Abstract
Mixl1 is thought to play important roles in formation of mesoderm and endoderm. Previously, Mixl1 expression was reported in the posterior primitive streak and allantois, but the precise spatiotemporal whereabouts of Mixl1 protein throughout gastrulation have not been elucidated. To localize Mixl1 protein, immunohistochemistry was carried out at 2-4 hour intervals on mouse gastrulae between primitive streak and 16-somite pair stages (~E6.5-9.5). Mixl1 localized to the entire primitive streak early in gastrulation. However, by headfold stages (~E7.75-8.0), Mixl1 diminished within the mid-streak but remained concentrated at either end of the streak, and localizing throughout midline posterior visceral endoderm. At the streak's anterior end, Mixl1 was confined to the posterior crown cells of Hensen's node, which contribute to dorsal hindgut endoderm and the posterior notochord. In the posterior streak, Mixl1 localized to the Allantoic Core Domain (ACD), which is the source of most of the allantois and contributes to the posterior embryonic-extraembryonic interface. In addition, Mix1 co-localized with the early hematopoietic marker, Runx1, in the allantois and visceral yolk sac blood islands. During hindgut invagination (4-16s, ~E8.5-9.5), Mixl1 localized to the hindgut lip, becoming concentrated within the midline anastomosis of the splanchnopleure, which appears to create the ventral component of the hindgut and omphalomesenteric artery. Surrounding the distal hindgut, Mixl1 identified midline cells within tailbud mesoderm. Mixl1 was also found in the posterior notochord. These findings provide a critical systematic, tissue-level understanding of embryonic Mixl1 localization, and support its role in regulation of crucial posterior axial mesendodermal stem cell niches during embryogenesis.
Keywords: allantois, blood island, gastrula, hematopoiesis, hindgut, mesendoderm, Mixl1, primitive streak, Runx1, stem cells, vasculogenesis
1. INTRODUCTION
The primitive streak is the overt manifestation of the antero-posterior axis of amniote embryos. Its appearance within the midline of the mouse epiblast foretells the posterior end of the embryo, and heralds the onset of gastrulation. The anterior end of the primitive streak condenses into the node, a stem/progenitor cell pool that elaborates midline patterning via formation of the notochord (Davidson et al., 1999), and contributes to embryonic endoderm (Beddington, 1994). Recent investigation into the posterior end of the primitive streak has demonstrated that, once the exocoelom forms, the streak extends further caudally and, together with associated posterior visceral endoderm (PVE), creates the Allantoic Core Domain (ACD; Downs, 2009; Downs et al., 2009). Ablation studies have demonstrated that the ACD, defined by localization of Brachyury (T), is crucial for appropriate elongation of the allantois to the chorion, enabling formation of the chorio-allantoic placenta (Downs et al., 2009). The ACD also exhibits multiple proteins involved in pluripotency, including Oct-3/4 (Downs, 2008), FGFs (Crossley and Martin, 1995; Ciruna and Rossant, 2001), Stella (Saitou et al., 2002; Mikedis and Downs, 2012), and c-myc (Downs et al., 1989). In addition, the ACD contributes to derivatives of all three embryonic primary germ layers, including surface ectoderm, hindgut endoderm, the allantois and posterior hemogenic vasculature, as well as to putative primordial germ cells (Mikedis and Downs, 2012). Thus, in addition to physical and other experimental evidence (Downs et al., 2009), the ACD exhibits hallmarks of the axial primitive streak. How the ACD differentiates into posterior cell lineages, particularly mesoderm and endoderm, its major derivatives (Downs et al., 2009; Mikedis and Downs, 2012), is not known.
A presumptive regulator of posterior progenitor mesendodermal cells is Mixl1 (Pereira et al., 2012a), a paired-type homeodomain transcription factor originally isolated in Xenopus, where it was expressed in mesoderm and blood in animal cap induction assays (Rosa, 1989; Mead et al., 1996). The human homologue of Mixl1 shares conserved Mix family domains; it can also induce expression of the hematopoietic α-globin gene in Xenopus animal caps (Guo, 2002). Mixl1 is also implicated in the development of hematopoietic malignancies. RT-PCR analysis revealed expression of human Mixl1 in tissues with hematopoietic expansion (e.g., lymph node germinal centers and spleen), as well as in T and B lymphocyte progenitors, but not in mature lymphocytes (Guo, 2002). While differentiated blood cells do not normally express Mixl1, poorly differentiated and rapidly proliferative human leukemias and lymphomas exhibit increased Mixl1 (Guo, 2002; Drakos et al., 2007). Further, irradiated mice that received donor hematopoietic stem cells constitutively expressing Mixl1 developed acute myeloid leukemia with anemia, thrombocytopenia, organomegaly and circulating myeloid blasts (Glaser et al., 2006; Metcalf et al., 2007). These findings suggest that aberrant Mixl1 may interfere with appropriate differentiation of hematopoietic stem cells.
Analysis of Mixl1 in embryonic stem cell (ESC) models has provided insight into its role in mesoderm/endoderm specification and hematopoiesis. Mixl1-GFP reporter in human ESCs under BMP-4 stimulation revealed early GFP expression, followed closely by co-expression with PDGFRα; later this subpopulation of cells expressed CD34, a more definitive hematopoietic marker (Davis et al., 2008). In cell culture assays, loss of Mixl1 resulted in loss of definitive endoderm and derangement of essential mesodermal structures; conversely, constitutive expression of Mixl1 in culture suppressed hematopoiesis and yielded a dramatic increase in expression of endodermal markers (Lim et al., 2009). These observations suggest that the quantity and/or timing of Mixl1 exposure within a progenitor population may impact descendants’ differentiation into ventral mesoderm (i.e., blood) or definitive endoderm. Therefore, based on available in vitro data, Mixl1 plays a role in the poorly understood events of mesendodermal differentiation within the posterior embryo and allantois, possibly through maintenance and specification of putative mesendodermal stem cell populations derived from the posterior primitive streak.
Mouse Mix-like 1 (Mixl1, also called mMix or mml) is the mouse homologue of Xenopus Mix.1 (Pearce and Evans, 1999). In mouse conceptuses, Mixl1 mRNA was first observed throughout the visceral endoderm prior to gastrulation, after which it became most prominent in the primitive streak and nascent mesoderm, with later restriction to the allantois and posterior primitive streak by headfold stages (Pearce and Evans, 1999; Robb et al., 2000; Mohn et al., 2003; staging of Downs and Davies, 1993). Weak expression in the tail bud then persisted through E11.5 (Pearce and Evans, 1999). Mixl1−/− embryos appeared unaffected until primitive streak stages when streak and node defects were observed; embryos ultimately exhibited shortening of the antero-posterior axis, poor neural fold development, mesenchymal disorganization, absence of a heart tube, and gut defects (Hart et al., 2002). Although the components of the exocoelom, including the yolk sac blood islands, appeared undisturbed, the allantois, which arises shortly after exocoelom formation, appeared unusually enlarged; Mixl1−/− embryos ultimately arrested in development around E9.0 (Hart et al., 2002).
In light of recent new findings on the relationship of the primitive streak to the allantois, we set out to characterize systematically, at the tissue level, localization of Mixl1 protein in the posterior region of the mouse conceptus, from formation of the primitive streak (~E6.5) through the completion of embryonic turning (~E9.5). Evaluation of co-localization with Runx1 has further allowed us to establish the relationship between Mixl1 and nascent blood-forming tissues, tailbud vasculature, and formation of the hindgut.
2. RESULTS
2.1. Specificity of Mixl1 antibody
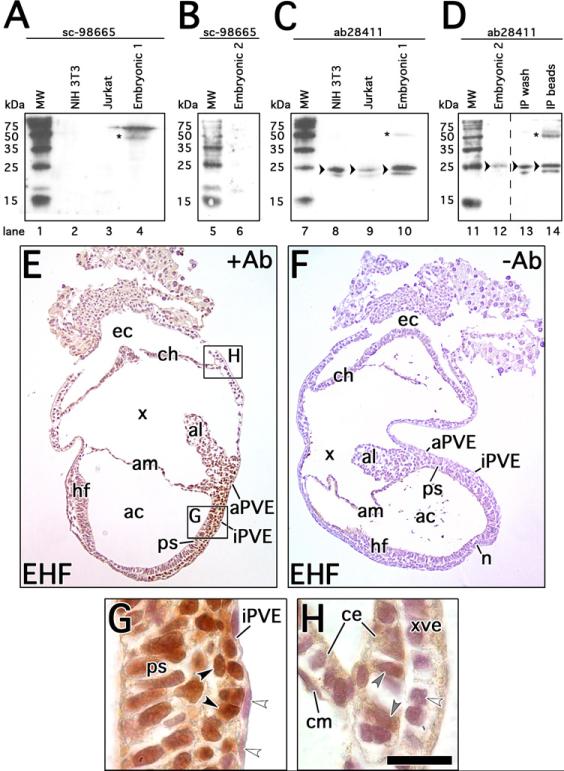
Two commercially available Mixl1 antibodies were compared by WB and IHC (see Section 4.3). The sc-98665 antibody did not identify a predicted band at 25kDa (Abcam, technical communication) in control NIH 3T3 or Jurkat cell lysates, nor in embryonic lysates 1 (denatured protein; Fig. 1A) or 2 (immunoprecipitated protein, Fig. 1B). Rather, sc-98665 identified two bands ≥50kDa (Fig. 1A), one of which correlated with immunoglobulin heavy chain, and which were not identified in embyronic lysate 2 (Fig. 1B). By contrast, the ab28411 antibody identified the predicted 25kDa band in control NIH 3T3 and Jurkat lysates, as well as in both types of embryonic lysates (Fig. 1C, D). It also identified a fainter band in each lysate at ~20kDa, as reported by the manufacturer on their WB (Abcam, technical communication). Ab28411 also identified a ~50kDa band, which correlated with immunoglobulin heavy chain, in embryonic lysate 1 (Fig. 1C) but which was absent in embryonic lysate 2 (Fig. 1D).
