Abstract
Rationale
Hemodynamic disturbed flow is associated with susceptibility to atherosclerosis. Endothelial KLF4 is an important anti-inflammatory atheroprotective transcription factor that is suppressed in regions of disturbed flow.
Objective
The plasticity of epigenomic KLF4 transcriptional regulation by flow-mediated DNA methylation was investigated in vitro and in arterial tissue.
Methods and Results
To recapitulate dominant flow characteristics of atheroprotected and atherosusceptible arteries, human aortic endothelial cells (HAEC) were subjected to pulsatile undisturbed flow (UF) or oscillatory disturbed flow (DF) containing a flow-reversing phase. Differential CpG site methylation was measured by methylation specific PCR, bisulfite pyrosequencing and restriction enzyme-PCR. The methylation profiles of endothelium from disturbed and undisturbed flow sites of adult swine aortas were also investigated. In vitro, DF increased DNA methylation of CpG islands within the KLF4 promoter that significantly contributed to suppression of KLF4 transcription; the effects were mitigated by DNA methyltransferase (DNMT) inhibitors and knock-down of DNMT3A. Contributory mechanisms included DF-induced increase of DNMT3A protein (1.7 fold), DNMT3A enrichment (11-fold) on the KLF4 promoter, and competitive blocking of a MEF2 binding site in the KLF4 promoter near the TSS. DF also induced DNMT-sensitive pro-pathological expression of downstream KLF4 transcription targets NOS3, thrombomodulin (THBD) and MCP-1. In support of the in vitro findings, swine aortic endothelium isolated from DF regions expressed significantly lower KLF4 and NOS3, and bisulfite sequencing of KLF4 promoter identified a hypermethylated MEF2 binding site.
Conclusions
Hemodynamics influence endothelial KLF4 expression through DNMT enrichment/MEF2 inhibition mechanisms of KLF4 promoter CpG methylation with regional consequences for atherosusceptibility.
Keywords: Atherosusceptible endothelium, hemodynamics, epigenetics, DNA methylation, KLF4, athergenesis
INTRODUCTION
Although associated with systemic environmental risk factors, genetic defects and aging1, atherosclerotic lesions typically originate locally, developing preferentially at predictable sites of curvature, branching, and bifurcation in large arteries where disturbed blood flow (DF) is a prominent characteristic2. In contrast, regions dominated by pulsatile unidirectional laminar undisturbed flow (UF) are relatively atheroresistant. Hemodynamic DF, although usually also laminar, is characterized by flow separation, transient flow reversals, and average low shear forces that define the atherosusceptible regional environment3.
The endothelium plays a central role in the initiation and development of inflammatory atherosclerosis. Endothelial phenotypes in prelesional atherosusceptible regions are subtly different from those located at nearby athero-resistant sites where UF is prevalent. In prelesional DF regions of mouse and swine, differential transcriptional profiling has identified endothelia that are sensitized for pro-inflammatory pathways, coagulation, and redox3-6, and the chronic low level activation of endoplasmic reticulum stress and the unfolded protein response7. When the principal characteristics of UF and DF are recapitulated in vitro, important protective pathways are suppressed by DF including the expression and activity of Kruppel-like factor 4 (KLF4) and nitric oxide synthase 3 (NOS3)5,8. The expression of NOS3, essential for regulation of vascular tone and maintenance of the quiescent state of endothelium8,9, is regulated by hemodynamic shear stress linked to a series of upstream transcription factors that includes KLF4, KLF2 and RelA/p6510-12. KLF4 and KLF2 are zinc-finger regulatory transcription factors for gene networks that confer atheroresistant anti-inflammatory and anti-thrombotic properties to the endothelium; localized dysfunction or suppression of KLF4 is therefore pro-pathological10,13,14. Dysregulation of KLF4 and NOS3 by genetic manipulation of the endothelium of mice significantly contributes to the development and progression of atherosclerosis10,15,16.
The mechanisms linking hemodynamics characteristics such as UF and DF to endothelial phenotype, function and pathosusceptibility are under intensive study at multiple levels of regulation, most recently epigenetic. Flow-induced histone modification and miRNAs have been shown to shape endothelial phenotype identities17-21 but differential DNA methylation responses to different flow profiles encountered in vivo and their recapitulation in vitro have not been addressed.
DNA methylation is one of the critical epigenetic mechanisms controlling gene expression22. In vertebrates, DNA methylation occurs at carbon 5 of cytosine in CpG dinucleotides (5mC). When occurring within the promoter regions of genes, it dramatically suppresses transcription by direct inhibition of transcription factor binding and recruitment of methyl-CpG-binding proteins, which further hinder access to the recognition site of transcription factors or modulate chromatin structure by the recruitment of histone-modifying proteins23-25. The DNA methylation landscape of the genome is established by methylation and demethylation enzymes. DNA (cytosine-5-)-methyltransferase 1 (DNMT1) maintains tissue-specific DNA methylation patterns via methylation of a hemi-methylated nascent DNA strand during cell proliferation26. DNMT3A and 3B are required for genome-wide de novo methylation and play crucial roles in the establishment of DNA methylation patterns during development26. Methylation by DNMTs is counterbalanced by passive and/or active DNA demethylation in which the TET (ten-eleven-translocation) genes pathway has been suggested to play a central role in oxidizing 5mC to 5-hydroxymethylcytosine (5hmC)24.
An appreciation of DNA methylation dynamics in physiological and pathological gene regulation is emerging22. Although the post-development DNA methylation status associated with many genes tends to remain stable and is often linked to the maintenance of cell identity, epigenetic plasticity including DNA methylation/demethylation dynamics may be important for cellular adaptation responses including endothelial phenotype identity in different arterial hemodynamic environments. Here we demonstrate the plasticity of endothelial DNA methylation within the promoter of the important atheroprotective transcription factor KLF4. We show that DF-induced hypermethylation significantly suppresses KLF4 transcription and regulates its downstream targets NOS3, thrombomodulin (THBD) and MCP-1. As far as we are aware these data are the first demonstrated changes in DNA methylation induced by physiological characteristics of flow and are supported by steady state measurements in endothelial cells isolated from in vivo regions of hemodynamic DF and UF in swine aorta.
METHODS
Reagents and detailed molecular biology procedures are described in detail in Online Data Supplement.
Cell culture and flow experiments
Human aortic endothelial cells (HAEC; passage 4-6; Lonza, Allendale NJ) were cultured in complete EGM-2 medium to confluence on 0.1% gelatin coated glass slides (75×38 mm). The flow experiments were conducted as previously described27. Post-confluent HAEC were subjected to pulsatile UF or DF in a parallel-plate flow chamber for 2 days. UF waveform is characterized by a higher mean wall shear stress (WSS) and fully antegrade flow (Figure 1A). In contrast, the DF waveform exposes cells to lower mean WSS and a retrograde flow for one third of each cycle. The flow waveforms capture the dominant characteristics of human arterial hemodynamics flow behavior in UF and DF arterial sites. All flow in large arteries is unsteady (pulsatile). The defining feature of DF regions is that there is flow reversal during the cardiac cycle whereas in UF, the flow is always unidirectional. Waveforms were generated digitally and converted to analog signals by a data acquisition card (USB-6229, National Instruments, Austin, TX) that controlled a 520U Watson-Marlow peristaltic pump (Cornwall, England). Flow was measured with an ultrasonic flow meter (Transonic Systems, Inc., Ithaca, NY) to ensure experimental repeatability. Both waveforms were sinusoidal while differing in amplitude, mean WSS and oscillatory shear index (OSI) values. WSS values for the UF waveform ranged from 9.6 to 1.5 dyne/cm2 (mean 5.1 dyne/cm2) and for DF from 2 to −1.2 dyne/cm2 (mean of 0.4 dyne/cm2). The OSI for UF equaled 0, while for DF it was 0.32. An OSI value of 0 corresponds to fully antegrade flow and 1 to fully retrograde flow.
