Abstract
Summary
Metabolic labeling with tritiated palmitate is a direct method for monitoring post-translational modification of Ras proteins with this fatty acid. Advances in intensifying screens have allowed for the easy visualization of tritium without the need for extended exposure times. While more energetic radioisotopes are easier to visualize, the lack of commercial source and need for shielding make them more difficult to work with. Since radiolabeled palmitate is directly incorporated into Ras, its loss can be monitored by traditional pulse-chase experiments that cannot be accomplished with the method of acyl exchange chemistry. As such, tritiated palmitate remains a readily accessible and direct method for monitoring the palmitoylation status of Ras proteins under a multitude of conditions.
Keywords: Ras, palmitoylation, pulse-chase, tritium, Transcreen
1. Introduction
Ras proteins must associate with cellular membranes in order to regulate signaling events [1]. Ras undergoes a series of post-translational modifications that result in a lipidated protein with affinity for phospholipid bilayers. A CAAX sequence at the C-terminus of Ras proteins signals for these modifications. The CAAX sequence of nascent Ras proteins is modified in three steps. First, a farnesyl lipid is added to the CAAX cysteine by farnesyltransferase. Next, the C-terminal AAX peptide is cleaved by Rce1. Finally, the newly C-terminal farnesyl cysteine is methyl esterified by Icmt [2]. CAAX processing produces relatively weak membrane affinity and is insufficient to properly localize Ras to the plasma membrane [3]. A second signal immediately upstream of the CAAX sequence affords added affinity. In the case of K-Ras4B, the second signal consists of a polybasic sequence that can interact with the negatively charged headgroups of the phospholipids in the inner leaflet of the plasma membrane. For all other Ras isoforms, including N-Ras, H-Ras, and K-Ras4A, the second signal consists of palmitoylation of one or two cysteines in proximity with the farnesylcysteine. Whereas CAAX processing is irreversible, the second signals can be modulated. In the case of the polybasic region of K-Ras4B, phosphorylation of a nearby serine partially neutralizes the positive charge [4]. For the palmitoylated Ras isoforms, the palmitates can be readily removed by hydrolysis under physiologic conditions [5,6]. The dynamic nature of palmitoylation and its necessity for oncogenic function have made it an active area of investigation in Ras biology.
The most direct way to monitor protein palmitoylation is by metabolic labeling with palmitic acid that incorporates a radioisotope. Because tritiated [3H] palmitate is commercially available, this has been the reagent of choice for more than three decades. Following metabolic labeling, Ras can be readily immunoprecipitated and analyzed by SDS-PAGE and the radiolabeled protein observed by fluorography using a scintillation reagent [7]. The challenge of working with [3H]palmitate has been the low energy of the β particle that is emitted when tritium naturally decays into 3He. The low energy is insufficient to allow the β particle to pass out of the polyacrylamide gel or to penetrate the emulsion of x-ray film, which necessitates impregnating the gel with a fluor such as 2,5 diphenyl oxazole in order to convert the β particles into photons and thereby allow for exposure of film. While this method reliably produces images of labeled proteins, it often requires long exposures of up to several weeks to attain a clear image. Alternatively, the gel can be cut into slices and digested to release the labeled proteins that can be quantified by scintillation counting [8]. This method affords direct quantification but is cumbersome and time-consuming. Some groups have solved the problem of low signal by labeling Ras with 16-[125I]iodohexadecanoic acid (IC), an analog of palmitate [9,10]. The gamma rays emitted upon decay of 125I are much more penetrating, allowing for direct autoradiography of dried gels with overnight exposures. However, the lack of a commercial source of 16-[125I]IC makes its use much more complicated since it must be synthesized by the investigator. Moreover, organic synthesis involving sodium [125I]iodide requires far more precautions than does working with tritium.
Recently, several new methods have been developed to analyze protein acylation without using radioisotopes. The most widely used of these is known as acyl-exchange chemistry. This method is applied ex vivo to lysates of cells or tissues. It entails a three-step procedure that includes blocking free thiols with N-ethylmaleimide (NEM), hydrolyzing thioester-linked lipids (e.g. palmitate) with hydroxylamine, and labeling the free sulfhydryls produced with a sulfhydryl-reactive reagent that is biotinylated, thereby allowing for purification and/or detection with streptavidin [11]. The great advantage of this method is the ability to assay the level of palmitoylation in samples that were not or could not be labeled with radioactive palmitate [12]. Perhaps the most powerful use of this method has been to survey global acylation of proteins in complex lysates of cells [13,14]. However, the disadvantages of this method include the need to optimize each step to obtain reproducible signals and the inability to conduct pulse-chase experiments since the method is always ex vivo. Another recently developed method utilizes bioorthogonal chemical ligation, or “click chemistry.” This two-step process involves metabolically labeling cells with alkynyl-analogues of palmitic acid; the labeled proteins are then reacted with azido-modified detection markers, such as fluorescent tags for visualization or biotin for visualization and/or purification [15]. The applicability of this method for examining palmitoylation in vivo, e.g. with pulse-chase experiments, and improvements in detection and use with newer generations of chemical reporters may result in click chemistry supplanting radioisotope labeling in the future [16].