Figure 1. Specificity and staining intensities of Mixl1 antibody.
A-D, Western blots comparing sc-98665 and ab28411 Mixl1 antibodies. All molecular weight lanes (MW; lanes 1, 5, 7, 11) loaded with 2.5μL Trail Mix standards. A, lane 2, 5μg NIH 3T3 lysate; lane 3, 5μg Jurkat lysate; and lane 4, 23μg embryonic lysate 1, probed with 0.2μg/mL sc-98665. Asterisk indicates ≥50kDa bands in lane 4. B, lane 6, 20μg embryonic lysate 2, probed with 0.4μg/mL sc-98665. Note in A, B an additional faint band in MW lanes 1 and 5 at approximately 18kDa, which was not seen on control Western blot performed without primary antibody (not shown). C, lane 8, 5μg NIH 3T3 lysate; lane 9, 5μg Jurkat lysate; and lane 10, 23μg embryonic lysate 1, probed with 1μg/mL ab28411. Arrowheads indicate 25kDa band at predicted size for Mixl1 in lanes 8-10. Asterisk indicates ~50kDa band in lane 10. D, lane 12, 20μg embryonic lysate 2; lane 13, 20μg post-immunopreciptiation (IP) wash; and lane 14, post-IP beads boiled in loading buffer, probed with 1μg/mL ab28411. Arrowheads indicate 25kDa band at predicted size for Mixl1 in lanes 12-14. Asterisk indicates ≥50kDa band in lane 14. Dashed line indicates site where lanes of this blot were digitally juxtaposed for clarity; vertical positioning with respect to the MW markers was maintained. E-F, mid-sagittal section through EHF conceptuses with (E, +Ab) and without primary antibody (F, -Ab) added during immunostaining, respectively. G-H, examples of strong and faint staining intensities, at locations indicated in E. G, magnification at posterior primitive streak (ps), illustrating strong Mixl1 localization (black arrowheads); no signal is seen in the intraembryonic primitive streak-associated posterior visceral endoderm (iPVE; white arrowheads). H, magnification at posterior chorion-yolk sac junction, illustrating faint Mixl1 localization (gray arrowheads) in the chorionic ectoderm (ce); individual cells of chorionic mesoderm (cm) and extraembryonic visceral endoderm (xve) are Mixl1-negative (white arrowhead). All conceptuses shown before mid-turning (≤12s stage, ~E9.0) are oriented with anterior/dorsal to the left, posterior/ventral to the right, chorionic pole upward and embryonic pole downward. Scale bar in H: 166μm (E-F); 20μm (G-H). Abbreviations: ac, amnionic cavity; al, allantois; am, amnion; aPVE, allantois-associated posterior visceral endoderm; ch, chorion; ec, ectoplacental cavity; hf, headfold; n, node; x, exocoelom.
Based on these results, we predicted that sc-98665 did not effectively recognize Mixl1 primary sequence (i.e., denatured protein). To test this, sc-98665 was conjugated to protein G-coupled sepharose beads and used to immunoprecipitate Mixl1 from embryonic lysate 2 (see Section 4.3). Sc-98665 antibody was unable to identify Mixl1 from the denatured immunoprecipitate in a subsequent WB (data not shown), while ab28411 recognized Mixl1 in the IP bead wash supernatant (Fig. 1D). When the beads were subsequently boiled with denaturing gel loading buffer, ab28411 was then able to identify intensified Mixl1 as well as a band ~50kDa, consistent with immunoglobulin released from the beads during denaturation (Fig. 1D).
In immunostaining, sc-98665 exhibited specific tissue localization, whereas at similar stages no consistent localization was seen with ab28411 (data not shown; please see Section 4.5 for numbers of specimens examined). These results accorded with our finding, above, that ab28411 best recognizes primary sequence of Mixl1 (i.e., denatured protein), while sc-98665 best recognizes either the secondary or tertiary structure of Mixl1 (i.e., that found in whole mount IHC specimens). Further, ab28411 was not listed as recommended for IHC by the manufacturer (Abcam, technical communication). Therefore, all IHC results presented in this study utilized the sc-98665 antibody.
Specificity of sc-98665 was further supported by comparing immunostained whole conceptuses at EHF stage (“+Ab”, Fig. 1E) with similar-staged ones in which the primary antibody was eliminated from the reaction (“-Ab”, Fig. 1F). At this stage, prominent Mixl1 signal was noted in the primitive streak, allantois, and PVE (Fig. 1E, G; and data not shown), consistent with previously published mRNA localization data (Pearce and Evans, 1999). No signal was observed in the -Ab controls (Fig. 1F). In addition, other areas exhibited an intermediate intensity, which was reproducible across the many specimens examined, such as that seen in the chorionic ectoderm (Fig. 1H). Both the greater and lesser intensity staining patterns will be referred to throughout this work.
2.2. Mixl1 localized to the nascent primitive streak
The original description of Mixl1 expression by whole mount in situ hybridization (ISH) reported that Mixl1 mRNA was restricted to the posterior midline of embryonic visceral endoderm (EVE) and to the nascent primitive streak (~E6-6.5 conceptus; Pearce and Evans, 1999). As ISH can be a less sensitive detection method than IHC (Speel et al., 1999; Rumballe et al., 2008; Smith et al., 2008), we systematically localized Mixl1 protein to the mouse gastrula at short time intervals, starting at the time of emergence of the streak.
Mixl1 appeared to be confined to the nucleus in all cell types. At ES through LS stages (~E6.5-7.0), Mixl1 was identified throughout the primitive streak as it extended cranially (Fig 2A) Transverse orientations confirmed Mixl1's localization within the streak (Fig. 2B-C). Nascent mesoderm was also Mixl1-positive during these stages (Fig. 2B-C). The epiblast had no demonstrable Mixl1 (Fig. 2A-C). Mixl1 was also found throughout the EVE at MS stage (Fig. 2B); localization became biased toward the posterior midline EVE by the LS stage, with occasional positive cells still present in the anterior and lateral EVE (Fig. 2C). By the LS stage, the ventral and posterior cells of the node, which was now morphologically visible (Downs and Davies, 1993), were Mixl1-positive and remained so through the EHF stage (see Section 2.3).
Figure 2. Mixl1 localization in the primitive streak through neural plate stages.
A, mid-sagittal section through MS stage conceptus, with Mixl1 localized throughout the primitive streak (ps). Black line indicates approximate plane of section in B,C. B-C, transverse sections through MS and LS stage conceptuses at the middle of the ps, respectively, illustrating Mixl1 localization to the ps and the presumptive laterally-migrating embryonic mesoderm (me). Arrowheads indicate Mixl1-positive embryo-associated visceral endoderm (eve) cells. D, mid-sagittal section through an EB stage conceptus in the posterior half, exhibiting the Mixl1-positive ps, extraembryonic ps (xps), allantoic bud (al) and ventral node (n), as well as the visceral yolk sac (ys) and posterior chorion (ch). E, magnified view of the n at site indicated in D. Strong Mixl1 signal is noted in the ventral n and posterior crown (pc). F-G, magnified views of the ps and posterior visceral endoderm at sites indicated in D. F, allantois-associated posterior visceral endoderm (aPVE) and adjacent xps are strongly Mixl1-positive. G, many cells of the midline eve localize Mixl1 (arrowheads). H, mid-sagittal section of the al and posterior ps at LB stage, exhibiting the contiguous Mixl1 localization from the ps into the xps and al. I-J, magnified views of the ps and posterior visceral endoderm at sites indicated in H. I, aPVE (arrowheads) and xps are strongly Mixl1-positive. J, intraembryonic primitive streak-associated visceral endoderm (iPVE) has intermittent Mixl1-positive cells at the midline (arrowheads). Scale bar in J: 56μm (A); 33μm (B-C); 119μm (D), 34μm (E), 20μm (F-G), 50μm (H), 20μm (I-J). Other abbreviations: ac, amnionic cavity; am, amnion; e, epiblast; ec, ectoplacental cavity; pac, proamnionic cavity; pe, parietal endoderm; x, exocoelom; xve, extraembryonic visceral endoderm.
By the neural plate stages (~E7.0-7.5), Mixl1 localization included the full extent of the streak, including the extraembryonic primitive streak (XPS) that extends into the exocoelom during this time period (Fig. 2D; Downs et al., 2009), as well as the ventral node and posterior crown (Fig. 2D, E). Once it appeared, the entire allantoic bud was also Mixl1-positive (Fig. 2D).Within the visceral endoderm at the EB stage, the allantois-associated posterior visceral endoderm (aPVE) was consistently Mixl1-positive (Fig. 2F), as was the posterior EVE (Fig. 2G). By the LB stage, Mixl1 remained prominent in the posterior streak, XPS, allantoic bud and aPVE (Fig. 2H-I), but was only intermittently positive in the posterior EVE (Fig. 2J).
2.3. Headfold stage Mixl1 localized to the postero-ventral primitive streak and ventral node
During the EHF stage (~E7.75), the breadth of Mixl1 within the primitive streak became reduced. In some specimens, only the anterior and posterior ends of the streak were positive (Fig. 3A), while in others, Mixl1 persisted throughout the streak, without apparent ventral bias (Fig. 3B). There was also variability of Mixl1 within the node, with some specimens continuing to demonstrate Mixl1 throughout its ventral region (Fig. 3A) and others showing a refinement of Mixl1 to the posterior crown only (Fig. 3B). By the LHF stage (~E8.0), node-associated Mixl1 was found only in the posterior crown and all streak-associated Mixl1 was ventrally localized except for the posteriormost aspect at the embryonic-extraembryonic junction, where Mixl1 continued to localize to the full dorso-ventral extent of the streak (data not shown).
