Abstract
Peripheral blood mononuclear cells (PBMCs) are stimulated by exercise and contribute not only to host defense, but also to growth, repair, and disease pathogenesis. Whether PBMC gene expression is altered by exercise in children is not known. 10 early pubertal boys (8-12 y) and 10 late pubertal boys (15-18 y) performed 10, 2-min bouts of strenuous, constant work rate exercise with 1-min rest intervals. PBMCs were isolated before and after exercise and microarray (Affymetrix U133+2 chips) analyzed. Statistical criterion to identify gene expression changes was less than 5% false discovery rate with 95% confidence. 1246 genes were altered in older boys (517 up, 729 down), but only 109 in the younger group (79 up, 30 down). In older boys, 13 gene pathways (using EASE p<0.05) were found (e.g., natural killer cell cytoxicity, apoptosis). Epiregulin gene expression (EREG, a growth factor involved in wound healing) increased in older boys. In older boys exercise altered genes such as TBX21, GZMA, PGTDR, and CCL5 that also play roles in pediatric inflammatory diseases like asthma. 66 genes changed significantly in both groups. The pattern of PBMC gene expression suggests the initiation of an immunological “danger” signal associated with a sudden change in energy expenditure.
Keywords: leukocytes, puberty, microarrays, inflammation, immune system
There is mounting data that leukocytes are not only involved in eradicating pathogens, but also play a role in wound repair, muscle growth, other key developmental processes, and in childhood diseases like asthma and arthritis (1-4). In both adults and children, a leukocytosis occurs following brief, heavy exercise (5). In addition, we now know from recent studies from this and other laboratories that in adults substantial changes in the gene expression profile pattern of the circulating leukocytes rapidly accompany exercise (6, 7). The impact of exercise on the immune response is increasingly seen as one of the novel mechanisms through which levels of physical activity modulates health, growth, and disease risk in both children and adults.
No studies have examined the leukocyte gene response to individual bouts of exercise in children, consequently, we hypothesized: first, that exercise would stimulate gene expression in peripheral blood mononuclear cells (PBMC); and secondly, that the genomic response would include alterations in pro and anti-inflammatory cytokines, stress factors, and growth mediators. Finally, we compared PBMC genomic response to exercise in both early and late-pubertal boys because we hypothesized that the impact of exercise on leukocyte gene responses would be altered by maturational status, reflecting the fact that immune function changes across puberty (8).
MATERIALS AND METHODS
Subjects
Twenty healthy males (age range 8-18 y) participated in this study (Table 1). We selected 10 participants (age range 8 to 12 y) who were classified as in early puberty and 10 participants (age range 15 to 18 y) who were late pubertal. We used a validated self-administered questionnaire which has been widely used to assess pubertal status (9, 10). We recognized the limitations of self reported assessment of pubertal status, and consequently, compared readily appreciated physical characteristics (such as facial hair or voice tonality) with the written response and planned to exclude those subjects in whom there was a clear disparity. Fortunately, we did not have to do this. In the present study, we focused on boys because gender influences many physiologic responses even in early puberty. Parallel studies are ongoing in females in our laboratory. Individuals participating in competitive sports and with a history of any chronic medical conditions or use of any medications were excluded from participation. The Institutional Review Board at the University of California, Irvine approved the study, and written informed assent and consent was obtained from all participants and their parents upon enrollment.
Table 1.
Anthropometrical and physiological characteristics of the 20 subjects
| Early Pubertal Boys (N=10) | Late Pubertal Boys (N=10) | |
|---|---|---|
| Age (yr) | 10.5 ± 0.4 | 17.4 ± 0.4* |
| Height (cm) | 141.7 ± 3.5 | 175.1 ± 3.4* |
| Weight (kg) | 36.2 ± 2.8 | 67.4± 3.7* |
| BMI (kg/m2) | 17.8 ± 0.9 | 21.9 ± 0.8* |
| BMI (%ile) | 58.1 ± 9.0 | 52.3 ± 8.8 |
| Peak VO2 (ml/min/kg) | 38.7 ± 2.8 | 44.6 ± 2.6 |
Values are means ± SE. BMI, body mass index; Peak VO2, peak oxygen Consumption.
Significant difference between early and late pubertal boys (p<0.005).
Anthropometric measurements
Standard, calibrated scales and stadiometers were used to determine height, body mass, and body mass index (BMI =wt/ht2). Since BMI changes with age, we also calculated BMI percentile for each child using the recently published standards from the Centers for Disease Control, National Center for Health Statistics (11).
