Abstract
Recently, there have been great interests for computer-aided diagnosis of Alzheimer’s disease (AD) and its prodromal stage, mild cognitive impairment (MCI). Unlike the previous methods that considered simple low-level features such as gray matter tissue volumes from MRI, and mean signal intensities from PET, in this paper, we propose a deep learning-based latent feature representation with a stacked auto-encoder (SAE). We believe that there exist latent non-linear complicated patterns inherent in the low-level features such as relations among features. Combining the latent information with the original features helps build a robust model in AD/MCI classification, with high diagnostic accuracy. Furthermore, thanks to the unsupervised characteristic of the pre-training in deep learning, we can benefit from the target-unrelated samples to initialize parameters of SAE, thus finding optimal parameters in fine-tuning with the target-related samples, and further enhancing the classification performances across four binary classification problems: AD vs. healthy normal control (HC), MCI vs. HC, AD vs. MCI, and MCI converter (MCI-C) vs. MCI non-converter (MCI-NC). In our experiments on ADNI dataset, we validated the effectiveness of the proposed method, showing the accuracies of 98.8, 90.7, 83.7, and 83.3 % for AD/HC, MCI/HC, AD/MCI, and MCI-C/MCI-NC classification, respectively. We believe that deep learning can shed new light on the neuroimaging data analysis, and our work presented the applicability of this method to brain disease diagnosis.
Keywords: Alzheimer’s disease (AD), Mild cognitive impairment (MCI), Multi-modal classification, Deep learning, Latent feature representation
Introduction
Alzheimer’s disease (AD), characterized by progressive impairment of cognitive and memory functions, is the most prevalent cause of dementia in elderly people and is recognized as one of the major challenges to global health care systems. A recent research by Alzheimer’s association reports that AD is the sixth-leading cause of death in the United States, rising significantly every year in terms of the proportion of cause of death (Alzheimer’s 2012). It is also indicated that 10–20 % of people aged 65 or older have mild cognitive impairment (MCI), a prodromal stage of AD (Alzheimer’s 2012), and situated in the spectrum between normal cognition and dementia (Cui et al. 2011). Due to the limited periods for which the symptomatic treatments are available, it has been of great importance for early diagnosis and prognosis of AD/MCI in the clinic.
To this end, researchers in many scientific fields have devoted their efforts to understand the underlying mechanism that causes these diseases and to identify pathological biomarkers for diagnosis or prognosis of AD/MCI by analyzing different types of neuroimaging modalities, such as magnetic resonance imaging (MRI) (Davatzikos et al. 2011; Wee et al. 2011), positron emission tomography (PET) (Nordberg et al. 2010), functional MRI (fMRI) (Greicius et al. 2004; Suk et al. 2013), cerebrospinal fluid (CSF) (Nettiksimmons et al. 2010), etc. In terms of clinical diagnosis, structural MRI provides visual information regarding the macroscopic tissue atrophy, which results from the cellular changes underlying AD/MCI, and PET can be used for the investigation of the cerebral glucose metabolism (Nordberg et al. 2010), which reflects the functional brain activity.
While these neuroimaging techniques have contributed substantially to our observation of the brain, significant breakthroughs in how we can efficiently understand and analyze the observed information have been of great concerns for the last decades. In that respect, machine learning has provided nice tools to tackle these challenges. Specifically, it has proved for their efficacy in multivariate pattern analysis and feature selection for clinical diagnosis. It is also impressive that they offered a new leverage strategy to efficiently fuse complementary information from different modalities including MRI, PET, biological and neurological data for discriminating AD/MCI patients from healthy normal controls (HC) (Fan et al. 2007; Perrin et al. 2009; Kohannim et al. 2010; Walhovd et al. 2010; Cui et al. 2011; Hinrichs et al. 2011; Zhang et al. 2011; Wee et al. 2012; Westman et al. 2012; Yuan et al. 2012; Zhang and Shen 2012). Kohannim et al. (2010) concatenated features from modalities into a vector and used a support vector machine (SVM) classifier. Walhovd et al. (2010) applied multi-method stepwise logistic regression analyses, and Westman et al. (2012) exploited a hierarchical modeling of orthogonal partial least squares to latent structures. Hinrichs et al. (2011) and Zhang et al. (2011), independently, utilized a kernel-based machine learning technique. There have been also attempts to select features by means of sparse learning, which jointly learns the tasks of clinical label identification and clinical scores prediction (Yuan et al. 2012; Zhang and Shen 2012).
Although these researches presented the effectiveness of their methods in their own experiments on multi-modal AD/MCI classification, the main limitation of the previous work is that they considered only simple low-level features such as cortical thickness and/or gray matter tissue volumes from MRI (Klöppel et al. 2008; Gray et al. 2013; Zhang et al. 2011; Zhang and Shen 2012; Cui et al. 2011; Desikan et al. 2009; Walhovd et al. 2010; Yao et al. 2012; Westman et al. 2012; Ewers et al. 2012; Zhou et al. 2011; Li et al. 2012; Liu et al. 2012), mean signal intensities from PET (Mosconi et al. 2008; Walhovd et al. 2010; Nordberg et al. 2010; Zhang et al. 2011; Zhang and Shen 2012; Gray et al. 2013), and t-tau, p-tau, and β-amyloid 42 (Aβ42) from CSF (Cui et al. 2011; Yuan et al. 2012; Zhang et al. 2011; Zhang and Shen 2012; Walhovd et al. 2010; Westman et al. 2012; Ewers et al. 2012; Tapiola et al. 2009). In this paper, we assume that there exists hidden or latent high-level information, inherent in those low-level features such as relations among them, which can be helpful to build a more robust model for AD/MCI diagnosis and prognosis.
To tackle this problem, we exploit a deep learning framework, which has been efficiently used to discover visual features in computer vision (Hinton and Salakhutdinov 2006; Bengio et al. 2007; Lee et al. 2011; Yu et al. 2011). The main concept of the deep learning is that deep architectures can be much more efficient than shallow architectures in terms of computational elements and parameters required to represent unknown functions (Bengio et al. 2007). Furthermore, one of the key features of the deep learning is that the low-layer represents low-level features and the high-layer abstracts those low-level features. In the case of our neuroimaging and biological data, the deep or hierarchical architecture can be efficiently used to discover latent or hidden representation, inherent in the low-level features from modalities, and ultimately to enhance classification accuracy. Specifically, ‘stacked auto-encoder’ (SAE) is considered to discover latent representations from the original neuroimaging and biological features. It is also noteworthy that thanks to the unsupervised characteristic of the pre-training in deep learning, the SAE model allows us to benefit from the target-unrelated samples to discover general latent feature representations, and hence to leverage for further enhancement of the classification accuracy.
The main contributions of our work can be summarized as follows: (1) To our best knowledge, this is the first work that considers a deep learning for feature representation in brain disease diagnosis and prognosis. (2) Unlike the previous work in the literature, we consider complicated nonlinear latent feature representation, which can be discovered from data in self-taught learning. (3) By constructing an augmented feature vector via a concatenation of the original low-level features and the SAE-learned latent feature representation, we can improve diagnostic accuracy on the public ADNI dataset. (4) By means of pre-training of SAE in an unsupervised manner with the target-unrelated samples and then fine-tuning with target-related samples, the proposed method further enhances the classification performance.
Materials and image processing
Subjects
In this work, we use the ADNI dataset publicly available on the web1. Specifically, we consider only the baseline MRI, 18-fluoro-deoxyglucose (FDG) PET, and CSF data acquired from 51 AD patients, 99 MCI patients (43 MCI converters, who progressed to AD, and 56 MCI non-converters, who did not progress to AD in 18 months), and 52 HC subjects2. The demographics of the subjects are detailed in Table 1. Along with the neuroimaging and biological data, two types of clinical scores, mini-mental state examination (MMSE) and Alzheimer’s disease assessment scale-cognitive subscale (ADAS-Cog), are also provided for each subject.
Table 1.