Figure 3. Mixl1 localization at EHF stage, compared with T and Oct-3/4.
A, midsagittal section, exhibiting Mixl1 within the ventral node (n) and posterior crown (pc), posterior primitive streak (ps) and allantoic core domain (ACD). B, midsagittal section of a specimen demonstrating Mixl1 within the pc and throughout the ps, and excluded from ventral n. C, midsagittal section at the allantois (al), with Mixl1 most prominently localized to the posterior ps and ventral ACD, and intermittently within allantois-associated posterior visceral endoderm (aPVE, white arrowheads). Individual mesenchymal cells in the al and allantoic mesothelium (m) are faintly Mixl1-positive (black arrowheads). D, schematic midsagittal view of EHF conceptus. Hashmarks indicate the ps, while black shading indicates the notochord (nc), n and ACD. Black horizontal lines indicate approximate positions of transverse sections shown in E-J. E+H, F+I and G+J are paired images taken from the same specimens, to facilitate comparison of staining intensity. E-G, transverse sections at the embryonic-extraembryonic junction exhibiting T, Oct-3/4 and Mixl1 localization, respectively. E, T labels the ACD as well as aPVE and nc. F, Oct-3/4 labels ACD and aPVE. Note that this is a frontally oblique transverse section in which the head folds and notochord are not visible. G, Mixl1 labels the ventral ACD and aPVE. Inset magnifies ventral ACD and aPVE; arrowheads indicate example Mixl1-positive aPVE cells. H-J, distal transverse sections exhibiting T, Oct-3/4 and Mixl1 localization, respectively. H, T labels the ps, embryonic mesoderm (me) and nc. I, Oct-3/4 labels ps, me and head folds (hf). J, Mixl1 is faintly detected in the ps and me, as highlighted in the inset; arrowheads indicate faintly Mixl1-positive embryonic visceral endoderm (eve) cells. Scale bar in J = 104μm (A), 35μm (B), 30μm (C), 111μm (E-J), 50μm (G inset), 31μm (J inset). Other abbreviations: ac, amnionic cavity; am, amnion; ec, ectoplacental cavity; iPVE, intraembryonic primitive streak-associated posterior visceral endoderm; x, exocoelom; ys, visceral yolk sac.
Within the allantois, intense Mixl1 was found in the ventral component of the ACD, adjacent to aPVE, the latter of which contained punctate, faintly Mixl1-positive cells (Fig. 3C). Distal allantoic mesenchyme and overlying mesothelium also exhibited punctate, faintly Mixl1-positive cells (Fig. 3C).
To better understand the pattern of Mixl1 at the EHF stage within the context of other proteins previously reported in the primitive streak, we compared Mixl1 localization with that of Brachyury (T) and Oct-3/4. Recent studies have established a direct interaction between T and Mixl1 proteins (Pereira et al., 2011). T localizes to nuclei within the primitive streak, node, notochord (Herrmann et al., 1990; Wilkinson et al., 1990), ACD (Inman and Downs, 2006b; Downs et al., 2009), nascent mesoderm, both embryonic and extraembryonic (Wilkinson et al., 1990; Inman and Downs, 2006b), endothelium (Inman and Downs, 2006b), hindgut endoderm (Wilkinson et al., 1990; Inman and Downs, 2006b) and chorionic ectoderm (Inman and Downs, 2006b). Oct-3/4, found in pluripotent stem cells (Pan et al., 2002; Sterneckert et al., 2012), localizes to the streak and its derivatives but in more dynamic antero-posterior patterns than T, diminishing anteriorly in advance of posterior down-regulation (Downs, 2008; Downs et al., 2009).
At the embryonic-extraembryonic junction into the allantois, T, Oct-3/4 and Mixl1 exhibited similar nuclear localization within the ACD (Fig. 3D-G). Punctate Mixl1 was also found within the aPVE (Fig. 3G). T and Oct-3/4 robustly identified the anterior aspect of the streak and nascent mesoderm (Fig. 3H-I), while Mixl1 was diminished there (compare Fig. 3J vs. 3H-I). Unlike T, Mixl1 was not appreciated in the notochord (compare Fig. 3E vs. 3G; Fig. 3H vs. 3J). EVE toward the anterior aspect of the streak was negative for Mixl1, save for occasional, faintly Mixl1-positive cells (compare insets, Fig. 3G vs. 3J) and Mixl1 was not detectable in the headfolds (Fig. 3J vs. 3I).
2.4. Mixl1 was maintained in the node posterior crown cells after neural plate stages
While mid-primitive streak Mixl1 waned during headfold stages (Fig. 3A), the posterior crown cells of the node, midline embryonic-extraembryonic junction, and ACD retained prominent Mixl1 at early somite stages (e.g., 2-6s; Fig. 4A-C). Neither the cells of the node's pit (compare Figs. 4A, B vs. 2E), the nearby streak, nor lateral mesoderm (Fig. 4B-D) exhibited Mixl1. Individual Mixl1-positive cells could be seen within mesoderm at the midline posterior to the node at each stage examined (e.g., Fig. 4A,E).
Figure 4. Mixl1 localization in the posterior crown and primitive streak at early somite stages.
A, mid-sagittal section at the posterior region at 2s stage. Mixl1 is present within the posterior crown (pc) of the node (n), posterior primitive streak (ps) and allantoic core domain (ACD). Occasional Mixl1 positive cells are seen within the more anterior aspects of the ps (arrowheads). Black lines indicate approximate planes of section in B-D. B-D, transverse sections at and posterior to the node at 4s stage. Mixl1 is not seen in the epiblast (e), ps or embryonic mesoderm (me) at this anatomic location. B, Mixl1 is not detected in the n. C, Mixl1 becomes evident and tightly clustered within the pc. D, immediately posterior to the pc, Mixl1 is not seen in the ps. E, mid-sagittal section through the n and anterior ps at 6s stage, exhibiting expanding midline Mixl1 posteriorly from the pc and scattered positive cells appearing within the nascent me (arrowheads). Scale bar in E = 100μm (A), 113μm (B-D), 50μm (E). Other abbreviations: al, allantois; am, amnion; eve, embryonic visceral endoderm; ys, visceral yolk sac.
2.5. Mixl1 was found at both ends of the elongating hindgut, and in the closely associated nascent posterior vasculature
To better understand Mixl1 localization during formation of the hindgut and posterior vasculature (9-16s), we evaluated Mixl1 in sagittal, oblique, transverse and frontal sections. By these stages, the major posterior arterial vessels have become distinct; these consist of the paired dorsal aortae (DA), omphalomesenteric artery (OA), and nascent umbilical artery (UA). These vessels anastomose at the vessel of confluence (VOC), located within the tailbud and immediately postero-ventral to the terminus of the hindgut (Fig. 5A; Downs, 1998; Daane and Downs, 2011).
Figure 5. Mixl1 localization within the posterior region at 13-16s stages.
A, whole mount of 16s stage tailbud region at the location indicated by the box in the inset. Major posterior vessels (da, dorsal aortae; oa, omphalomesenteric artery; vv, vitelline vessels; ua, umbilical artery; VOC, vessel of confluence) and the hindgut (hg) are diagrammed at their approximate locations. Black lines indicate approximate planes of section in panels D-H. As these specimens have undergone turning, the dorsal (rightward) and ventral (leftward) positions are reversed in this figure with respect to previous figures. B, mid-sagittal section at 16s stage tailbud. Mixl1 localizes to the tailbud mesoderm (tm) and posterior notochord (nc), while being excluded from the oa and most of the hg endoderm. Mixl1-positive cells can be seen peppering the mesoderm between the nc and tm Mixl1-positive populations (white arrowheads). Mixl1 is also noted throughout the allantois (al). C, magnified distal/dorsal region of the tailbud and hg at an adjacent section to B, illustrating punctate Mixl1-positive cells between the tm and nc populations (white arrowheads) and others interspersed within the dorsal hg endoderm (black arrowheads). Occasional Mixl1-positive cells are noted in the embryonic surface ectoderm (ee; hatched arrowhead). D-H, transverse sections through 13s stage embryo. D, posterior splanchnopleure (sp) exhibits Mixl1 as it folds toward the midline, as do the nc and visceral yolk sac (ys). E, immediately posterior to the midline (asterisk) joining of the sp, the hg endoderm remains Mixl1-positive, as does the nc. F, as the vv join at the midline (asterisk) to form the oa, these vessels exhibit Mixl1 and the nc remains positive. G, posterior to fusion of the vv, Mixl1 wanes within the hg and oa, and is limited to punctate positive cells in the nc (arrowheads). H, site anterior to hg closure, illustrating minimal Mixl1 localization outside of the ys. Scale bar in H = 64μm (A), 255μm (A inset), 88μm (B), 56μm (C), 44μm (D-H). Other abbreviations: nt, neural tube; pm, paraxial mesoderm.
During turning stages, Mixl1-positive cells were clustered within the tailbud mesoderm adjacent to the posterior terminus of the hindgut (Fig. 5B). Additionally, the putative posterior notochord population of axial mesodermal cells exhibited Mixl1 (Fig. 5B). The mesoderm spanning these two Mixl1-positive clusters, adjacent to the dorsal hindgut, also contained scattered Mixl1 positive cells (Fig. 5B-C). Intermittently, Mixl1-positive cells were interspersed within the dorsal hindgut endoderm (Fig. 5C). Mixl1 signal was also appreciated diffusely throughout the allantois (Fig. 5B), and in occasional cells along the embryonic surface ectoderm adjacent to the Mixl1-positive tailbud mesoderm (Fig. 5C).