Measurement of fitness
Each subject performed a ramp-type progressive cycle ergometer using the SensorMedics metabolic system (Ergoline 800S, Yorba Linda, CA). Subjects were vigorously encouraged during the high-intensity phases of the exercise protocol. Gas exchange was measured breath-by-breath and the anaerobic (lactate) threshold and peak oxygen consumption (V̇O2) was calculated using a standard methods (12).
Exercise Protocol
At least 48 hours, but not exceeding seven days following the completion of the ramp test, each subject performed exercise consisting of 10 two-minute bouts of constant work rate cycle ergometry, with one-minute rest interval between each bout of exercise. The work rate was individualized for each child and was calculated to be equivalent to the work rate corresponding roughly to 50% of the work rate between the anaerobic threshold and the peak oxygen uptake (as determined non-invasively from the ramp-type test). This resulted in a relative work rate that was equivalent between study subjects. We have used this protocol in the past first, to more closely mimic the “stop-start” nature of spontaneous physical activity (13) and secondly to ensure that the exercise input was standardized to physiological indicators of each subject’s exercise capacity (14).
Blood sampling and analysis
An indwelling catheter was inserted into the antecubital vein. A baseline sample was taken 30 min after the placement of the catheter and before the onset of exercise. We waited 30 min to ensure that measurable physiological parameters of stress (e.g., heart rate and blood pressure) were at baseline levels. Subjects then completed the 10 two-minute bouts of constant work rate, and additional blood samples were obtained immediately after exercise (total of 40 samples). The plasma was separated and stored at −80°C and thawed only once for analysis. Complete blood counts (CBC) for white blood cell analysis were obtained by standard methods from the clinical hematology laboratory at UC Irvine.
PBMC separation
PBMCs were isolated using OptiPrep® Density Gradient Medium (SIGMA). Standard and consistent practices were employed in an effort to minimize any potential changes in mRNA expression levels due to manipulation of PBMCs. The duration from blood draw to stabilization of RNA never exceeded 60 min.
RNA extraction
Total RNA was extracted using TRIzol® (Gibco BRL Life Technologies, Rockville, MD) reagent and purified using the RNeasy Midi columns method (Qiagen, Valencia, CA). RNA pellets were resuspended in diethyl pyrocarbonate-treated water. RNA integrity was assessed by running out a small amount of each sample (typically 25-250 ng/well) onto a RNA Lab-On-A-Chip (Caliper Technologies Corp., Mountain View, CA) that was evaluated on an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Palo Alto, CA, USA).
Preparation of labeled cRNA
The detailed protocol for preparation and microarray processing was performed as recommended by the manufacturer and is available in the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix Inc., Santa Clara, CA). Briefly, 4 μg total RNA was used as a template for double-stranded cDNA synthesis. Single-stranded then double-stranded cDNA was synthesized. A portion of the resulting double-stranded cDNA was used as a template to generate biotin-tagged cRNA from an in vitro transcription reaction (IVT), using the Affymetrix GeneChip® IVT Labeling Kit.
Hybridization to microarray
A total of 15μg of the resulting biotin-tagged cRNA was fragmented to an average strand length of 100 bases (range 35-200 bases) following prescribed protocols (Affymetrix GeneChip® Expression Analysis Technical Manual). Subsequently, 10 μg of this fragmented target cRNA was hybridized at 45°C with rotation for 16 hours (Affymetrix GeneChip® Hybridization Oven 640) on an Affymetrix U133+2 arrays. The GeneChip® arrays were washed and then stained (SAPE, streptavidin-phycoerythrin) on an Affymetrix Fluidics Station 450, followed by scanning on a GeneChip® Scanner 3000.
Data Analysis
Microarray Analysis
The results were quantified and analyzed using GCOS 1.4 software (Affymetrix, Inc.) using default values (Scaling Target Signal Intensity=500). The microarray data was analyzed using ArrayAssist® version 5.2.2 (STRATAGENE®). We normalized the data using GC-RMA. Only probe sets that reached a signal value ≥20 in at least one array and a present call by MAS5 criteria in at least 30 arrays were selected for further analysis. Overall, 20065 out of 54675 probe sets represented on the array met these criteria. The microarray CEL files and GC-RMA normalized data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11761). We further applied BRB-ArrayTools software Version 3.6.0 (http//linus.nci.nih.gov/~brb/tool.htm) to determine significantly changed probe sets from before to after exercise for early-pubertal and late-pubertal boys separately. Traditional Student’s paired t-test was first applied to each probe set and then significantly changed probe sets were identified with permutation tests (15). With 95% confidence, the final list of significantly changed probe sets in each group has less than a 5% false discovery rate (FDR). The mean fold change of genes that were significantly changed in both lists was further evaluated using Pearson correlation for similarity.