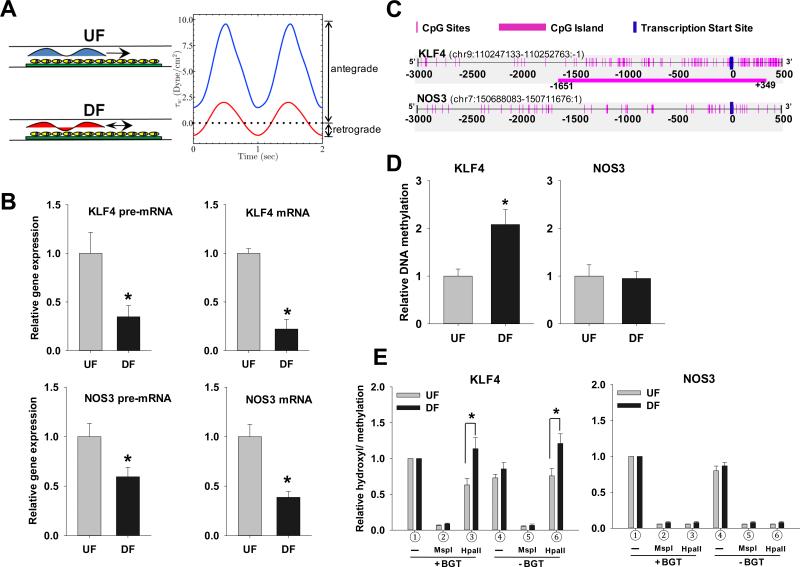
Figure 1. UF and DF regulation of KLF4 and NOS3 promoter methylation and gene transcription.
(A) Schematic illustration of the parallel plate flow apparatus and the arterial flow waveforms. Confluent HAEC were subjected to UF or DF for 2 days. Flow characteristics were confirmed by ultrasonic flow meter. (B) DF-induced suppression of KLF4 transcription. mRNA and pre-mRNA of KLF4 and NOS3 were determined by qPCR. The relative expression of DF to UF is computed following normalization of UF or DF to no-flow cells. All qPCR are normalized to ubiquitin B (UBB) and expressed as mean ± SEM fold of UF. (C) Schematic illustration of CpG island and CpG sites in human KLF4 and NOS3 promoters. (D) DF differentially regulates KLF4 and NOS3 promoter methylation. DNA methylation was determined by MSP using primers targeting KLF4 (−152/+9) and NOS3 (−369/−196) promoter region. Data are normalized to UBB promoter and expressed as mean ± SEM fold of UF. (E) Methylation and hydroxymethylation of KLF4 and NOS3 promoters. After glucosylation by T4 β-glucosyltransferase (BGT) and restriction enzyme MspI and HpaII digestion, the digested genomic DNA were used for qPCR with gene specific primers targeting CCGG-9 of the KLF4 promoter and CCGG-137 of the NOS3 promoter.  input,
input,  5hmC,
5hmC,  5mC and 5hmC,
5mC and 5hmC,  input,
input,  background,
background,  5mC and 5hmC. Data are normalized to
5mC and 5hmC. Data are normalized to  uncut DNA and expressed as mean ± SEM. *P < 0.05. n= 4.
uncut DNA and expressed as mean ± SEM. *P < 0.05. n= 4.
Animal studies
Endothelia were obtained from adult pigs (6-month-old; ~250 lb) immediately after euthanasia at a local slaughterhouse (Clemens Foods, Hatfield, PA). Aortas were harvested, and the vessel lumen was rinsed with ice-cold PBS. Endothelial cells were freshly harvested by gentle scraping of 1 cm2 regions located at the inner curvature of the aortic arch (AA) and nearby descending thoracic aorta (DT) representing DF and UF respectively. Cells were transferred directly to lysis buffer for DNA or RNA extraction. Endothelial purity was routinely assessed with antibodies against platelet endothelial cell adhesion molecule (PECAM-1) and alpha smooth muscle actin (ACTA2). Endothelial purity was also monitored by examining ACTA2 promoter hypermethylation (Online Figure VIII).
Statistical analysis
Results are expressed as mean ± SEM. Statistical analysis was performed by using an independent Student t test for two groups of data and ANOVA. If a normality test failed, data were compared by Mann-Whitney Rank Sum test. P value < 0.05 was considered significant.
RESULTS
Induction of endothelial KLF4 promoter methylation by disturbed flow in vitro
UF and DF characteristics were monitored in real-time. The pulsatile flow was always antegrade in UF whereas a brief reverse flow phase (negative shear stress, retrograde flow) lasting one third of the cycle provided an oscillatory shear index in DF (Figure 1A).
Gene expression
Figure 1B (upper panel) demonstrates that, when referenced to UF, DF significantly inhibited the expression of both KLF4 pre-mature mRNA (pre-mRNA) and mature mRNA by 65% and 75% respectively. The introns are spliced out in mature mRNA which is composed of exons only. DF also inhibited NOS3 pre-mRNA and mRNA by 41% and 61% respectively (Figure 1B lower panels). These data agree with previous reports of DF suppression of endothelial KLF4 and NOS35,8.
DNA methylation
A CpG island (CGI) exists at the transcription start site (TSS) of human KLF4 but not at the TSS of NOS3 (Figure 1C). The KLF4 promoter CGI is 2000 bp in length and the CpG observed/ expected (CpGo/e) = 0.74. Promoter methylation is usually associated with gene regulation22; therefore, its association with the suppression of transcription was interrogated. Methylation status was determined by methylation specific PCR (MSP) using specific primers targeting the KLF4 promoter. With reference to UF, DF significantly enhanced the KLF4 promoter methylation to 2.0 fold in CpG-rich regions after 2 days (Figure 1D left panel). NOS3, an athero-protective and KLF4 down-stream target gene, was also evaluated. In contrast to KLF4, the human NOS3 promoter has poor CpG content (CpGo/e < 0.4). Importantly, methylation of NOS3 promoter was unchanged by DF (Figure 1D right panel) despite suppression of NOS3 transcript expression. Thus, DF-suppressed KLF4 gene transcription was directly associated with hypermethylation of the KLF4 promoter, a status lacking in the NOS3 promoter.
Minimal contribution by hydroxymethylation
Hydroxymethylation at CpG sites is suggested to be associated with a DNA demethylation pathway that influences gene transcription regulation24,28. MSP does not discriminate between methylation and hydroxymethylation of CpG29. Therefore we quantified hydroxymethylation and methylation by using restriction enzyme-PCR. Two CCGG sites in the KLF4 promoter (804 bp and 9 bp upstream of TSS) were interrogated for DNA methylation and hydroxymethylation after exposure to UF or DF for 2 days (Figure 1E and Online Figure IA). At both sites, T4-BGT and MspI enzyme treatment followed by qPCR demonstrated that hydroxymethylation at the KLF4 promoter was very low and remained unchanged by either flow treatment. In contrast, HpaII treatment followed by qPCR confirmed that DF significantly enhanced KLF4 promoter methylation in the same experiments (Figure 1E and Online Figure IA). Three CCGG sites (−745, −194 and −137) in the NOS3 promoter were also tested (Figure 1E and Online Figure I). T4-BGT and MspI enzyme treatment followed by qPCR demonstrated that hydroxymethylation of NOS3 promoter was almost undetectable, while methylation remained very low and not significantly different between UF and DF. We conclude that DNA methylation of the KLF4 - but not NOS3 - promoter region is influenced by flow characteristics and that the contribution by hydroxymethylation is not significant.
