Abstract
Climatic extremes threaten agricultural sustainability worldwide. One approach to increase plant water-use efficiency is to introduce crassulacean acid metabolism (CAM) into C3 crops. Such a task requires comprehensive systems-level understanding of the enzymatic and regulatory pathways underpinning this temporal CO2 pump. Here, we review the progress that has been made in achieving this goal. Given that CAM arose through multiple independent evolutionary origins, comparative transcriptomics and genomics of taxonomically diverse CAM species are being used to define the genetic ‘parts list’ required to operate the core CAM functional modules of nocturnal carboxylation, daytime decarboxylation, and inverse stomatal regulation. Engineered CAM offers the potential to sustain plant productivity for food, feed, fiber, and biofuel production in hotter and drier climates.
Keywords: Crassulacean acid metabolism, water-use efficiency (WUE), engineering CAM into C3 plants, biodesign, bioenergy
Photosynthesis for a parched planet
Earth’s population is projected to exceed 9 billion by 2050. The consequent demands on agriculture for food, feed, fiber, and fuels, coupled with decreasing arable land area and increasing nitrogen and phosphate fertilizer requirements for crop production all point to the need to produce more plant-derived biomass with reduced resource inputs [1]. Approximately 40% of the world’s land area is considered arid, semi-arid, or dry sub-humid, with precipitation amounts that are inadequate for most conventional agriculturally important C3 or C4 crops [2]. Although water is the most crucial resource for sustainable agriculture, projections regarding global warming suggest that there could be a gradual increase in the severity and frequency of extreme weather conditions, including higher temperatures and drought conditions [3-5]. Prolonged drought and overreliance on groundwater for crop irrigation has led to the depletion of aquifers in the USA [6, 7] and in other regions of the world [8, 9].
Multiple strategies have been proposed to improve agricultural productivity via enhancing photosynthesis [1, 10, 11]. These include, but are not limited to, the introduction of: (i) either single-cell or two-celled C4 photosynthesis into C3 plants [12-14]; (ii) a CO2-transporting aquaporin [15]; (iii) inorganic carbon-concentrating mechanisms (CCMs) from cyanobacteria [16, 17]; (iv) a CCM shared by all eukaryotic photosynthetic organisms for active recycling of photorespiratory CO2 from mitochondria to chloroplasts [18]; (v) synthetic carbon fixation pathways [1, 19, 20]; and (vi) strategies to reduce photorespiration during carbon fixation [21]. By contrast, relatively little attention has been paid to the potential of moving crassulacean acid metabolism (CAM) into C3 plants. CAM is a temporally controlled plant inorganic CCM that maximizes water-use efficiency (WUE) by shifting all or part of the CO2 uptake to the nighttime, when evapotranspiration rates are reduced compared with the daytime. This review assesses the progress that has been made in defining the genetic requirements and strategies for the assembly and operation of CAM in C3 plants via synthetic biology.
CAM – a strategic target for synthetic biology
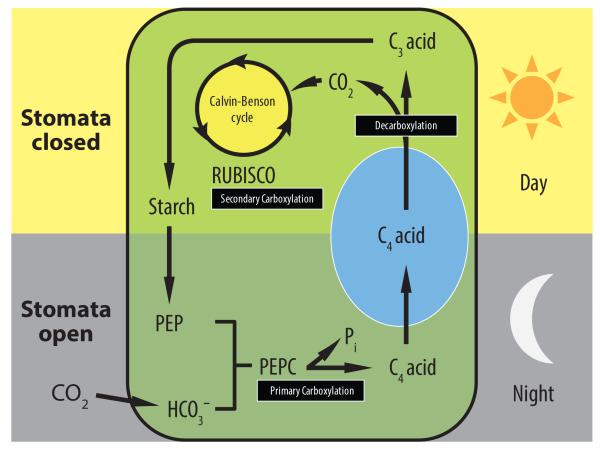
The major plant inorganic CCMs in terrestrial vascular plants are CAM and C4 photosynthesis [22]. CAM arose through multiple, independent evolutionary origins in at least 343 genera across 36 plant families representing >6% of higher plant species [23]. CAM resembles C4 photosynthesis in its use of C4 organic acids as storage intermediates during carbon fixation, but exploits a temporal separation of primary and secondary CO2 fixation. Furthermore, CAM maximizes WUE by concentrating CO2 around ribulose-1-5-bisphosphate carboxylase/oxygenase (RUBISCO), favoring carboxylase activity (Figure 1). The two most distinctive features of CAM are: (i) nocturnal CO2 uptake and fixation by phosphoenolpyruvate carboxylase (PEPC) in the cytosol, which leads to the formation of C4 organic acids that are stored in the vacuole and (ii) an inverse stomatal behavior, in which stomata are closed during part of or all of the day and are open at night. The organic acids accumulated overnight are subsequently decarboxylated during the day to release CO2, which is refixed by RUBISCO in the chloroplast, leading to carbohydrate production via the C3 Calvin–Benson cycle (Figure 1).
Figure 1.
A simplified view of crassulacean acid metabolism (CAM). Stomata open at night allowing atmospheric CO2 to enter the cell to be captured as bicarbonate (HCO3−) by cytosolic phosphoenolpyruvate carboxylase (PEPC) leading to the formation of C4 acid, which undergoes protonation and is stored in the vacuole as malic acid. Stomata are closed for all or part of the subsequent day owing to malic acid efflux from the vacuole and decarboxylation by the malic enzyme to release CO2, which is then refixed by plastidic ribulose-1,5- bisphosphate carboxylase/oxygenase (RUBISCO) through the Calvin–Benson photosynthetic carbon reduction cycle. C3 acids produced in the cytosol serve as substrates for the gluconeogenic pathway during which carbohydrates are regenerated and may be stored as starch (shown) or other storage carbohydrates.
For convenience, the CAM day-night cycle is defined classically as four separate phases of gas exchange, the onset or duration of which can be reduced or eliminated based upon prevailing environmental conditions [24, 25] or upon the degree of tissue succulence as shown by system dynamics modeling [26]. Phase I is the period of high stomatal conductance and nocturnal CO2 uptake and assimilation of atmospheric and respiratory CO2 by PEPC into oxaloacetate (OAA). OAA is subsequently reduced to malate by NAD(P)-malate dehydrogenase and stored in the vacuole as malic acid. Under natural conditions, this phase results in improved WUE as the leaf-air vapor pressure deficit (VPD) is usually lower than during the day [25]. The rate of nocturnal CO2 uptake is limited by mesophyll processes, such as carboxylation capacity derived from storage carbohydrates [27, 28] or vacuolar storage capacity, instead of by stomatal conductance [29]. Phase II is defined as the transition from dark C4 carboxylation by PEPC to daytime RUBISCO-mediated carboxylation as photosynthetically active radiation (PAR) increases in the early morning [25]. During phase II, a combination of CO2 from the atmosphere and that released from organic acid decarboxylation is fixed. Phase III is the period of daytime C3 photosynthesis when net CO2 uptake falls to zero [24] and of major decarboxylation of C4 acids by either NAD(P)-malic enzyme or PEP carboxykinase depending on the species [29]. CO2 liberated during this phase is concentrated behind closed stomata from 2- to 60-fold in the vicinity of RUBISCO [30], essentially creating a ‘CO2 pump’, which can potentially reduce photorespiration, a process that can decrease photosynthesis by up to 40% in C3 plants [31]. The 3-carbon compounds (e.g., pyruvate or PEP) released by malate decarboxylation are converted into storage carbohydrates, which reach peak accumulation at the end of the light period. Phase IV occurs towards the end of the daylight period when partial net CO2 uptake recommences as a result of the depletion of nocturnally accumulated organic acids, and leads to a decline in leaf internal partial pressure of CO2 (pi) and an increase in stomatal conductance and transpiration [25]. As in phase II, both C3 and C4 carboxylation reactions can occur although C4 carboxylation increases as the dark period nears [25]. In summary, CAM can dramatically limit water loss and enhance the magnitude and duration of net CO2 uptake over a 24-h cycle in resource-limited environments [32]. Requirements for optimal CAM operation are summarized in Box 1.
Box 1. Requirements for CAM.
Nocturnal CO2 uptake: depending on the species and environmental conditions, all, most, or the majority of CO2 uptake by CAM plants occurs during the nighttime when leaf-air VPD is low [25].
Inverse day–night pattern of stomatal closure: to accommodate nocturnal CO2 uptake, stomata open in the dark (phase I) and for variable periods of time at the start (phase II) and end (phase IV) of the photoperiod, depending on plant water status and species, but close during the daytime (phase III) [25].
Diel accumulation–depletion of organic acids: the primary carboxylation reactions result in the nocturnal accumulation of C4 organic acids (mainly malate), which are subsequently degraded to provide an internal CO2 source during the subsequent photoperiod [28].
Reciprocal turnover of storage carbohydrates: storage carbohydrates, such as starch, glucans, soluble hexoses and/or disaccharides, exhibit accumulation patterns that are reciprocal to the pattern of C4 organic acid accumulation [28]. Up to 20% of leaf dry weight can be committed to carbohydrates for supplying phosphoenolpyruvate (PEP) to CAM [136].