Gene annotation
The final list of significantly changed probe sets was then additionally analyzed using the functional annotation tools provided by DAVID-- Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov) in order to classify the genes into pathways using the KEGG database. Only pathways with EASE score<0.05 are presented in this analysis. EASE score (Expression Analysis Systematic Explorer) is a modified Fisher Exact p-value in DAVID system used for gene-enrichment analysis. EASE queries the KEGG pathways database and determines functional enrichment by calculating a Fisher’s exact test p-value for each pathway. EASE score P-Value = 0 represents perfect enrichment. P-Value ≤ 0.05 is considered as gene-enrichment in a specific annotation category (http://david.abcc.ncifcrf.gov/helps/functional_annotation.html#summary).
Physiological Data
The physiological data are presented as mean and standard error mean (SEM). The two-sided paired t-test was applied for testing changes from before to after the exercise within each group and the two-sample t-test was applied for examining group difference. All analyses were done using SAS9 (Cary, NC) and the significance level was set at 0.05.
Real-Time Polymerase Chain Reaction (RT-PCR)
For confirmation of microarray gene expression findings, RT-PCR was carried out on 5 genes that we felt were of particular physiological significance in the context of leukocyte function and inflammation (HSPA1B, CTSW, CASP6, GZMB, and TBX21) in the late pubertal boys, and one gene (HSPA1B) in the early pubertal boys. One μg of RNA was reverse transcribed using the Reverse Transcription Reagents kit (Applied Biosystems) according to the manufacturer’s instructions, using random primers in a 100-μl reaction. The RT-PCR analysis was performed with the ABI PRISM 7000 Sequence Detection System (TaqMan; Applied Biosystems) by using TaqMan Universal PCR Master Mix and Assays-on-Demand Gene Expression probes (Applied Biosystems) (HSPA1B: Assay ID: Hs00271244_s1, CTSW: Assay ID: Hs00175160_m1, CASP6: Assay ID: Hs00154250_m1, GZMB: Assay ID: Hs00188051_m1, TBX21: Assay ID: Hs00203436_m1). Human Actin beta was used as an endogenous control. The RT-PCR results were compared with the microarray results on the directional change (up- or down-regulated) from before-exercise to after-exercise using Fisher’s exact test.
RESULTS
Anthropometric and physiological characteristics
The anthropometric and physiologic characteristics of the 20 subjects appear in Table 1. The average BMI was within normal limits in both groups. The distribution of BMI percentile was similar between the two groups; the median and range were 68.2 and 82.4 for early-pubertal boys and were 51.8 and 74.4 for late-pubertal boys.
Exercise intensity and lactic acid levels
Relative exercise intensity as determined by HR, WR, and V̇O2 was similar between the two groups (Figure 1). In the older boys, lactate increased from 1.44±0.19 mmol/l to 7.28±0.87 mmol/l, p<0.0001 (+445.4±59.3%). Although lactate also increased significantly in the early pubertal boys [(from 1.96±0.18 mmol/l to 3.89±0.52 mmol/l, p=0.0061 (+112.1±33.8%)], the increase was smaller in the younger compared with the older boys (p=0.0005).
Figure.1.
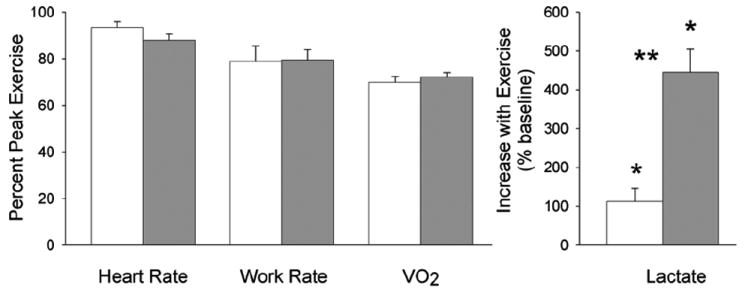
Assessment of exercise intensity during the 10, 2-min bouts used to gage the PBMC genomic response. The white bars represent the early pubertal boys. The gray bars represent the late pubertal boys. The values represent the means ± SEM of heart rate, work rate, and VO2 expressed as percent of the individual participant’s peak values obtained from the progressive exercise protocol. As can be seen, the relative exercise intensity was virtually the same in the two groups. Lactate increased in both groups (*p<0.05); however, to a significantly greater degree in the older boys (**p<0.005).