In the meantime, we have taken advantage of the advances in x-ray film sensitivity and specialized intensifying screens for tritium to optimize conventional metabolic labeling with [3H]palmitate. Our method consists of [3H]palmitate labeling, immunoprecipitation, SDS-PAGE, transfer onto PVDF membranes and high sensitivity fluorography using the Kodak BioMax Transcreen-LE intensifying screen and BioMax MS film. Our method allows for visualization of fluorograms in 1–3 days, depending on the Ras isoform being investigated. Moreover, since the exposed substrate is bound to PVDF membrane, protein levels can also be directly assessed by immunoblots quantified with a Li-Cor imaging system. This has allowed us to monitor changes in the palmitoylation status of H-Ras caused by peptidyl-prolyl isomerase inhibitors [17]. The method also allows for pulse-chase experiments that are crucial for determining how alterations to the steady state palmitoylation of Ras are achieved. In this chapter, we outline our protocol for such assays.
2. Materials
2.1 Cell culture
Labeling medium: Dulbecco’s Modified Eagle Medium (DMEM) + 10% dialyzed fetal bovine serum (FBS), 5 mM sodium pyruvate (to inhibit beta oxidation of palmitate), 3.6 mg/mL fatty acid-free bovine serum albumin (BSA). Make fresh prior to experiment. For 10 mL, weigh 36 mg of fatty acid-free BSA. Add 1 mL of dialyzed FBS, 0.5 mL of sodium pyruvate (from 100 mM stock), 8.5 mL of DMEM, and mix. Sterile-filter using 0.45 µm filter (see Note 1).
Chase medium: DMEM + 10% dialyzed FBS, 5 mM sodium pyruvate, 3.6 mg/mL fatty acid-free BSA, 200 µM palmitic acid. Prepare medium as indicated for labeling media, then add 200 µM palmitic acid (1:1000 dilution from 200 mM stock in ethanol).
Palmitic Acid, [9,10–3H(N)]- (Perkin Elmer, Waltham, MA, USA) (tritiated) Palmitic Acid (Sigma Aldrich, St. Louis, MO, USA) (untritiated): make 200 mM stock in absolute ethanol.
Transfection reagent: Lipofectamine 2000 (Invitrogen).
2.2 Immunoprecipitation
RIPA buffer: 20 mM Tris, pH 7.5, 137 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 10% glycerol, protease inhibitors (Roche cOmplete, Mini, EDTA-free protease inhibitor cocktail tablets), 2 mM Pefabloc SC. Add protease inhibitors fresh before use; store at 4°C.
Immunoprecipitation of Ras: Y13-259 pan-Ras conjugated antibody (Santa Cruz Biotechnology)
2.3 SDS-PAGE analysis
SDS-PAGE running buffer: 0.3% Tris (w/v), 1.44% glycine (w/v), 0.1% SDS (w/v). Laemmli buffer (SDS-PAGE sample buffer): 120 mM Tris-HCl, pH 6.8, 4% SDS (w/v), 20% glycerol, 0.02% bromophenol blue (w/v)
Dithiothreitol (DTT)
14% acrylamide gel
Towbin buffer (Tris-glycine transfer buffer): 0.3% Tris (w/v), 1.44% glycine (w/v)
Semi-dry transfer: PVDF membrane, semi-dry transfer apparatus, Whatman filter paper.
Immunoblot detection of Ras: RAS10 (Calbiochem), anti-GFP (Invitrogen) Secondary antibodies: LiCor IRDye 680/800 conjugated goat anti-mouse/rabbit IgG LiCor Odyssey Imaging System for development and quantification of immunoblot
2.4 Tritium fluorography
X-ray film cassette
Kodak BioMax Transcreen-LE intensifying screen
Kodak BioMax MS Film, Maximum Sensitivity – Radioisotope
Kodak X-Omat or other film developer system
3. Methods
3.1 Steady-state monitoring of H-Ras palmitoylation
Seed 106 COS-1 cells in each well of a 6-well dish.
The next day, transfect cells with GFP-H-Ras using Lipofectamine 2000, according to the manufacturer’s instructions.
