol. Cell. Biol. 2005;25:1238–1257. doi: 10.1128/MCB.25.4.1238-1257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Askew EB, Bai S, Hnat AT, Minges JT, Wilson EM. Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators. J. Biol. Chem. 2009;284:34793–34808. doi: 10.1074/jbc.M109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai S, Wilson EM. Epidermal growth factor-dependent phosphorylation and ubiquitinylation of MAGE-11 regulates its interaction with the androgen receptor. Mol. Cell. Biol. 2008;28:1947–1963. doi: 10.1128/MCB.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkel T, Duc J, Fearon ER, Dang CV, Tomaselli GF. Detection and modulation in vivo of helix–loop–helix protein–protein interactions. J. Biol. Chem. 1993;268:5–8. [PubMed] [Google Scholar]
- 28.Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol. Cell. Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raivio T, Palvimo JJ, Dunkel L, Wick-man S, Jänne OA. Novel assay for determination of androgen bioactivity in human serum. J. Clin. Endocrinol. Metab. 2001;86:1539–1544. doi: 10.1210/jcem.86.4.7329. [DOI] [PubMed] [Google Scholar]
- 30.Kemppainen JA, Wilson EM. Agonist and antagonist activities of hydroxyflutamide and Casodex relate to androgen receptor stabilization. Urology. 1996;48:157–163. doi: 10.1016/s0090-4295(96)00117-3. [DOI] [PubMed] [Google Scholar]
- 31.Berrevoets CA, Doesburg P, Steketee K, Trapman J, Brinkmann AO. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol. Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 32.He B, Minges JT, Lee LW, Wilson EM. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem. 2002;277:10226–10235. doi: 10.1074/jbc.M111975200. [DOI] [PubMed] [Google Scholar]
- 33.Doesburg P, Kuil CW, Berrevoets CA, Steketee K, Faber PW, Mulder E, Brinkmann AO, Trapman J. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 34.Lubahn DB, Joseph DR, Sar M, Tan J, Higgs HN, Larson RE, French FS, Wilson EM. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol. Endocrinol. 1988;2:1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- 35.Choong CS, Kemppainen JA, Zhou ZX, Wilson EM. Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol. Endocrinol. 1996;10:1527–1535. doi: 10.1210/mend.10.12.8961263. [DOI] [PubMed] [Google Scholar]
- 36.Choong CS, Kemppainen JA, Wilson EM. Evolution of the primate androgen receptor: a structural basis for disease. J. Mol. Evol. 1998;47:334–342. doi: 10.1007/pl00006391. [DOI] [PubMed] [Google Scholar]
- 37.Choong CS, Wilson EM. Trinucleotide repeats in the human androgen receptor: a molecular basis for disease. J. Mol. Endocrinol. 1998;21:235–257. doi: 10.1677/jme.0.0210235. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Lu J, Yong EL. Ligand- and coactivator-mediated transactivation function (AF2) of the androgen receptor ligand-binding domain is inhibited by the cognate hinge region. J. Biol. Chem. 2001;276:7493–7499. doi: 10.1074/jbc.M009916200. [DOI] [PubMed] [Google Scholar]
- 39.Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67:4514–4523. doi: 10.1158/0008-5472.CAN-06-1701. [DOI] [PubMed] [Google Scholar]
- 40.Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J. Biol. Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]
- 41.Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
- 42.Chandra S, Shao J, Li JX, Li M, Longo FM, Diamond MI. A common motif targets huntingtin and the androgen receptor to the proteasome. J. Biol. Chem. 2008;283:23950–23955. doi: 10.1074/jbc.M800467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehm SM, Regan KM, Schmidt LJ, Tindall DJ. Selective role of an NH2-terminal WxxLF motif for aberrant androgen receptor activation in androgen depletion independent prostate cancer cells. Cancer Res. 2007;67:10067–10077. doi: 10.1158/0008-5472.CAN-07-1267. [DOI] [PubMed] [Google Scholar]
- 44.Hsu CL, Chen YL, Yeh S, Ting HJ, Hu YC, Lin H, Wang X, Chang C. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J. Biol. Chem. 2003;278:23691–23698. doi: 10.1074/jbc.M211908200. [DOI] [PubMed] [Google Scholar]
- 45.van de Wijngaart DJ, Dubbink HJ, Molier M, de Vos C, Trapman J, Jenster G. Functional screening of FxxLF-like peptide motifs identifies SMARCD1/BAF60a as an androgen receptor cofactor that modulates TMPRSS2 expression. Mol. Endocrinol. 2009;23:1776–1786. doi: 10.1210/me.2008-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van de Wijngaart DJ, van Royen ME, Hersmus R, Pike AC, Houtsmuller AB, Jenster G, Trapman J, Dubbink HJ. Novel FXXFF and FXXMF motifs in androgen receptor cofactors mediate high affinity and specific interactions with the ligand-binding domain. J. Biol. Chem. 2006;281:19407–19416. doi: 10.1074/jbc.M602567200. [DOI] [PubMed] [Google Scholar]