PBMC response to exercise
As shown in Figure 2, the number of total WBCs, lymphocytes, and monocytes was found to be significantly elevated at Peak exercise in both early pubertal boys [WBC increases 1680±274.8 (34%, p=0.0002), lymphocytes 600±210.8 (30%, p=0.019), and monocytes 120±35.9 (37%, p=0.0086)] and late pubertal boys [WBC 2930±330.3 (65%, p<0.0001), lymphocytes 1140±212 (66.5%, p=0.0004), monocytes 390±58.6 (158%, p<0.0001)]. The increase was significantly greater in late- compared with early pubertal boys for total WBCs (p=0.0096) and monocytes (p=0.0014).
Figure.2.
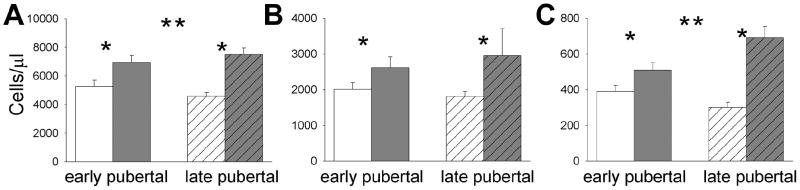
Effect of exercise on PBMCs in the early and late pubertal boys. The data represent the mean±SEM before (white bars) and immediately after exercise (gray bars) for (A) all leukocytes, (B) lymphocytes, and (C) monocytes. * indicates a significant (p<0.05) increase from before to after exercise. ** indicates an increase that was significantly (p<0.05) greater in the late compared with early pubertal boys.
The effect of exercise on gene expression
Exercise induced significant changes in PBMC gene expression in both the early and late pubertal boys. There were differences in the PBMC gene response between the two groups: in late pubertal boys, exercise altered 1246 genes (517 up, 729 down) (FDR<0.05) compared to only 109 (79 up, 30 down) in the early pubertal boys. A complete detailed lists of genes that were altered by exercise in early- and late pubertal boys are shown in Tables S1-S2 respectively (supplementary material , online at www.pedresearch.org). Figure 3 schematically compares the magnitude of the PBMC gene change in the two groups, and in Figure 4, we show the correlation in fold-change of the 66 common exercise-sensitive genes in the two groups. Of these 66 genes, the direction of change was similar in 64 (Table 2). Moreover, in the 37 common exercise-sensitive upregulated genes, the average fold change was virtually the same in the two groups (early pubertal, 1.55±0.11 vs. late pubertal, 1.55±0.07). Similarly, in the 27 common down-regulated genes, the average fold-change values were 0.63±0.02 in early pubertal boys vs. 0.63±0.03 in the late).
Figure.3.
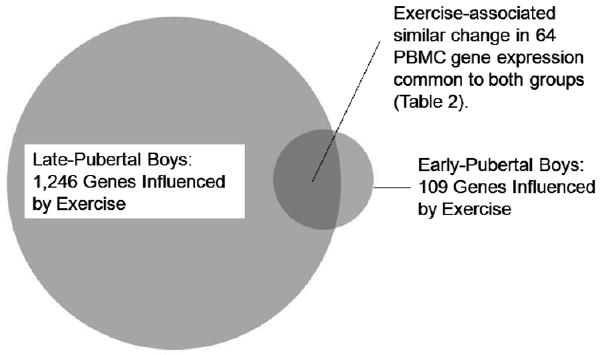
Comparison of the effect of exercise on PBMC genes in early- and late pubertal boys. The figure shows the relative magnitude of the effect (circles) and the size of the overlap (shaded area).
Figure.4.
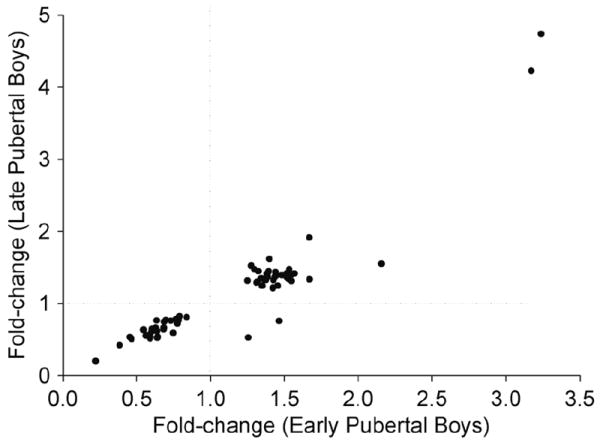
Comparison of fold-change in PBMC genes that were significantly affected by exercise in both the younger- and older boys. The data points represent the mean fold-change for each of the exercise-sensitive genes in both groups. The dotted lines demarcate up-regulation (fold change > 1) from down-regulation (fold-change < 1). The fold-change in the two groups was highly correlated (r=0.92). In only two of the common exercise-sensitive genes were the direction of fold-change different in the two groups.