To prepare tritiated labeling medium for 6 samples, in a sterile tissue culture hood add 1.2 mCi of [3H]palmitic acid to a 10 cm plate (for a final concentration of 0.2 mCi/mL). Allow ethanol solvent to evaporate completely (see Note 2).
Add 6 mL of labeling medium on top of the 10 cm plate containing the [3H]palmitate and incubate at 37°C for 5 minutes.
Change medium for the transfected cells using the prepared labeling medium containing [3H]palmitate. If applicable, apply the appropriate concentration of pharmacologic agent, e.g. 1 µM FK506, 50 ng/mL cycloheximide, 500 nM rapamycin, or 25 µM 2-BP. Incubate at 37°C overnight (see Note 3).
The next day, place cells on ice. Manually remove medium from cells and wash once with ice-cold PBS. Treat all media and washes as liquid radioactive waste.
Lyse cells in ice-cold RIPA buffer for 5 minutes on ice.
Collect cells using a cell scraper into microfuge tubes and clarify lysate by spinning at top speed (~20,000 × g) in a 4°C pre-chilled tabletop centrifuge for 10 minutes.
Immunoprecipitate Ras from the clarified lysate by adding clarified lysate to a fresh tube containing 10 µL of Y13-259 beads (anti-pan-Ras). Rotate for 1 hour at 4°C.
Spin down beads at 2,500 × g for 5 minutes at 4°C in a pre-chilled tabletop centrifuge.
Manually remove supernatant and wash the beads once with ice-cold RIPA buffer (see Note 4).
Manually remove supernatant and resuspend beads in 20 µL of 2X Laemmli sample buffer + 5 mM DTT (see Note 5).
Perform SDS-PAGE on samples, followed by transfer onto PVDF membrane using a semi-dry transfer apparatus (see Note 6).
Block membrane with 5% milk in PBS for 20 minutes, then blot for Ras or GFP with the antibodies described above for 1 hr. Wash 3x with PBS + 1% Triton X-100 for 5 minutes each, then blot with LiCor IRDye 680/800 conjugated secondary antibodies for 1 hr. Visualize and quantify Ras by LiCor Odyssey Imaging.
After immunoblotting, dry membranes overnight on Whatman paper.
Affix membranes with tape to the Kodak BioMax intensifying screen that is mounted in an x-ray film cassette.
In a darkroom, place Kodak BioMax MS film into the cassette containing the membrane. This must be done in complete darkness.
Place cassette in a −80°C freezer for 24–48 hrs (the cold temperature increases the sensitivity of the film).
When ready to develop film, remove cassette from −80°C freezer and allow to thaw to room temperature (roughly half an hour or more). Wipe away condensation from outside of cassette to prevent water from getting onto film or membrane.
In a darkroom (again, in complete darkness), develop film using a Kodak X-mat film developer or alternative. If exposure is too low, additional pieces of film can be put down for longer exposures (2 weeks or less is usually sufficient).
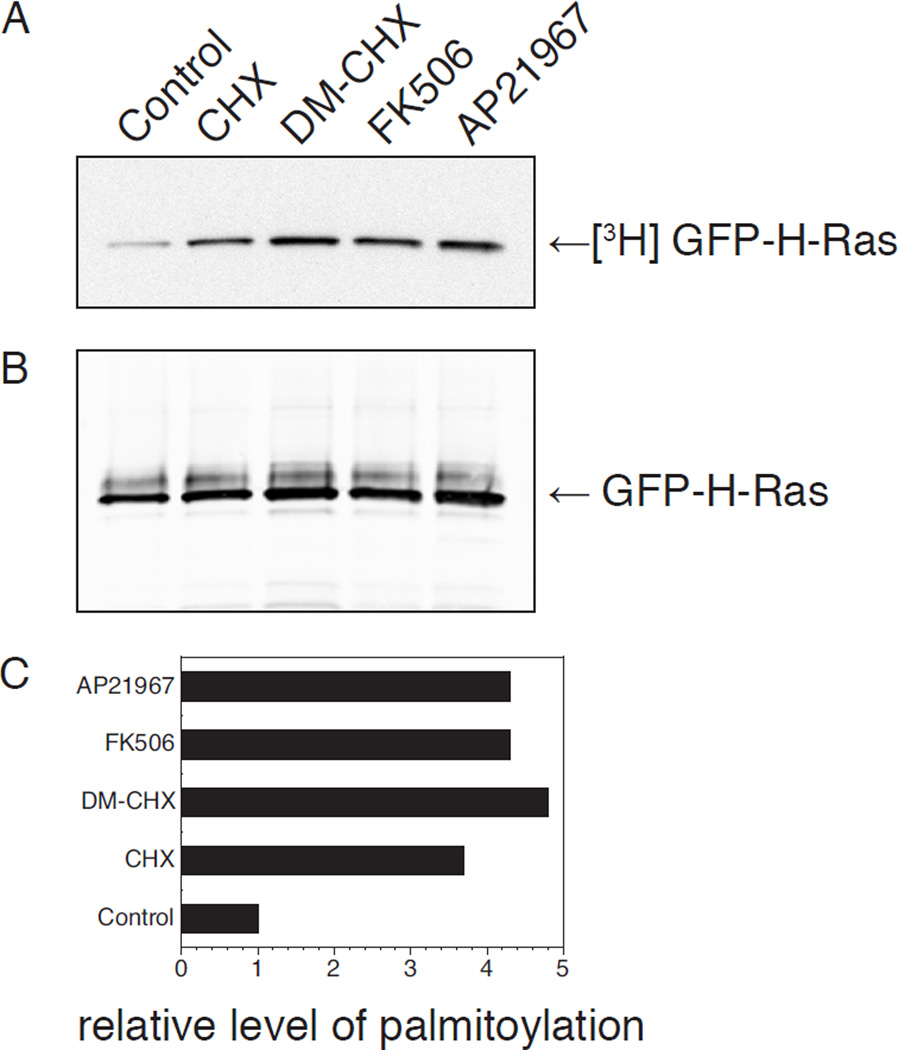
The bands representing palmitoylated proteins on the film can be scanned using a flatbed scanner with a transillumination mode and their optical density quantified using ImageJ software (Figure 1).