Enhanced expression of C4 anabolism enzymes: carbonic anhydrase (CA), which converts CO2 to HCO3−; the primary carboxylation enzyme phosphoenolpyruvate carboxylase (PEPC), which converts HCO3− to oxaloacetate (OAA); and malate dehydrogenase (NADP+- and NAD+-MDH), which converts OAA to malate, each exhibit increased mRNA and protein expression and enzyme activities relative to those in C3-performing plants [58, 76].
Enhanced expression of C4 catabolism enzymes: the primary decarboxylation enzymes NADP+-/NAD+-malic enzyme (ME), which convert malate to pyruvate with the release of CO2 in the cytosol with subsequent conversion of pyruvate to PEP by pyruvate orthophosphate dikinase (PPDK), or decarboxylation of malate to PEP by phosphoenolpyruvate carboxykinase (PEPCK) depending on the species, each exhibit increased mRNA and protein expression and enzyme activities relative to those in C3-performing plants [57, 58, 76].
Enhanced expression of glycolytic–gluconeogenic pathway enzymes: coordinated expression of a suite of glycolysis and gluconeogenesis pathway enzymes to supply substrates for nocturnal primary carboxylation and daytime decarboxylation (e.g., PEP, pyruvate) reactions, respectively, is essential for CAM [76, 136-138].
Transport activities between subcellular organelles: the operation of the metabolic sequence of CAM requires the orchestration of transporters at the tonoplast (e.g., voltage-gated inward-rectifying malate channel, tonoplast dicarboxylate transporter, vacuolar H+-ATPase), mitochondrial, and chloroplast envelope membranes [83, 139, 140].
Leaf or stem succulence: some degree of leaf succulence, characterized by increased mesophyll cell size owing to large storage vacuoles and increased mesophyll tissue and leaf thickness to ensure a high capacity for nocturnal organic acid storage [107]. Large cell size, tightly packed cells, reduced intercellular air spaces, and reduced surface area exposure to these air spaces are likely to limit CO2 diffusion within these tissues.
Circadian clock control of CO2 fixation: CAM plants show robust rhythms of CO2 uptake that persist under constant light and temperature conditions, indicating that these rhythms are under circadian clock control. Expression of mRNAs and post-translational regulatory events, such as the reversible phosphorylation of PEPC by PEPC kinase, are also clock controlled [59, 81, 137].
The inherently high WUE of CAM plants signifies their potential for sustainable production of biomass in a warmer and drier world [33, 34]. Highly succulent CAM species, such as Agave spp. or Opuntia ficus-indica, have been grown commercially for centuries as sources of fiber, sugars for alcohol-containing beverages, and as food or animal forage and fodder in semi-arid and arid regions of the world. More recently, the recognition of the ability of these species to operate at near-maximum productivity with relatively low requirements for water [33, 34] and nutrient inputs has engendered scientific interest in their use as sustainable bioenergy feedstocks [35-39]. These CAM crops avoid competition for existing land resources because they can be grown on marginal or degraded land with poor soil conditions, where precipitation totals or frequency are insufficient to support traditional C3 or C4 crops [35-37, 40]. Furthermore, CAM plants could be used for sustainable production on irrigated lands using up to 80% less water to produce similar amounts of biomass compared with C3 species [32, 33]. Thus, research into expanding the agricultural uses of CAM species should be a high priority to ensure that adequate food, feed, and fiber needs are met in future warmer climates with diminishing arable land and water resources.
Engineering of CAM
The development of bioenergy feedstocks and food crops engineered with the improved WUE of CAM plants complements the direct use of CAM species to supply human needs. CAM and C4 photosynthesis have been described as products of either parallel or convergent evolution of a complex trait with the implication that many, if not all, of the genes and some regulatory elements necessary for these photosynthetic specializations are already present in C3 species [23, 41-43]. Such reasoning also underpins the ambitious aims and rationale for transferring C4 properties to C3 plants as a means of enhancing plant productivity to achieve food security [12, 14]. Whereas optimal performance of C4 photosynthesis requires specialized anatomy, including bundle sheath and mesophyll cells to accommodate the spatially separated reactions of C3 and C4 carboxylation [41], CAM might prove more tractable because it is a single-cell adaptation requiring only mesophyll cells in contrast to two-cell C4 photosynthesis. Many permutations of CAM have been described within the evolutionary continuum of CAM species including CAM idling, CAM cycling, weak CAM, latent CAM, facultative or inducible CAM, and obligate or constitutive CAM [23] . In facultative CAM species, CAM expression is readily modified by salinity, water deficit, and high light [29]. Furthermore, some CAM species are capable of switching from C3 to CAM and back to C3 [34], which implies that there are no metabolic incompatibilities between C3 photosynthesis and the water-conserving adaptation. Thus, there appears to be no a priori reason why the pathway cannot be engineered into non-CAM crops as a means of enhancing water-use efficiency or increasing carbon balance.
A logical goal for engineering CAM is the installation of a complete, obligate CAM pathway as this would likely maximize WUE. However, intermediate steps in CAM engineering might also be considered beneficial. Some CAM-cycling or facultative CAM species are thought to benefit from a partial commitment to CAM, not by increasing net carbon gain, but by simply maintaining a positive carbon balance by reducing respiratory CO2 losses. Such variants of CAM increase WUE and water absorption, resulting in an extension of the plant’s life cycle and thus improve reproductive success under water-deficit stress [44]. Thus, strategies to incrementally engineer a partial CAM pathway might provide partial benefit to C3 plants. For example, expression of an engineered Solanum tuberosum PEPC under the control of a dark-induced promoter from Arabidopsis thaliana resulted in Arabidopsis plants with greater stomatal conductance, respiration, and transpiration in dark-adapted leaves, and increased CO2 assimilation rates under different external CO2 concentrations (Ca) and light intensities compared to wild-type plants [45].
Probing phylogenetically diverse lineages to enable comparative CAM genomics
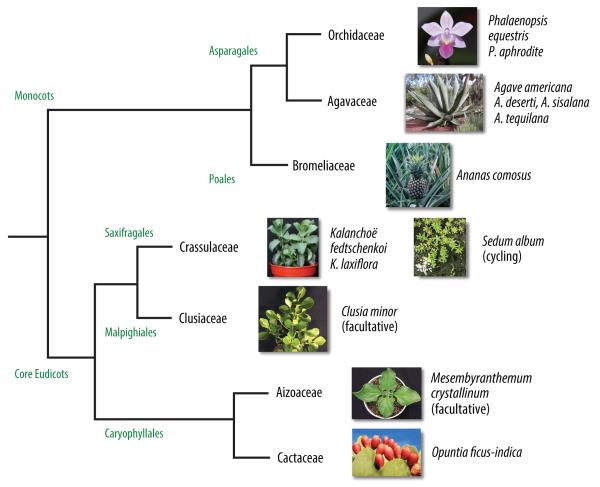
A fundamental requirement for engineered CAM is to first understand the minimal set of genes and proteins required for its efficient establishment and operation. Until recently, there has been a paucity of genome or transcriptome information available for CAM species. However, the genomic sequences and transcriptome atlases available from cycling, facultative, or obligate CAM species sampled from diverse phylogenetic origins should rapidly redefine our understanding of the molecular genetics of CAM (Figure 2).
Figure 2.
Phylogenetic relationships among major model and agronomically important CAM species for which genomic-scale transcriptomic or genomic sequence datasets are available. Phylogeny derived from the Angiosperm Phylogeny Website (http://www.mobot.org/MOBOT/research/APweb/). Phalaenopsis equestris image © 2008 AltonX. Ananas comosus image © 2009 Enviromantic. Opuntia ficus-indica image © 2005 martinalfaro. Sedum album image © 2007 Frank Vincentz.
Among monocots, the partial transcriptomes or genomes of CAM orchids in the genus Phalaenopsis have been characterized [46-50]. The transcriptome [51] and genome (R. Ming, personal communication) of pineapple (Ananas comosus) are also being sequenced. RNA-sequencing (RNA-seq) and quantitative whole-transcriptome analysis has been performed in young (C3) and mature (CAM) leaves of Agave americana (X. Yang, unpublished) and across the leaf developmental gradient in several Agave species, including Agave deserti [52], Agave sisalana [53] (J. Hartwell, unpublished), and Agave tequilana [52, 54]. Systematic analysis of gene expression patterns along such developmental gradients are expected to reveal putative regulatory factors involved in the establishment and daily optimization of CAM, in a manner similar to the establishment of the developmental profile of C4 photosynthesis in maize (Zea mays) leaves [55, 56].
Among core eudicots, several species have emerged as genetic and ecophysiological models for CAM, including the common ice plant (Mesembryanthemum crystallinum), in which CAM is induced following the imposition of salinity or water-deficit stress and Clusia minor, in which CAM is rapidly inducible and reversible [57]. Comparative messenger RNA (mRNA) expression profiling experiments in M. crystallinum performing C3 and CAM have revealed changes in the abundance of mRNAs encoding key enzymes for CAM [58]. However, the use of facultative CAM species requires that mRNA responses to the stresses used to induce CAM (e.g., high salinity or water deficit) must be distinguished from those associated specifically with CAM.