Table 2.
Sixty four common exercise-sensitive genes in which fold-change was congruent
| Up-Regulated |
Down-Regulated |
||
|---|---|---|---|
| HSPA1B | FBXO30 | TAF9 | PSMD7 |
| FASLG | EDG1 | YPEL5 | EPC1 |
| ZBTB6 | KIAA1632 | HMGB1 | WTAP |
| ZNF200 | SLC38A2 | HIST1H2BD | YME1L1 |
| EOMES | CCDC117 | UBE2J1 | BTG1 |
| GPR18 | ARNTL | CPNE5 | SP3 |
| ANKRD46 | JARID1A | WIBG | SNORA28 |
| CSE1L | MIER3 | RHBDD1 | FUS |
| WDR20 | PPP1R10 | CLIC4 | |
| MFAP3 | DNAJB4 | EAF2 | |
| ZNF443 | ARHGEF3 | KRR1 | |
| TAGAP | ZNF644 | ZCCHC10 | |
| DLEU2 | TMEM2 | ATF7IP2 | |
| MFAP1 | KCTD6 | H3F3B | |
| FEM1B | IVNS1ABP | USP15 | |
| GOSR1 | PHF23 | LCN2 | |
| FBXO8 | TGFBR1 | ARF1 | |
| WDR33 | HIPK1 | SNRPA1 | |
| MEX3C | EIF1 | ||
We characterized the genes affected by exercise using gene pathways. In the late pubertal boys, we found 13 gene pathways classified by Kegg that were significantly (Ease score <0.05) altered by exercise (Table 3). Many of the pathways identified were involved in innate or early immune responses. A complete detailed list of the individual genes that were altered by exercise, within each pathway is shown in Table S3 (supplementary material , online at www.pedresearch.org).
Table 3.
Thirteen pathways which were altered by exercise, in late pubertal boys, classified by Kegg
| Pathway | # of Genes | P-Value (Ease Score) |
|---|---|---|
| Natural killer cell mediated cytotoxicity | 27 | 1.30E-06 |
| Hematopoietic cell lineage | 21 | 1.80E-06 |
| Antigen processing and presentation | 16 | 5.50E-04 |
| Cell adhesion molecules (CAMs) | 20 | 2.10E-03 |
| Cytokine-cytokine receptor interaction | 31 | 5.90E-03 |
| B cell receptor signaling pathway | 12 | 6.70E-03 |
| Glycan structures - biosynthesis 1 | 17 | 1.30E-02 |
| T cell receptor signaling pathway | 14 | 1.80E-02 |
| Apoptosis | 13 | 1.80E-02 |
| Type I diabetes mellitus | 8 | 2.10E-02 |
| Prostate cancer | 13 | 2.30E-02 |
| Calcium signaling pathway | 21 | 2.70E-02 |
| Adherens junction | 11 | 4.10E-02 |
RT-PCR verification of specific genes
We verified the change from before to after the exercise of 5 genes in late pubertal boys (HSPA1B, CTSW, CASP6, GZMB, and TBX21), and one gene in early pubertal boys (HSPA1B), using RT-PCR. The results showed 58 out of the 60 comparisons are consistent between RT-PCR and microarray (50 up-regulated and 8 down-regulated, Fisher Exact Test p-value<0.0001). Only the CSP6 gene of two late boys showed inconsistency, where RT-PCR showed up regulated but microarray showed down regulated.