Demographic and clinical information of the subjects
| AD (N = 51) |
MCI converter (N = 43) |
MCI non- converter (N = 56) |
HC (N = 52) |
|
|---|---|---|---|---|
| Female/male | 18/33 | 15/28 | 17/39 | 18/34 |
| Age (mean ± SD) | 75.2 ± 7.4 [59–88] | 75.7 ± 6.9 [58–88] | 75.0 ± 7.1 [55–89] | 75.3 ± 5.2 [62–85] |
| Education (mean ± SD) | 14.7 ± 3.6 [4–20] | 15.4 ± 2.7 [10–20] | 14.9 ± 3.3 [8–20] | 15.8 ± 3.2 [8–20] |
| MMSE (mean ± SD) | 23.8 ± 2.0 [20–26] | 26.9 ± 2.7 [20–30] | 27.0 ± 3.2 [18–30] | 29 ± 1.2 [25–30] |
| CDR (mean ± SD) | 0.7 ± 0.3 [0.5–1] | 0.5 ± 0 [0.5–0.5] | 0.5 ± 0 [0.5–0.5] | 0 ± 0 [0–0] |
N number of subjects, SD standard deviation, [min–max]
With regard to the general eligibility criteria in ADNI, subjects were in the age of between 55 and 90 with a study partner, who could provide an independent evaluation of functioning. General inclusion/exclusion criteria3 are as follows: (1) healthy subjects: MMSE scores between 24 and 30 (inclusive), a clinical dementia rating (CDR) of 0, non-depressed, non-MCI, and non-demented; (2) MCI subjects: MMSE scores between 24 and 30 (inclusive), a memory complaint, objective memory loss measured by education adjusted scores on Wechsler memory scale logical memory II, a CDR of 0.5, absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, and an absence of dementia; and (3) mild AD: MMSE scores between 20 and 26 (inclusive), CDR of 0.5 or 1.0, and meets the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/AD-RDA) criteria for probable AD.
MRI and PET scanning
The structural MR images were acquired from 1.5 T scanners. We downloaded data in Neuroimaging Informatics Technology Initiative (NIfTI) format, which had been pre-processed for spatial distortion correction caused by gradient nonlinearity and B1 field inhomogeneity. The FDG-PET images were acquired 30–60 min post-injection, averaged, spatially aligned, interpolated to a standard voxel size, normalized in intensity, and smoothed to a common resolution of 8 mm full width at half maximum. CSF data were collected in the morning after an overnight fast using a 20- or 24-gauge spinal needle, frozen within 1 h of collection, and transported on dry ice to the ADNI Biomarker Core Laboratory at the University of Pennsylvania Medical Center.
Image processing and feature extraction
The MR images were preprocessed by applying the typical procedures of anterior commissure (AC)–posterior commissure (PC) correction, skull-stripping, and cerebellum removal. Specifically, we used MIPAV software4 for AC-PC correction, resampled images to 256 × 256 × 256, and applied N3 algorithm (Sled et al. 1998) to correct intensity inhomogeneity. An accurate and robust skull stripping (Wang et al. 2011) was performed, followed by cerebellum removal. We further manually reviewed the skull-stripped images to ensure clean and dura removal. Then, FAST in FSL package5 (Zhang et al. 2001) was used for structural MR image segmentation into three tissue types of gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF). We finally pacellated them into 93 regions of interests (ROIs) by warping Kabani et al.’s (1998) atlas to each subject’s space via HAMMER (Shen and Davatzikos 2002), although other advanced registration methods can also be applied for this process (Friston et al. 1995; Evans and Collins 1997; Rueckert et al. 1999; Shen et al. 1999; Wu et al. 2006; Xue et al. 2006a, b; Avants et al. 2008; Yang et al. 2008; Tang et al. 2009; Vercauteren et al. 2009; Jia et al. 2010). In this work, we only considered GM for classification, because of its relatively high relatedness to AD/MCI compared to WM and CSF (Liu et al. 2012).
Regarding FDG-PET images, they were rigidly aligned to the respective MR images, and then applied parcellation propagated from the atlas by registration. For each ROI, we used the GM tissue volume from MRI, and the mean intensity from FDG-PET as features6, which are most widely used in the field for AD/MCI diagnosis (Davatzikos et al. 2011; Hinrichs et al. 2011; Zhang and Shen 2012; Liu et al. 2013). Therefore, we have 93 features from a MR image and the same dimensional features from FDG-PET image. Here, we should note that although it is known that the regions of medial temporal and superior parietal lobes are mainly affected by the disease, we assume that other brain regions, although their relatedness to AD is not clearly investigated yet, may also contribute to the diagnosis of AD/MCI and thus we consider 93 ROIs in our study. In addition, we have three CSF biomarkers of Aβ42, t-tau, and p-tau as features.
Stacked auto-encoder for latent feature representation
In this section, we describe the proposed method for AD/MCI classification. Figure 1 illustrates a schematic diagram of the proposed method. Given multi-modal data along with the class-label and clinical scores, we first extract low-level features from MRI and FDG-PET as explained in “Image processing and feature extraction”. We then discover a latent feature representation from the low-level features in MRI, FDG-PET, and CSF, individually, by deep learning with SAE. In deep learning, we perform two steps sequentially: (1) We first pre-train the SAE in a greedy layer-wise manner to obtain good initial parameters. (2) We then fine-tune the deep network to find the optimal parameters. A sparse learning on the augmented feature vectors, i.e., a concatenation of the original low-level features and the SAE-learned features, is applied to select features that efficiently regress the target values, e.g., class-label and/or clinical scores. Finally, we fuse the selected multi-modal feature information via a multi-kernel SVM (MK-SVM) for diagnosis. Note that the latent feature representation and feature selection are performed for each modality individually. Hereafter, we do not explicitly indicate the modality of samples, unless specified, in order for simplicity. Basically, the method described below can be applied for each modality individually, but also applicable to the concatenated feature vectors of three modalities in terms of information fusion, which is considered later in our experiments for comparison.
Fig. 1.
An illustration of the proposed method for AD/MCI diagnosis
Sparse auto-encoder
An auto-encoder, also called as auto-associator, is one type of artificial neural networks structurally defined by three layers: input layer, hidden layer, and output layer. The input layer is fully connected to the hidden layer, which is further fully connected to the output layer as illustrated in Fig. 2. The aim of the auto-encoder is to learn a latent or compressed representation of the input, by minimizing the reconstruction error between the input and the reconstructed one from the learned representation.
Fig. 2.
Illustration of an auto-encoder and its parameters. (The bias parameters b1 and b2 are omitted for clarity.)
Let DH and DI denote, respectively, the number of hidden and input units in a neural network. Given a set of training samples from N subjects, an auto-encoder maps xi to a latent representation yi ∈ ℝDH through a linear deterministic mapping and a nonlinear activation function f as follows:
| (1) |
where W1 ∈ ℝDH×DI is an encoding weight matrix and b1 ∈ ℝDH is a bias vector. Regarding the activation function, in this study, we consider a logistic sigmoid function for f(a) = 1/(1 + exp(−a)), which is the most widely used in the field of pattern recognition or machine learning (Bengio et al. 2007; Lee et al. 2008; Bengio 2009; Larochelle et al. 2009; Ngiam et al. 2011; Shin et al. 2013). The representation yi of the hidden layer is then mapped to a vector zi ∈ ℝDI, which approximately reconstructs the input vector xi by another linear mapping as follows:
| (2) |
where W2 ∈ ℝDI×DH and b2 ∈ ℝDI are a decoding weight matrix and a bias vector, respectively.
Structurally, the number of input and output units are fixed to the dimension of an input vector. Meanwhile, the number of hidden units can be determined based on the nature of the data. If the number of hidden units is less than the dimension of the input data, then the auto-encoder can be used for dimensionality reduction. However, it is noteworthy that in order for obtaining complicated non-linear relations among neuroimaging features, we can allow the number of hidden units to be even larger than the input dimension, from which we can still find an interesting structure by imposing a sparsity constraint (Lee et al. 2008; Larochelle et al. 2009).
From a learning perspective, we aim to minimize the reconstruction error between the input xi and the output zi with respect to the parameters. Let and denote a reconstruction error. In order for the sparseness of the hidden units, we further consider a Kullback-Leibler (KL) divergence between the average activation ρ̂j of the jth hidden unit over the training samples and the target average activation ρ defined as follows (Shin et al. 2013):
| (3) |
where ρ and ρ̂j are Bernoulli random variables. Then our objective function can be written as follows:
| (4) |
With the introduction of the KL divergence weighted by a sparsity control parameter γ to the target objective function, we penalize a large average activation of a hidden unit over the training samples by setting ρ small7. This penalization drives many of the hidden units’ activation to be equal or close to zero, resulting in sparse connections between layers.
Note that the output from the hidden layer determines the latent representation of the input vector. However, due to its simple shallow structural characteristic, the representational power of a single-layer auto-encoder is known to be very limited.
Stacked auto-encoder
Inspired from the biological model of the human visual cortex (Fukushima 1980; Serre et al. 2005), recent studies in machine learning have shown that a deep or hierarchical architecture is useful to find highly non-linear and complex patterns in data (Bengio 2009). Motivated by the studies, in this paper, we consider a SAE (Bengio et al. 2007), in which an auto-encoder becomes a building block, for a latent feature representation in neuroimaging or biological data. Specifically, as the name says, we stack auto-encoders one after another taking the outputs from the hidden units of the lower layer as the input to the upper layer’s input units, and so on. Figure 3 shows an example of a SAE model with three auto-encoders stacked hierarchically. Note that the number of units in the input layer is equal to the dimension of the input feature vector. But the number of hidden units in the upper layers can be determined according to the nature of the input, i.e., even larger than the input dimension.