Viewed transversely, the relationship of Mixl1 to hindgut and vascular formation was elucidated at mid-turning (~13s). Hindgut development appears to proceed in caudo-cranial sequence, with the lateral edges of the yolk sac (splanchnopleure) joining at the midline (Daane and Downs, 2011) to create the ventral component of the hindgut and OA (Fig. 5A, 5D-G). Mixl1 marked the midline splanchnopleure (Fig. 5D, E). Once the ventral hindgut was enclosed, Mixl1 was found throughout the circumference of the hindgut (Fig. 5E). Posterior to the site of OA formation and hindgut closure, midline Mixl1 in the hindgut, OA and vitelline vessels faded to undetectable (Fig. 5F, G). The notochord was strongly positive for Mixl1 adjacent to sites of hindgut closure and OA formation, fading to fainter, punctate signal at a more posterior level (Figs 5D,F vs. 5G). At sites anterior to the closing hindgut portal, Mixl1 was less prominent, found with relatively faint intensity in the visceral yolk sac endoderm (Fig. 5H).
2.6. Mixl1 co-localized with Runx1expression in multiple posterior sites
Previously, Mixl1 was implicated in hematopoiesis (Willey et al., 2006). Given Mixl1's localization to the allantois, which has hematopoietic potential (Zeigler et al., 2006; Corbel et al., 2007), and since some cells of the ACD give rise to hematopoietic cells (Mikedis and Downs, 2012), we assessed Mixl1's relation to expression of Runx1, a major early hematopoietic transcription factor (North et al., 2002) that has been identified in the allantois beginning at EHF stage (Zeigler et al., 2006; Daane and Downs, 2011).
Mixl1 and Runx1 co-localized throughout the EHF stage allantois (Fig. 6A-C). While Runx1 was prominently expressed throughout the distal allantois, in both the core and mesothelium, Mixl1 was faint but detectable (Fig. 6A, B). By contrast, while Runx1 expression was relatively less intense but still detectable in the ventral ACD, these cells exhibited the greatest Mixl1 intensity (Fig. 6C; see also Fig. 3C). Mixl1 and Runx1 co-localized with the greatest combined intensity along the aPVE (Fig. 6C), near the junction with the yolk sac and adjacent to the putative site of VOC formation, also called the “nook” (Rhee and Iannaccone, 2012). By 6s, Mixl1 co-localized with Runx1 at ventral sites in the proximal allantois (Fig. 6D), most prominently in presumptive budding blood cells and endothelium of the nascent UA (Fig. 6E), in ventral allantoic mesothelium (data not shown), and along the ventral hindgut lip, contiguously joining the hindgut with the visceral endoderm (Fig. 6F, G).
Figure 6. Mixl1 co-localization with Runx1.
Specimens shown are Runx1:lacZ reporter specimens; blue coloration indicates Runx1, and brown represents Mixl1 immunostaining. A-C, EHF stage, mid-sagittal view. A, Allantois (al) and posterior primitive streak (ps). Runx1 is seen throughout the al and visceral yolk sac (ys) endoderm; Mixl1 exhibits greatest intensity within the allantoic core domain (ACD). Boxes indicate approximate locations of panels B-C. B, magnified view of distal al, illustrating faint Mixl1 co-localization with prominent Runx1 expression throughout the distal al and mesothelium (m). C, magnified view of proximo-ventral al and ventral ACD; intense Mixl1 is seen throughout the ACD (black arrowheads), colocalizing with Runx1 most intensely in ventral cells (white arrowheads) near the site of the presumptive vessel of confluence (asterisk), as well as within the allantois-associated posterior visceral endoderm (aPVE). D-G, 6s stage, mid-sagittal view. D, posterior embryo and al. Mixl1 is noted within the proximal al and the presumptive posterior crown population (arrowhead). Boxes indicate approximate locations of panels E-G. E, magnified view of nascent umbilical vasculature (asterisks) in the proximal al, exhibiting strong Mixl1 localization in more proximal cells (black arrowhead) and Mixl1-Runx1 colocalization within endothelium-associated cells (white arrowheads). F-G, magnified view of adjacent sections at the embryonic-extraembryonic junction. Mixl1 is most intensely localized to primitive streak (ps)-associated cells adjacent to the hindgut (hg) invagination (black arrowheads), which do not exhibit Runx1. Runx1 is noted in the nascent vasculature (asterisks) and vitelline vessels (vv). Faint co-localization is noted within the ventral hg lip (hgl) and proximo-ventral m (white arrowheads). H, 10s stage, midsagittal section at site of hg invagination. Mixl1 is again noted at the hgl, and excluded from hg endoderm more posterior to the hgl. Box indicates area of magnification in I. I, magnified view of blood island (bi) in the ys, illustrating Mixl1 throughout the ys/bi mesoderm (ysm) and extraembryonic visceral endoderm (xve); the latter strongly co-localizes Runx1. Presumptive erythroblasts (eb) also co-localize Mixl1 and Runx1 (arrowheads). J, sagittal section at 9s stage, magnified to show the postero-lateral chorionic-ys junction. Mixl1 is noted at the lateral chorionic ectoderm (ce), adjacent to the site of contiguity with the ys (black arrowheads). Mixl1 and Runx1 co-localize within the xve, and, to a lesser extent, within the lateral ce. K, 10s stage, anterior transverse section, treated with brief X-gal exposure (see Materials and Methods), illustrating hazy Mixl1 signal throughout the amnion (am) and embryonic surface ectoderm (ee). The ys, bi and eb (black arrowhead) are Mixl1-positive, including some, but not all, circulating eb within the embryonic vasculature (inset, black vs. white arrowheads). L, 9s stage, anterior transverse section, treated with brief X-gal exposure, exhibiting no demonstrable Mixl1 signal within any of the indicated structures. Scale bar in L = 50μm (A), 35μm (B-C), 133μm (D), 49μm (E), 63μm (F-G), 78μm (H), 21μm (I), 47μm (J), 100μm (K-L), 18μm (K inset). Other abbreviations: ac, amnionic cavity; ch, chorion; cm, chorionic mesoderm; da, dorsal aorta; ec, ectoplacental cavity; fg, foregut; ht, heart; iPVE, intraembryonic primitive streak-associated posterior visceral endoderm; nc, notochord; nt, neural tube; oa, omphalomesenteric artery; pm, paraxial mesoderm; x, exocoelom.
Within the yolk sac blood islands, Mixl1 was initially found in a sparse, salt-and-pepper distribution as early as LB stage; as blood islands increased in size and number through 3s stage, Mixl1 localization was found in some – although not all – cells of each blood island (data not shown). This pattern of yolk sac blood island Mixl1 localization persisted through turning stages. Viewing the vitelline vessels and OA contiguously (Fig. 6H), Mixl1 localized to the yolk sac blood islands and developing blood cells of the OA as the latter became established within the embryo. Within the visceral yolk sac, both endothelium and blood cells co-localized Mixl1 and Runx1, with Mixl1 being strongest in the endothelium and Runx1 strongest in the blood cells, also beginning at bud stages (data not shown). In accord with results of a previous study (North et al., 1999), Runx1 was prominently expressed in the yolk sac endoderm (Fig. 6I).
2.7. Chorionic ectoderm exhibited hazy but persistent Mixl1
The chorion, which contains a trophoblast stem cell population (Uy et al., 2002), consists of a bilayer of extraembryonic mesoderm, which lines the exocoelomic cavity, and extraembryonic ectoderm, which faces the ectoplacental cavity. At its lateral edges, the chorion's contiguous extraembryonic ectoderm is apposed to distal extraembryonic endoderm, both of which contain Brachyury (Inman and Downs, 2006b). Prior to chorio-allantoic fusion (CAF), which begins in most conceptuses at the 4-5s stage (Downs and Gardner, 1995), Mixl1 localization was hazy and faint but consistent throughout the chorionic ectoderm (Fig. 1H, 2D), reminiscent of Brachyury (Inman and Downs, 2006b). This signal was not noted in -Ab controls (Fig. 1F). After CAF, Mixl1 localized to the lateral chorionic ectoderm, extending along the ectoplacental extraembryonic ectoderm, and including the distal visceral endoderm contiguous with the yolk sac (Fig. 6J). Runx1 co-localized with Mixl1 within the distal visceral endoderm and lateral chorionic ectoderm, which are ultimately thought to invade the lateral placenta with parietal endoderm to form the potentially hematopoietic Sinuses of Duval (Ogura et al., 1998; Daane and Downs, 2011).
2.8. Mixl1 localization within anterior and dorsal organs was minimal
Consistent with its not being found in epiblast or headfolds (Fig. 2A-C, 3G,J), Mixl1 was also not detectable later in the neural tube or developing brain (Fig. 6K-L). At and after early somite stages (i.e., 3-4s), anterior mesoderm and endoderm derivatives variably exhibited no, or faint, Mixl1, which was at a relatively minimal intensity when compared with the posterior positive structures within the same specimens. These organs included the heart, anterior DA, paraxial mesoderm, and foregut (Fig. 6K-L). Mixl1 also was found in some, but not all, circulating erythroblasts (Fig. 6K).
3. DISCUSSION
We have systematically evaluated Mixl1 localization throughout the mouse conceptus from the onset of gastrulation through embryonic turning, and have expanded on previous ISH-based expression data. Similar to previous results (Pearce and Evans, 1999; Robb et al., 2000; Mohn et al., 2003), we found that Mixl1 marked the early primitive streak. We have also elucidated tissue-level Mixl1 localization within the anterior and posterior remnants of the streak: the node posterior crown cells and the ACD. By contrast to previous results, Mixl1 was found in a variety of hematopoietic and vasculogenic mesodermal derivatives, including the allantois, VOC, tailbud, nascent OA and UA, blood islands, and chorion, as well as in the forming hindgut, despite a previous report that Mixl1 is not expressed in definitive endoderm (Mohn et al., 2003). We attribute these differences in localization to differences in sensitivity between ISH, used previously, and IHC, which we have employed; in particular, ISH signal loss has been documented when whole mount specimens are paraffin-embedded and sectioned (Speel et al., 1999; Rumballe et al., 2008; Smith et al., 2008), possibly complicating previous authors’ ability to discern tissue-level Mixl1 expression. Fig. 7 summarizes Mixl1 localization during mouse gastrulation and elaborates upon Mixl1 expression previously presented (Pereira et al., 2012a).