- 47.Burd CJ, Petre CE, Moghadam H, Wilson EM, Knudsen KE. Cyclin D1 binding to the androgen receptor NH2-terminal domain inhibits AF2 association and reveals dual roles for AR corepression. Mol. Endocrinol. 2005;19:607–620. doi: 10.1210/me.2004-0266. [DOI] [PubMed] [Google Scholar]
- 48.Shenk JL, Fisher CJ, Chen SY, Zhou XF, Tillman K, Shemshedini L. p53 represses androgen-induced transactivation of prostate-specific antigen by disrupting hAR amino- to carboxyl-terminal interaction. J. Biol. Chem. 2001;276:38472–38479. doi: 10.1074/jbc.M103652200. [DOI] [PubMed] [Google Scholar]
- 49.Uesugi M, Verdine GL. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA. 1999;96:14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Hsu CL, Ni J, Wang PH, Yeh S, Keng P, Chang C. Human checkpoint protein hRad9 functions as a negative coregulator to repress androgen receptor transactivation in prostate cancer cells. Mol. Cell. Biol. 2004;24:2202–2213. doi: 10.1128/MCB.24.5.2202-2213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Q, Fu W, Li P, Nicosia SV, Jenster G, Zhang X, Bai W. FoxO1 mediates PTEN suppression of androgen receptor N- and C-terminal interactions and coactivator recruitment. Mol. Endocrinol. 2009;23:213–225. doi: 10.1210/me.2008-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dotzlaw H, Moehren U, Mink S, Cato AC, Iñiguez Lluhí JA, Baniahmad A. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol. Endocrinol. 2002;16:661–673. doi: 10.1210/mend.16.4.0798. [DOI] [PubMed] [Google Scholar]
- 53.Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, Ghosh JC, Li H, Chen JD. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 2003;278:5052–5061. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 54.Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 2002;16:1492–1501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, Kawate H, Ohnaka K, Nawata H, Takayanagi R. Nuclear compartmentalization of N-CoR and its interactions with steroid receptors. Mol. Cell. Biol. 2006;26:6633–6655. doi: 10.1128/MCB.01534-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bubulya A, Chen SY, Fisher CJ, Zheng Z, Shen XQ, Shemshedini L. c-Jun potentiates the functional interaction between the amino and carboxyl ter-mini of the androgen receptor. J. Biol. Chem. 2001;276:44704–44711. doi: 10.1074/jbc.M107346200. [DOI] [PubMed] [Google Scholar]
- 57.Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AA, Miner JN, Diamond MI. The structural basis of androgen receptor activation: intramolecular and intermolecular amino-carboxy interactions. Proc. Natl. Acad. Sci. USA. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB. Compartmentalization of androgen receptor protein-protein interactions in living cells. J. Cell Biol. 2007;177:63–72. doi: 10.1083/jcb.200609178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang CY, Hager GL, Saatcioglu F. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol. Cell. Biol. 2007;27:1823–1843. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc. Natl. Acad. Sci. USA. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 62.Tetel MJ, Giangrande PH, Leonhardt SA, McDonnell DP, Edwards DP. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol. Endocrinol. 1999;13:910–924. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- 63.Kraus WL, McInerney EM, Katzenellenbogen BS. Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc. Natl. Acad. Sci. USA. 1995;92:12314–12318. doi: 10.1073/pnas.92.26.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rogerson FM, Fuller PJ. Interdomain interactions in the mineralocorticoid receptor. Mol. Cell. Endocrinol. 2003;200:45–55. doi: 10.1016/s0303-7207(02)00413-6. [DOI] [PubMed] [Google Scholar]
- 65.Tung L, Abdel-Hafiz H, Shen T, Harvell DM, Nitao LK, Richer JK, Sartorius CA, Takimoto GS, Horwitz KB. Progesterone receptors (PR)-B and -A regulate transcription by different mechanisms: AF-3 exerts regulatory control over coactivator binding to PR-B. Mol. Endocrinol. 2006;20:2656–2670. doi: 10.1210/me.2006-0105. [DOI] [PubMed] [Google Scholar]
- 66.Pippal JB, Yao Y, Rogerson FM, Fuller PJ. Structural and functional characterization of the interdomain interaction in the mineralocorticoid receptor. Mol. Endocrinol. 2009;23:1360–1370. doi: 10.1210/me.2009-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murai-Takeda A, Shibata H, Kurihara I, Kobayashi S, Yokota K, Suda N, Mitsuishi Y, Jo R, Kitagawa H, Kato S, Saruta T, Itoh H. NF-YC functions as a corepressor of agonist-bound mineralocorticoid receptor. J. Biol. Chem. 2010;285:8084–8093. doi: 10.1074/jbc.M109.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]