Fig. 3.
A deep architecture of our stacked auto-encoder and the two-step (unsupervised greedy layer-wise pretraining and supervised fine-tuning) parameter optimization scheme. (The black arrows denote the parameters to be optimized in the current stage). a Pre-training of the first hidden layer with the training samples as inputs, b pre-training of the second hidden layer with the outputs from the first hidden layer as inputs, c pre-training of the third hidden layer with the output from the second hidden layer as inputs, d fine-tuning of the whole network with an additional label-output layer, taking the pre-trained parameters as the starting point in optimization
Thanks to the hierarchical nature in structure, one of the most important characteristics of the SAE is to learn or discover highly non-linear and complicated patterns such as the relations among input features. Another important characteristic of the deep learning is that the latent representation can be learned directly from the data. Utilizing its representational and self-taught learning properties, we can find a latent representation of the original low-level features directly extracted from neuroimaging or biological data. When an input sample is presented to a SAE model, the different layers of the network represent different levels of information. That is, the lower the layer in the network, the simpler patterns (e.g., linear relations of features); the higher the layer, the more complicated or abstract patterns inherent in the input feature vector (e.g., non-linear relations among features).
With regard to training parameters of the weight matrices and the biases in the deep network of our SAE model, a straightforward way is to apply back-propagation with the gradient-based optimization technique starting from random initialization taking the deep network as a conventional multi-layer neural network. Unfortunately, it is generally known that deep networks trained in that manner perform worse than networks with a shallow architecture, suffering from falling into a poor local optimum (Larochelle et al. 2009). However, recently, Hinton et al. introduced a greedy layer-wise unsupervised learning algorithm and showed its success to learn a deep belief network (Hinton et al. 2006). The key idea in a greedy layer-wise learning is to train one layer at a time by maximizing the variational lower bound (Hinton et al. 2006). That is, we first train the first hidden layer with the training data as input, and then train the second hidden layer with the outputs from the first hidden layer as input, and so on. That is, the representation of the lth hidden layer is used as input for the (l + 1)-th hidden layer. This greedy layer-wise learning is called ‘pre-training’ (Fig. 3a–c). The pre-training is performed in an unsupervised manner with a standard back-propagation algorithm (Bishop 1995). Later in our experiments, we utilize this unsupervised characteristic in pre-training to further find optimal parameters to discover a latent representation in the neuroimaging or biological data, taking benefits from target-unrelated samples.
Focusing on the ultimate goal of our work to improve diagnostic performance in AD/MCI identification, we further optimize the deep network in a supervised manner. In order for that, we stack another output layer on top of the SAE (Fig. 3d). This top output layer is used to represent the class-label of an input sample. We set the number of units in the output layer to be equal to the number of classes of interest. This extended network can be considered as a multi-layer neural network and, in this paper, we call it ‘SAE-classifier’. Therefore, it is straightforward to optimize the deep network by back-propagation with gradient descent, having parameters, except for the last classification network, initialized by the pre-trained ones. Note that the initialization of the parameters via pre-training makes the deep network different from the conventional neural network, and it helps the supervised optimization, called ‘finetuning’, reduce the risk of falling into local poor optima (Hinton et al. 2006; Larochelle et al. 2009). We summarize the deep learning of the SAE in Algorithm 1. Besides the fine-tuning of the parameters, we also utilize the SAE-classifier to determine the optimal SAE structure.
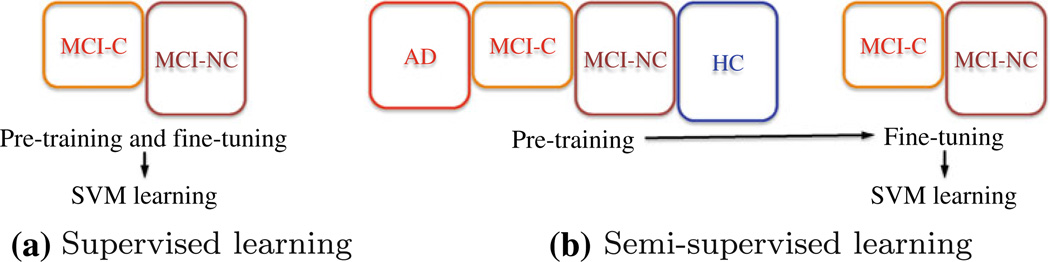
Later in our experiments, we consider the following two learning schemes, in which the main difference lies in the way of utilizing the training samples available: (1) The supervised approach learns the parameters of SAE from solely the target-related training samples. For example, in the task of classifying MCI converter (MCI-C) and MCI non-converter (MCI-NC), we use the target-related training samples of MCI-C and MCI-NC for both pre-training and fine-tuning in deep learning, and for the SVM classifier learning (Fig. 4a). (2) The semi-supervised approach first performs pre-training using both target-related and target-unrelated samples, and then fine-tune the model with only the target-related samples. For example, in the task of discriminating MCI-C from MCI-NC, we first perform pretraining with the samples of AD and HC as well as those of MCI-C and MCI-NC, and then fine-tuning with only the MCI-C and MCI-NC training samples (Fig. 4b). Finally, the representation of the target-related MCI-C and MCI-NC training samples are used for SVM learning. The motivation of applying this learning scheme in our work is that the more samples we use in pre-training of the deep architecture, the better good initialization of the parameters we can obtain, and thus the better latent representation inherent in the low-level features we can discover (Larochelle et al. 2009). Hereafter, we use the terms of ‘supervised’ and ‘semi-supervised’, respectively, to specify the strategy of learning parameters of a SAE model as described above throughout the paper.
Algorithm 1.
Deep learning of a stacked auto-encoder
| Input: train feature samples: X ∈ ℝDI × N, train labels: L ∈ ℝN, sparsity control parameter: γ, target average activation: ρ | |
| Output: weight matrices: , biases: | |
| /* H: number of hidden layers */ | |
Step 1) Pre-training hidden layers:
| |
Step 2) Fine-tuning the whole network:
| |
Fig. 4.
An example of SAE model training schemes for MCI converter (MCI-C) and MCI non-converter (MCI-NC) classification. The colored-boxes denote the samples used for training during the specified step. The size of a rectangle represents the number of training samples available for each class
Once we determine the structure of a SAE model, we consider the outputs from the top hidden layer as our latent feature representation, i.e., , where and denote, respectively, the trained weight matrix and bias of the top Hth hidden layer, and YH−1 is the outputs from the (H−1)-th hidden layer. To utilize both the low-level simple features and the high-level latent representation, we construct an augmented feature vector X̂ by concatenating the SAE-learned feature representation YH with the original low-level features X, i.e., , which is then fed into the sparse learning for feature selection as described below.
Feature selection with sparse representation learning
Earlier, Zhang and Shen showed the efficacy of sparse representation for feature selection in AD/MCI diagnosis (Zhang and Shen 2012). Here, we consider two sparse representation methods, namely, least absolute shrinkage and selection operator (lasso) (Tibshirani 1996) and group lasso (Yuan and Lin 2006), which penalize a linear regression model with l1-norm and l21-norm, respectively. In this work, we select features for each modality individually and defer the multi-modal information fusion to MK-SVM learning. The rationale for the modality-specific feature selection is that we believe it would be helpful to find the discriminative features in a low dimension rather than in a high dimension of the modality-concatenated feature vectors.
Let m ∈ {1, …, M} denote an index of modalities and X̂(m) ∈ ℝN×D denote a set of the augmented feature vectors, where N and D(= DH + DI) are, respectively, the number of samples and the dimension of the augmented feature vector. In lasso, we focus on finding optimal weight coefficients a(m) to regress the target response vector t(m) ∈ ℝN by a combination of the features in X̂(m) with a sparsity constraint as follows:
| (5) |
where λ1 is a sparsity control parameter. In our work, the target response vector corresponds to the target clinical labels. The l1-norm penalty to linear regression imposes sparsity to the solution of a(m), which means that many of the elements are to be zero. By the application of the lasso, we can select features whose weight coefficients are non-zero.
Meanwhile, unlike the lasso that considers a single target response vector, the group lasso can accommodate multiple target response vectors, where each target response vector can be regarded as one task, and impose a constraint that encourages the correlated features to be jointly selected for multiple tasks in a data-driven manner.
| (6) |
where s ∈ {1, …, S} denotes an index of tasks8, , and λ2 is a group-sparsity control parameter. In Eq. 6, , where A(m)[d, :] denotes the dth row of the matrix A(m). This l2,1-norm imposes to select features that are jointly used to represent the target response vector across tasks9. We can select features whose absolute weight coefficient is larger than zero.