Figure 7. Schematic representation of Mixl1 localization in the mouse conceptus through ~E9.0.
Mixl1 localization is represented by violet shading. Gradient shading indicates diffuse or fading localization; hatched shading indicates punctate expression. Structures are not necessarily drawn to scale. A-E represent mid-sagittal positions within the conceptus. Solid black lines in A-B indicate embryonic-extraembryonic interface (arrowhead). A, MS stage: Mixl1 identifies the primitive streak (ps) and embryo-associated visceral endoderm (eve). Arrow indicates ps progression toward the distal egg cylinder and concomitant Mixl1 within the expanding ps. B, OB stage: Mixl1 is found throughout the ps and into the now-developing extraembryonic extension of the streak (xps), ventral node (n), and in a punctate pattern within posterior eve. C, EB-EHF stages: Mixl1 within the ps recedes toward the posterior (gradient arrow), although ventral n, posterior crown (pc) and ventral ps cells still exhibit Mixl1. Mixl1 localizes throughout the allantoic (al) bud during neural plate stages, and labels the allantoic core domain (ACD) starting at headfold stages (dashed line). The allantois-associated posterior visceral endoderm (asterisk) remains Mixl1-positive. D, 4s-6s stages: Mixl1 has become largely confined to populations at either end of the ps: the pc as it progresses posteriorly (gradient arrow), and a collection of posterior/ventral cells at the embryonic-extraembryonic junction surrounding the site of hindgut (hg) invagination. Definitive hg endoderm at the invagination, adjacent visceral endoderm, lateral chorionic ectoderm (ce) and yolk sac (ys) visceral endoderm also localize Mixl1. E, 9s-11s: Magnified view of the posterior conceptus. The Mixl1-positive cells of the pc, at the position of presumptive posterior notochord (nc), have retracted to the proximal dorsal hg. Mixl1 also localizes to the mesoderm surrounding the distal hg, as well as to the endoderm surrounding the site of closure of the proximal hg; this includes the ventral hg lip (hgl). Black lines indicate planes of transverse views in F-I. F-I, progressive anterior-to-posterior transverse views of the posterior region at 11s, illustrating Mixl1 localization within the posterior notochord (nc) and at the closing edges of the forming omphalomesenteric artery (oa) and hg. Other abbreviations: ac, amnionic cavity; am, amnion; bw, body wall; ch, chorion; da, dorsal aorta; e, epiblast; hf, head fold; nf, neural fold; pac, proamnionic cavity; ua, umbilical artery; VOC, vessel of confluence; vv, vitelline vessels; ve, visceral endoderm; x, exocoelom.
3.1. During early gastrulation, Mixl1 localizes to visceral endoderm and embryonic endoderm
As the primitive streak formed, Mixl1 was observed throughout the embryonic visceral endoderm, in salt-and-pepper distribution, ultimately becoming confined to the posterior midline and yolk sac visceral endoderm by headfold stages (Fig. 7A-C). The initial formation of definitive endoderm has been shown to involve intercalation of primitive streak-derived cells into the visceral endoderm layer (Kwon et al., 2008), likely causing displacement of much of the visceral endoderm to the yolk sac (Lawson et al., 1986; Lawson et al., 1991), although some visceral endoderm-derived cells persist in the posterior midline (Kwon et al., 2008). Interestingly, labeled Mixl1−/− cells grafted to the posterior primitive streak exhibit reduced contribution to endoderm and increased contribution to proliferative but disorganized mesoderm when compared with Mixl1+/- cells; consequently, visceral endoderm cells exhibit poor displacement posteriorly (Hart et al., 2002; Tam et al., 2007). Our findings of persistent Mixl1 in the posterior visceral endoderm may represent displaced visceral endoderm (Fig. 7B-C; see below), while punctate endodermal Mixl1 is reminiscent of the intercalation of gastrulating embryonic endoderm cells (Kwon et al., 2008), consistent with reports that Mixl1 is a marker of early endodermal differentiation in ESCs (Tada et al., 2005; Pereira et al., 2012a).
3.2. During early gastrulation, Mixl1 localizes to primitive streak-associated organizer tissue
During ES-LS stages, Mixl1 localized to the entire streak, including the anteriormost aspect (Fig. 7A). This region of the elongating streak during ES and MS stages, respectively called the early- and mid-gastrula organizer (EGO, MGO; Lawson et al., 1991; Kinder et al., 2001), contributes mesodermal cells that migrate anteriorly and build the anterior head process (Robb and Tam, 2004; Yamanaka et al., 2007). This structure, in turn, contributes to the most cranial region of the notochord, and elaborates signals for induction of brain, cranial mesenchyme, heart and foregut development (Robb and Tam, 2004). Intriguingly, the mesodermal derivatives of the EGO, MGO and anterior streak, specifically the anterior axial and paraxial mesoderm and heart mesenchyme, did not contain detectable Mixl1 (Fig. 6K-L), although the Mixl1−/− embryo exhibits dramatic defects in heart, head and brain development (Hart et al., 2002). Thus, Mixl1 in the early anterior primitive streak may be necessary for maintenance and/or specification of EGO/MGO populations before they migrate anteriorly.
3.3. Anterior streak-associated Mixl1 becomes refined to the node posterior crown cells and their descendants
By the LS stage, Mixl1 was observed throughout the ventral node (Fig. 7B), a structure that forms from the anterior condensation of the streak at LS stage, and has been described as the “late organizer” (Robb and Tam, 2004; Yamanaka et al., 2007). Node formation depends on the forkhead transcription factor FoxH1 (Hoodless et al., 2001), which is part of the complex that promotes Mixl1 expression (Hart et al., 2005). Cells arising in the node give rise to trunk notochord (Yamanaka et al., 2007), and induce proper dorsoventral neural patterning (Davidson et al., 1999; Lee and Anderson, 2008); the Mixl1−/− mouse lacks a morphologically distinct node, has grossly disorganized axial mesoderm, and develops a malformed neural plate (Hart et al., 2002), supporting a role for Mixl1 in development or maintenance of the node.
From LS through headfold stages, Mixl1 localization waned from the ventral node and was retained in the posterior crown cells thereafter (Fig. 7C-D). The ventral node gives rise to the posterior crown cells, also described as notochord progenitor cells (Kinder et al., 2001; Ukita et al., 2009). These cells prominently express Oct-3/4 (Downs, 2008), which is associated with stem cell maintenance and developmental potency (Pan et al., 2002; Sterneckert et al., 2012). While they appear mitotically quiescent from headfold stages through E8.25 (Bellomo et al., 1996), posterior crown cells become markedly more proliferative from E9.25-E10.5 (Ukita et al., 2009). Throughout early somitogenesis stages, they also express Noto and migrate to the tailbud, forming the posterior extension of the notochord (Robb and Tam, 2004; Yamanaka et al., 2007; Ukita et al., 2009). We found intermittent Mixl1-positive cells within the axial mesoderm posterior to the posterior crown as early as the 2s stage (Fig. 4A,E), reminiscent of Noto-expressing cells migrating to the tailbud to form the tail notochord-forming cluster (Yamanaka et al., 2007). The posterior crown region has also been fate-mapped to dorsal hindgut endoderm (Lawson et al., 1986; Wilson and Beddington, 1996), and our findings of Mixl1 in this region during hindgut elongation correlate closely with a fate map of cells originating from the posterior crown at LS stage (compare Lawson et al. 1986 Fig. 2 Zone IV vs. Fig. 7E). Finally, posterior crown derivatives also fate map to the tailbud mesoderm, a progenitor pool thought to support posterior axial elongation (Tam and Tan, 1992; Wilson and Beddington, 1996).
Therefore, the cells of the ventral node-derived posterior crown, which localize Mixl1, act as progenitors for embryonic mesoderm (e.g., notochord, tailbud) and endoderm (e.g., dorsal hindgut). This finding is mirrored in the posterior remnant of the streak.
3.4. Posterior streak-associated Mixl1 becomes refined to the ACD and tailbud mesoderm
During neural plate stages, Mixl1 identified the posterior extension of the streak, the XPS (Fig. 7B), and became refined to the posterior streak and proximal ACD during headfold stages (Fig. 7C). Recently, the posterior end of the streak has been found to extend into the exocoelom during neural plate stages (Downs et al., 2009). There, with overlying posterior visceral endoderm, the XPS establishes the ACD within the base of the allantois by EHF stage (Downs, 2009; Downs et al., 2009). Genetic and microsurgical deletion of this domain has revealed its critical role as a progenitor pool for elongation of the allantois to the chorion (Inman and Downs, 2006b; Downs et al., 2009).
Synchronous orthotopic grafting experiments revealed that the ACD can robustly contribute to mesoderm, especially the hemogenic posterior vasculature, as well as to ventral hindgut endoderm and the putative primordial germ cells (Mikedis and Downs, 2012). We found that Mixl1 waned within the allantois and ACD from headfold through early somite stages, and by 6s stage was found in a relatively small population of cells at the base of the allantois, associated with the hindgut invagination and VOC (Fig. 7D), consistent with known proximal ACD descendants (Mikedis and Downs, 2012). Through turning stages (e.g., 9-16s), Mixl1 was retained in the allantoic vasculature (Fig. 5B), to which the ACD contributes (Mikedis and Downs, 2012), and in the tailbud mesoderm (Fig. 5B-C), to which the posterior crown (see Section 3.3) and ACD also contribute (Tam and Tan, 1992; Wilson and Beddington, 1996; Mikedis and Downs, 2012). Thus, Mixl1-localizing cells of the proximal ACD exhibit contributions to both mesoderm (e.g., vasculature, tailbud), and endoderm (e.g., ventral hindgut).