DNA methylation inhibits the transcriptional activity of myocyte enhancer factor-2 (MEF2)
It has been suggested that tumor necrosis factor α (TNFα) and/or resveratrol induction of the KLF4 gene can be regulated by myocyte enhancer factor 2 (MEF2) transcription factors13,14,19,20. In silico analysis suggested only one MEF2 binding site TATTTAAAGTA (−64/−55) in the human KLF4 promoter. To test if MEF2 can bind to KLF4 promoter in cells, the MEF2 enrichment of chromatin was tested by ChIP-PCR assay using 4 primers targeting the promoter region of KLF4. MEF2 was dramatically enriched in the region from −161 to −25 (MEF2 binding site), but not in other regions of the KLF4 promoter (Online Figure II).
To confirm the ability of MEF2 to bind to the KLF4 promoter, nuclear protein extract from flow-acclimated HAEC was incubated with fluorescent labeled oligonucleotide containing MEF2 binding site. Gel mobility assay showed one shifted band which was abolished by anti-MEF2 antibody directed to the C-terminus of MEF2, suggesting that C-terminus of MEF2 is required in the formation of MEF2 complex and its association with the KLF4 promoter (Online Figure III). This was not a test of differential flow; protein extracts from DF and UF cells induced similar binding capacity of MEF2 to the unmethylated oligonucleotide.
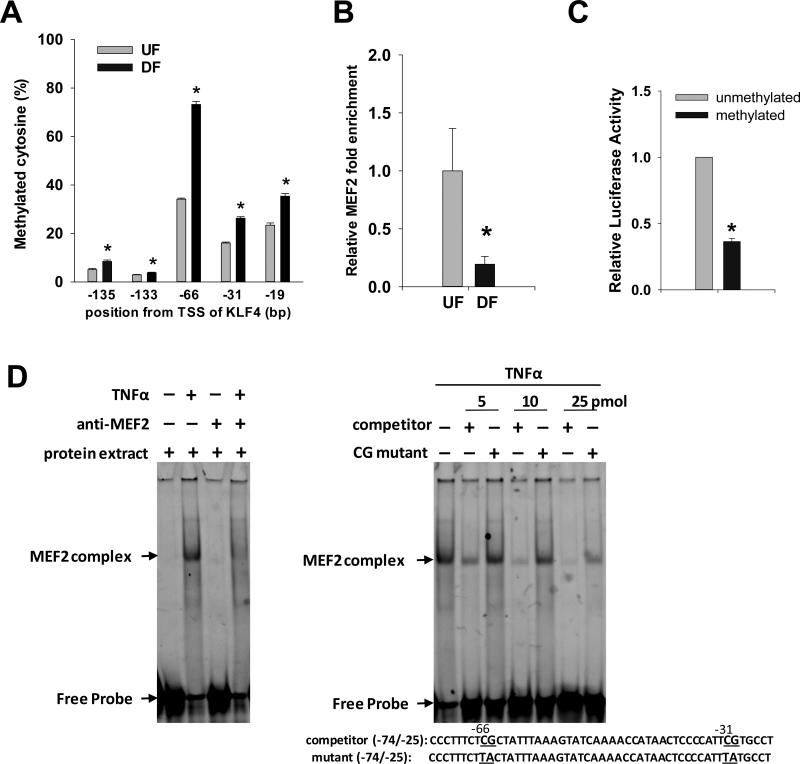
To test if hemodynamic forces could regulate the methylation status of CpG near the MEF2 binding sequence, bisulfite pyrosequencing was used to quantify the methylation levels at individual CpG sites (−135, −133, −66, −31 and −19bp from KLF4 TSS). Consistent with MSP analysis (Figure 1D), DF further enhanced CpG methylation close to the MEF2 binding sequence. Methylation of CpG site −66 was enhanced from 34% to 73% and methylation of CpG site −31 was enhanced from 16% to 26% (Figure 2A). Consistent with robust DF-enhanced methylation near the MEF2 binding site, DF reduced the chromatin loading of MEF2 protein to KLF4 promoter by 80% (Figure 2B), confirming that MEF2-enhanced KLF4 gene transcription is impeded by methylation of KLF4 promoter.
Figure 2. KLF4 promoter methylation blocks MEF2-mediated transcriptional activation of KLF4.
(A) Genomic DNA were isolated from confluent HAEC which were subjected to UF or DF for 2 days. Bisulfite pyrosequencing quantified the methylation levels of individual CpG sites near the MEF2 binding sequence in the KLF4 promoter. (B) Two-day UF and DF induction of MEF2 loading at the KLF4 promoter (−161/−25) was analyzed by ChIP-qPCR. Data are normalized to chromatin enrichment of the ACTA2 promoter and are expressed as mean ± SEM fold of UF. (C) Methylated and unmethylated KLF4 promoter construct containing MEF2 binding sequence was transfected into HAEC. KLF4 promoter transcription activity was measured by luciferase enzyme activity. Data are expressed as mean ± SEM fold of unmethylation. (D) TNFα induction of MEF2 binding to KLF4 promoter sequence. Confluent HAEC were treated with TNFα for 6 hours. 10 μg of nuclear protein was incubated with FAM labeled oligonucleotides with MEF2 binding sequence (TATTTAAAGTA) of the KLF4 promoter for 30 min. Anti-MEF2 antibody was pre-incubated with protein before adding the oligonucleotides (left panel). Competition experiments were carried out by pre-incubations of the protein extract with anti-MEF2 antibody, competitor or mutant oligonucleotides (right panel). Two CpG sites (−66 and −31bp from KLF4 TSS) and the corresponding mutants flanking the core MEF2 binding sequence are underlined. *P < 0.05. n= 4-6.
To further test if methylation of KLF4 promoter can impede KLF4 transcription, an in vitro luciferase reporter assay was carried out. After successfully methylating a KLF4 promoter construct that contains MEF2 binding sequence (Online Figure IV), HAEC were transiently transfected with methylated and mock-methylated constructs. Methylation of KLF4 promoter construct significantly decreased its promoter activity by 64%, compared with the unmethylated construct (Figure 2C), further demonstrating inhibition of MEF2-enhanced KLF4 gene transcription by methylation of KLF4 promoter.
Endothelia in prelesional DF regions of artery are sensitized for pro-inflammatory pathways3-7. To test if pro-inflammatory cytokine TNFα can induce similar interactions of MEF2 with KLF4 promoter, oligonucleotide containing MEF2 binding sequence was incubated with nuclear protein extract from HAEC treated with or without TNFα. Gel mobility assay showed a prominent shifted band after TNFα treatment that was completely abolished by MEF2 antibody (Figure 2D left panel), while no shifted band was observed in cells without TNFα treatment. The specificity of MEF2 site in the KLF4 promoter to bind endogenous MEF2 factors was then confirmed by progressive abolition of MEF2 protein binding by wild-type competitor (5 - 50 fold molar excess) as the molar concentration increased (Figure 2D right panel). The importance of CpG in mediating MEF2 binding to KLF4 promoter was tested by mutation of two CpG dinucleotides (−66 and −33) flanking the MEF2 binding sequence. These mutations resulted in a mild effect on competing MEF2 binding, suggesting that the CpG sites flanking the core binding sequence are critical in mediating MEF2 binding. Taken together, these data suggest that DF-induced hypermethylation and/or mutation of CpG sites flanking MEF2 binding sequence (Figure 2A) can suppress the chromatin loading of MEF2 to KLF4 promoter (Figure 2B and 2D) and inhibit transcriptional activity (Figure 2C).