DISCUSSION
We found that an acute bout of exercise induced a remarkable change in PBMC gene expression in healthy boys. These observations were made using a stringent statistical analysis of the microarray data to limit the possibility of false-positive results. Moreover, we found that a different technique, RT-PCR in a small group of genes, corroborated the microarray data. In the late pubertal participants, the relatively brief but heavy-exercise protocol not only influenced more than a thousand individual genes, but also significantly affected thirteen known gene pathways like natural killer cell mediated cytotoxicity, antigen processing and presentation, cytokine-cytokine receptor interaction, and apoptosis all of which have relevance for health and disease in children (Table 3). Exercise is a complex and profound physiologic perturbation in which, not surprisingly, the sudden insult to cellular homeostasis leads to a systemic “danger-type” response (16), one reflecting the increased vulnerability of an organism required to flee a predator or pursue prey. Our data indicate that brief exercise is associated not only with increased numbers of immune cells in the circulation, but also may serve as a “wake-up” call in which key gene pathways are activated preparing PBMCs for fighting infection, wound repair, and, at the same time, setting the stage for apoptosis should these functions not be needed and the activated cells be eliminated.
Previous investigators have examined, predominately in adults, immune consequences of exercise primarily by measuring the impact of physical activity on circulating immune mediators and on leukocyte numbers and function (e.g., (17)). Very few studies have focused on the earliest steps of immune cell activation, namely gene expression. The robust response we observed in this study reflects previous work in this laboratory in young adult males (6). Using a different approach, 30-min of continuous, heavy exercise and pooling blood samples by groups of three from a total of fifteen participants, we found that 311 PBMC genes/probe sets were influenced by exercise in the men of which 163 (52.4%) were also found in the late pubertal boys in the present study. Among the leukocyte genes whose expression was altered by exercise in both the young men of the Connolly study (6) and the late-pubertal participants in the current study were, for example, CCL4, FASLG, GZMA, PRF1, HSPA1B, consistent with the emerging concept that brief exercise stimulates inflammatory, stress, and apoptotic pathways in leukocytes.
Intriguingly, the gene for epiregulin (EREG, an epidermal growth factor) was stimulated to a greater degree by exercise in PBMCs in both young adult men (3.5 fold) and late pubertal boys (3.6 fold) than almost any other gene. Although epiregulin can promote wound healing (18, 19), it may also play a role in inflammation as epiregulin gene expression in leukocytes is upregulated in in vitro models of rhinovirus infection (20). Systemic stress prior to injury activates leukocyte trafficking in a manner that could promote subsequent would healing (21). Our data supports this potentially beneficial effect of exercise stress by showing in adolescents an increased gene expression of specific tissue growth mediators like epiregulin in PBMCs.
Exercise is known to play a role, sometimes deleterious and sometimes beneficial in a variety of childhood diseases in which immune cells are involved. Exercise-induced bronchoconstriction, for example, is a common and distressing clinical feature of most children with asthma (22) characterized by airway inflammation. We found that exercise affected a number of asthma-related PBMC genes (e.g., TBX21, CCL5, PTGDR, GZMA), which could play a role in asthma pathogenesis ranging from altering susceptibility to atopy in general to factors that govern the recruitment and activation of inflammatory cells (23-26). Whether levels of physical activity and fitness can alter the PBMC function in health and disease in children by influencing gene expression and translation has not been determined.
Although many PBMC genes significantly changed in response to exercise in both groups, far fewer PBMC genes responded to exercise in the younger boys (Figure 3). Of the 109 exercise-sensitive genes in the early pubertal participants, 66 (60%) were also exercise-sensitive in the older participants. There were a number of revealing patterns in this set of common genes. For example, Fas ligand (TNF superfamily, member 6, FASLG) increased expression by 3.2-fold in the younger boys and by 4.2 in the older boys. Interaction of Fas with this ligand is critical in triggering apoptosis of some types of cells such as lymphocytes (27). Another pleiotropic PBMC gene that was exercise sensitive in both the older and younger subjects was heat shock 70kDa protein 1B (HSPA1B). Heat shock protein-70 (HSP70) is one member of a family of highly conserved intracellular proteins synthesized to resist stress-induced damage by assisting with the proper folding and transport of proteins and known to be responsive to exhaustive exercise in adults (28). Increasingly, HSP are seen as playing a role in juvenile autoimmune diseases (29, 30) and, possibly, allergy and asthma (31).
We can only speculate as to the mechanisms responsible for the differences in PBMC gene expression between the younger and older boys. When we focused solely on the PBMC genes that were responsive to exercise in both groups, we found remarkable congruity in the direction and magnitude of the changes (Figure 4). These data provide indirect evidence for the existence of a number of PBMC genes that are sensitive to the perturbation caused by exercise, yet independent of the pubertal status of the participants.