Within the Crassulaceae, obligate CAM species, such as Kalanchoë fedtschenkoi, Kalanchoë daigremontiana, and Kalanchoë laxiflora, develop CAM as leaves expand from the shoot apical meristem [59], even under well-watered conditions [60]. Quantitative RNA-seq comparisons are underway between developmentally immature, C3-performing leaves and mature, CAM-performing leaves in these species to discover CAM-specific genes (J. Hartwell, unpublished). Kalanchoë species are readily transformable [61, 62], and thus represent a powerful model system for probing the function of genes and regulatory elements essential for CAM via transgenic RNA interference and overexpression approaches. The genome and transcriptome sequence data being generated for K. fedtschenkoi and K. laxiflora should dramatically expand the possibilities for CAM functional genomics within Kalanchoë. Genome sequence information has been generated for another member of the Crassulaceae, the inducible CAM-cycling species Sedum album [63] (T.P. Michael, unpublished). Lastly, partial transcriptome information is available for a second member of the Caryophyllales, Opuntia ficus-indica [64], which is the most widely cultivated member of the cactus family [65].
Comparative CAM genomics
Comparative transcriptomic and genomic approaches can be used to discern CAM gene function by comparing the expression patterns of known CAM enzymes and transporters among closely related C3 and CAM species within the same genus or family that show C3, weak CAM, and strong CAM [23]. Comparative genome and transcriptome sequencing studies of C3 and C4 model species from diverse taxonomic origins do not support the duplication or expansion of C4 pathway genes or the long-standing hypothesis that C4 photosynthesis has been facilitated by gene duplications followed by neofunctionalization [42]. Similarly, comparative analysis among CAM species is expected to provide informative examples of either parallel or convergent evolutionary events leading to CAM as transcriptome and genome sequence information becomes available (Figure 2).
Another strategy for resolving CAM gene function is to study DNA polymorphisms associated with CAM phenotypes among diverse genotypes arising from either natural or artificial populations within the same CAM species. Ideally, a reference genome sequence should be available so that re-sequencing of hundreds of individual genotypes is feasible and affordable. Embracing this strategy, the research community studying CAM is working with he DOE Joint Genome Institute to establish K. laxiflora as an Arabidopsis-like CAM model species. K. laxiflora has several traits that make it useful as a model, including that it: (i) is diploid (n = 17) with a relatively small genome (~250 Mb), (ii) has relatively small stature (30 to 45 cm tall), (iii) has a short life cycle (~ 6 months), (iv) is easily transformed, and (v) is self-compatible, with the ability to produce many thousands of small seeds per plant. Although M. crystallinum has been studied extensively as a facultative CAM model, its genome is larger (~390 Mb) [66] than that of K. laxiflora and M. crystallinum is not readily transformed.
Coexpression network modeling of CAM
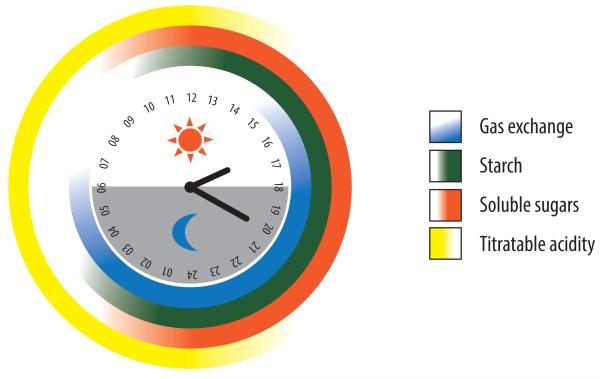
A key challenge for the engineering of CAM into C3 plant species lies in understanding the temporal regulatory events controlling not only the core carboxylation–decarboxylation of C4 acids, but also the coincident metabolic fluxes through glycolysis–gluconeogenesis, storage carbohydrate synthesis and breakdown, as well as stomatal control, over the course of the day-night cycle as illustrated in Figure 3A. A multi-layer approach incorporating transcriptional data, functional genomics annotation, and genetics within an integrative modeling framework might afford the best means to discover genes comprising a functional CAM module (i.e., a set of gene or gene products related by their participation in a common biological process) and then to translate this information into well-informed biodesign strategies.
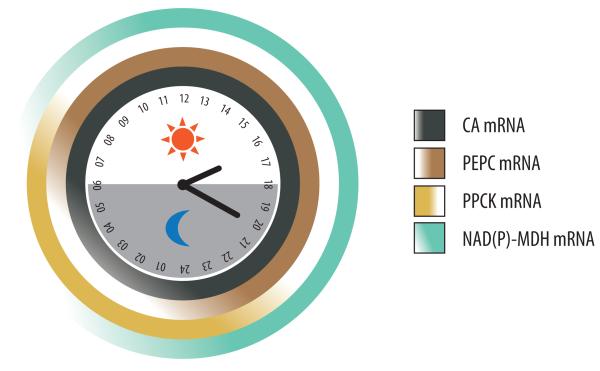
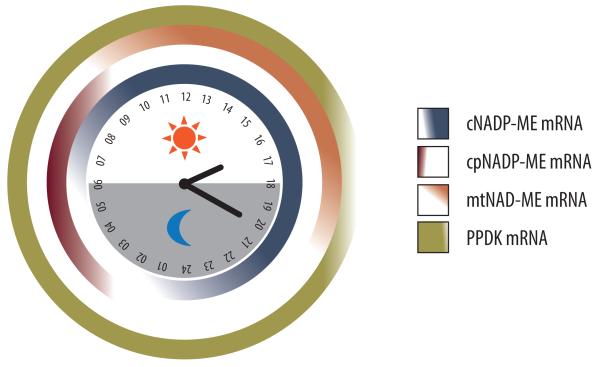
Figure 3.
Temporal dynamics of the major physiological and biochemical processes and key mRNA expression patterns of CAM. (A) Gas exchange, which occurs mostly at night resulting in the peak accumulation of C4 acids at dawn, is measured as titratable acidity, and reciprocal accumulation of starch or soluble sugars, which peak at dusk [28]. (B) Carboxylation module showing the accumulation of steady-state mRNA for chloroplastic (and possibly cytosolic) β-carbonic anhydrase (CA), cytosolic phosphoenolpyruvate carboxylase (PEPC), cytosolic PEPC kinase (PPCK), and multiple NAD(P) malate dehydrogenase [NAD(P)-MDH] genes, which show similar mRNA expression patterns in M. crystallinum and thus are represented together [58, 81]. (C) Decarboxylation module showing the accumulation of steady-state mRNA for cytosolic NADP-dependent malic enzyme (cNADP-ME), chloroplastic NADP-dependent malic enzyme (cpNADP-ME), mitochondrial NAD-dependent malic enzyme (mtNAD-ME), and plastidic pyruvate orthophosphate dikinase (PPDK). Expression data summarized from CAM-performing M. crystallinum [58, 81] with the exception of mtNAD-ME (J.C. Cushman, unpublished).
Putative functional gene groups and their associated regulation can be inferred by combining multiple layers of functional genomics data into gene network models [67, 68]. Network models are constructed as simple node-edge graphs in which genes and proteins are connected by an edge if they are deemed to be associated or similar to one another in some manner using experimental data. Network gene modules are loosely defined as densely connected structures in a node-edge graph. Gene coexpression networks created solely from relative mRNA abundance data can declare a gene pair or cohort to be associated based on user-defined similarity metrics and expression thresholds. This process results in the construction of coexpression networks of genes that are closely connected through potential co-regulation or functional association.
Protein–protein interaction networks offer other approaches to identify genes with related functions or shared regulation [68, 69]. Such networks are built upon interactions that are either known experimentally [70] or predicted by data mining methods that detect protein-protein interologs, which are interacting pairs of homologous proteins conserved across different species [69, 71, 72]. Integration of protein–protein interaction and coexpression network modules with Gene Ontology [68] or transcription factor regulatory interaction data [69] are approaches well-suited for extracting gene modules of similar function. The integration of functional genomics data, particularly gene coexpression network data with phylogenetic information, referred to as ‘systems genetics’, is an emerging discipline aimed at understanding the molecular mechanisms governing complex traits [73]. Although promising, the integration of functional genomic data alone does not provide evolutionary insights from phylogenomics data. The establishment of complete genomes and transcriptomes for phylogenetically ordered CAM species (Figure 2), along with numerous genetic resources for various C3 species, now make systems genetics a viable strategy for CAM biodesign. Lastly, integration of phenotypic or genotypic data with gene networks using machine-learning algorithms has been shown to improve the identification of genetic elements contributing to mouse weight [74] and human disease-causing alleles [75]. With increasing genetic and genomic resources for CAM species, exploration of such approaches is expected to be fruitful in the context of biodesign efforts.
Biodesign of CAM modules
Although network-modeling approaches can provide information about new candidate genes by virtue of their association with known genes within a functional module, empirical testing of minimal functional modules, coupled with information from loss-of-function studies of individual enzymes, regulatory proteins, or transcription factors can provide important empirical information about the basic genetic requirements for CAM biodesign. For simplicity, a set of discrete functional modules for carboxylation and decarboxylation, and stomatal control, as well as anatomical requirements for CAM, can be designed and tested as a set of minimal functional modules rather than on a gene-by-gene basis to accelerate the empirical testing process. The rationale for engineering a complete module at one time is that single-enzyme engineering is unlikely to result in a functional CAM pathway with the desired improvements in WUE. This is illustrated by attempts to engineer constitutive, nocturnal expression of PEPC in Arabidopsis, which resulted in only incremental improvements in CO2 assimilation rates compared to wild type plants [45].