3.5. Mixl1 identifies putative axial mesendodermal progenitors derived from either end of the primitive streak
On the basis of the above observations, Mixl1 may identify populations of bipotential mesendodermal stem cells (MSCs) at either end of the primitive streak. The existence of MSCs has been supported by molecular signature in ESC culture, in which Gsc, E-cadherin, Foxa2 and PDGFRα identified the MSCs (Tada et al., 2005), but proving their existence in vivo has been elusive. Supportive of this role, analysis of Mixl1+ and Mixl1-null mouse ESC transcriptomes revealed that loss of Mixl1 yielded decreases in multiple gene products often associated with both endoderm (e.g., Gsc, Gata6, Cxcr4) and mesoderm (specifically, blood-and vasculature-forming, e.g., Flk1, PDGFRα) specification (Pereira et al., 2012b).
Differential dosage of Mixl1 may govern differentiation of progenitors into mesoderm or endoderm. Willey and colleagues (2006) used a conditional promoter to drive Mixl1 in embryoid bodies, and showed an increase in cells with mesodermal potential, specifically primitive and definitive hematopoietic cells. Similarly, descendants of labeled human ESCs expressing Mixl1 and PDGFRα, cultured under mesoderm-biasing conditions, were found to express CD34, a marker of pluripotent hematopoietic stem cells (Davis et al., 2008). Conversely, others have shown that mouse Mixl1 overexpression induced transcription of endoderm-associated genes, such as Foxa2, Gsc, Sox17 and E-cadherin, without apparent hematopoietic descendants (Lim et al., 2009). Thus, Mixl1 can specify mesodermal or endodermal fates in progenitor cells, potentially dependent on the quantity or duration of exposure (proposed by Lim et al., 2009), or on available binding partners in situ (Pereira et al., 2012a).
3.6. Mix1 and T coincide in progenitor populations and at sites of gut formation, primitive hematopoiesis and vasculogenesis
As with many homeodomain proteins, Mixl1 is capable of dimerization (Zhang et al., 2009; Pereira et al., 2012b), and T has recently been shown to bind Mixl1 and attenuate its activity (Pereira et al., 2011). Conversely, Mixl1−/− embryos exhibit an expanded domain of T (Hart et al., 2002), suggesting that mouse Mixl1 suppresses T, just as Xenopus Mix.1 represses Xbra expression (Latinkić and Smith, 1999). The most striking posterior morphogenetic defect reported in Mixl1−/− was an enlarged allantois (Hart et al., 2002). One possible explanation for this observation is that Mixl1 maintains but limits the scope of the ACD progenitor population, and that this population experiences poorly-regulated proliferation when Mixl1 is lost, perhaps through expanded activity of T. As we have found sites of coincident localization patterns of Mixl1 and T in the posterior notochord and ACD, these transcription factors may help to balance proliferation and differentiation through reciprocal inhibition in these progenitor populations.
Maintenance of the node-derived posterior crown population is considered essential for proper tailbud outgrowth and neurectodermal and mesodermal patterning (Yamanaka et al., 2007), and depends on T. Brachyury mutants likely exhibit tail truncation phenotypes due to loss of the posterior extension of the notochord (Herrmann and Kispert, 1994). Similarly, in the posterior conceptus, a subregion of extraembryonic visceral endoderm, the aPVE (formerly “allantois-associated extraembryonic visceral endoderm,” AX; Downs et al., 2009) has recently been identified. This strip of visceral endoderm becomes T- and Oct-3/4-positive only once the ACD has formed (Fig. 3E, F; Downs, 2008; Downs et al., 2009). Presumably, the Oct-3/4 and T-positive cells enter the aPVE as part of the hindgut's cellular endowment from the streak. By contrast, the aPVE was Mixl1-positive prior to ACD formation (Fig. 2F, 2I, 7C). This region of endoderm at the embryonic-extraembryonic junction, corresponding to the posterior visceral endoderm tissue formed by earlier displacement of visceral endoderm by intercalating definitive endoderm described above (Lawson et al., 1991; Kwon et al., 2008), may anticipate the future site of hindgut invagination (Hara et al., 2009). This process depends on both Mixl1 and T, especially as mutants of both genes exhibit malformations of the hindgut (Hart et al., 2002; Inman and Downs, 2006a; Tam et al., 2007).
We have also found that Mixl1 and T localization coincides in sites of primitive hematopoiesis and placentation. Mixl1 was found throughout the blood islands, co-localized with Runx1 in subjacent yolk sac visceral endoderm (Fig. 6I), coincident with T localization to the yolk sac visceral endoderm between the EB and 10s stages (Inman and Downs, 2006b). During these same stages, blood islands arise within the yolk sac mesoderm in response to inductive signals from visceral endoderm (Belaoussoff et al., 1998; Palis and Yoder, 2001). Blood islands can form endothelium and primitive erythroblasts, representing the earliest rudiments of the vitelline vasculature.
Within the chorion, Mixl1 ultimately localized to the lateral chorionic ectoderm, straddling the junction with the yolk sac (Fig. 6J, 7D), and T has been found to identify the chorionic ectoderm prior to its fusion with the ectoplacental cone (Inman and Downs, 2006b). The unfused lateral chorionic ectoderm exhibits increased mitotic activity and greater trophoblast stem cell potential than the central fusing chorion (Uy et al., 2002). Mouse and rat conceptuses develop an intraplacental yolk sac organ (also called Sinuses of Duval), consisting of invagination of yolk sac endoderm into the placental disc, such that the chorion-yolk sac interface becomes adjacent to the parietal endoderm at the placental pole (Duval, 1892; Ogura et al., 1998). Runx1 has been found at sites of yolk sac invagination into the developing placenta, suggestive of a role in hematopoiesis (Daane and Downs, 2011). T and Mixl1 may interact within the yolk sac and lateral chorion, sites of extraembryonic mesoderm and endoderm or ectoderm apposition respectively, to specify primitive hematopoiesis and vasculogenesis. Since Mixl1 is found in tissues with early hematopoietic and vasculogenic potential, it may identify the earliest-known hemangioblast precursors (Huber et al., 2004). Surprisingly, the exocoelom, visceral yolk sac and blood islands were reportedly normal in the Mixl1−/− embryo, and no perturbation in T expression at these sites was described (Hart et al., 2002). Nevertheless, T could be up-regulated in Mixl1−/− extraembryonic membranes as it is in the embryo proper, which appears consistent in the whole mount ISH specimens previously described (compare Hart et al., 2002 Fig. 5B vs. 5A).
4. MATERIALS AND METHODS
4.1. Mouse husbandry, dissections, and embryo staging
Mice were treated in accordance with Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals (Public Law 99–158) as enforced by the University of Wisconsin-Madison. Standard F2 embryological specimens were obtained by intercrossing inbred hybrids B6CBAF1/J (Jackson Laboratory, Bar Harbor, ME; Downs, 2006). Individual estrous females (Champlin et al., 1973) and stud males were placed together just before lights went off (13.00 hours/01.00 hours or 21.00/09.00 lights off/on); copulation plugs were identified up to 12 hours later. Runx-1:lacZ specimens were obtained by mating heterozygous males (Runx1tm2Spe; North et al., 1999; North et al., 2002) bred onto the B6CBAF1/J background (Daane and Downs, 2011). Dissection and staging of conceptuses were as described by Downs and Davies (1993).
4.2. Fixation, storage and X-gal staining
Staged conceptuses were fixed in 4% paraformaldehyde (2 hours, 4°C) and rinsed in phosphate-buffered saline (PBS, Sigma-Aldrich, St. Louis, MO; Downs, 2008). Runx1:lacZ+/− conceptuses were stained with X-gal (below), after which they, as well as B6CBAF1/J F2 specimens were dehydrated in increasing methanols/PBS through absolute methanol and stored at –20°C for at least three days in anticipation of immunostaining. To ensure equivalent penetration of X-gal solution and immunostaining reagents, all specimens from the 9s stage onward were pierced with a beveled insulin needle (28-gauge) at the lateral yolk sac and amnion after fixation.
X-gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside; B4252, Sigma-Aldrich) staining for Runx1 expression utilized a preparation of 10 mM of K4Fe(CN)6•3H2O, 2mM MgCl2 and 1mg/ml X-gal (stock concentration 40 mg/mL DMSO; Daane and Downs, 2011) and was carried out on fixed conceptuses for 13-17 hours at 37°C; to reduce X-gal intensity, selected specimens at 9-12s employed X-gal incubation times of 4-6 hours. No background staining suggestive of endogenous β-galactosidase activity was observed in non-Runx1/lacZ+ littermates (data not shown).
4.3. Immunoprecipitation (IP) and Western blotting (WB)
Two primary Mixl1 antibodies were used throughout this study: (1) sc-98665, rabbit polyclonal antibody raised against the N-terminal 134 amino acid epitope of mouse Mixl1, (Santa Cruz Biotechnology, Santa Cruz, CA), and (2) ab28411, rabbit polyclonal antibody raised against keyhole limpet hemocyanin-conjugated peptide hapten synthesized from the C-terminal 32 amino acid residues of mouse Mixl1 (Abcam, Cambridge, MA).
Embryonic lysate 1 consisted of denatured protein extracted from EHF-5s (~E7.75-8.25) conceptuses (gift of M. Mikedis; Mikedis and Downs, 2013). Embryonic lysate 2 was prepared from seven litters of late streak through 8 somite pair (~E7.0-8.75) staged conceptuses, divested of trophoblast giant cells, parietal endoderm and ectoplacental cone, then flash frozen, homogenized with a Tissue Miser (OM02012, Fisher Scientific, Pittsburgh, PA) in complete RIPA buffer (sc-24948, Santa Cruz) to facilitate nuclear extraction and to avoid ionic detergent exposure. The supernatant protein lysate was recovered as previously described (Abcam, technical communication).