Moreover, to avoid exercise “dose-response” differences in children of different sizes and exercise capacity, we employed careful techniques to ensure that the magnitude of the exercise dose was comparable between the two groups. HR, WR, and VO2 data showed that we achieved parity between the groups (Figure 1). There were significant differences in the effect of exercise on the number of circulating PBMCs (Figure 2), but the effect was so small that it is unlikely related to the very large difference in the number of genes affected in the early and late pubertal boys (Figure 3). Although lactate increased significantly in both groups, the magnitude of the increase was substantially larger in the older participants (Figure 1). This is a well-known maturational difference (32) related, most likely to lower levels of muscle lactate dehydrogenase (33) and/or a greater dependence on aerobic rather than anaerobic metabolism in the performance of muscular work (34) in the younger participants. Whether the smaller lactate responses (and the accompanying changes in tissue and systemic pH) to exercise in younger compared with older children could explain the difference in PBMC gene expression between the early and late pubertal boys is not known.
In summary, this is the first study to show significant alteration in PBMC gene expression in response to exercise in children. The exercise protocol, while strenuous, was brief and not unrepresentative of actual patterns of physical activity encountered in the daily lives of many children. Marked differences, perhaps related to pubertal status, in the gene response were observed. In the late pubertal boys, 1246 genes and 13 established gene pathways were significantly altered by exercise. The genes involved included growth mediators, cytotoxic killing factors, and agents involved in apoptosis, reinforcing the notion that exercise can initially lead to the initiation in PBMCs of a number of protective physiologic responses that might be of benefit to the organism during or following the perceived “danger” associated with a sudden change in energy expenditure. These data shed new light on possible immune-cell pathways linking exercise with health and disease in the growing child, but the extent to which changes in gene expression in PBMCs lead to functional or physiological changes in these cells has yet to be determined.
Supplementary Material
Acknowledgments
Financial Support: This work was supported in part by National Institutes of Health grants RO1-HL080947, P01HD-048721 and the UCI Satellite GCRC:MO1 RR00827.
ABBREVIATIONS
- GZMA
granzyme A
- PBMCs
peripheral blood mononuclear cells
- TBX21
T-box 21
- V̇O2
oxygen consumption
Footnotes
This article contains supplementary material available online at www.pedresearch.org.
References
- 1.Tidball JG, Wehling-Henricks M. Macrophages promote muscle membrane repair and muscle fibre growth and regeneration during modified muscle loading in mice in vivo. J Physiol. 2007;578:327–336. doi: 10.1113/jphysiol.2006.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarvis JN, Petty HR, Tang Y, Frank MB, Tessier PA, Dozmorov I, Jiang K, Kindzelski A, Chen Y, Cadwell C, Turner M, Szodoray P, McGhee JL, Centola M. Evidence for chronic, peripheral activation of neutrophils in polyarticular juvenile rheumatoid arthritis. Arthritis Res Ther. 2006;8:R154. doi: 10.1186/ar2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabroe I, Whyte MK. Toll-like receptor (TLR)-based networks regulate neutrophilic inflammation in respiratory disease. Biochem Soc Trans. 2007;35:1492–1495. doi: 10.1042/BST0351492. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, Gazit R, Yutkin V, Benharroch D, Porgador A, Keshet E, Yagel S, Mandelboim O. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 5.Schwindt CD, Zaldivar F, Wilson L, Leu SY, Wang-Rodriguez J, Mills PJ, Cooper DM. Do circulating leucocytes and lymphocyte subtypes increase in response to brief exercise in children with and without asthma? Br J Sports Med. 2007;41:34–40. doi: 10.1136/bjsm.2006.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly PH, Caiozzo VJ, Zaldivar F, Nemet D, Larson J, Hung SP, Heck JD, Hatfield GW, Cooper DM. Effects of exercise on gene expression in human peripheral blood mononuclear cells. J Appl Physiol. 2004;97:1461–1469. doi: 10.1152/japplphysiol.00316.2004. [DOI] [PubMed] [Google Scholar]