Nocturnal carboxylation module
Nocturnal CO2 uptake via open stomata and primary fixation as HCO3− in the dark (phase I) requires the coordinated action of a set of CAM-specific enzymes, some of which have multiple subcellular locations, along with a regulatory kinase [i.e., phosphoenolpyruvate carboxylase (PEPC) kinase (PPCK)] (Figures 3A,B and 4). In the facultative CAM model M. crystallinum, the induction of CAM results in a 2- to 30-fold increase in the transcript abundance of CAM-specific enzymes depending on the particular isogene in question [58]. Corresponding enzyme activities have also been shown to increase relative to those in C3-performing plants [76]. However, evidence for rhythmic patterns in the abundance of corresponding proteins (e.g. PEPC and RUBISCO) over the light–dark cycle is lacking [77, 78]. Although RUBISCO is activated by RUBISCO activase (RCA) in response to light and circadian clock signals [79], PEPC is activated as a result of allosteric regulation modulated via N-terminal phosphorylation of the enzyme [59]. In CAM plants, the circadian clock activates PEPC kinase (PPCK) transcription and translation in the dark, leading to enhanced phosphorylation of PEPC. Phospho-PEPC has reduced sensitivity to feedback inhibition by malate, and this phosphorylation helps to sustain CO2 fixation for the majority of the dark period [59, 80, 81]. Thus, engineering an efficient CAM carboxylation module will require the introduction of a clock-controlled, dark-phased PPCK, which should help to prevent a futile cycle of malate synthesis and decarboxylation. The temporal phasing of requisite mRNA expression patterns is illustrated in Figure 3B. It is generally thought that CAM-specific enzymes would possess the kinetic properties required for CAM, which may not be present in C3 non-photosynthetic isoforms, as has been described in C4 plants [82]. Correct temporal expression in the target host C3 species will require cis-regulatory expression patterns with circadian clock control to drive expression of these enzymes only during the dark period.
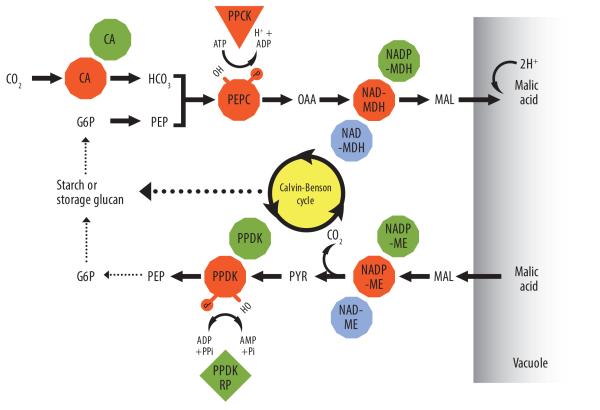
Figure 4.
Proposed model of a minimum gene set encoding enzymes of core CAM carboxylation–decarboxylation modules and their known regulation based upon enzyme activity [76, 141, 142] and mRNA expression studies [58, 59, 81]. Key metabolites include: G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; OAA, oxaolacetate; MAL, malate; PYR, pyruvate. Broken arrows indicate multiple metabolic steps. Key enzymes include: CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PPCK, PEPC kinase; NAD(P)-MDH, NAD or NADP-dependent malate dehydrogenase; NAD(P)-ME, NAD- or NADP-dependent malic enzyme; PPDK, pyruvate orthophosphate dikinase; PPDK-RP, PPDK regulatory protein. The involvement of PPDK-RP is inferred from studies in C3 and C4 species [91]. Colors indicate subcellular locations of enzymes: cytosol (orange), chloroplasts (green), and mitochondria (blue).
Another key consideration for a fully functional carboxylation module is the possible need to engineer enhanced malate transport into the vacuole during phase I, if insufficient nocturnal vacuolar malate accumulation is observed following testing of the core carboxylation module (Figure 4). Malate influx is presumably mediated by one or more voltage-gated inward rectifying malate channels [83] each with functional redundancy and discrete regulatory responses for controlling malate influx and efflux [84]. Such channels have been characterized in Arabidopsis mesophyll (AtALMT9) [85] and guard cells (AtALMT6) [84]. Furthermore, malate transport in mesophyll vacuoles of Arabidopsis is accomplished by a tonoplast dicarboxylate transporter AttDT [86, 87]. However, the genes or gene products encoding these channels and transporters have not been described functionally for any CAM species to date. Therefore, an important goal of ongoing transcriptomic and genomic sequencing efforts is the molecular identification of these malate channels and transporters.
Daytime decarboxylation module
Daytime CO2 release from decarboxylation of stored malate and refixation via the Calvin cycle (phase III) also requires the coordinated action of a set of CAM-specific enzymes. The release of CO2 from malate can be catalyzed by one of several NAD(P)-malic enzymes (MEs), some of which exhibit elevated and circadian clock-controlled mRNA expression patterns (Figure 3C) and distinct subcellular locations (Figure 4). In M. crystallinum, these CAM-specific enzymes show up to 8-fold increases in mRNA abundance [58] depending on the isogene, along with corresponding increases in enzyme activities relative to C3-performing plants [76]. In CAM plants, pyruvate orthophosphate dikinase (PPDK) is required to recycle pyruvate from malate decarboxylation to PEP. PPDK can be localized to the chloroplast, to the cytosol, or to both compartments depending on the genus or species [88, 89]. PPDK mRNA and protein expression are increased during C3-to-CAM induction in M. crystallinum [58, 76, 90]. PPDK-regulatory protein (RP) is a bifunctional kinase/phosphatase that catalyzes the reversible phosphorylation–dephosphorylation of PPDK over the light–dark cycle, leading to inactivation–activation of PPDK in C3 and C4 plants [91].
As in the design of the carboxylation module, a key consideration for a fully functional decarboxylation module is the possible need to engineer enhanced malate efflux from the vacuole during phase III (Figure 4). However, the process of malic acid efflux from the vacuole is not well understood. Efflux might occur by passive diffusion, by a proton-linked symporter, or by a tonoplast dicarboxylate transporter (AttDT) [86, 87] potentially regulated by reversible phosphorylation events [83]. Therefore, the molecular identification and functional characterization of putative vacuolar efflux transporters is needed.
Stomatal regulation module
In CAM plants, the inverse day–night pattern of stomatal closure and opening that underpins the high WUE of the pathway has been linked to the substantial changes in the leaf pi generated during the diel process of malate turnover [92, 93]. At night, stomata are thought to open in response to the drawdown in leaf pi as PEPC is activated. Furthermore, the expression of a PEPC engineered to exhibit reduced malate sensitivity indicated that both constitutive and dark-induced expression of this ‘CAM-like’ PEPC in Arabidopsis resulted in enhanced stomatal opening and transpiration rates at night [45]. Such efforts illustrate the need for engineering coordinated activation–deactivation of carboxylases–decarboxylases over the diel cycle to ensure CAM-like stomatal regulation. During the day in the CAM leaf when stomata close, up to 10,000 μmol CO2 mol−1 may be generated via malate decarboxylation [92]. Stomatal closure is thought to be driven by a signaling cascade wherein CO2 is sensed either directly by guard cells, by leaf mesophyll cells, or both cell types [94].
Defining the cell-type specificity of CO2 signaling components will require more research. In brief, guard cell-specific β-CARBONIC ANHYDRASE (βCA1, βCA4) and the HIGH LEAF TEMPERATURE 1 (HT1) kinase mediate early stomatal closure signaling events that converge with downstream ABA- and Ca2+-signaling transduction networks that act on SLOW ANION CHANNEL ASSOCIATED 1 (SLAC1) S-type (and R-type) anion channels to trigger anion efflux from the guard cells. This anion efflux results in membrane depolarization, which drives K+ efflux from guard cells via outward-rectifying K+ out channels, resulting in stomatal closure [94]. The idea that changes in the concentration of malate in the apoplast of C3 plants might mediate guard cell responses to [CO2] by activating H +-out efflux channels and anion channels in guard cells and thereby facilitate the efflux of the osmoregulatory anions Cl− and malate2− during stomatal closure [95, 96] was first proposed in 1993-1994 [97, 98]. More recent evidence from C3 plants with genetically altered flux through the TCA cycle supports the concept of mesophyll-derived, apoplastic [malate] as a reporter of leaf pi, and thus, a key effector for linking mesophyll and stomatal function [99, 100].
The diel variation in stomatal responsiveness to leaf pi that has been reported for some Kalanchoë species [101] implies that circadian gating of guard cell responsiveness to apoplastic [malate] might be required to provide a further layer of control over CAM stomata. An important task that needs to be undertaken as part of CAM biodesign will be to identify the genes and proteins that regulate the transport of malate between the cytosol and the vacuole, and between the cytosol and the apoplast, and to establish the appropriate level of circadian control over these potential checkpoints for linking stomatal regulation with mesophyll metabolism.
The ‘malate as CO2 sensor’ idea supports the concept that the mesophyll regulates guard cell function [102], and that it should thus be possible to bioengineer the CAM-defining nocturnal opening and daytime closure of stomata in C3 plants by installing the enzymatic machinery responsible for diel turnover of malate in the leaf mesophyll. However, alterations in C3 guard cell metabolism or signaling processes might also be required.