DF-induced DNMT3A enrichment in the KLF4 promoter
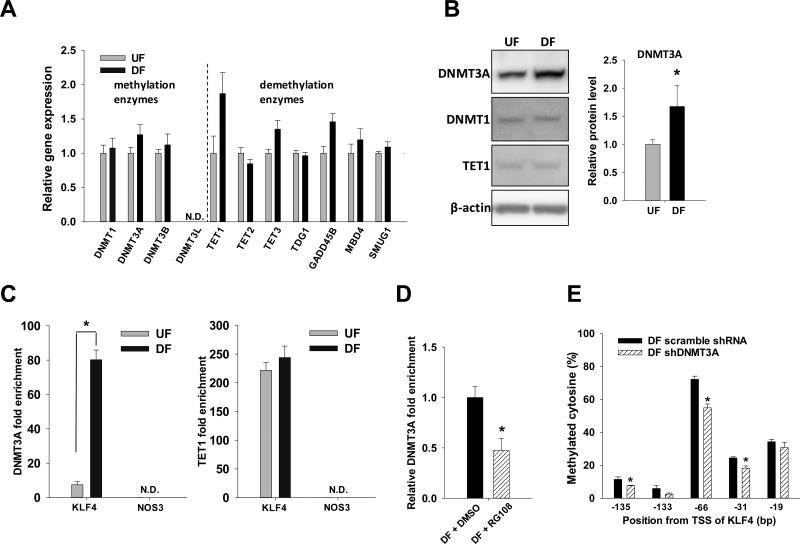
DNA methylation at CpG sites is the net balance of methylation and demethylation dynamics. To test if DF can change the net methylation equilibrium, mRNA of the enzymes involved in methylation and demethylation processes were examined by qPCR (Figure 3A). The relative transcript expressions of methylation and demethylation enzymes were not significantly different between UF and DF; the mRNA of DNMT3L was not detectable by qPCR.
Figure 3. UF and DF regulation of methylation and demethylation enzymes.
Confluent HAEC were subjected to UF or DF for 2 days. (A) mRNA of methylation enzymes (DNMT1, 3A, 3B, 3L) and enzymes involved in demethylation pathways24 (TET1, 2, 3; TDG1; GADD45B; MBD4; SMUG1) were determined by qPCR. Data are normalized to UBB and expressed as mean ± SEM fold of UF. (B) Proteins of DNMT1, DNMT3A and TET1 were analyzed by Western blot. Optical density of DNMT3A normalized to β-actin is expressed as mean ± SEM fold of UF. (C) UF- and DF-induced DNMT3A and TET1 enrichment in KLF4 (−161/−25) and NOS3 (−167/−15) promoter were analyzed by ChIP-qPCR. Data are normalized to the chromatin loading to ACTA2 promoter and are expressed as mean ± SEM. (D) Confluent HAEC were subjected to DF with or without RG108 for 2 days. Effect of RG108 on DF-induced DNMT3A loading to KLF4 promoter (−161/−25) was analyzed by ChIP-qPCR. Data are normalized to chromatin loading of DNMT3A to ACTA2 promoter and expressed as mean ± SEM fold of UF. (E) Confluent HAEC on glass slide post-transfected with DNMT3A-specific shRNA or scramble shRNA were subjected to DF for 2 days. Bisulfite pyrosequencing was used to quantify the methylation level of individual CpG sites at the KLF4 promoter. *P < 0.05. n= 4.
DNMT3A protein was 1.7 fold enhanced in DF (p<0.05, Figure 3B). An increase in DNMT3A protein without changing the mRNA levels may suggest a potential post-transcriptional and/or post-translational mechanism (eg. sumoylation)30 in regulating DNMT3A by flow. While DNMT1 and TET1 protein levels were comparable between UF and DF, DNMT3B protein was not detectable in HAEC by Western blot using two different antibodies from two vendors.
Chromatin loading of DNMT3A enzyme at the KLF4 promoter and NOS3 promoter was examined by ChIP-PCR assay (Figure 3C left panel). DF significantly enhanced (11-fold; p<0.00002) the enrichment of DNMT3A protein at the KLF4 promoter (−161/−25) but not at the NOS3 promoter (−167/−15) where DNMT3A was undetectable. The chromatin loading of TET1 enzyme at the KLF4 promoter remained unchanged between UF and DF, while TET1 at the NOS3 promoter was undetectable (Figure 3C right panel). Pre-treatment with RG108, a specific DNMT inhibitor, significantly suppressed (by 55%; p<0.05) DF-induced chromatin loading of DNMT3A to the KLF4 promoter (Figure 3D). Specific knockdown of DNMT3A significantly inhibited the DF-induced methylation of CpG sites near the MEF2 binding sequence (Online Figure V and Figure 3E). These data demonstrate that DF induced the chromatin enrichment of DNMT3A, leading to the hypermethylation of KLF4 promoter. They also suggest that methylation/ demethylation enzymes DNMT3A and TET1 do not regulate NOS3 promoter methylation and hydroxymethylation in HAEC (Figure 1D right panel and Online Figure I) under hemodynamic forces.
DF-enhanced, DNMT-mediated KLF4 promoter methylation and gene silencing
To determine if DF-enhanced methylation of KLF4 promoter is mediated by the activation of DNMTs, 5-azacytidine (5-Aza) and RG108 were used to block DNMT activity. 5-Aza can incorporate into DNA and covalently trap and inhibit DNMTs. RG108 has been shown to specifically bind to DNMTs and inhibit the enzyme activity with long half-life (20 days) and without significantly inducing apoptosis, cytotoxicity and genotoxicity31. In HAEC cultured under static conditions (no flow), dose-response curves showed that RG108 up to 100 μM and 5-azacytidine up to 1 μM did not significantly inhibit the methylation of the KLF4 promoter (Online Figure VIA).
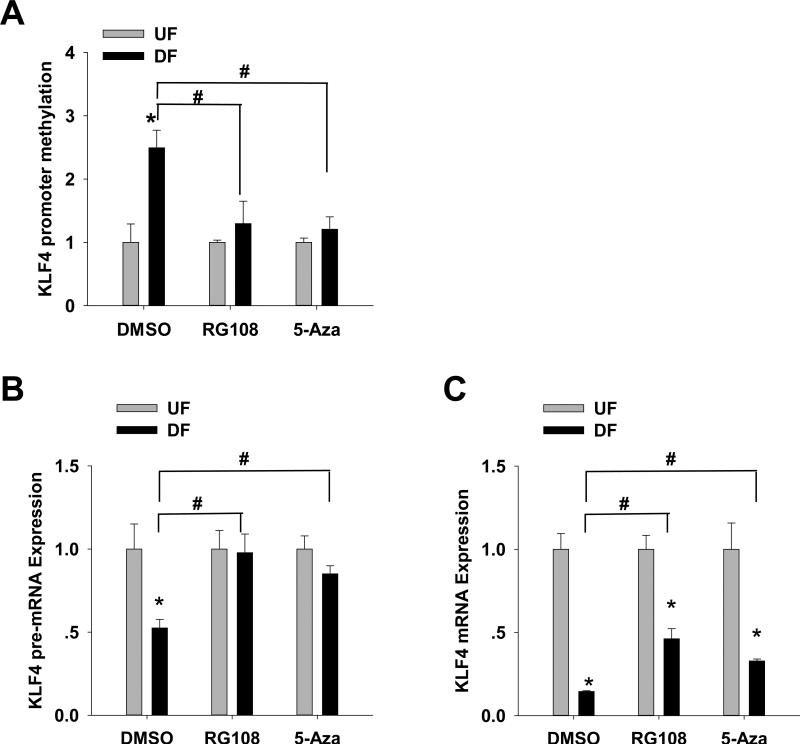
DF-mediated methylation was then measured following treatment with DNMT inhibitors. RG108 (20 μM) and/or 5-Aza (1 μM) completely prevented DF-specific (vs UF) methylation of the KLF4 promoter (−152/+9, Figure 4A), consistent with DF-induced KLF4 promoter hypermethylation mediated by DNMT. We also tested if DF-induced hypermethylation in other regions of the KLF4 promoter could be blocked by DNMT inhibitors. Consistent with the findings in −152/+9, DF-induced hypermethylation in regions −1520/−1328, 1339/−1141 and −808/−648 was completely blocked by RG108 (Online Figure VIB) presumably through (non-MEF2) DNMT global inhibition.
Figure 4. Effects of RG108 and 5-azacytidine on UF- and DF-induced KLF4 promoter methylation and gene expression.