From the inspection of Eqs. 5 and 6, we can see that the group lasso is a generalized form of the lasso in terms of the number of tasks involved in regression. That is, if we have information for a single task, then the group lasso becomes the conventional lasso. Later in our experiments, we consider both of these sparse representation learning and observe their effects on selecting features as well as classification performance. We use a set of class-labels in lasso, and clinical scores as well as class-labels in group lasso. The hyper-parameters of λ1 and λ2 in Eqs. 5 and 6, respectively, are determined by a grid search within a space of [0.001, 0.005, 0.01, 0.05, 0.1, 0.2, 0.3]. For the optimization, we use a SLEP toolbox10.
Multi-kernel SVM learning
It is witnessed in the previous studies that the biomarkers from different modalities can provide complementary information in AD/MCI diagnosis (Perrin et al. 2009). In this paper, we combine the complementary information from modalities of MRI, FDG-PET, and CSF in the feature kernel space with linear SVM, which has proved its efficacy in many fields (Wee et al. 2012; Han and Davis 2012; Suk and Lee 2013). Given the dimension-reduced training samples through the sparse representation learning as described in “Feature selection with sparse representation learning”, and the test sample of x̃(m), where m ∈ {1, …, M} denotes an index of modalities, the decision function of the MK-SVM is defined as follows:
| (7) |
where ζi is the class-label of the ith sample, αi and b are, respectively, a Lagrangian multiplier and a bias, is a kernel function, ϕ(m) is a kernel-induced mapping function, and β(m) ≥ 0 is a weight coefficient of the mth modality with the constraint of . Refer to Gönen (2011) for a detailed explanation on the MK-SVM.
Experimental results
Experimental setup
In this section, we evaluate the effectiveness of the proposed method for a non-linear latent feature representation by deep learning with SAE, considering four binary classification problems: AD vs. HC, MCI vs. HC, AD vs. MCI, and MCI-C vs. MCI-NC. In the classifications of MCI vs. HC, and AD vs. MCI, both MCI-C and MCI-NC data were used as the MCI class. For each classification problem, we applied a tenfold cross validation technique. That is, we randomly partitioned the dataset into 10 subsets, each of which included 10 % of the total dataset, and then used 9 out of 10 subsets for training and the remaining one for testing. We repeated these whole process 10 times for unbiased evaluation.
To show the validity of the proposed method of combining SAE-learned feature representation with the original low-level features, we compared the results of the proposed method with those from the original low-level features and SAE-learned feature representation, respectively, by applying the same strategies of feature selection and MK-SVM learning. Hereafter, we denote LLF, SAEF, and LLF + SAEF, respectively, for the cases of using the original low-level features, SAE-learned features, and the concatenation of LLF and SAEF. It is noteworthy that we use the same training and test samples over the competing methods for fair comparison.
Determination of the structure of a SAE model
With regard to the structure of a SAE model, we considered three hidden layers for MRI, FDG-PET, and CONCAT11, and two hidden layers for CSF, by taking into account the dimensionality of the low-level features in each modality. To determine the number of hidden units, we performed classification with a SAE-classifier by a grid search12. Due to the possibility of over-fitting with a small number of training samples, we early stopped the fine-tuning step by setting a small number for iteration. The optimal structure of the SAE models and the respective performance are presented in Table 2. For example, in classification of AD and HC, we obtained the best accuracy of 85.7 % with MRI from a SAE-classifier of 500-50-10 (from bottom to top) hidden units in supervised learning13. We used a DeepLearnToolbox14 to train our SAE model.
Table 2.
Classification accuracies (mean ± standard deviation) obtained from SAE-classifiers and their corresponding structures in terms of the number of hidden units
| Supervised | Semi-supervised | |||
|---|---|---|---|---|
| # Hidden units | Accuracy | # Hidden units | Accuracy | |
| AD vs. HC | ||||
| MRI | 500-50-10 | 0.857 ± 0.018 | 300-50-20 | 0.844 ± 0.025 |
| PET | 1,000-50-30 | 0.859 ± 0.021 | 100-50-10 | 0.834 ± 0.020 |
| CSF | 50-3 | 0.831 ± 0.016 | 10-3 | 0.757 ± 0.048 |
| CONCAT | 500-100-20 | 0.899 ± 0.014 | 1,000-100-10 | 0.888 ± 0.009 |
| MCI vs. HC | ||||
| MRI | 100-100-20 | 0.706 ± 0.021 | 500-50-20 | 0.697 ± 0.032 |
| PET | 300-50-10 | 0.670 ± 0.018 | 500-50-20 | 0.673 ± 0.021 |
| CSF | 10-3 | 0.683 ± 0.020 | 10-2 | 0.664 ± 0.021 |
| CONCAT | 100-50-20 | 0.737 ± 0.025 | 100-50-20 | 0.752 ± 0.025 |
| AD vs. MCI | ||||
| MRI | 1,000-50-30 | 0.645 ± 0.024 | 1,000-50-10 | 0.655 ± 0.027 |
| PET | 100-50-10 | 0.659 ± 0.017 | 500-50-10 | 0.655 ± 0.026 |
| CSF | 10-1 | 0.661 ± 0.009 | 10-1 | 0.660 ± 0.013 |
| CONCAT | 100-100-20 | 0.689 ± 0.023 | 1,000-100-20 | 0.672 ± 0.025 |
| MCI-C vs. MCI-NC | ||||
| MRI | 100-100-10 | 0.549 ± 0.037 | 300-100-30 | 0.571 ± 0.036 |
| PET | 100-100-10 | 0.595 ± 0.044 | 100-50-30 | 0.581 ± 0.045 |
| CSF | 30-2 | 0.589 ± 0.026 | 30-1 | 0.562 ± 0.020 |
| CONCAT | 500-50-20 | 0.602 ± 0.031 | 300-100-30 | 0.613 ± 0.042 |
Refer to Fig. 4a, b, and the contexts for explanation of ‘supervised’ and ‘semi-supervised’
Classification results
Regarding the feature selection, we observed that the lasso-based method showed better classification performance compared to the group lasso-based one. Here, we present the classification results obtained by lasso-based feature selection method.
Table 3 shows the mean accuracies of the competing methods in the classification of AD and HC. Although the proposed method of LLF + SAEF with a single-modality was outperformed for a couple of cases by the LLF-based one, e.g., 89 % (LLF) vs. 88.2 % (LLF + SAEF) with MRI, 93.7 % (LLF) vs. 93.5 % (LLF + SAEF) with CONCAT, those from multi-modality fusion via MK-SVM showed the best accuracies of 97.9 and 98.8 % in supervised and semi-supervised learning, respectively. Compared to the accuracy of 97 % with a LLF-based method, the proposed method improved the accuracy by 0.9 and 1.8 %, in supervised and semi-supervised learning, respectively.
Table 3.
Performance comparison of different feature sets with lasso-based feature selection in AD vs. HC classification
| AD vs. HC | |||
|---|---|---|---|
| LLF | SAEF | LLF + SAEF | |
| Supervised | |||
| MRI | 0.890 ± 0.018 | 0.821 ± 0.026 | 0.882 ± 0.019 |
| PET | 0.848 ± 0.026 | 0.832 ± 0.020 | 0.850 ± 0.018 |
| CSF | 0.797 ± 0.014 | 0.802 ± 0.017 | 0.801 ± 0.018 |
| CONCAT | 0.937 ± 0.013 | 0.835 ± 0.018 | 0.935 ± 0.012 |
| MK-SVM | 0.970 ± 0.010 | 0.947 ± 0.018 | 0.979 ± 0.007 |
| Semi-supervised | |||
| MRI | – | 0.838 ± 0.028 | 0.924 ± 0.015 |
| PET | – | 0.827 ± 0.023 | 0.887 ± 0.027 |
| CSF | – | 0.785 ± 0.033 | 0.797 ± 0.014 |
| CONCAT | – | 0.889 ± 0.018 | 0.960 ± 0.014 |
| MK-SVM | – | 0.945 ± 0.017 | 0.988 ± 0.004 |
Bold best performance across both the feature types and the learning schemes, italics best performance across the feature types in the same learning scheme
In the classification of MCI and HC, as presented in Table 4, the proposed method showed the best classification accuracies of 88.8 and 90.7 % with supervised and semi-supervised learning schemes, respectively. The performance improvements compared to the classification accuracy of 84.8 % with the LLF-based method were 4 and 5.9 %, respectively.
Table 4.