Possible divergence of signaling networks between C3 and CAM guard cells is suggested by reports of the insensitivity of CAM stomata to blue light, inferred from studies with individual leaves or epidermal peels from the facultative CAM species Portulacaria afra and M. crystallinum [103-105]. Defining the guard cell transcriptome, proteome, and metabolome in a CAM model species might reveal whether the signaling or metabolic hubs and networks of C3 guard cells need to be re-engineered to achieve the inverse day–night pattern of stomatal conductance that typifies CAM.
Anatomical requirements for CAM
Leaf or stem succulence, a concomitant anatomical trait with CAM, results from the presence of large cell vacuoles that are presumed to be a prerequisite for the storage of malic acid during the night [106, 107]. Within the relatively undifferentiated leaves that typify most CAM species, increased succulence tends to reduce internal air space and the surface area of chloroplast-containing mesophyll cells directly exposed to intercellular air spaces [107, 108]. These anatomical traits reduce leaf internal conductance to CO2 and direct uptake of atmospheric CO2, but enhance carbon economy during decarboxylation in phase III of CAM because net CO2 efflux from the leaf is minimized [109]. At first glance, this ‘trade-off’ between the optimal leaf anatomy for CAM and the ideal internal structure for C3 photosynthesis would appear to present a challenge for engineering CAM in C3 crops without incurring significant penalties in yield potential. However, a study of functional leaf anatomy within the genus Clusia, which includes C3, C3–CAM, and constitutive CAM species, indicates that distinct layers of palisade and spongy mesophyll present further options for accommodating both direct uptake of CO2 via RUBISCO and CAM [34]. The relatively well-aerated spongy mesophyll of Clusia helps to optimize direct C3-mediated CO2 fixation, whereas the enlarged and densely packed palisade cells accommodate the potential for C4 carboxylation and nocturnal storage of organic acids [34]. These findings indicate that selecting genotypes of C3 target crops with increased ploidy level, which is correlated expediently with increased cell size and biomass productivity [110], succulence [66], and a well-developed palisade mesophyll, in principle, should expedite engineering of CAM into C3 crops. However, if such target genotypes are unavailable, the mesophyll cells could be selectively enlarged by introducing basic helix–loop–helix transcription factors that increase cell size [111]. Alternatively, expression of a putative xyloglucan endotransglucosylase hydrolase gene could result in an increase in leaf water storage and succulence, alongside an increase in the number of mesophyll cells and reduced intracellular air space without incurring an increase in leaf thickness [112].
Identifying target host species
Initial CAM biodesign efforts will target the genetic model Arabidopsis, or close relatives, owing to its rapid growth rate and ease of transformation. With regard to bioenergy feedstocks, preferred targets are rapid-cycling C3 crops, such as members of the Brassicaceae, particularly oilseed crops and fast-growing woody plants within the Populus genus, which are used extensively in the timber, pulp, and paper industries, and more recently as a bioenergy crop. There are many genetic and genomic resources available for Populus [113], including several sequenced genomes [114, 115] and well-developed transformation systems [116, 117]. There is also substantial variability in leaf anatomy and morphology among Populus spp. [118, 119]. Moreover, leaf cell size in Populus is stimulated by free-air CO2 enrichment [120]. These results indicate that Populus leaf cell size has the potential to be increased through genetic modification strategies, as outlined above. Improvement of the WUE of Populus by the introduction of CAM would enable sustainable agroforestry production and possibly expansion of production acreage into more arid regions.
Moving complex traits into target host species
Many biological processes are controlled by gene modules composed of an array of genes [121, 122]. Similarly, complex trait engineering, such as the proposed CAM engineering project outlined here, will require the use of multigene stacking technologies. Most plant genetic engineering attempts have been limited to the introduction of one or a few genes at a time [123]. To address this limitation, new methods have been developed recently to assemble multigene plant transformation vectors that include a zinc-finger nuclease and homing endonuclease [123], in vivo site-specific assembly [124], recombination-assisted multifunctional DNA assembly [125], or a standardized assembly system based on type IIS restriction enzymes that allows the indefinite expansion of reusable gene modules made from standardized DNA components [126, 127]. The multigene plant transformation vector approach has one major drawback: the maximum number of genes in each vector is limited by the cloning capacity of the recipient vectors. Although the TAC vector, derived from the bacteriophage P1 cloning system, is capable of accepting DNA fragments of up to 100 kb [128] and the BIBAC vector, derived from the bacterial artificial chromosome system, is capable of maintaining a genomic DNA fragment of 150–300 kb [129, 130], the assembly of such large constructs with a large number of genes remains a challenge [131]. Multigene plant transformation vector approaches have successfully transferred fewer than 10 genes at a time [123, 124]. However, in planta gene stacking by site-specific recombination has advantages for multigene transfer because of its ability to effectively resolve complex transgenes into precise, single-copy insertions at known genomic target sites [132]. In planta site-specific recombination is limited to stacking one gene at a time and would be time-consuming to use for stacking more than 10 genes. Mini-chromosomes (or artificial chromosomes) provide another vehicle for stacking multiple genes and offer several advantages. Transgenes are grouped into a closely linked block, which avoids linkage drag [131]. Both ‘top-down’ (i.e., involving engineering of the existing chromosomes within a cell) and ‘bottom-up’ approaches (i.e., de novo assembly of chromosomes from centromeric arrays, telomere repeats, and replication origins) can be used to create mini-chromosomes [131, 133]. Although mini-chromosomes have been constructed for genetic transformation in maize [134], Arabidopsis [135], and rice (Oryza sativa) [131], this new technology is not yet ready for routine gene stacking in Populus [133]. Efforts are being made to develop mini-chromosome technology in Populus, and the centromeric regions of several chromosomes have been identified in the Populus genome assembly (X. Yang, unpublished). Implementation of such large-scale cloning and multigene stacking technologies will ensure that each CAM module will be assembled and expressed efficiently within host genomes.
Conclusions and future directions
The ability of succulent CAM species, such as Agave and Opuntia, to maintain high biomass productivities with water input of only 20% of that required by C3 or C4 crops has drawn attention to their possible use as feedstocks for biofuel production. Indeed, such CAM species can be grown in areas where precipitation is typically insufficient to support C3 or C4 crops. Thus, exploring the agricultural uses of CAM species should bear fruit as global warming continues to erode finite arable land and water resources.
The transfer of the WUE of the CAM to C3 crops represents an exciting alternative to the direct use of CAM crops, thereby creating the theoretical potential for sustainable agricultural production on semi-arid lands and avoiding competition with current food and biofuel production systems. One limitation for engineering CAM is the lack of a comprehensive ‘parts list’ required for the design of functional nocturnal carboxylation, daytime decarboxylation, and inverse stomatal regulation modules. However, the rapid expansion of comparative ‘omics’ resources across a phylogenetically diverse set of monocot and eudicot CAM species is imminent. Comparative genomic analysis of this wealth of information is expected to aid in the identification of evolutionarily conserved enzymes and regulatory factors that constitute the minimal requirements for a functional CAM system. Such information will in turn aid the design and transfer of CAM to C3 crops.
Key challenges for successfully engineering a functional CAM system into C3 crops in the near future will include improved understanding of: (i) temporal or circadian clock-controlled regulation of enzyme activities besides those known for PEPC and PPDK in the carboxylation and decarboxylation modules, respectively; (ii) the functional characterization and temporal control of malate channels and transporters that mediate the nighttime influx and daytime efflux of malate into and out of the vacuole; (iii) the signaling pathways that control nighttime opening and daytime closing of stomatal guard cells; (iv) the temporal regulation of the glycolytic and gluconeogenic pathways and associated transporter activities, which feed C3 substrates to carboxylation and from the decarboxylation modules, respectively; (v) the regulation of the hydrolytic and phosphorolytic pathways of starch degradation and starch biosynthesis enzyme activities, which supply the glycolytic and gluconeogenic pathways, respectively; (vi) in soluble sugar-accumulating CAM species, regulation of the activity of tonoplast sugar transporters, which move sugars into and out of the vacuole; (vii) RNAi-mediated transcriptional and posttranscriptional regulatory circuits, if present; and (viii) posttranscriptional and posttranslational regulation of the synthesis and turnover of protein components of the CAM machinery. CAM module design and implementation will also need to be coupled with leaf tissue anatomy and succulence to obtain a fine balance between CO2 diffusion into the leaf and CO2 trapping and concentration within the leaf to optimize the efficient operation of CAM. Lastly, cloning technologies for the efficient assembly or stacking and transfer of multigene constructs into target host species are improving rapidly, and are expected to enable the successful engineering of CAM into Arabidopsis and Populus, a highly productive bioenergy feedstock, as well as other food and fiber species in the near future.
Highlights.
Crassulacean acid metabolism (CAM) species exhibit high water-use efficiency (WUE).
Introduction of CAM into crops should serve to enhance water-use efficiency and increase carbon balance.
Genome and transcriptome sequencing of diverse taxa of CAM species is in progress.
Coexpression network modelling can help define the genetic requirements for CAM biodesign.