Embryonic lysate 2 was pre-cleared using 582 μg of raw lysate mixed with 4 μg of rabbit IgG (sc-2027, Santa Cruz) and precipitated with 25 μL of protein G-sepharose conjugate (101241, Invitrogen, Life Technologies, Grand Island, NY) as previously described (Abcam, technical communication). Additionally, 65 μL of protein G-sepharose beads were bound to 5 μg of Mixl1 antibody (sc-98665, Santa Cruz), then crosslinked in 6.5 mg/mL dimethyl pimelimidate and 0.1 M triethanolamine in three successive treatments. Quenching was performed with 50 mM ethanolamine in two washes, followed by removal of unlinked antibody by two washes in 1 M glycine pH 3, as previously described (Abcam, technical communication). IP was performed with 296 μg pre-cleared lysate 2 and 65 μL sc-98665–crosslinked sepharose beads, then washed gently in complete RIPA buffer, followed by a second elution in WB loading buffer plus β-mercaptoethanol and boiling for 10 min (Abcam, technical communication).
To detect Mixl1 protein, 20-23 μg of each embryonic lysate and the post-IP wash were subjected to SDS-PAGE under reducing conditions in a 15% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane (IPVH00010 Immobilon-P Transfer Membrane, EMD Millipore, Darmstadt, Germany), and blocked in 5.0% milk/Tris-buffered saline (TBS)-Tween overnight at 4°C. The blot was then probed with either: sc-98665 at 0.2-0.4 μg/mL (Santa Cruz, technical communication), or ab28411 at 1 μg/mL (Abcam, technical communication) in 2.5% milk/TBS-Tween overnight at 4°C, washed three times in TBS-Tween, probed with horseradish peroxidase-conjugated donkey anti-rabbit secondary antibody (sc-2313, Santa Cruz) at 1/5,000 dilution (80ng/mL) in 2.5% milk/TBS-Tween for 1h at room temperature, washed three times, and visualized via chemiluminescent detection (RPN2134, Amersham ECL Western blotting detection reagents, GE Healthcare, Madison, WI) on Amersham Hyperfilm ECL (28906835; GE Healthcare). NIH 3T3 whole cell lysate (sc-2210, Santa Cruz, 5 μg/lane) and Jurkat whole cell lysate (sc-2204, Santa Cruz, 5μg/lane) were used as positive controls for Mixl1 (Drakos et al., 2007; Zhang et al., 2009). Novagen Trail Mix Protein Molecular Weight Markers (71048-3, EMD Millipore) were used according to manufacturer's instructions.
4.4. Immunohistochemistry (IHC) and histological analysis
IHC using primary antibodies against Mixl1 was carried out in whole conceptus preparations as previously described (Downs, 2008). Two antibodies were applied to conceptuses for comparison (see above). Primary antibody sc-98665 (stock 200 μg/mL) was optimized at 1:100 (2 μg/mL); in addition, ab28411 (stock 200 μg/mL) was evaluated at a range of concentrations from 1:200 (1 μg/mL) through 1:50 (4 μg/mL) but an optimal concentration was not achieved; nevertheless this antibody provided valuable information about antibody specificity (please see Results). Negative control immunostaining followed the same protocol, with omission of the primary antibody. No control peptide is available for sc-98665 (Santa Cruz Biotechnology, technical communication). T and Oct-3/4 immunostaining had been carried out previously (Downs, 2008; Downs et al., 2009).
Immunostained whole mount conceptuses were dehydrated and embedded in paraffin as previously described (Downs, 2008). Specimens were sectioned at a thickness of 6 micrometers (microns, μm), prepared for histology, and counterstained with Gill's hematoxylin (Sigma-Aldrich). Slides were viewed using a Nikon Microphot-SA compound microscope (Nikon Instruments Inc., Melville, NY) and photographed with an attached Spot RTKE (Spot Diagnostic Instruments Inc., Sterling Heights, MI) or QImaging Retiga 2000R digital camera (QImaging, Surrey BC, Canada). Whole mount specimens were evaluated in a Nikon SMZ1500 dissection microscope and photographed as above. Image processing was performed in Adobe Photoshop CS5 (Adobe Systems Inc., San Jose, CA), utilizing level and saturation adjustments and a sharpening filter.
4.5. Number of specimens evaluated
A total of 140 F2 conceptuses was evaluated for Mixl1 localization using sc-98665 antibody: 22 × early streak (ES) – late streak (LS), 1 × no bud (OB), 5 × early bud (EB), 11 × late bud (LB), 26 × early headfold (EHF), 10 × late headfold (LHF), 5 × 1 somite pair (1s), 9 × 2s, 8 × 3s, 5 × 4s, 3 × 5s, 1 × 6s, 3 × 8s, 17 × 9-12s, and 14 × 13-16s. A total of 116 X-Gal-treated Runx1/lacZ+/− conceptuses was evaluated for co-localization of Mixl1 and Runx1: 3 × ES-LS, 1 × OB, 2 × EB, 2 × LB, 8 × EHF, 16 × LHF, 5 × 1s, 9 × 2s, 17 × 3s, 8 × 4s, 5 × 6s, 2 × 7s, 4 × 8s, and 34 × 9-12s. A total of 30 F2 conceptuses was evaluated with ab28411 at similar stages but without positive results (see Results): 2 × LB, 13 × EHF, 1 × 3s, 4 × 5s, 2 × 6s, 2 × 9s, and 6 × 10s. The above totals reflect a combination of specimens sectioned primarily in sagittal and transverse orientations; some frontal and oblique orientations were also prepared from selected 9-16s stage conceptuses.
Mixl1 protein has been localized to the mouse gastrula from ~E6.5-E9.5.
Mixl1 may identify putative mesendodermal progenitors within the axial midline.
Mixl1 localizes to early hemogenic/vasculogenic sites in the posterior conceptus.
Mixl1 localizes to the invaginating hindgut.
Results support Mixl1 in mesendodermal stem cell niches in early organogenesis.
ACKNOWLEDGMENTS
The authors are grateful to Maria M. Mikedis for help with Western blotting, and for critical reading of parts of this manuscript, and to Dexter Jin, Erin Willing and Lauren Wierenga for technical assistance. This study was supported by grants from the March of Dimes (1-FY09-511) and National Institutes of Child Health and Development (R01 HD042706) (K.M.D.). A.D.W. is further supported by NIH National Heart, Lung, and Blood Institute Grant “Research Training in Hematology” (T32 HL07899), and is a current recipient of an NIH Extramural Pediatric Research Loan Repayment Program grant (NOT-OD-11-086).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beddington RS. Induction of a second neural axis by the mouse node. Development. 1994;120:613–620. doi: 10.1242/dev.120.3.613. [DOI] [PubMed] [Google Scholar]
- Belaoussoff M, Farrington SM, Baron MH. Hematopoietic induction and respecification of A-P identity by visceral endoderm signaling in the mouse embryo. Development. 1998;125:5009–5018. doi: 10.1242/dev.125.24.5009. [DOI] [PubMed] [Google Scholar]
- Bellomo D, Lander A, Harragan I, Brown NA. Cell proliferation in mammalian gastrulation: The ventral node and notochord are relatively quiescent. Developmental Dynamics. 1996;205:471–485. doi: 10.1002/(SICI)1097-0177(199604)205:4<471::AID-AJA10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Champlin AK, Dorr DL, Gates AH. Determining the stage of the estrous cycle in the mouse by the appearance of the vagina. Biology of reproduction. 1973;8:491–494. doi: 10.1093/biolreprod/8.4.491. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Corbel C, Salaun J, Belo-Diabangouaya P, Dieterlen-Lievre F. Hematopoietic potential of the pre-fusion allantois. Developmental biology. 2007;301:478–488. doi: 10.1016/j.ydbio.2006.08.069. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Daane JM, Downs KM. Hedgehog signaling in the posterior region of the mouse gastrula suggests manifold roles in the fetal-umbilical connection and posterior morphogenesis. Developmental Dynamics. 2011;240:2175–2193. doi: 10.1002/dvdy.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BP, Kinder SJ, Steiner K, Schoenwolf GC, Tam PP. Impact of node ablation on the morphogenesis of the body axis and the lateral asymmetry of the mouse embryo during early organogenesis. Developmental biology. 1999;211:11–26. doi: 10.1006/dbio.1999.9276. [DOI] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Downs KM. The murine allantois. Current topics in developmental biology. 1998;39:1–33. doi: 10.1016/s0070-2153(08)60451-2. [DOI] [PubMed] [Google Scholar]
- Downs KM. In vitro methods for studying vascularization of the murine allantois and allantoic union with the chorion. Methods in molecular medicine. 2006;121:241–272. doi: 10.1385/1-59259-983-4:239. [DOI] [PubMed] [Google Scholar]
- Downs KM. Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:464–475. doi: 10.1002/dvdy.21438. [DOI] [PubMed] [Google Scholar]
- Downs KM. The enigmatic primitive streak: prevailing notions and challenges concerning the body axis of mammals. BioEssays. 2009;31:892–902. doi: 10.1002/bies.200900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Downs KM, Gardner RL. An investigation into early placental ontogeny: allantoic attachment to the chorion is selective and developmentally regulated. Development. 1995;121:407–416. doi: 10.1242/dev.121.2.407. [DOI] [PubMed] [Google Scholar]
- Downs KM, Inman KE, Jin DX, Enders AC. The Allantoic Core Domain: new insights into development of the murine allantois and its relation to the primitive streak. Developmental Dynamics. 2009;238:532–553. doi: 10.1002/dvdy.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes & development. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Drakos E, Rassidakis GZ, Leventaki V, Guo W, Medeiros LJ, Nagarajan L. Differential expression of the human MIXL1 gene product in non-Hodgkin and Hodgkin lymphomas. Human pathology. 2007;38:500–507. doi: 10.1016/j.humpath.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Duval M. Le placenta des rongeurs. Felix Alcan; Paris, France: 1892. p. 640. [Google Scholar]
- Glaser S, Metcalf D, Wu L, Hart AH, DiRago L, Mifsud S, D'Amico A, Dagger S, Campo C, Chan AC, Izon DJ, Robb L. Enforced expression of the homeobox gene Mixl1 impairs hematopoietic differentiation and results in acute myeloid leukemia. PNAS. 2006;103:16460–16465. doi: 10.1073/pnas.0607776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. A human Mix-like homeobox gene MIXL shows functional similarity to Xenopus Mix.1. Blood. 2002;100:89–95. doi: 10.1182/blood.v100.1.89. [DOI] [PubMed] [Google Scholar]