Confluent HAEC were subjected to UF and DF with RG108 (20 μM), 5-azacytidine (5-Aza, 1 μM) or vehicle (DMSO) for 2 days. (A) KLF4 promoter methylation was analyzed by MSP. Data are normalized to UBB promoter and expressed as mean ± SEM fold of UF. (B) pre-mRNA and (C) mRNA of KLF4 were determined by qPCR. Data are normalized to UBB and expressed as mean ± SEM fold of UF. *,#P < 0.05. n= 4-6. *Different from UF. #Different from vehicle control.
The effects of RG108 and 5-Aza on gene transcription were then examined. KLF4 pre-mRNA was completely restored to UF levels by RG108 and to 90% UF levels by 5-Aza (Figure 4B), consistent with the release of DF-suppressed KLF4 transcription by inhibition of DNMT. Mature KLF4 mRNA was rescued from 10% to 50% and 30% of UF levels by RG108 and 5-Aza respectively (Figure 4C) indicating a post-transcriptional partial inhibition of KLF4 mRNA by DF. Our recent finding21 that intronic miRNA-92a could decrease KLF4 (and KLF2) mRNA stability may be related to this finding.
Implications for the regulation of KLF4 target genes
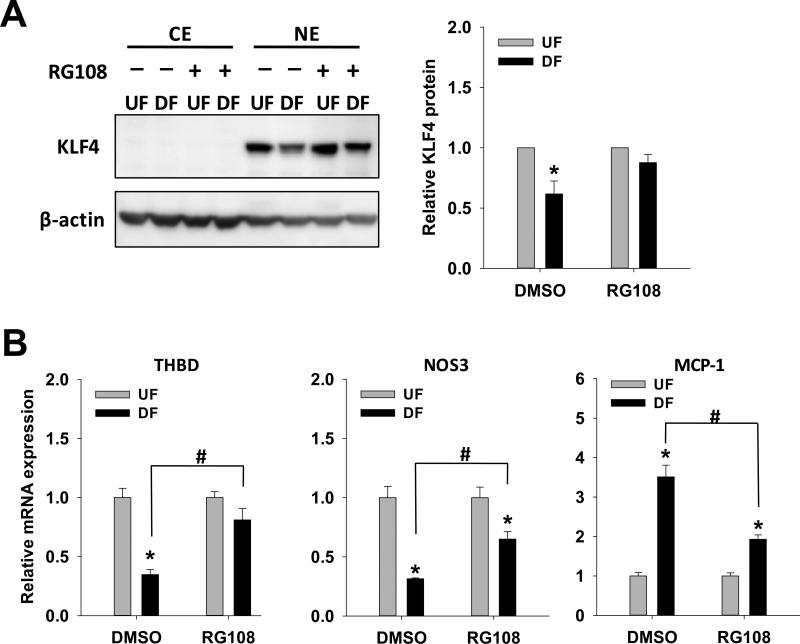
KLF4 protein expression was inhibited by 38% (p<0.05) in DF (Figure 5A), consistent with in vivo data that endothelial KLF4 protein is down-regulated in the DF region of aortic arch21. DF-suppressed KLF4 protein levels in HAEC were rescued by the DNMT inhibitor RG108 which selectively increased KLF4 protein levels in DF without affecting UF levels (Figure 5A). As an atheroprotective transcription factor, KLF4 up-regulates anti-inflammatory and anti-thrombotic factors such as NOS3 and thrombomodulin (THBD), whereas it inhibits the expression of pro-inflammatory factor, monocyte chemoattractant protein-1 (MCP-1)10,21. To show the effects of DF on the transcript expressions of these molecules and to test if blocking DNA methylation pathway could potentially restore the atheroprotective phenotypes of these KLF4 gene targets, the expression of NOS3, THBD and MCP-1 were evaluated by qPCR in the absence and presence of DNMT inhibitor RG108 (Figure 5B). In controls (DMSO vehicle) DF significantly inhibited NOS3 and THBD gene expression, and enhanced MCP-1 gene expression (Figure 5B). This atherosusceptible profile was partially rescued by RG108. Suppression of THBD by DF was completely reversed by RG108 while inhibition of NOS3 and enhancement of MCP-1 by DF were both two-thirds reversed by RG108. The suppression and restoration of these KLF4 target genes by RG108 was not associated with changes in DNA methylation of low CpG promoter of NOS3 or high CpG promoter of THBD (Online Figure VII). These results demonstrate that DF-induced pro-inflammatory and pro-thrombotic profiles in HAEC can be rescued by blocking upstream DNA methylation pathways in KLF4.
Figure 5. Effect of RG108 on KLF4 protein and KLF4-downstream genes transcription.
Confluent HAEC were subjected to UF and DF with RG108 (20 μM) or vehicle control (DMSO) for 2 days. (A) KLF4 proteins in cytoplasmic extract (CE) and nuclear extract (NE) were determined by Western blot. Data normalized to β-actin are expressed as fold of UF. (B) thrombomodulin (THBD), MCP-1 and NOS3 mRNA were determined by qPCR. Data are normalized to UBB and expressed as mean ± SEM fold of UF *,#P < 0.05. n= 4.*Different from UF. #Different from vehicle control.
KLF4 and NOS3 in the swine genome
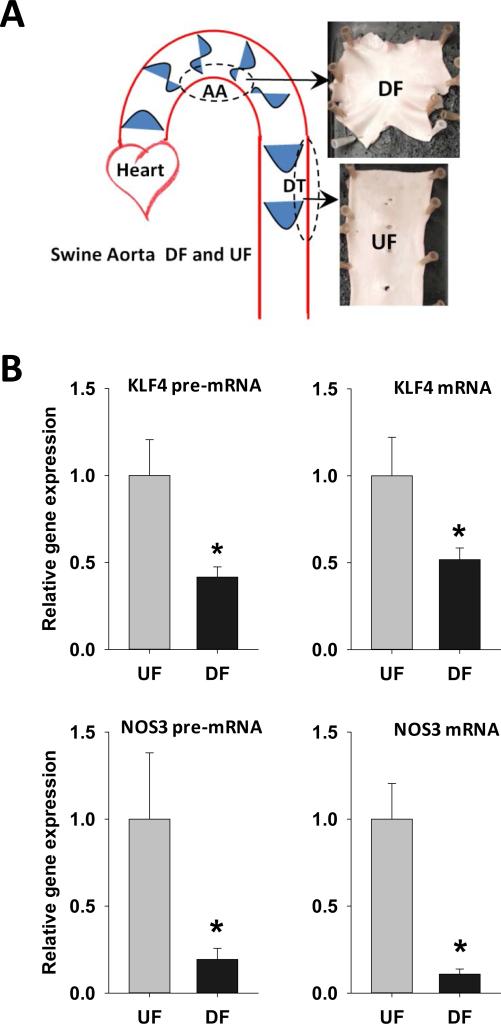
The hemodynamic environment, transcriptome and atherosclerosis susceptibility in the arterial tree are similar between humans and swine4. To explore parallel in vivo/in vitro epigenomic regulatory mechanisms, endothelial cells were isolated from distinct hemodynamic sites of DF (aortic arch; atherosusceptible) and UF (descending thoracic aorta; atheroresistant) in adult swine (Figure 6A). Consistent with the in vitro data, KLF4 and NOS3 mRNA and pre-mRNA in vivo were elevated in the UF region and suppressed in the DF region (Figure 6B).
Figure 6. Differential gene expression and promoter methylation of swine arterial endothelial KLF4and NOS3 in vivo.