Carboxylation, decarboxylation, and stomatal regulation module engineering will be required.
Acknowledgements
This material is based upon work supported by the Department of Energy (DOE), Office of Science, Genomic Science Program under Award Number DE-SC0008834. The Mesembryanthemum crystallinum transcriptome and mRNA expression data were supported by the National Science Foundation, USA (IOS-084373 awarded to K.A.S. and J.C.C.). The Kalanchoë fedtschenkoi sequencing and CAM functional genomics project was supported by the Biotechnology and Biological Sciences Research Council, UK (BB/F009313/1 awarded to J.H.). This publication was also made possible by grants from the National Center for Research Resources (5P20RR016464-11) and the National Institute of General Medical Sciences (8 P20 GM103440-11) from the National Institutes of Health (NIH) through its support of the Nevada Genomics Center and the Nevada Center for Bioinformatics. The contents of this review are solely the responsibility of the authors and do not necessarily represent the official views of DOE or NIH. We acknowledge the contributions of all the members of the CAM Biodesign research team, including: Jin-Gui Chen, Enrique De Paoli, Nancy Engle, Lee Gunter, Sara Jawdy, Guruprasad H. Kora, Kaitlin Palla, Hengfu Yin, Zack Moore, and Heather Tran (ORNL); Susie Boxall and Louisa Dever (University of Liverpool); Rebecca Albion, Travis Garcia, Jungmin Ha, Sung Lim, Jesse Mayer, Bernard Wone, and Won Yim (University of Nevada); Hong Guo (University of Tennessee). We also thank Mary Ann Cushman for critical review and clarifying comments on the manuscript and Lori Kunder for figure presentation assistance. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE–AC05–00OR22725.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maurino V, Weber A. Engineering photosynthesis in plants and synthetic microorganisms. J. Exp. Bot. 2013;64:743–751. doi: 10.1093/jxb/ers263. [DOI] [PubMed] [Google Scholar]
- 2.United Nations Environment Programme (UNEP) Global Environment Outlook: Environment for Development. 2012.
- 3.Intergovernmental Panel on Climate Change (IPCC) Climate Change 2007: The Physical Science Basis. Cambridge University Press; 2007. [Google Scholar]
- 4.Carnicer J, et al. Widespread crown condition decline, food web disruption, and amplified tree mortalinity with increased climate change-type drought. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1474–1478. doi: 10.1073/pnas.1010070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treberth K. Changes in precipitation with climate change. Climate Res. 2011;47:123–138. [Google Scholar]
- 6.Scanlon B, et al. Groundwater depletion and sustainability of irrigation in the US High Plains and Central Valley. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9320–9325. doi: 10.1073/pnas.1200311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famiglietti J, Rodell M. Water in the balance. Science. 2013;340:1300–1301. doi: 10.1126/science.1236460. [DOI] [PubMed] [Google Scholar]
- 8.Voss K, et al. Groundwater depletion in the Middle East from GRACE with implications for transboundary water management in the Tigris-Euphrates-Western Iran region. Water Resour. Res. 2013;49:904–914. doi: 10.1002/wrcr.20078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moiwo J, et al. GRACE, GLDAS and measured groundwater data products show water storage loss in Western Jilin, China. Water Sci. Technol. 2012;65:1606–1614. doi: 10.2166/wst.2012.053. [DOI] [PubMed] [Google Scholar]
- 10.Ducat D, Silver P. Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 2012;16:337–344. doi: 10.1016/j.cbpa.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans J. Improving photosynthesis. Plant Physiol. 2013;162:1780–1793. doi: 10.1104/pp.113.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covshoff S, Hibberd J. Integrating C4 photosynthesis into C3 crops to increase yield potential. Curr. Opin. Biotechnol. 2012;23:209–214. doi: 10.1016/j.copbio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Ruan C, et al. A critical review on the improvement of photosynthetic carbon assimilation in C3 plants using genetic engineering. Crit. Rev. Biotechnol. 2012;32:1–21. doi: 10.3109/07388551.2010.533119. [DOI] [PubMed] [Google Scholar]
- 14.Leegood R. Strategies for engineering C4 photosynthesis. J. Plant Physiol. 2013;170:378–388. doi: 10.1016/j.jplph.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Sade N, et al. The role of tobacco Aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Plant Physiol. 2010;152:245–254. doi: 10.1104/pp.109.145854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price G, et al. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 2013;64:753–768. doi: 10.1093/jxb/ers257. [DOI] [PubMed] [Google Scholar]
- 17.Zarzycki J, et al. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 2013;64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- 18.Zabaleta E, et al. A basal carbon concentrating mechanism in plants? Plant Sci. 2012;187:97–104. doi: 10.1016/j.plantsci.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Bar-Even A, et al. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-Even A, et al. A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 2012;63:2325–2342. doi: 10.1093/jxb/err417. [DOI] [PubMed] [Google Scholar]
- 21.Peterhansel C, et al. Engineering photorespiration: current state and future possibilities. Plant Biol. (Stuttg) 2013;15:754–758. doi: 10.1111/j.1438-8677.2012.00681.x. [DOI] [PubMed] [Google Scholar]
- 22.Raven J, et al. The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Philos. Trans. R Soc. Lond. B: Biol. Sci. 2008;363:2641–2650. doi: 10.1098/rstb.2008.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvera K, et al. Evolution along the crassulacan acid metabolism continuum. Funct. Plant Biol. 2010;37:995–1010. [Google Scholar]
- 24.Osmond C. Crassulacean acid metabolism: a curiosity in context. Annu. Rev. Plant Physiol. 1978;29:379–414. [Google Scholar]
- 25.Griffiths H. Crassulacean acid metabolism: a re-appraisal of physiological plasticity in form and function. Adv. Bot. Res. 1988;15:43–92. [Google Scholar]
- 26.Owen N, Griffiths H. A system dynamics model integrating physiology and biochemical regulation predicts extent of crassulacean acid metabolism (CAM) phases. New Phytol. 2013 Aug 29; doi: 10.1111/nph.12461. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Borland A, et al. Metabolite control overrides circadian regulation of phosphoenolpyruvate carboxylase kinase and CO2 fixation in crassulacean acid metabolism. Plant Physiol. 1999;121:889–896. doi: 10.1104/pp.121.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cushman JC, et al. Isolation and characterization of mutants of common ice plant deficient in Crassulacean acid metabolism. Plant Physiol. 2008;147:228–238. doi: 10.1104/pp.108.116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter K. Crassulacean acid metabolism. In: Barber J, Baker N, editors. Photosynthetic mechanisms and the environment. Elsevier; 1985. pp. 329–387. [Google Scholar]
- 30.Lüttge U. CO2-concentrating: consequences in crassulacean acid metabolism. J. Exp. Bot. 2002;53:2131–2142. doi: 10.1093/jxb/erf081. [DOI] [PubMed] [Google Scholar]
- 31.Ehleringer J, Monson R. Evolutionary and ecological aspects of photosynthetic pathway variation. Annu. Rev. Ecol. Syst. 1993;24:411–439. [Google Scholar]
- 32.Nobel P. High productivities of certain agronomic CAM species. In: Winter K, Smith J, editors. Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution. Springer-Verlag; 1996. pp. 255–265. [Google Scholar]
- 33.Borland A, et al. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. Journal of Experimental Botany. 2009;60:2879–2896. doi: 10.1093/jxb/erp118. [DOI] [PubMed] [Google Scholar]
- 34.Borland A, et al. The photosynthetic plasticity of crassulacean acid metabolism: an evolutionary innovation for sustainable productivity in a changing world. New Phytol. 2011;191:619–633. doi: 10.1111/j.1469-8137.2011.03781.x. [DOI] [PubMed] [Google Scholar]
- 35.Somerville C, et al. Feedstocks for lignocellulosic biofuels. Science. 2010;329:790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 36.Davis S, et al. The global potential for Agave as a biofuel feedstock. Global Change Biol. Bioenergy. 2011;3:68–78. [Google Scholar]
- 37.Owen N, Griffiths H. Marginal land bioethanol yield potential of four crassulacean acid metabolism candidates (Agave fourcroydes, Agave salmiana, Agave tequilana and Opuntia ficus-indica) in Australia. Global Change Biol. Bioenergy. 2013 [Google Scholar]
- 38.Holtum J, et al. Agave as a biofuel feedstock in Australia. Global Change Biol. Bioenergy. 2011;3:58–67. [Google Scholar]
- 39.Yan X, et al. Life cycle energy and greenhouse gas analysis for agave-derived bioethanol. Energy Environ. Sci. 2011;4:3110–3121. [Google Scholar]
- 40.Nobel P. Environmental Biology of Agaves and Cacti. Cambridge University Press; 1988. [Google Scholar]
- 41.Sage R, et al. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- 42.Williams B, et al. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 2012;17:213–220. doi: 10.1016/j.tplants.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Gowik U, et al. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell. 2011;23:2087–2105. doi: 10.1105/tpc.111.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrera A. Crassulacean acid metabolism and fitness under water deficit stress: if not for carbon gain, what is facultative CAM good for? Annals Bot. 2009;103:645–653. doi: 10.1093/aob/mcn145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kebeish R, et al. Constitutive and dark-induced expression of Solanum tuberosum phosphoenolpyruvate carboxylase enhances stomatal opening and photosynthetic performance of Arabidopsis thaliana. Biotech. Bioeng. 2012;109:536–544. doi: 10.1002/bit.23344. [DOI] [PubMed] [Google Scholar]