Figure 1. Steady-state metabolic labeling of GFP-H-Ras with various drug treatments.
COS-1 cells were transfected with GFP-H-Ras and labeled with [3H]palmitate while being treated either with DMSO (control), 50 ng/mL cyclohexamide (CHX), 10 µM DM-CHX, 1 µM FK506, or 2 µM AP21967. Cells were lysed the following day in RIPA buffer. Cell lysates were immunoprecipitated for Ras and subjected to SDS-PAGE analysis. (A) Fluorogram for [3H]palmitate labeling, 1 day exposure. (B) Immunoblot for Ras. (C) [3H]GFP-H-Ras bands were quantified using ImageJ software (for fluorogram) and LiCor software (for immunoblot) and are plotted as normalized ratio. Levels indicate amount of GFP-H-Ras incorporating [3H]palmitate relative to total protein amount.
3.2 Pulse-chase depalmitoylation assay
Seed 106 COS-1 cells in each well of a 6-well dish.
The next day, transfect cells with GFP-H-Ras using Lipofectamine 2000, according to the manufacturer’s instructions.
On the following day (post-transfection), prepare [3H]palmitic acid labeling medium for the pulse. For 6 samples, in a sterile tissue culture hood add 6 mCi of [3H]palmitic acid to a 10 cm plate (for a final concentration of 1 mCi/mL). Allow ethanol solvent to evaporate completely before adding 6 mL of labeling medium on top of the [3H]palmitate in the 10 cm plate. Incubate at 37°C for 5 minutes (see Note 7).
To begin pulse, add 500 µL of labeling medium containing [3H]palmitate to the well corresponding to the maximum chase timepoint (= 60 min). If applicable, add pharmacologic agents. Incubate for 5 min (see Note 8).
Remove [3H] medium and wash cells twice with room-temperature PBS. Add 1 mL of chase medium and incubate for 60 min.
Ten minutes after step 3, aspirate media from cells in the adjacent well and repeat steps 2–3, but incubate in chase medium for 45 min.
Repeat step 4 for each successive well, decreasing length of chase by 15 min. Timepoints may be adjusted as desired (see Note 9).
Once all timepoints are complete, place cells on ice and manually remove medium. Wash once with ice-cold PBS and proceed to lyse samples for SDS-PAGE, Western analysis, and film exposure as described above starting from step 7 of steady-state labeling protocol (Section 3.1).
Footnotes
We have found that to avoid confusion between labeling and chase media it is advantageous to use phenol red-free DMEM in making labeling medium.
Treat this 10 cm plate, pipette tips, and all other [3H]palmitate-containing materials as solid waste. When working with radioisotopes, care must be taken to minimize both solid and liquid waste. All forms of waste must be carefully contained and monitored to prevent contamination of lab materials and surfaces. This is particularly important due to the long half-life of tritium (12.3 years). Monitoring potential tritium contamination should be performed by wipe testing and scintillation counting for all lab surfaces after every experiment and decontamination should be performed promptly, if necessary.