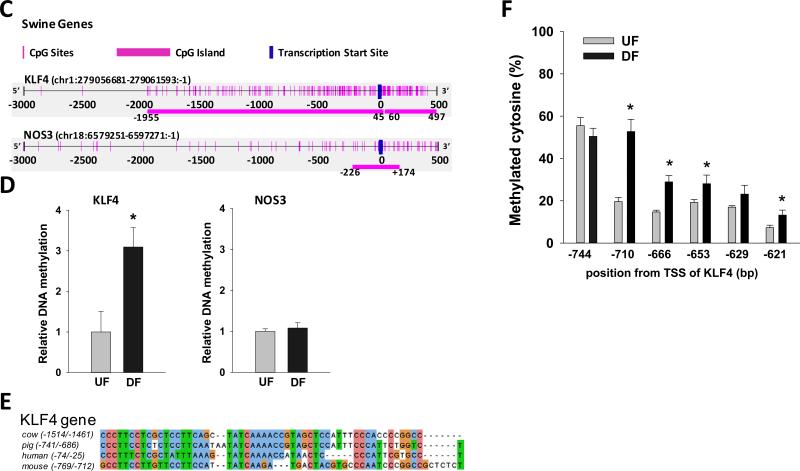
(A) Schematic illustration of targeted UF and DF regions in swine aorta. Endothelial cells were scraped gently from the descending thoracic aorta (DT) where undisturbed flow (UF) is dominant, and from the inner curvature of aortic arch (AA) where disturbed flow (DF) is dominant. (B) mRNA and pre-mRNA of swine KLF4 and NOS3 were determined by qPCR. Data are normalized to the geometric mean of GAPDH and PECAM1 and are expressed as mean ± SEM fold of UF. (C) Schematic illustration of the CpG island and CpG sites in swine KLF4 and NOS3 promoters. (D) Methylation of KLF4 promoter and NOS3 promoter in UF and DF region of swine aorta was determined by using MSP targeting CpG-rich region. Data are normalized to UBB promoter without CpG sites and expressed as mean ± SEM fold of UF. (E) Alignment of KLF4 promoter region in multiple mammalian species showing a highly conserved region of MEF2 binding sequence. (F) Methylation level of individual CpG sites in swine KLF4 promoter determined by bisulfite-pyrosequencing. *Different from UF. P < 0.05. n= 6.
Alignment of human and swine protein sequences showed homologies of 94.9% for KLF4 protein and 96.0% for NOS3 protein, and 89.3% similarity for KLF4 mRNA and 88.5% for NOS3 mRNA. Analysis of swine KLF4 promoter indicated two CGIs with CpGo/e 0.80 and 1.0 (Figure 6C), which are higher than that in human (0.74). In contrast to human NOS3 promoter which does not contain CGIs (Figure 1C, −500/+101 CpGo/e = 0.23; −200/+101 CpGo/e = 0.36), swine NOS3 promoter contains a CpG island at the TSS (400 bp, −226/+174 CpGo/e = 0.61).
DNA methylation in swine endothelium isolated from UF and DF aortic regions was therefore measured in CpG rich regions of KLF4 and in NOS3. As shown in Figure 6D (left panel), while DNA methylation remained unchanged in NOS3 promoter, in the KLF4 promoter there was significantly increased methylation in DF (3-fold; p<0.05). Multiple sequence alignment of human, mouse, cow and pig revealed one highly conserved region of MEF2 binding sequence in the KLF4 gene promoter (Figure 6E); for swine this site was −741/−686. Bisulfite pyrosequencing demonstrated hypermethylation of CpG sites inside and flanking the MEF2 binding sequence (−710, −666,−653,−621) in swine arterial endothelium isolated from DF sites (Figure 6F). These 4 CpG sites were hypomethylated (about 20%) in UF; while the methylation level was increased 1.5-2.7 fold in DF, consistent with the MSP analysis (Figure 6D). Thus the swine KLF4 promoter methylation findings in vivo were in agreement with the in vitro HAEC profile of CGIs. Furthermore, the DNA methylation of NOS3 promoter remained unchanged between UF and DF (Figure 6D right panel) in agreement with our in vitro HAEC data, despite the presence of a CGI near the TSS of swine NOS3.
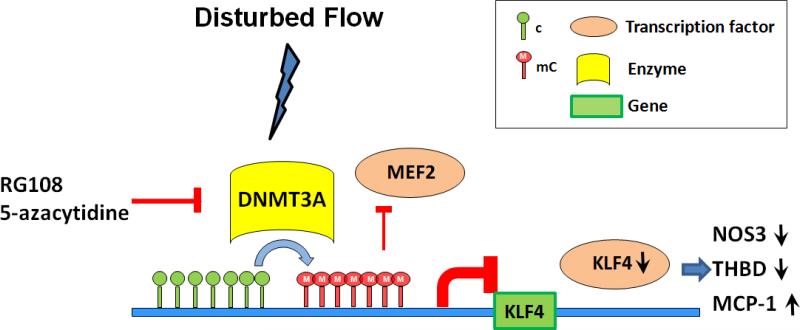
A suggested mechanism of enhanced DNA methylation that is part of the dynamic regulation of KLF4 transcription is outlined in Figure 7. Disturbed flow, a recapitulation of the in vivo hemodynamic regions susceptible to atherosclerosis, induced KLF4 promoter hypermethylation of cytosine. The broad-acting DNMT inhibitors RG108 and 5-Aza suggested that DNMT balance is changed by DF. Our data show that the promoter becomes enriched in methylation enzyme DNMT3A while the demethylation side of the equilibrium, reflected by TET1, remains unchanged. The resulting hypermethylation of KLF4 promoter induced gene silencing by preventing the chromatin binding of MEF2 to the KLF4 promoter.
Figure 7. Summary schematic of KLF4 promoter methylation mechanisms contributing to suppression of transcription.
DF-induced DNMT3A enrichment of endothelial KLF4 promoter near the TSS increased CpG methylation. Hypermethylation prevented MEF2-complex binding resulting in inhibition of KLF4 transcription. Decreased KLF4 expression can lower the interaction of KLF4 with its transcription targets independently of their methylation status leading to a pro-inflammatory, pro-atherosclerosis phenotype. Intervention by DNMT inhibitors (RG108; 5-Aza) can rescue this pathway.
DISCUSSION
As noted in a recent review22, DNA methylation can be evaluated in different genomic contexts that result in functional purposes that range from cell identity to splicing to dynamic - as well as fixed -transcriptional regulation. Here we have demonstrated that KLF4 promoter CpG methylation is responsive to different physiological flow profiles of pathological importance to human arterial endothelial function and we present a plausible DNMT3A/MEF2 mechanism for CpG methylation of promoter sequence near the TSS. In vivo patterns of steady state KLF4 promoter methylation in aortic endothelium from UF and DF regions in swine supported the in vitro interpretation. The characteristics of DF may therefore contribute to the atherosusceptibility of regions associated with branches, curvatures and bifurcations via inducible methylation of the KLF4 promoter that results in its transcriptional suppression with downstream effects upon expression of its targets.
KLF4 has been characterized as an essential transcription factor in the regulation of inflammation and maintenance of a quiescent endothelium. The consequences of enhanced DNA methylation by hemodynamic DF include inhibition of KLF4 expression that removes a degree of protection against the pro-inflammatory pathways that lead to atherogenesis. Among its targets is NOS3 which is also suppressed in regions of DF.
Although histone modification has been reported to be involved in shear stress-induced NOS3 transcription37, DF did not affect methylation, hydroxymethylation and chromatin-enrichment of DNMT3A and TET1 in promoter regions of human NOS3. However, we noted that DF also failed to methylate a CpG island proximal to the TSS of swine NOS3 promoter suggesting resistance of NOS3 to hypermethylation. This is broadly consistent with the report by Chan et al (2004)38 who noted the resilience of the human NOS3 gene to hypermethylation, attributing it to a critical determinant of endothelial cell identity established during development. The same study38 established the principle of a role for DNA methylation in NOS3 transcription by demonstrating reduced NOS3 promoter activity following in vitro transfection of methylated promoter/reporter constructs. However, as demonstrated in this study, neither human nor swine NOS3 promoter methylation was directly affected by DF and it appears that at least part of the NOS3 downregulation may be secondary to DNMT-sensitive DNA methylation effects on upstream transcription factors including, but not necessarily exclusive to, KLF4.