Performance comparison of different feature sets with lasso-based feature selection in MCI vs. HC classification
| MCI vs. HC | |||
|---|---|---|---|
| LLF | SAEF | LLF + SAEF | |
| Supervised | |||
| MRI | 0.736 ± 0.013 | 0.674 ± 0.020 | 0.802 ± 0.016 |
| PET | 0.683 ± 0.016 | 0.672 ± 0.027 | 0.745 ± 0.018 |
| CSF | 0.678 ± 0.020 | 0.662 ± 0.023 | 0.679 ± 0.022 |
| CONCAT | 0.756 ± 0.022 | 0.726 ± 0.031 | 0.836 ± 0.005 |
| MK-SVM | 0.848 ± 0.014 | 0.799 ± 0.024 | 0.888 ± 0.012 |
| Semi-supervised | |||
| MRI | – | 0.709 ± 0.022 | 0.794 ± 0.019 |
| PET | – | 0.682 ± 0.021 | 0.749 ± 0.025 |
| CSF | – | 0.664 ± 0.019 | 0.682 ± 0.013 |
| CONCAT | – | 0.724 ± 0.033 | 0.833 ± 0.020 |
| MK-SVM | – | 0.808 ± 0.017 | 0.907 ± 0.012 |
Bold best performance across both the feature types and the learning schemes, italics best performance across the feature types in the same learning scheme
In the classification of AD and MCI, as shown in Table 5, the proposed method showed the best classification accuracies of 82.7 and 83.7 % with supervised and semi-supervised learning schemes, respectively. We could enhance the classification accuracy by 3.9 and 4.9 % with supervised and semi-supervised learning schemes, respectively, compared to the LLF-based method, whose accuracy was 78.8 %.
Table 5.
Performance comparison of different feature sets with lasso-based feature selection in AD vs. MCI classification
| AD vs. MCI | |||
|---|---|---|---|
| LLF | SAEF | LLF + SAEF | |
| Supervised | |||
| MRI | 0.617 ± 0.020 | 0.631 ± 0.023 | 0.704 ± 0.026 |
| PET | 0.667 ± 0.023 | 0.645 ± 0.015 | 0.711 ± 0.025 |
| CSF | 0.659 ± 0.004 | 0.661 ± 0.002 | 0.655 ± 0.009 |
| CONCAT | 0.693 ± 0.019 | 0.681 ± 0.023 | 0.752 ± 0.030 |
| MK-SVM | 0.788 ± 0.018 | 0.759 ± 0.019 | 0.827 ± 0.025 |
| Semi-supervised | |||
| MRI | – | 0.659 ± 0.025 | 0.721 ± 0.039 |
| PET | – | 0.640 ± 0.021 | 0.715 ± 0.024 |
| CSF | – | 0.659 ± 0.002 | 0.659 ± 0.005 |
| CONCAT | – | 0.682 ± 0.022 | 0.781 ± 0.028 |
| MK-SVM | – | 0.757 ± 0.017 | 0.837 ± 0.015 |
Bold best performance across both the feature types and the learning schemes, italics best performance across the feature types in the same learning scheme
In discriminating MCI-C from MCI-NC, the proposed method also outperformed the LLF-based method as presented in Table 6. While the LLF-based method showed the classification accuracy of 76 % with multi-modality fusion via MK-SVM, we could obtain the classification accuracies of 77.9 and 83.3 % in supervised and semi-supervised learning, respectively. It is remarkable that the semi-supervised learning scheme enhanced the performance by 7.3 % compared to that of the LLF-based method.
Table 6.
Performance comparison of different feature sets with lasso-based feature selection in MCI converter (MCI-C) vs. MCI non-converter (MCI-NC) classification
| MCI-C vs. MCI-NC | |||
|---|---|---|---|
| LLF | SAEF | LLF + SAEF | |
| Supervised | |||
| MRI | 0.541 ± 0.042 | 0.544 ± 0.026 | 0.561 ± 0.037 |
| PET | 0.573 ± 0.025 | 0.598 ± 0.048 | 0.611 ± 0.039 |
| CSF | 0.569 ± 0.017 | 0.581 ± 0.028 | 0.576 ± 0.032 |
| CONCAT | 0.597 ± 0.034 | 0.596 ± 0.030 | 0.713 ± 0.030 |
| MK-SVM | 0.760 ± 0.020 | 0.733 ± 0.035 | 0.779 ± 0.027 |
| Semi-supervised | |||
| MRI | – | 0.559 ± 0.060 | 0.693 ± 0.020 |
| PET | – | 0.573 ± 0.029 | 0.689 ± 0.038 |
| CSF | – | 0.548 ± 0.024 | 0.577 ± 0.030 |
| CONCAT | – | 0.596 ± 0.048 | 0.786 ± 0.032 |
| MK-SVM | – | 0.737 ± 0.031 | 0.833 ± 0.021 |
Bold best performance across both the feature types and the learning schemes, italics best performance across the feature types in the same learning scheme
We also plotted the best performances of the competing methods, regardless of the model training schemes, for four binary classification problems with their sensitivity and specificity given in Fig. 5. From the figure, we can clearly see that the proposed method outperforms the competing methods. It is noteworthy that there is a tendency of the improvement increase in the order of AD vs. HC, AD vs. MCI, MCI vs. HC, and MCI-C vs. MCI-NC. That is, we made higher improvements in the more challengeable and important tasks, e.g., classifying between MCI-C and MCI-NC, for early diagnosis and treatment.
Fig. 5.
Comparison of the best performances of the competing methods, regardless of the learning schemes for a SAE model
Discussions
Deep learning-based latent feature representation
In our method of discovering a latent feature representation, we built a SAE-classifier for a means of determining the optimal SAE-structure. It is worth noting that, across classification tasks, different numbers of hidden units for the same modality were determined, e.g., 500-50-10 in AD vs. HC, 100-100-20 in MCI vs. HC, 1000-50-30 in AD vs. MCI, and 100-100-10 in MCI-C vs. MCI-NC for MRI in supervised learning. We believe that this reflects the necessity of considering different high-level non-linear relations inherent in LLF for different classification problems.
In terms of the model architecture, the SAE-classifier can be considered as a simple logistic regression model taking the SAE-learned feature representation as input. Despite the simple architecture, it presented classification accuracies higher than or comparable to those from the SVM classifier, into which SAE-learned features were fed after feature selection. This is resulted from the fact that the SAE-learned features were optimized to the SAE-classifier, not to the SVM classifier.
In the meantime, when we constructed an augmented feature vector via a concatenation of LLF and SAEF, we could greatly improve the accuracies. That is, the original low-level features are still informative for brain disease diagnosis along with the latent feature representations.
In comparison with the LLF-based method, the proposed method of LLF + SAEF, greatly improved the diagnostic accuracy over all the classification problems considered in this work. Specifically, the proposed method consistently outperformed the competing methods over uni-modality and multi-modality with semi-supervised learning.
In deep learning, it is an important issue for the size of training samples. While there is a limited number of samples available in ADNI dataset, we would like to note that under the circumstance of a small sample size, there is an empirical proof that the unsupervised pre-training helps deep learning find better optimal parameters for reducing errors (Erhan et al. 2010). In the same perspective, we could also obtain the best performances in four binary classification problems from the semi-supervised learning, which means that we could benefit from the target-unrelated samples for pre-training and learning the optimal parameters for the deep network, and hence enhance the classification accuracy. This is one of the most prominent and important characteristics of deep learning in SAE, compared to the conventional neural network. In the conventional neural network, we find the optimal parameters starting from random initialization in a supervised manner, which means that we can only use limited number of target-related samples in learning. Therefore, it is restricted for the application of neural networks with only a small number of layers in structure. Meanwhile, the deep learning allows to utilize the unlabeled or target-unrelated samples in learning. From a practical point of view, it is of great importance to exploit information from unlabeled or target-unrelated data, which we have much more available in the reality.
It is also important for the interpretation of the trained weights and the latent feature representations. We can regard the trained weights as filters that can find different types of relations among the inputs. For example, each hidden unit in the first hidden layer captures a different representation via the non-linear transformation of the weighted linear combination of the input low-level features. Note that each unit has a different weight set and the weights of the input low-level features can be positive, negative, or zero. That is, by assigning different weights to each low-level feature, e.g., GM tissue volume from MRI or mean intensity from FDG-PET, the model discovers different latent relations among the low-level features from hidden units. From a neuroscience perspective, the hidden layer can discover the structural non-linear relations from MRI features and the functional non-linear relations from FDG-PET features. The outputs of the first hidden layer are further combined in the upper hidden layer capturing even more complicated relations. In this way, the SAE hierarchically captures latent complicated information inherent in the input low-level features, which are helpful to classify patients and healthy normal controls. Theoretically, to date, there is no standard way to visualize or interpret the trained weights in an intuitive way, but it still remains a challenging issue also in the field of pattern recognition or machine learning. We would like to mention that while it is not straightforward to interpret the meaning of the trained weights or the latent feature representations, it is clear from our experiments that the latent complicated information is useful in AD/MCI diagnosis.
To further validate the effectiveness of the proposed method, we also presented a statistical significance of the results with paired t test in Table 7. The test was performed with the results obtained from LLF and LLF + SAEF with MK-SVM. The lasso-based feature selection was considered for both methods, and, for LLF + SAEF, the SAE model was learned in a semi-supervised manner. The proposed method statistically outperformed the LLF-based method across all cases, except for CSF, rejecting the null hypothesis beyond the 95 % of confidence level. We believe that due to the low dimensionality of the original features from CSF, the SAE-learned latent feature representation was not much informative in classification.