Treatment with pharmacologic agents must be titrated for both dose and duration to allow for sufficient pharmacologic activity while at the same time minimizing cell toxicity. For the pharmacologic agents used here to inhibit peptidyl-prolyl isomerization, overnight treatments are suitable for steady-state labeling, while much shorter treatments are used for pulse-chase labeling.
To facilitate removal of supernatant from beads, use flat-orifice gel loading tips.
Dithiothreitol (DTT) is used at 5 mM as reducing agent for SDS-PAGE sample preparation instead of β-mercaptoethanol to avoid cleavage of thioester linkages. Samples in Laemmli buffer may be stored at −80°C prior to SDS-PAGE analysis.
PVDF affords a greater affinity for hydrophobic and lipidated proteins than nitrocellulose and, as such, is the preferred membrane to use; however, nitrocellulose membranes can sufficiently bind and be used to detect palmitoylated Ras species. A semi-dry transfer system is used to minimize production of liquid radioactive waste.
A greater concentration of [3H]palmitate is required for pulse-chase analysis than for steady-state labeling. Here we increase the concentration five fold.
Our pulse-chase assay utilizes a pulse of only 5 minutes. Previous efforts to monitor the latency of H-Ras palmitoylation used pulse intervals of one hour or more [18], but these methods are confounded by the kinetics of palmitate metabolism. The rate of accumulation of exogenous palmitic acid in cells is controlled by saturable fatty acid transporters. Once unlabeled palmitic acid has gained access to the cytosol, it must be conjugated with Coenzyme A in order to compete for the labeling of proteins with the [3H]palmitoyl Co-A that was produced during the pulse. Thus, an instantaneous chase using even a great excess of unlabeled palmitate is not possible. Further confounding the ability to achieve a rapid and complete chase is the fact that labeled fatty acids can be recycled [19] such that a pool of [3H]palmitoyl Co-A can be produced even after [3H]palmitate is removed from the growth medium. In our experience, a characteristic increase in the incorporation of the label was routinely seen between 15 and 30 minutes following chase, necessitating both a shorter pulse and longer chase timepoints [17].
In our pulse-chase protocol, we perform timepoints of 15 minute intervals, which corresponds to the time elapsed between treatment of each well (steps 2–3). Performing each chase timepoint counting backwards allows us to harvest cells at the end of each chase duration simultaneously, minimizing timing errors in our pulse-chase. The length of time between steps 3 and 4 can be adjusted to allow for different chase intervals, e.g. 5 or 10 min differences, as required.
References
- 1.Willumsen BM, Norris K, Papageorge AG, et al. Harvey murine sarcoma virus p21 ras protein: biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J. 1984;3:2581–2585. doi: 10.1002/j.1460-2075.1984.tb02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Choy E, Chiu VK, Silletti J, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 4.Bivona TG, Quatela SE, Bodemann BO, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin JS, Drake KR, Rogers C, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 7.Sefton BM, Trowbridge IS, Cooper JA, et al. The transforming proteins of Rous sarcoma virus, Harvey sarcoma virus and Abelson virus contain tightly bound lipid. Cell. 1982;31:465–474. doi: 10.1016/0092-8674(82)90139-8. [DOI] [PubMed] [Google Scholar]
- 8.Hancock JF. Prenylation and palmitoylation analysis. Methods Enzymol. 1995;255:237–245. doi: 10.1016/s0076-6879(95)55026-7. [DOI] [PubMed] [Google Scholar]
- 9.Peseckis SM, Deichaite I, Resh MD. Iodinated fatty acids as probes for myristate processing and function. Incorporation into pp60v-src. J Biol Chem. 1993;268:5107–5114. [PubMed] [Google Scholar]
- 10.Liang X, Nazarian A, Erdjument-Bromage H, et al. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276:30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 11.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 12.Drisdel RC, Alexander JK, Sayeed A, et al. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Kang R, Wan J, Arstikaitis P, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth AF, Wan J, Bailey AO, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charron G, Zhang MM, Yount JS, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 16.Yang YY, Ascano JM, Hang HC. Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahearn IM, Tsai FD, Court H, et al. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41:173–185. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veit M, Ponimaskin E, Schmidt MF. Analysis of S-acylation of proteins. Methods Mol Biol. 2008;446:163–182. doi: 10.1007/978-1-60327-084-7_12. [DOI] [PubMed] [Google Scholar]
- 19.Qanbar R, Bouvier M. Determination of protein-bound palmitate turnover rates using a three-compartment model that formally incorporates [3H]palmitate recycling. Biochemistry. 2004;43:12275–12288. doi: 10.1021/bi049176u. [DOI] [PubMed] [Google Scholar]