- 46.Fu C, et al. OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 2011;52:238–243. doi: 10.1093/pcp/pcq201. [DOI] [PubMed] [Google Scholar]
- 47.Su C, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52:1501–1514. doi: 10.1093/pcp/pcr097. [DOI] [PubMed] [Google Scholar]
- 48.Tsai W, et al. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 2013;54:e7. doi: 10.1093/pcp/pcs187. [DOI] [PubMed] [Google Scholar]
- 49.Hsu C-C, et al. An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biology. 2011;11:3. doi: 10.1186/1471-2229-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsiao Y, et al. Gene discovery using next-generation pyrosequencing to develop ESTs for Phalaenopsis orchids. BMC Genomics. 2011;12:360. doi: 10.1186/1471-2164-12-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong W, et al. De novo assemby, characterization and functional annotation of pineapple fruit transcriptome through massively parallel sequencing. PLoS One. 2012;7:e46937. doi: 10.1371/journal.pone.0046937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross S, et al. De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics. 2013;14:563. doi: 10.1186/1471-2164-14-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou W-Z, et al. Construction and evaluation of normalized cDNA libraries enriched with full-length sequences for rapid discovery of new genes from Sisal (Agave sisalana Perr.) different developmental stages. Int. J. Mol. Sci. 2012;13:13150–13168. doi: 10.3390/ijms131013150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpson J, et al. Genomic resources and transcriptome mining in Agave tequilana. Global Change Biol. Bioenergy. 2011;3:25–36. [Google Scholar]
- 55.Pick T, et al. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell. 2011;23:4208–4220. doi: 10.1105/tpc.111.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li P, et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 57.Borland A, et al. Inducibility of crassulacean acid metabolism (CAM) in Clusia species; physiological/biochemical characterisation and intercellular localisation of carboxylation processes in three species which show different degrees of CAM. Planta. 1998;205:342–351. [Google Scholar]
- 58.Cushman J, et al. Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM) J. Exp. Bot. 2008;59:1875–1894. doi: 10.1093/jxb/ern008. [DOI] [PubMed] [Google Scholar]
- 59.Hartwell J, et al. Phosphoenolpyruvate carboxylase kinase is a novel protein kinase is a novel protein kinase regulated at the level of gene expression. Plant J. 1999;20:333–342. doi: 10.1046/j.1365-313x.1999.t01-1-00609.x. [DOI] [PubMed] [Google Scholar]
- 60.Winter K, et al. On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchoë, and Opuntia. J. Exp. Bot. 2008;59:1829–1840. doi: 10.1093/jxb/ern080. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Sogo B, et al. Efficient transformation of Kalanchoe blossfeldiana and production of male-sterile plants by engineered anther ablation. Plant Cell Rep. 2010;29:61–77. doi: 10.1007/s00299-009-0798-8. [DOI] [PubMed] [Google Scholar]
- 62.Garcés H, et al. Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15578–15583. doi: 10.1073/pnas.0704105104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castillo F. Antioxidant protection in the inducible CAM plant Sedum album L. following the imposition of severe water stress and recovery. Oecologia. 1996;107:469–477. doi: 10.1007/BF00333937. [DOI] [PubMed] [Google Scholar]
- 64.Mallona I, et al. Conserved and divergent rhythms of crassulacean acid metabolism-related and core clock gene expression in the cactus Opuntia ficus-indica. Plant Physiol. 2011;156:1978–1989. doi: 10.1104/pp.111.179275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paterson A, et al. The fruits of tropical plant genomics. Trop. Plant Biol. 2008;1:3–19. [Google Scholar]
- 66.De Rocher E, et al. Developmentally regulated systemic endopolyploid in succulents with small genomes. Science. 1990;250:99–101. doi: 10.1126/science.250.4977.99. [DOI] [PubMed] [Google Scholar]
- 67.Usadel B, et al. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32:1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 68.Heyndrickx K, Vandepoele K. Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 2012;159:884–901. doi: 10.1104/pp.112.196725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Bodt S, et al. CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 2012;195:707–720. doi: 10.1111/j.1469-8137.2012.04184.x. [DOI] [PubMed] [Google Scholar]
- 70.Consortium, A.I.M. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Bodt S, et al. CORNET: a user-friendly tool for data mining and integration. Plant Physiol. 2010;152:1167–1179. doi: 10.1104/pp.109.147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Haagen H, et al. Novel protein-protein interactions inferred from literature context. PLoS One. 2009;4:e7894. doi: 10.1371/journal.pone.0007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H. System genetics in “-omics” era: current and future development. Theory Biosci. 2013;132:1–16. doi: 10.1007/s12064-012-0168-x. [DOI] [PubMed] [Google Scholar]
- 74.Chen Z, Zhang W. Integrative analysis using module-guided random forest reveals correlated genetic factors related to mouse weight. PLoS Comput. Biol. 2013;9:e1002956. doi: 10.1371/journal.pcbi.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Itan Y, et al. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5558–5563. doi: 10.1073/pnas.1218167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holtum J, Winter K. Activities of enzymes of carbon metabolism during the induction of crassulacean acid metabolism in Mesembryanthemum crystallinum L. Planta. 1982;155:8–16. doi: 10.1007/BF00402925. [DOI] [PubMed] [Google Scholar]
- 77.Winter K, et al. Activity and quantity of ribulose bisphosphate carboxylase- and phosphoenolpyruvate carboxylase-protein in two Crassulacean acid metabolism plants in relation to leaf age, nitrogen nutrition, and point in time during a day/night cycle. Planta. 1982;154:309–317. doi: 10.1007/BF00393908. [DOI] [PubMed] [Google Scholar]
- 78.Davies B, Griffiths H. Competing carboxylases: circadian and metabolic regulation of Rubisco in C3 and CAM Mesembryanthemum crystallinum L. Plant Cell Environ. 2012;35:1211–1220. doi: 10.1111/j.1365-3040.2012.02483.x. [DOI] [PubMed] [Google Scholar]
- 79.Portis A. Rubisco activase – Rubisco’s catalytic chaperone. Photosyn. Res. 2003;75:11–27. doi: 10.1023/A:1022458108678. [DOI] [PubMed] [Google Scholar]
- 80.Taybi T, et al. Expression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase genes. Implications for genotypic capacity and phenotypic plasticity in the expression of crassulacean acid metabolism. Plant Physiol. 2004;135:587–598. doi: 10.1104/pp.103.036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taybi T, et al. A minimal Ser/Thr protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in CAM-induced leaves of Mesembryanthemum crystallinum. Plant Physiol. 2000;123:1471–1482. doi: 10.1104/pp.123.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Engelmann S, et al. Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria – a gradual increase from C3 to C4 characteristics. Planta. 2003;217:717–725. doi: 10.1007/s00425-003-1045-0. [DOI] [PubMed] [Google Scholar]
- 83.Holtum J, et al. Intracellular transport and pathways of carbon flow in plants with crassulacean acid metabolism. Funct. Plant Biol. 2005;32:429–449. doi: 10.1071/FP04189. [DOI] [PubMed] [Google Scholar]
- 84.Meyer S, et al. Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 2011;67:247–257. doi: 10.1111/j.1365-313X.2011.04587.x. [DOI] [PubMed] [Google Scholar]
- 85.Kovermann P, et al. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007;52:1169–1180. doi: 10.1111/j.1365-313X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- 86.Emmerlich V, et al. The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11122–11126. doi: 10.1073/pnas.1832002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hurth M, et al. Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 2005;137:901–910. doi: 10.1104/pp.104.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kondo A, et al. Species variation in the intracellular localization of pyruvate, Pi dikinase in leaves of crassulacean-acid-metabolism plants: an immunogold electron-microscope study. Planta. 2000;210:611–621. doi: 10.1007/s004250050051. [DOI] [PubMed] [Google Scholar]
- 89.Kondo A, et al. Leaf inner structure and immunogold localization of some key enzymes involved in carbon metabolism in CAM plants. J. Exp. Bot. 1998;49:1953–1961. [Google Scholar]
- 90.Fißlthaler B, et al. Age-dependent induction of pyruvate, orthophosphate dikinase in Mesembryanthemum crystallinum L. Planta. 1995;196:492–500. doi: 10.1007/BF00203649. [DOI] [PubMed] [Google Scholar]
- 91.Astley H, et al. The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis are both bifunctional and interact with the catalytic and nucleotide-binding domains of pyruvate, orthophosphate dikinase. Plant J. 2011;68:1070–1080. doi: 10.1111/j.1365-313X.2011.04759.x. [DOI] [PubMed] [Google Scholar]
- 92.Cockburn W, et al. Relationships between stomatal behavior and internal carbon-dioxide concentrations in crassulacean acid metabolism plants. Plant Physiol. 1979;63:1029–1032. doi: 10.1104/pp.63.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spalding M, et al. Crassulacean acid metabolism and diurnal variations in internal CO2 and O2 concentrations in Sedum praealtum DC. Aust. J. Plant Physiol. 1979;6:557–567. [Google Scholar]