Table 7.
Statistical significance (paired t test) between the classification accuracies obtained from LLF and LLF + SAEF, which used supervised and semi-supervised learning schemes, respectively
| AD vs. HC |
MCI vs. HC |
AD vs. MCI |
MCI-C vs. MCI-NC |
|
|---|---|---|---|---|
| MRI | 0.0014 | 4.18e–06 | 2.32e–06 | 2.67e–06 |
| PET | 0.0025 | 3.26e–05 | 2.51e–04 | 2.51e–06 |
| CSF | 0.8673 | 0.4031 | 0.9955 | 0.2357 |
| CONCAT | 0.0014 | 5.74e–07 | 2.53e–05 | 2.18e–06 |
| MK-SVM | 8.45e–04 | 5.78e–07 | 5.36e–05 | 9.28e–06 |
| All | 0.005 | 6.70e–16 | 2.80e–12 | 3.43e–11 |
Lasso vs. group lasso for feature selection
Here, we compare the performances with lasso- and group lasso-based feature selection methods. In group lasso, we considered the clinical labels and clinical scores of MMSE and ADAS-cog as the target responses. In conclusion, we observed that the method of lasso-based feature selection outperformed that of group lasso-based one as presented in Fig. 6. The reason for this result is that, we believe, although the l21-norm-based multi-task learning can be used to take the advantage of richer information, it focuses on the target regression instead of the classification. Therefore, it finds features that most accurately regress the target values, i.e., clinical labels and clinical scores, regardless of the discriminative power of the selected features between classes. Moreover, the MMSE scores for different groups were highly overlapped, which means it provided mere information and might act as a potential confounding in discriminative feature selection. Meanwhile, in l1-norm-based single-task learning, the clinical labels, the prediction of which is our main goal, are used as the target response. That is, the selected features to regress the target clinical labels can be class-discriminative in some sense. However, we should note that the multi-task learning is a generalized form of the single-task sparse learning. Therefore, if there exists other class-related information, we should utilize the information in the framework of multi-task learning and it should thus produce better performance.
Fig. 6.
Comparison of the best performances between lasso- and group lasso-based feature selection methods
Comparison with the state-of-the-art method
We also compared the performance of the proposed method with that of the multi-task multi-modal learning (M3T) method (Zhang and Shen 2012), which first performs multi-task learning, i.e., group lasso, on LLF for feature selection and then fuses multi-modal information via MK-SVM. For fair comparison, we used the same training and test samples for M3T. Compared to the accuracies of M3T, which were 94.5 ± 0.8, 84 ± 1.1, 78.8 ± 1.8, and 71.8 ± 2.6 % for AD vs. HC, MCI vs. HC, AD vs. MCI, and MCI-C vs. MCI-NC classification, respectively, the proposed method with LLF + SAEF made a performance improvement of 3.4, 4.8, 3.9, and 6.1 % using a supervised learning scheme, and 4.3, 6.7, 4.9, and 11.5 % using a semi-supervised learning scheme, both of which used a l1-norm based feature selection.
Selected region of interests
From Figs. 7, 8, 9 and 10, we can see that the SAE-learned latent features did not show high frequency of being selected for classification. However, based on the classification accuracies and the fewer number of high frequency ROIs in the graphs, we assume that the SAE-learned latent features affected to filter out the original low-level features, which were not discriminative in classification, during feature selection. But, in classification of MCI vs. HC, a larger number of ROIs were involved for discrimination in the proposed method. Our understanding for this phenomenon is that due to its subtlety of the involved cognitive impairment in MCI compared to AD, we need to consider a larger number of ROIs and also the relations among them for more accurate diagnosis.
Fig. 7.
Frequencies of the selected ROIs in AD vs. HC classification. Blue and red bars correspond, respectively, to the original low-level features and the SAE-learned feature representations
Fig. 8.
Frequencies of the selected ROIs in MCI vs. HC classification. Blue and red bars correspond, respectively, to the original low-level features and the SAE-learned feature representations
Fig. 9.
Frequencies of the selected ROIs in AD vs. MCI classification. Blue and red bars correspond, respectively, to the original low-level features and the SAE-learned feature representations
Fig. 10.
Frequencies of the selected ROIs in MCI-C vs. MCI-NC classification. Blue and red bars correspond, respectively, to the original low-level features and the SAE-learned feature representations
The selected ROIs included medial temporal lobe that involves a system of anatomically related structures that are vital for declarative or long-term memory: amygdala, hippocampal formation, entorhinal cortex, hippocampal region, and the perirhinal, entorhinal, and parahippocampal cortices (Braak and Braak 1991; Visser et al. 2002; Mosconi 2005; Lee et al. 2006; Devanand et al. 2007; Burton et al. 2009; Desikan et al. 2009; Ewers et al. 2012; Walhovd et al. 2010), and also the regions of supramarginal gyrus (Buckner et al. 2005; Desikan et al. 2009; Dickerson et al. 2009; Schroeter et al. 2009), angular gyrus (Schroeter et al. 2009; Nobili et al. 2013; Yao et al. 2012), superior parietal lobule, precuneus, cuneus (Bokde et al. 2006; Singh et al. 2006; Davatzikos et al. 2011), cingulate region (Mosconi 2005), anterior limb of internal capsule (Zhang et al. 2009), caudate nucleus (Dai et al. 2009), fornix (Copenhaver et al. 2006).
Limitations of the current work
Although we could achieve performance enhancements in four different classification problems, there exist some limitations and disadvantages of the proposed method.
First, in PET imaging, it is known that the partial volume effect, caused by a combination of the limited resolution of PET and image sampling, can lead to underestimation or overestimation of regional concentrations of radioactivity in the reconstructed images and further errors in statistical parametric images (Aston et al. 2002). However, in this work, we did not apply a procedure for partial volume correction. Therefore, there is a possibility of resulting in mixed combination of multiple tissue values in each voxel, reducing the differences between GM and WM. On the other hand, since we are using the ROI-based features for our classification, the performance of our method is less affected by this partial volume effect.
Second, as for the computational complexity, once the model was built by determining the network structure, learning the model parameters, and selecting the features, it took less than a minute to get the result for a given patient in our system of Mac OSX with 3.2GHz Intel Core i5 and 16 GB of memory. However, as stated in “Deep learning-based latent feature representation”, to date, there is no general or intuitive method for visualization of the trained weights or for interpretation of the latent feature representations. The problem of efficient visualization or interpretation of the latent feature representation is another big challenge that should be tackled by the communities of machine learning and clinical neuroscience, collaboratively. Furthermore, we used a relatively small data samples (51 AD, 43 MCI-C, 56 MCI-NC, and 52 HC). Therefore, the network structures used to discover latent information in our experiments are not necessarily optimal for other datasets. We believe that it needs more intensive studies such as learning the optimal network structure from big data for practical use of deep learning in clinical settings.
Third, according to a recent broad spectrum of studies, there are increasing evidences that subjective cognitive complaints are one of the important genetic risk factors increasing the risk of progression to MCI or AD (Loewenstein et al. 2012; Mark and Sitskoorn 2013). That is, among cognitively normal elderly individuals, who have subjective cognitive impairments, there exists a high possibility for some of them to be in the stage of ‘pre-MCI’. However, in the ADNI dataset, there is no related information. Thus, in our experiments, the HC group could include both genuine controls and those with subjective cognitive complaints.
Lastly, we should mention that the data fusion in our deep learning is considered through a simple concatenation of the features from modalities into a vector, resulting in a low performance compared to that of the multi-kernel SVM. But, in terms of the network architecture, it is limited as a shallow model to discover the non-linear relations among features from multiple modalities. We believe that although the proposed SAE-based deep learning is successful to find latent information in this work, there is still a room to design a multi-modal deep network for the shared representation across modailities. Furthermore, inspired from the recent computer vision researches (Ngiam et al. 2011; Srivastava and Salakhutdinov 2012), we can efficiently handle the incomplete data problem (Yuan et al. 2012) with multi-modal deep learning. Therefore, it will be our forthcoming research issue to build a novel multimodal deep architecture that can efficiently model and combine complementary information in a unified framework. Besides that, while we used the complimentary information from three different modalities of MRI, FDG-PET, and CSF in this work, it will be also beneficiary to consider the genetic risk factor such as the presence of the allele ε4 in the Apoliopoprotein E (ApoE) for our future work.