DF-induced KLF4 hypermethylation in HAEC also inhibited the expression of THBD and upregulated MCP-1 (which is normally inhibited by KLF432). All 3 genes are KLF4-targets that contain or lack CpG islands in their promoters; their responses reflect a pro-inflammmatory pattern we previously observed in HAEC when siRNA was used to knock-down KLF421.
The observation that hemodynamic forces are capable of changing the methylation of KLF4, an important transcription factor for flow-sensitive genes13, sheds new light on how physical factors such as blood flow influence gene expression and disease susceptibility. By translating and integrating the effects of biochemical and biomechanical stimuli, KLF4 coordinates atheroprotective and atheroprone gene expression13. Together with other epigenetic mechanisms that relate to endothelial flow responses, e.g. chromatin and miRNA regulation, we show that DNA methylation is a potent contributor to the mechanistic link between the genome and environment that is important in the spatial distribution of atherogenesis. Indeed our earlier study on the regulation of KLF4 gene expression by miRNA-92a suggests that KLF4 is tightly regulated by multiple flow-related mechanisms21. The 50% rescue of KLF4 spliced mature mRNA by DNMT inhibitors whereas the recovery of pre-mRNA expression was complete (Figure 4) is consistent with KLF4 regulation by intronic miRNA mechanisms21 as well as by promoter methylation contributions.
Many experimental studies have reported the effect of undisturbed laminar flow on endothelial responses when referenced to no-flow. However, extrapolation of no-flow comparisons to physiological arterial flow is problematic because of the constantly changing blood velocity/shear stress throughout each cardiac cycle. In the present study pulsatile UF and DF, recapitulating dominant dynamic characteristics of in vivo arterial flow, were directly compared. DF and UF results were normalized to no-flow controls in each set of experiments only to control for unknown criteria unrelated to flow. The epigenomic measurements were conducted after 48h of in vitro flow in order to establish a degree of cell adaptation to the flow to better match the steady state in vivo environment. This time window avoided the first 24h of flow (UF or DF) when transient changes in gene expression occur caused by the shift from no-flow. At 48h much of this activity has subsided. In support of this choice is the correlative evidence for a similar pathway in vivo in site-specific swine aortic endothelial cells (Figure 6); these cells are in an adapted ‘steady-state’ at the time of harvest.
The association of hypermethylation of KLF4 promoter and down-regulation of KLF4 have been reported in lymphoma and epithelial tumors where KLF4 typically functions as a tumor suppressor33,34. In the cardiovascular system the association of genomic DNA methylation with cardiovascular diseases has been noted in peripheral blood mononuclear cells of hyperhomocysteinemia patients and in the whole aorta tissue of apoE-null mice35,36. In mammals, DNMTs use S-adenosyl methionine (SAM) as a methyl group donor for DNA methylation. Hyperhomocysteinemia and the subsequent decreased production or bioavailability of SAM is associated with an increased risk of cardiovascular disease35. Furthermore, atheroprone apoE-null mice show changes in DNA methylation patterns in the whole aorta before the appearance of histologically detectable vascular lesion36. In contrast, our work reveals DNA methylation plasticity in response to hemodynamics representative of atherosusceptible and protected locations in arteries. It demonstrates the first in vitro hypermethylation by spatially differential hemodynamic force characteristics in endothelial cells with strong corroborative evidence in vivo.
In endothelium exposed to DF (atherosusceptible), the kinetics of signaling pathways (KLFs, pro-inflammatory molecules; ER-stress/UPR etc) are set differently resulting in differential phenotype expression18,21,41. Indeed we consider DF itself as a risk factor in sensitizing the cell to pathological change and the basis of atherosusceptibility. Yet while these may be biomarkers of susceptibility they are also adaptive responses and no overt pathology is evident. We suggest that atherogenesis is a 2-hit initiation process where the addition of a second risk factor such as hypercholesterolemia stress may be required to initiate inflammatory pathological change.
The dynamic nature of DNA methylation and demethylation may offer opportunities for therapeutic intervention. Unlike DNA mutations, DNA methylation abnormalities are reversible by drugs in a laboratory setting and this reversal allows cancer cells to reactivate the silenced genes and produce normal proteins. In the present study the specific DNMT inhibitors 5-azacytidine (5-Aza) and RG108 could both rescue DF-suppressed atheroprotective gene expression and negatively regulate DF-induced atherosusceptible genes. 5-Aza has been approved (as Vidaza™) by the US Food and Drug Administration for the treatment of myelodysplastic syndrome, a pre-leukemic bone marrow disorder, by inhibiting DNA methylation and cell proliferation.
The physico-chemical mechanism(s) by which the endothelium distinguishes between UF and DF is unclear and may reside in subcellular spatio-temporal mechanotransduction criteria3,39. Transmission of flow-related deformation forces throughout the cytoskeleton is a plausible mechanical link to the nuclear membrane, the mechanics of which may influence gene regulation40. Furthermore, the local redox environment near the cell surface may influence DNA methylation via the presence of reactive oxygen species (ROS) that are significantly elevated in endothelium at atherosusceptible (DF) sites in normal swine41. In response to ROS the CpG-rich KLF4 promoter may recruit the silencing complex (DNMTs, SIRT1 and polycomb members)42. Since NFkB pathway is also more active in DF18 and Rel/p65 could recruit DNMTs to specific genome loci43, these pathways may influence DNMT3A enrichment. As reflected in Figure 7, mechanotransduction experiments aimed at the induction of DNMT3A may be a fruitful avenue of investigation.
The present studies, in which flow characteristics are the principal experimental variable leading to phenotype adaptation and increased susceptibility to atherosclerosis, were conducted in normal human cells and normocholesterolemic swine. The added introduction of hypercholesterolemia to initiate atherogenesis and associated cytokine-stimulated inflammatory responses will facilitate further evaluation of the epigenetic and epigenomic regulation of endothelial phenotype adaptation during early pathological change.
Supplementary Material
Novelty and Significance.
What Is Known?
Atherosclerotic plaques develop preferentially in regions of disturbed flow.
Irreversible epigenetic DNA methylation patterns are established during development and cell differentiation.
Suppression of endothelial expression of the key athero-protective transcription factor Kruppel-like factor 4 (KLF4) predisposes to atherogenesis. DNA hypermethylation is proposed as a potent inhibitor of KLF4 transcription.
What new information does this article contribute?
Disturbed flow induces DNA methylation of CpG islands in the KLF4 promoter.
Hypermethylation of KLF4 promoter is due to an imbalance in methylation/demethylation activities involving competition for myocyte enhancer factor-2 (MEF-2) binding sites.
Change in the biophysical environment can influence pre-transcriptional endothelial gene expression, a mechanism that may represent epigenomic adaptive physiological regulation.
The distribution of atherosclerotic lesions has been linked to arterial branches, bifurcations and curvatures where the blood vessel geometry causes the flow to create vortices and eddies, referred to as disturbed flow. In the absence of disease at these sites the endothelium nevertheless expresses pro-inflammatory genes that collectively sensitize it to the initiation of atherosclerosis. These cells are therefore considered to be ‘athero-susceptible’, a pre-pathological state of stress adaptation to the local hemodynamic environment. Regulatory mechanisms that link disturbed flow to the susceptible endothelial phenotype include inhibition of KLF4 transcription and translation by disturbed flow. KLF4 is suppressed in endothelium of regions with disturbed flow in vivo where we found DNA hypermethylation of its promoter, the fingerprint of a potent epigenetic suppression mechanism. Using human aortic endothelial cells and controlled flow environments in vitro we show that disturbed flow -induced KLF4 hypermethylation to be a dynamic epigenomic response with pro-atherogenic consequences. This analysis of flow-related epigenomic plasticity of an atheroprotective gene may be representative of a broader adaptive epigenomic response to environmental conditions, some of which may be truly epigenetic (inheritable).