- 94.Kim T, et al. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brearley J, et al. The effect of elevated CO2 concentrations on K+ and anion channels of Vicia faba L. guard cells. Planta. 1997;203:145–154. [Google Scholar]
- 96.Raschke K. Alternation of the slow with the quick anion conductance in whole guard cells effected by external malate. Planta. 2003;217:651–657. doi: 10.1007/s00425-003-1034-3. [DOI] [PubMed] [Google Scholar]
- 97.Hedrich R, et al. Malate-sensitive anion channels enable guard-cells to sense changes in the ambient CO2 concentration. Plant J. 1994;6:741–748. [Google Scholar]
- 98.Hedrich R, Marten I. Malate-induced feedback regulation of plasma membrane anion channels could provide a CO2 sensor to guard cells. EMBO J. 1993;12:897–901. doi: 10.1002/j.1460-2075.1993.tb05730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Araújo W, et al. Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell. 2011;23:600–627. doi: 10.1105/tpc.110.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Araújo W, et al. Control of stomatal aperture: a renaissance of the old guard. Plant Signal. Behav. 2011;6:1305–1311. doi: 10.4161/psb.6.9.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Von Caemmerer S, Griffiths H. Stomatal responses to CO2 during a diel Crassulacean acid metabolism cycle in Kalanchoe daigremontiana and Kalanchoe pinnata. Plant Cell and Environment. 2009;32:567–576. doi: 10.1111/j.1365-3040.2009.01951.x. [DOI] [PubMed] [Google Scholar]
- 102.Mott K, et al. The role of the mesophyll in stomatal responses to light and CO2. Plant Cell Environ. 2008;31:1299–1306. doi: 10.1111/j.1365-3040.2008.01845.x. [DOI] [PubMed] [Google Scholar]
- 103.Lee D, Assmann S. Stomatal responses to light in the facultative crassulacean acid metabolism species, Portulacaria afra. Physiol. Plant. 1992;85:35–42. [Google Scholar]
- 104.Mawson B, Zaugg M. Modulation of light-dependent stomatal opening in isolated epidermis following induction of crassulacean acid metabolism in Mesembryanthemum crystallinum L. J. Plant Physiol. 1994;144:740–746. [Google Scholar]
- 105.Tallman G, et al. Induction of CAM in Mesembryanthemum crystallinum abolishes the stomatal response to blue light and light-dependent zeaxanthin formation in guard cell chloroplasts. Plant Cell Physiol. 1997;38:236–242. [Google Scholar]
- 106.Nelson E, Sage R. Functional constraints of CAM leaf anatomy: tight cell packing is associated with increased CAM function across a gradient of CAM expression. J. Exp. Bot. 2008;59:1841–1850. doi: 10.1093/jxb/erm346. [DOI] [PubMed] [Google Scholar]
- 107.Nelson E, et al. Functional leaf anatomy of plants with crassulacean acid metabolism. Funct. Plant Biol. 2005;32:409–419. doi: 10.1071/FP04195. [DOI] [PubMed] [Google Scholar]
- 108.Nelson E, Sage R. Functional contraints of CAM leaf anatomy: tight cell packing is associated with increased CAM function across a gradient of CAM expression. J. Exp. Bot. 2008;59:1841–1850. doi: 10.1093/jxb/erm346. [DOI] [PubMed] [Google Scholar]
- 109.Maxwell K, et al. Is a low internal conductance to CO2 diffusion a consequence of succulence in plants with crassulacean acid metabolism? Aust. J. Plant Physiol. 1997;24:777–786. [Google Scholar]
- 110.De Veylder L, et al. Molecular control and function of endoreduplication in development and physiology. Trends Plant Sci. 2011;16:624–634. doi: 10.1016/j.tplants.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 111.Nicolas P, et al. The grape berry-specific basic helix-loop-helix transcription factor VvCEB1 affects cell size. J. Exp. Bot. 2013;64:991–1003. doi: 10.1093/jxb/ers374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han Y, et al. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J. Exp. Bot. 2013;64:4225–4238. doi: 10.1093/jxb/ert229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang X, et al. Poplar genomics: state of the science. Crit. Rev. Plant Sci. 2009;28:285–308. [Google Scholar]
- 114.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 115.Ma T, et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013;4:2797. doi: 10.1038/ncomms3797. [DOI] [PubMed] [Google Scholar]
- 116.Song J, et al. Genetic transformation of Populus trichocarpa genotype Nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol. 2006;47:1582–1589. doi: 10.1093/pcp/pcl018. [DOI] [PubMed] [Google Scholar]
- 117.Cseke L, et al. High efficiency poplar transformation. Plant Cell Rep. 2007;26:1529–1538. doi: 10.1007/s00299-007-0365-0. [DOI] [PubMed] [Google Scholar]
- 118.Al Afas N, et al. Variability in Populus leaf anatomy and morphology in relation to canopy position, biomass production, and varietal taxon. Annals Forest Sci. 2007;64:521–532. [Google Scholar]
- 119.Ridge C, et al. Leaf growth characteristics of fast-growing poplar hybrids Populus trichocarpa × P. deltoides. Tree Physiol. 1986;1:209–216. doi: 10.1093/treephys/1.2.209. [DOI] [PubMed] [Google Scholar]
- 120.Ferris R, et al. Leaf area is stimulated in Populus by free air CO2 enrichment (POPFACE), through increased cell expansion and production. Plant Cell Environ. 2001;24:305–315. [Google Scholar]
- 121.Stuart JM, et al. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 122.Ogata Y, et al. CoP: a database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics. 2010;26:1267–1268. doi: 10.1093/bioinformatics/btq121. [DOI] [PubMed] [Google Scholar]
- 123.Zeevi V, et al. Zinc finger nuclease and homing endonuclease-mediated assembly of multigene plant transformation vectors. Plant Physiol. 2012;158:132–144. doi: 10.1104/pp.111.184374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen Q, et al. MISSA Is a highly efficient in vivo DNA assembly method for plant multiple-gene transformation. Plant Physiol. 2010;153:41–51. doi: 10.1104/pp.109.152249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma L, et al. RMDAP: a versatile, ready-to-use toolbox for multigene genetic transformation. PLoS One. 2011;6:e19883. doi: 10.1371/journal.pone.0019883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sarrion-Perdigones A, et al. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. Plos One. 2011;6:e21622. doi: 10.1371/journal.pone.0021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sarrion-Perdigones A, et al. GoldenBrain 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013;162:1618–1631. doi: 10.1104/pp.113.217661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sternberg N. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc. Natl. Acad. Sci. U.S.A. 1990;87:103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shizuya H, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hamilton C, et al. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu C, et al. Construction of rice minichromosomes by telomere mediated chromosomal truncation. Plant J. 2012;70:1070–1079. doi: 10.1111/j.1365-313X.2012.04916.x. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, et al. Recombinase technology: applications and possibilities. Plant Cell Rep. 2011;30:267–285. doi: 10.1007/s00299-010-0938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dhar M, et al. Towards the development of better crops by genetic transformation using engineered plant chromosomes. Plant Cell Rep. 2011;30:799–806. doi: 10.1007/s00299-011-1001-6. [DOI] [PubMed] [Google Scholar]
- 134.Yu W, et al. Construction and behavior of engineered minichromosomes in maize. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8924. doi: 10.1073/pnas.0700932104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Teo C, et al. Induction of telomere-mediated chromosomal truncation and stability of truncated chromosomes in Arabidopsis thaliana. Plant J. 2011;68:28–39. doi: 10.1111/j.1365-313X.2011.04662.x. [DOI] [PubMed] [Google Scholar]
- 136.Borland A, Dodd A. Carbohydrate partitioning in crassulacean acid metabolism plants: reconciling potential conflicts of interest. J. Exp. Bot. 2002;29:707–716. doi: 10.1071/PP01221. [DOI] [PubMed] [Google Scholar]
- 137.Dodd A, et al. Integrating diel starch metabolism with the circadian and environmental regulation of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Planta. 2003;6:789–797. doi: 10.1007/s00425-002-0930-2. [DOI] [PubMed] [Google Scholar]
- 138.Paul M, et al. Starch-degrading enzymes during the induction of CAM in Mesembryanthemum crystallinum. Plant Cell Environ. 1993;16:531–538. [Google Scholar]
- 139.Häusler R, et al. Plastidic metabolite transporters and their physiological functions in the inducible crassulacean acid metabolism plant Mesembryanthemum crystallinum. Plant J. 2000;24:285–296. doi: 10.1046/j.1365-313x.2000.00876.x. [DOI] [PubMed] [Google Scholar]
- 140.Kore-eda S, et al. Transcriptional profiles of organellar metabolite transporters during induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Funct. Plant Biol. 2005;32:451–466. doi: 10.1071/FP04188. [DOI] [PubMed] [Google Scholar]
- 141.Spalding M, et al. Intracellular localization of some key enzymes of crassulacean acid metabolism in Sedum praealtum. Plant Physiol. 1979;63:738–743. doi: 10.1104/pp.63.4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Winter K, et al. Intracellular localization of enzymes of carbon metabolism in Mesembryanthemum crystallinum exhibiting C3 photosynthetic characteristics or performing Crassulacean acid metabolism. Plant Physiol. 1982;69:300–307. doi: 10.1104/pp.69.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]