Conclusions
Due to the increasing proportion of AD as the cause of death in elderly people, there have been great interests in early diagnosis and prognosis of the neurodegenerative disease in the clinic. Recent neuroimaging tools and machine learning techniques have greatly contributed for computer-aided brain disease diagnosis. However, the previous work in the literature considered only simple low-level features such as cortical thickness and/or gray matter tissue volumes from MRI, mean signal intensities from FDG-PET, and t-tau, p-tau, and Aβ42 from CSF.
The main motivation of our work is that there may exist hidden or latent high-level information inherent in the original low-level features, such as relations among features, which can be helpful to build a more robust diagnostic model. To this end, in this paper, we proposed to utilize a deep learning with SAE for a latent feature representation from the data for AD/MCI diagnosis.
While the SAE is a neural network in terms of the model structure, thanks to the two-step learning scheme of greedy layer-wise pre-training and the fine-tuning in deep learning, we could reduce the risk of falling into a poor local optimum, which is the main limitation of the conventional neural network. We believe that deep learning can shed new light on the analysis of neuroimaging data, and our paper presented the applicability of the method to brain disease diagnosis for the first time.
The contributions of our work are that (1) to our best knowledge, this is the first work that considers a deep learning for feature representation in brain disease diagnosis and prognosis, (2) unlike the previous work in the literature, we considered a complicated non-linear latent feature representation, which was directly discovered from data, (3) by constructing an augmented feature vector via a concatenation of the original low-level features and the SAE-learned latent feature representation, we could greatly improve diagnostic accuracy, and (4) thanks to the unsupervised characteristic of the pre-training in deep learning, the proposed method can utilize target-unrelated samples to discover a general feature representation, which helped to further enhance classification performance. Using the publicly available ADNI dataset, we evaluated the effectiveness of the proposed method and achieved the maximum accuracies of 98.8, 90.7, 83.7, and 83.3 %forADvs. NC, MCI vs. NC, AD vs. MCI, and MCI-C vs. MCI-NC classification, respectively, outperforming the competing methods.
Acknowledgments
This work was supported in part by NIH grants EB006733, EB008374, EB009634, AG041721, MH100217, and AG042599, and also by the National Research Foundation grant (No. 2012-005741) funded by the Korean government.
Footnotes
Although there exist in total more than 800 subjects in ADNI database, only 202 subjects have the baseline data including all the modalities of MRI, FDG-PET, and CSF.
Refer to http://www.adniinfo.org for the details.
While the low-level simple features should be the voxels in MRI and FDG-PET, due to high dimensionality and a small sample problem, in this paper, we take a ROI-based approach and consider the conical GM tissue volumes and the mean intensity for each ROI from MRI and FDG-PET, respectively, as the low-level features.
In this work, we set γ = 0.01 and ρ = 0.05.
In our case, the tasks are to regress class-label, and MMSE and ADAS-Cog scores.
In this work, .
CONCAT represents a concatenation of the features from MRI, FDG-PET, and CSF into a single vector, which is the most direct and intuitive way of combining multimodal information.
We considered [100, 300, 500, 1,000]–[50, 100]–[10, 20, 30] and [10, 20, 30]–[1, 2, 3] (bottom–top) for three-layer and two-layer networks, respectively.
Refer to “Sparse auto-encoder” for explanation of the supervised learning.
Contributor Information
Heung-Il Suk, Email: hsuk@med.unc.edu, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina, Chapel Hill, NC 27599, USA.
Seong-Whan Lee, Department of Brain and Cognitive Engineering, Korea University, Seoul 136-713, Republic of Korea.
Dinggang Shen, Email: dgshen@med.unc.edu, Biomedical Research Imaging Center (BRIC) and Department of Radiology, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Brain and Cognitive Engineering, Korea University, Seoul 136-713, Republic of Korea.
References
- Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2012;8(2):131–168. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Aston JAD, Cunningham VJ, Asselin MC, Hammers A, Evans AC, Gunn RN. Positron emission tomography partial volume correction: estimation and algorithms. J Cereb Blood Flow Metab. 2002;22(8):1019–1034. doi: 10.1097/00004647-200208000-00014. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengio Y. Learning deep architectures for AI. Found Trends Machine Learn. 2009;2(1):1–127. [Google Scholar]
- Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. In: Schölkopf B, Platt J, Hoffman T, editors. Advances in neural information processing systems. Vol. 19. Cambridge: MIT Press; 2007. pp. 153–160. [Google Scholar]
- Bishop CM. Neural networks for pattern recognition. New York: Oxford University Press, Inc.; 1995. [Google Scholar]
- Bokde ALW, Lopez-Bayo P, Meindl T, Pechler S, Born C, Faltraco F, Teipel SJ, Möller HJ, Hampel H. Functional connectivity of the fusiform gyrus during a face-matching task in subjects with mild cognitive impairment. Brain. 2006;129(5):1113–1124. doi: 10.1093/brain/awl051. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton EJ, Barber R, Mukaetova-Ladinska EB, Robson J, Perry RH, Jaros E, Kalaria RN, O’Brien JT. Medial temporal lobe atrophy on MRI differentiates Alzheimer’s disease from dementia with Lewy bodies and vascular cognitive impairment: a prospective study with pathological verification of diagnosis. Brain. 2009;132(1):195–203. doi: 10.1093/brain/awn298. [DOI] [PubMed] [Google Scholar]
- Copenhaver BR, Rabin LA, Saykin AJ, Roth RM, Wishart HA, Flashman LA, Santulli RB, McHugh TL, Mamourian AC. The fornix and mammillary bodies in older adults with Alzheimer’s disease, mild cognitive impairment, and cognitive complaints: a volumetric MRI study. Psychiatry Res Neuroimaging. 2006;147(2–3):93–103. doi: 10.1016/j.pscychresns.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin JS. The Alzheimer’s disease neuroimaging initiative: identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6(7):e21, 896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Lopez O, Carmichael O, Becker J, Kuller L, Gach H. Mild cognitive impairment and Alzheimer disease: patterns of altered cerebral blood flow at MR imaging. Radiology. 2009;250(3):856–866. doi: 10.1148/radiol.2503080751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32(12):2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Cabral H, Hess C, Dillon W, Salat D, Buckner R, Fischl B Initiative ADN. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132:2048–2057. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Hoing LS, Mayeux R, Stern Y, Tabert MH, de Leon JJ. Hippocampal and entorhinal atrophy in mild cognitive impairment. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL. The cortical signature of Alzheimer’s disease: Regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:828–836. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhan D, Bengio Y, Courville A, Manzagol PA, Vincent P, Bengio S. Why does unsupervised pre-training help deep learning. Int J Pattern Recognit Artif Intell. 2010;11:625–660. [Google Scholar]
- Evans AC, Collins DL. Animal: validation and applications of nonlinear registration-based segmentation. Int J Pattern Recognit Artif Intell. 1997;11(8):1271–1294. [Google Scholar]
- Ewers M, Walsh C, Trojanowski JQ, Shaw LM, Petersen RC, Jack CR, Jr, Feldman HH, Bokde AL, Alexander GE, Scheltens P, Vellas B, Dubois B, Weiner M, Hampel H. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. 2012;33(7):1203–1214. e2. doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Rao H, Hurt H, Giannetta J, Korczykowski M, Shera D, Avants BB, Gee JC, Wang J, Shen D. Multivariate examination of brain abnormality using both structural and functional MRI. NeuroImage. 2007;36(4):1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Poline JB, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Hum Brain Mapp. 1995;3(3):165–189. [Google Scholar]
- Fukushima K. Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980;36(4):93–202. doi: 10.1007/BF00344251. [DOI] [PubMed] [Google Scholar]
- Gönen M, Alpaydm E. Multiple kernel learning algorithms. J Machine Learn Res. 2011;12:2211–2268. [Google Scholar]
- Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Davis LS. Density-based multifeature background subtraction with support vector machine. IEEE Trans Pattern Anal Machine Intell. 2012;34(5):1017–1023. doi: 10.1109/TPAMI.2011.243. [DOI] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage. 2011;55(2):574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- Jia H, Wu G, Wang Q, Shen D. ABSORB: Atlas building by self-organized registration and bundling. NeuroImage. 2010;51(3):1057–1070. doi: 10.1016/j.neuroimage.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, MacDonald D, Holmes C, Evans A. A 3D atlas of the human brain. NeuroImage. 1998;7(4):S717. [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol Aging. 2010;31(8):1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle H, Bengio Y, Louradour J, Lamblin P. Exploring strategies for training deep neural networks. J Machine Learn Res. 2009;10:1–40. [Google Scholar]