- Hara K, Kanai-Azuma M, Uemura M, Shitara H, Taya C, Yonekawa H, Kawakami H, Tsunekawa N, Kurohmaru M, Kanai Y. Evidence for crucial role of hindgut expansion in directing proper migration of primordial germ cells in mouse early embryogenesis. Developmental biology. 2009;330:427–439. doi: 10.1016/j.ydbio.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PPL, Elefanty AG, Robb L. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development. 2002;129:3597–3608. doi: 10.1242/dev.129.15.3597. [DOI] [PubMed] [Google Scholar]
- Hart AH, Willson TA, Wong M, Parker K, Robb L. Transcriptional regulation of the homeobox gene Mixl1 by TGF-beta and FoxH1. Biochemical and biophysical research communications. 2005;333:1361–1369. doi: 10.1016/j.bbrc.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Kispert A. The T genes in embryogenesis. TIG. 1994;10:280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbe E, Attisano L, Rossant J, Wrana JL. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes & development. 2001;15:1257–1271. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Brachyury is required for elongation and vasculogenesis in the murine allantois. Development. 2006a;133:2947–2959. doi: 10.1242/dev.02454. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene expression patterns : GEP. 2006b;6:783–793. doi: 10.1016/j.modgep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PP. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Developmental Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkić BV, Smith JC. Goosecoid and Mix.1 repress Brachyury expression and are required for head formation in Xenopus. Development. 1999;126:1769–1779. doi: 10.1242/dev.126.8.1769. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Cell Fate and Cell Lineage in the Endoderm of the Presomite Mouse Embryo, Studied with an Intracellular Tracer. Developmental biology. 1986;115:325–339. doi: 10.1016/0012-1606(86)90253-8. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- Lee JD, Anderson KV. Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left-right asymmetry in the mouse. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:3464–3476. doi: 10.1002/dvdy.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SM, Pereira L, Wong MS, Hirst CE, Van Vranken BE, Pick M, Trounson A, Elefanty AG, Stanley EG. Enforced expression of Mixl1 during mouse ES cell differentiation suppresses hematopoietic mesoderm and promotes endoderm formation. Stem cells. 2009;27:363–374. doi: 10.1634/stemcells.2008-1008. [DOI] [PubMed] [Google Scholar]
- Mead PE, Brivanlou IH, Kelley CM, Zon LI. BMP-4-responsive regulation of dorsal-ventral patterning by the homeobox protein Mix.1. Nature. 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- Metcalf D, Glaser S, Mifsud S, Di Rago L, Robb L. The preleukemic state of mice reconstituted with Mixl1-transduced marrow cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20013–20018. doi: 10.1073/pnas.0710339104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. STELLA-positive subregions of the primitive streak contribute to posterior tissues of the mouse gastrula. Developmental biology. 2012;363:201–218. doi: 10.1016/j.ydbio.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. Widespread but tissue-specific patterns of interferon-induced transmembrane protein 3 (IFITM3, FRAGILIS, MIL-1) in the mouse gastrula. Gene expression patterns : GEP. 2013 doi: 10.1016/j.gep.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn D, Chen SW, Dias DC, Weinstein DC, Dyer MA, Sahr K, Ducker CE, Zahradka E, Keller G, Zaret KS, Gudas LJ, Baron MH. Mouse Mix gene is activated early during differentiation of ES and F9 stem cells and induces endoderm in frog embryos. Developmental dynamics : an official publication of the American Association of Anatomists. 2003;226:446–459. doi: 10.1002/dvdy.10263. [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Takakura N, Yoshida H, Nishikawa S-I. Essential Role of Platelet-Derived Growth Factor Receptor α in the Development of the Intraplacental Yolk Sac / Sinus of Duval in Mouse Placenta. Biology of reproduction. 1998;58:65–72. doi: 10.1095/biolreprod58.1.65. [DOI] [PubMed] [Google Scholar]
- Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Experimental Hematology. 2001;29:927. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- Pan GJ, Chang ZY, Scholer HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell research. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- Pearce JJH, Evans MJ. Mml, a mouse Mix-like gene expressed in the primitive streak. Mechanisms of Development. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Pereira LA, Wong MS, Lim SM, Sides A, Stanley EG, Elefanty AG. Brachyury and related Tbx proteins interact with the Mixl1 homeodomain protein and negatively regulate Mixl1 transcriptional activity. PLoS ONE. 2011;6:e28394. doi: 10.1371/journal.pone.0028394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LA, Wong MS, Mei Lim S, Stanley EG, Elefanty AG. The Mix family of homeobox genes--key regulators of mesendoderm formation during vertebrate development. Developmental biology. 2012a;367:163–177. doi: 10.1016/j.ydbio.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Pereira LA, Wong MS, Mossman AK, Sourris K, Janes ME, Knezevic K, Hirst CE, Lim SM, Pimanda JE, Stanley EG, Elefanty AG. Pdgfralpha and Flk1 are direct target genes of Mixl1 in differentiating embryonic stem cells. Stem cell research. 2012b;8:165–179. doi: 10.1016/j.scr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Rhee JM, Iannaccone PM. Mapping mouse hemangioblast maturation from headfold stages. Developmental biology. 2012;365:1–13. doi: 10.1016/j.ydbio.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L, Hartley L, Begley CG, Brodnicki TC, Copeland NG, Gilbert DJ, Jenkins NA, Elefanty AG. Cloning, expression analysis, and chromosomal localization of murine and human homologues of a Xenopus mix gene. Developmental dynamics : an official publication of the American Association of Anatomists. 2000;219:497–504. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1070>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Robb L, Tam PP. Gastrula organiser and embryonic patterning in the mouse. Seminars in cell & developmental biology. 2004;15:543–554. doi: 10.1016/j.semcdb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Rosa FM. Mix.1, a Homeobox mRNA Inducible by Mesoderm Inducers, Is Expressed Mostly in the Presumptive Endodermal Cells of Xenopus Embryos. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- Rumballe B, Georgas K, Little MH. High-Throughput Paraffin Section In Situ Hybridization and Dual Immunohistochemistry on Mouse Tissues. Cold Spring Harbor Protocols. 2008 doi: 10.1101/pdb.prot5030. 2008:pdb.prot5030-pdb.prot5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Smith A, Zhang J, Guay D, Quint E, Johnson A, Akimenko MA. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:417–425. doi: 10.1002/dvdy.21417. [DOI] [PubMed] [Google Scholar]
- Speel EJM, Hopman AHN, Komminoth P. Amplification Methods to Increase the Sensitivity of In Situ Hybridization: Play CARD(S). Journal of Histochemistry & Cytochemistry. 1999;47:281–288. doi: 10.1177/002215549904700302. [DOI] [PubMed] [Google Scholar]
- Sterneckert J, Hoing S, Scholer HR. Concise review: Oct4 and more: the reprogramming expressway. Stem cells. 2012;30:15–21. doi: 10.1002/stem.765. [DOI] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa S-I. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- Tam PP, Khoo PL, Lewis SL, Bildsoe H, Wong N, Tsang TE, Gad JM, Robb L. Sequential allocation and global pattern of movement of the definitive endoderm in the mouse embryo during gastrulation. Development. 2007;134:251–260. doi: 10.1242/dev.02724. [DOI] [PubMed] [Google Scholar]
- Tam PP, Tan SS. The somitogenetic potential of cells in the primitive streak and the tail bud of the organogenesis-stage mouse embryo. Development. 1992;115:703–715. doi: 10.1242/dev.115.3.703. [DOI] [PubMed] [Google Scholar]
- Ukita K, Hirahara S, Oshima N, Imuta Y, Yoshimoto A, Jang CW, Oginuma M, Saga Y, Behringer RR, Kondoh H, Sasaki H. Wnt signaling maintains the notochord fate for progenitor cells and supports the posterior extension of the notochord. Mechanisms of Development. 2009;126:791–803. doi: 10.1016/j.mod.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy GD, Downs KM, Gardner RL. Inhibition of trophoblast stem cell potential in chorionic ectoderm coincides with occlusion of the ectoplacental cavity in the mouse. Development. 2002;129:3913–3924. doi: 10.1242/dev.129.16.3913. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Willey S, Ayuso-Sacido A, Zhang H, Fraser ST, Sahr KE, Adlam MJ, Kyba M, Daley GQ, Keller G, Baron MH. Acceleration of mesoderm development and expansion of hematopoietic progenitors in differentiating ES cells by the mouse Mix-like homeodomain transcription factor. Blood. 2006;107:3122–3130. doi: 10.1182/blood-2005-10-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Beddington RSP. Cell fate and morphogenetic movement in the late mouse primitive streak. Mechanisms of Development. 1996;55:79–89. doi: 10.1016/0925-4773(95)00493-9. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Developmental Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Zeigler BM, Sugiyama D, Chen M, Guo Y, Downs KM, Speck NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- Zhang H, Fraser ST, Papazoglu C, Hoatlin ME, Baron MH. Transcriptional activation by the Mixl1 homeodomain protein in differentiating mouse embryonic stem cells. Stem cells. 2009;27:2884–2895. doi: 10.1002/stem.203. [DOI] [PMC free article] [PubMed] [Google Scholar]