ACKNOWLEDGMENTS
We thank Drs. Scott Diamond, Elisabetta Manduchi and Chris Stoeckert of the University of Pennsylvania for discussions.
SOURCES OF FUNDING
The research was supported by AHA Postdoctoral Fellowship 13POST14070010 (YJ), NIH grants P01 HL62250 (PFD), K25 HL107617 (JMJ), T32 HL07954 (MEM) and by the Biomedical Graduate Studies division of the University of Pennsylvania (KO).
Nonstandard Abbreviations and Acronyms
- 5-Aza
5-azacytidine
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- AA
aortic arch
- DF
disturbed flow
- DNMT
DNA methyltransferase
- DT
descending thoracic aorta
- HAEC
human aortic endothelial cell
- KLF4
Kruppel-like factor 4
- MEF2
myocyte enhancer factor-2
- NOS3
nitric oxide synthase 3
- TET
Tet methylcytosine dioxygenase
- UF
undisturbed flow
Footnotes
DISCLOSURES
None
REFERENCES
- 1.Lusis AJ. Genetics of atherosclerosis. Trends Genetics. 2012;28:267–275. doi: 10.1016/j.tig.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies PF. Endothelial transcriptome profiles in vivo in complex arterial flow fields. Annals Biomed Eng. 2008;36:563–570. doi: 10.1007/s10439-007-9400-0. [DOI] [PubMed] [Google Scholar]
- 3.Davies PF. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nature Clin Prac Cardiovasc Med. 2009;6:16–26. doi: 10.1038/ncpcardio1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Passerini AG, Polacek DC, Shi C, Francesco NM, Manduchi E, Grant GR, Pritchard WF, Powell S, Chang GY, Stoeckert CJ, Jr., Davies PF. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci U S A. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni CW, Qiu H, Rezvan A, Kwon K, Nam D, Son DJ, Visvader JE, Jo H. Discovery of novel mechanosensitive genes in vivo using mouse carotid artery endothelium exposed to disturbed flow. Blood. 2010;116:e66–73. doi: 10.1182/blood-2010-04-278192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C, van Haperen R, de Waard M, van Damme LC, Tempel D, Hanemaaijer L, van Cappellen GW, Bos J, Slager CJ, Duncker DJ, van der Steen AF, de Crom R, Krams R. Shear stress affects the intracellular distribution of eNOS: direct demonstration by a novel in vivo technique. Blood. 2005;106:3691–3698. doi: 10.1182/blood-2005-06-2326. [DOI] [PubMed] [Google Scholar]
- 7.Davies PF, Civelek M, Fang Y, Fleming I. The atherosusceptible endothelium: Endothelial phenotypes in complex hemodynamic shear stress regions in vivo. Cardiovasc Res. 2013;99:315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Won D, Zhu SN, Chen M, Teichert AM, Fish JE, Matouk CC, Bonert M, Ojha M, Marsden PA, Cybulsky MI. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–1704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawashima S, Yokoyama M. Dysfunction of endothelial nitric oxide synthase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:998–1005. doi: 10.1161/01.ATV.0000125114.88079.96. [DOI] [PubMed] [Google Scholar]
- 10.Zhou G, Hamik A, Nayak L, Tian H, Shi H, Lu Y, Sharma N, Liao X, Hale A, Boerboom L, Feaver RE, Gao H, Desai A, Schmaier A, Gerson SL, Wang Y, Atkins GB, Blackman BR, Simon DI, Jain MK. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest. 2012;122:4727–4731. doi: 10.1172/JCI66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis ME, Grumbach IM, Fukai T, Cutchins A, Harrison DG. Shear stress regulates endothelial nitric-oxide synthase promoter activity through nuclear factor kappaB binding. J Biol Chem. 2004;279:163–168. doi: 10.1074/jbc.M307528200. [DOI] [PubMed] [Google Scholar]
- 12.Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Jr, Balasubramanian V, García-Cardeña G, Jain MK. Kruppel-likefactor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96:e48–57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 13.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 14.Villarreal G, Jr, Zhang Y, Larman HB, Gracia-Sancho J, Koo A, García-Cardeña G. Defining the regulation of KLF4 expression and its downstream transcriptional targets in vascular endothelial cells. Biochem Biophys Res Comm. 2010;391:984–989. doi: 10.1016/j.bbrc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawashima S, Yokoyama M. Dysfunction of endothelial nitric oxide synthase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:998–1005. doi: 10.1161/01.ATV.0000125114.88079.96. [DOI] [PubMed] [Google Scholar]
- 16.Kuhlencordt PJ, Gyurko R, Han F, Scherrer-Crosbie M, Aretz TH, Hajjar R, Picard MH, Huang PL. Accelerated atherosclerosis, aortic aneurysm formation, and ischemic heart disease in apolipoprotein E/endothelial nitric oxide synthase double-knockout mice. Circulation. 2001;104:448–454. doi: 10.1161/hc2901.091399. [DOI] [PubMed] [Google Scholar]
- 17.Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in atherosusceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol Cell Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Ha CH, Jhun BS, Wong C, Jain MK, Jin ZG. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y, Davies PF. Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium. Arterioscler Thromb Vasc Biol. 2012;32:979–987. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Rev Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 23.Ordovás JM, Smith CE. Epigenetics and cardiovascular disease. Nature Rev Cardiol. 2010;7:510–519. doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirodkar AV, Marsden PA. Epigenetics in cardiovascular disease. Curr Opin Cardiol. 2011;26:209–215. doi: 10.1097/HCO.0b013e328345986e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez JM, Prasad V, Yu MD, Kampmeyer CP, Kaakour AH, Wang PJ, Maloney SF, Wright N, Johnston I, Jiang YZ, Davies PF. Macro- and microscale variables regulate stent haemodynamics, fibrin deposition and thrombomodulin expression. J R Soc Interface. 2014;11(94):20131079. doi: 10.1098/rsif.2013.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, Madigou T, Durand E, Froguel P, Staels B, Lefebvre P, Métivier R, Eeckhoute J, Salbert G. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–8265. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PloS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling Y1, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 32.Shen B, Smith RS, Jr, Hsu YT, Chao L, Chao J. Kruppel-like factor 4 is a novel mediator of Kallistatin in inhibiting endothelial inflammation via increased nitric-oxide synthase expression. J Biol Chem. 2009;284:35471–35478. doi: 10.1074/jbc.M109.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, Mitsuya H, Matsuoka M. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 34.Guan H, Xie L, Leithäuser F, Flossbach L, Möller P, Wirth T, Ushmorov A. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116:1469–1478. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 35.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Human Mol Genet. 2005;14:R139–147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 36.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Bacanamwo M, Harrison DG. Activation of p300 histone acetyltransferase activityis an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J Biol Chem. 2008;283:16293–16298. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan Y, Fish JE, D'Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 39.Davies PF, Helmke BP. Endothelial Mechaotransduction. In: Mofrad RK, Kamm RD, editors. ‘Cellular Mechanotransduction: Diverse Perspectives from Molecules to Tissue’. Cambridge University Press; 2010. pp. 20–60. Chapt. 2. [Google Scholar]
- 40.Gleni RS, Hendzel MJ. Mechanotransduction from the ECM to the genome: are the pieces now in place? J Cell Biochem. 2008;104:1964–1987. doi: 10.1002/jcb.21364. [DOI] [PubMed] [Google Scholar]
- 41.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Davies PF. Coronary artery endothelial transcriptome in vivo: identification of endoplasmic reticulum stress and enhanced reactive oxygen species by gene connectivity network analysis. Circulation Cardiovasc Genet. 2011;4:243–252. doi: 10.1161/CIRCGENETICS.110.958926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y1, Mayo MW, Nagji AS, Smith PW, Ramsey CS, Li D, Jones DR. Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene. 2012;31:1143–1154. doi: 10.1038/onc.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.