- Lee ACH, Buckley MJ, Gaffan D, Emery T, Hodges JR, Graham KS. Differentiating the roles of the hippocampus and perirhinal cortex in processes beyond long-term declarative memory: a double dissociation in dementia. J Neurosci. 2006;26(19):5198–5203. doi: 10.1523/JNEUROSCI.3157-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Ekanadham C, Ng A. Sparse deep belief net model for visual area v2. In: Platt J, Koller D, Singer Y, Roweis S, editors. Advances in neural information processing systems. Vol. 20. Cambridge: MIT Press; 2008. pp. 873–880. [Google Scholar]
- Lee H, Grosse R, Ranganath R, Ng AY. Unsupervised learning of hierarchical representations with convolutional deep belief networks. Commun ACM. 2011;54(10):95–103. [Google Scholar]
- Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. 2012;33(2):427.e15–427.e30. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer’s disease. NeuroImage. 2012;60(2):1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Suk HI, Wee CY, Chen H, Shen D. High-order graph matching based feature selection for Alzheimer’s disease identification. Proceedings of the 16th international conference on medical image computing and computer-assisted intevention; Springer; Berlin, Heidelberg. 2013. pp. 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenstein DA, Greig MT, Schinka JA, Barker W, Shen Q, Potter E, Raj A, Brooks L, Varon D, Schoenberg M, Banko J, Potter H, Duara R. An investigation of PreMCI: subtypes and longitudinal outcomes. Alzheimer’s Dementia. 2012;8(3):172–179. doi: 10.1016/j.jalz.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark RE, Sitskoorn MM. Are subjective cognitive complaints relevant in preclinical Alzheimer’s disease? A review and guidelines for healthcare professionals. Rev Clin Gerontol. 2013;23:61–74. [Google Scholar]
- Mosconi L. Brain glucose metabolism in the early and specific diagnosis of Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2005;32(4):486–510. doi: 10.1007/s00259-005-1762-7. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Tsui WH, Herholz K, Pupi A, Drzezga A, Lucignani G, Reiman EM, Holthoff V, Kalbe E, Sorbi S, Diehl-Schmid J, Perneczky R, Clerici F, Caselli R, Beuthien-Baumann B, Kurz A, Minoshima S, de Leon MJ. Multicenter standardized 18F-FDG PET diagnosis of mild cognitive impairment, Alzheimer’s disease, and other dementias. J Nucl Med. 2008;49(3):390–398. doi: 10.2967/jnumed.107.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettiksimmons J, Harvey D, Brewer J, Carmichael O, DeCarli C, Jack CR, Petersen R, Shaw LM, Trojanowski JQ, Weiner MW, Beckett L. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiol Aging. 2010;31(8):1419–1428. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY. Multimodal deep learning; Proceedings of the 28th International Conference on Machine Learning; 2011. pp. 689–696. [Google Scholar]
- Nobili F, Mazzei D, Dessi B, Morbelli S, Brugnolo A, Barbieri P, Girtler N, Sambuceti G, Rodriguez G, Pagani M. Unawareness of memory deficit in amnestic MCI: FDG-PET findings. J Alzheimer’s Dis. 2010;22(3):993–1003. doi: 10.3233/JAD-2010-100423. (2010) [DOI] [PubMed] [Google Scholar]
- Nordberg A, Rinne JO, Kadir A, Langstrom B. The use of PET in Alzheimer disease. Nat Rev Neurol. 2010;6(2):78–87. doi: 10.1038/nrneurol.2009.217. [DOI] [PubMed] [Google Scholar]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Non-rigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Schroeter ML, Stein T, Maslowski N, Neumann J. Neural correlates of Alzheimer’s disease and mild cognitive impairment: a systematic and quantitative meta-analysis involving 1351 patients. NeuroImage. 2009;47(4):1196–1206. doi: 10.1016/j.neuroimage.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre T, Wolf L, Poggio T. Object recognition with features inspired by visual cortex. Proceedings of the 2005 IEEE computer society conference on computer vision and pattern recognition. 2005;2:994–1000. [Google Scholar]
- Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shen D, Wong WH, Ip HH. Affine-invariant image retrieval by correspondence matching of shapes. Image Vis Comput. 1999;17(7):489–499. [Google Scholar]
- Shin HC, Orton MR, Collins DJ, Doran SJ, Leach MO. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. IEEE Trans Pattern Anal Machine Intell. 2013;35(8):1930–1943. doi: 10.1109/TPAMI.2012.277. [DOI] [PubMed] [Google Scholar]
- Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer’s disease. Brain. 2006;129(11):2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Srivastava N, Salakhutdinov R. Multimodal learning with deep Boltzmann machines. In: Bartlett P, Pereira F, Burges C, Bottou L, Weinberger K, editors. Advances in neural information processing systems. Vol. 25. 2012. pp. 2231–2239. [Google Scholar]
- Suk HI, Lee SW. A novel Bayesian framework for discriminative feature extraction in brain-computer interfaces. IEEE Trans Pattern Anal Machine Intell. 2013;35(2):286–299. doi: 10.1109/TPAMI.2012.69. [DOI] [PubMed] [Google Scholar]
- Suk HI, Wee CY, Shen D. Discriminative group sparse representation for mild cognitive impairment classification. Proceedings of the 4th international workshop on machine learning in medical imaging; Springer; Switzerland. 2013. pp. 131–138. [Google Scholar]
- Tang S, Fan Y, Wu G, Kim M, Shen D. RABBIT: rapid alignment of brains by building intermediate templates. NeuroImage. 2009;47(4):1277–1287. doi: 10.1016/j.neuroimage.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiola T, Alafuzoff I, Herukka SK, Parkkinen L, Hartikainen P, Soininen H, Pirttilä T. Cerebrospinal fluid β-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Archives Neurol. 2009;66(3):382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc. 1996;58(1):267–288. [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. NeuroImage. 2009;45(1) Suppl 1:S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Visser PJ, Verhey FRJ, Hofman PAM, Scheltens P, Jolles J. Medial temporal lobe atrophy predicts Alzheimer’s disease in patients with minor cognitive impairment. J Neurol Neurosurg Psychiatry. 2002;72:491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd K, Fjell A, Brewer J, McEvoy L, Fennema-Notestine C, Hagler DJ, Jr, Jennings R, Karow D, Dale A. The Alzheimer’s disease Neuroimaging Initiative Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nie J, Yap PT, Shi F, Guo L, Shen D. Robust deformable-surface-based skull-stripping for large-scale studies. Proceedings of the 14th international conference on medical image computing and computer-assisted intervention; Springer; Berlin, Heidelberg. 2011. pp. 635–642. [DOI] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Li W, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Enriched white matter connectivity networks for accurate identification of MCI patients. Neuroimage. 2011;54(3):1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Zhang D, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Identification of MCI individuals using structural and functional connectivity networks. Neuroimage. 2012;59(3):2045–2056. doi: 10.1016/j.neuroimage.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman E, Muehlboeck JS, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage. 2012;62(1):229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Wu G, Qi F, Shen D. Learning-based deformable registration of MR brain images. IEEE Trans Med Imaging. 2006;25(6):1145–1157. doi: 10.1109/tmi.2006.879320. [DOI] [PubMed] [Google Scholar]
- Xue Z, Shen D, Davatzikos C. Statistical representation of high-dimensional deformation fields with application to statistically constrained 3D warping. Med Image Anal. 2006a;10(5):740–751. doi: 10.1016/j.media.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Xue Z, Shen D, Karacali B, Stern J, Rottenberg D, Davatzikos C. Simulating deformations of MR brain images for validation of atlas-based segmentation and registration algorithms. NeuroImage. 2006b;33(3):855–866. doi: 10.1016/j.neuroimage.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Shen D, Davatzikos C, Verma R. Diffusion tensor image registration using tensor geometry and orientation features. Proceedings of the 11th international conference on medical image computing and computer-assisted intervention; Springer; Berlin, Heidelberg. 2008. pp. 905–913. [DOI] [PubMed] [Google Scholar]
- Yao Z, Hu B, Liang C, Zhao L, Jackson M. The Alzheimer’s disease neuroimaging initiative: a longitudinal study of atrophy in amnestic mild cognitive impairment and normal aging revealed by cortical thickness. PLoS One. 2012;7(11):e48, 973. doi: 10.1371/journal.pone.0048973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Lin Y, Lafferty J. Learning image representations from the pixel level via hierarchical sparse coding; Proceedings of the 2011 IEEE computer society conference on computer vision and pattern recognition, Providence; 2011. pp. 1713–1720. [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. NeuroImage. 2012;61(3):622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc Ser B. 2006;68(1):49–67. [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage. 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schuff N, Du AT, Rosen HJ, Kramer JH, Gorno-Tempini ML, Miller BL, Weiner MW. White matter damage in frontotemporal dementia and Alzheimer’s disease measured by diffusion MRI. Brain. 2009;132(9):2579–2592. doi: 10.1093/brain/awp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang Y, Li Y, Yap PT, Shen D ADNI. Hierarchical anatomical brain networks for MCI prediction: revisiting volumetric measures. PLoS ONE. 2011;6(7):e21935. doi: 10.1371/journal.pone.0021935. [DOI] [PMC free article] [PubMed] [Google Scholar]