- 7.Radom-Aizik S, Zaldivar F, Jr, Leu SY, Galassetti P, Cooper DM. Effects of 30 min of aerobic exercise on gene expression in human neutrophils. J Appl Physiol. 2008;104:236–243. doi: 10.1152/japplphysiol.00872.2007. [DOI] [PubMed] [Google Scholar]
- 8.Lamason R, Zhao P, Rawat R, Davis A, Hall JC, Chae JJ, Agarwal R, Cohen P, Rosen A, Hoffman EP, Nagaraju K. Sexual dimorphism in immune response genes as a function of puberty. BMC Immunol. 2006;7:2. doi: 10.1186/1471-2172-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson EB, Skinner ML, Love MM, Elder GH, Conger RD, Dubas JS, Petersen AC. The Pubertal Development Scale: a rural and suburban comparison. J Early Adolesc. 1992;12:174–186. [Google Scholar]
- 10.Petersen AC, Crockett L, Richards M, Boxer A. Self-Report Measure of Pubertal Status-Reliability, Vailidity, and Initial Norms. J Youth Adolesc. 1988;17:117–133. doi: 10.1007/BF01537962. [DOI] [PubMed] [Google Scholar]
- 11.Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR, Roche AF, Johnson CL. 2000 CDC Growth Charts for the United States: methods and development. Vital Health Stat. 2002;11:1–190. [PubMed] [Google Scholar]
- 12.Cooper DM, Weiler-Ravell D, Whipp BJ, Wasserman K. Aerobic parameters of exercise as a function of body size during growth in children. J Appl Physiol. 1984;56:628–634. doi: 10.1152/jappl.1984.56.3.628. [DOI] [PubMed] [Google Scholar]
- 13.Bailey RC, Olson J, Pepper SL, Barstow TJ, Porszsasz J, Cooper DM. The level and tempo of children’s physical activities: An observational study. Med Sci Sports Exerc. 1995;27:1033–1041. doi: 10.1249/00005768-199507000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Tirakitsoontorn P, Nussbaum E, Moser C, Hill M, Cooper DM. Fitness, acute exercise, and anabolic and catabolic mediators in cystic fibrosis. Am J Respir Crit Care Med. 2001;164:1432–1437. doi: 10.1164/ajrccm.164.8.2102045. [DOI] [PubMed] [Google Scholar]
- 15.Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and Analysis of DNA Microarray Investigations. Springer; New York: 2003. Class Prediction; pp. 65–94. [Google Scholar]
- 16.Matzinger P. Friendly and dangerous signals: is the tissue in control? Nat Immunol. 2007;8:11–13. doi: 10.1038/ni0107-11. [DOI] [PubMed] [Google Scholar]
- 17.Gleeson M, Bishop NC. The T cell and NK cell immune response to exercise. Ann Transplant. 2005;10:43–48. [PubMed] [Google Scholar]
- 18.Draper BK, Komurasaki T, Davidson MK, Nanney LB. Epiregulin is more potent than EGF or TGFalpha in promoting in vitro wound closure due to enhanced ERK/MAPK activation. J Cell Biochem. 2003;89:1126–1137. doi: 10.1002/jcb.10584. [DOI] [PubMed] [Google Scholar]
- 19.Roy S, Khanna S, Rink C, Biswas S, Sen CK. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics. 2008;34:162–184. doi: 10.1152/physiolgenomics.00045.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Gualano RC, Hibbs ML, Anderson GP, Bozinovski S. Epidermal growth factor receptor signaling to Erk1/2 and STATs control the intensity of the epithelial inflammatory responses to rhinovirus infection. J Biol Chem. 2008;283:9977–9985. doi: 10.1074/jbc.M710257200. [DOI] [PubMed] [Google Scholar]
- 21.Viswanathan K, Dhabhar FS. Stress-induced enhancement of leukocyte trafficking into sites of surgery or immune activation. Proc Natl Acad Sci USA. 2005;102:5808–5813. doi: 10.1073/pnas.0501650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storms WW. Asthma associated with exercise. Immunol Allergy Clin North Am. 2005;25:31–43. doi: 10.1016/j.iac.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Munthe-Kaas MC, Carlsen KH, Haland G, Devulapalli CS, Gervin K, Egeland T, Carlsen KL, Undlien D. T cell-specific T-box transcription factor haplotype is associated with allergic asthma in children. J Allergy Clin Immunol. 2008;121:51–56. doi: 10.1016/j.jaci.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 24.Steinke JW, Borish L. Genetics of allergic disease. Med Clin North Am. 2006;90:1–15. doi: 10.1016/j.mcna.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhu G, Vestbo J, Lenney W, Silverman M, Whyte M, Helms P, Anderson WH, Pillai SG. Association of PTGDR gene polymorphisms with asthma in two Caucasian populations. Genes Immun. 2007;8:398–403. doi: 10.1038/sj.gene.6364399. [DOI] [PubMed] [Google Scholar]
- 26.Bratke K, Haupt F, Kuepper M, Bade B, Faehndrich S, Luttmann W, Virchow JC., Jr Decrease of cytotoxic T cells in allergic asthma correlates with total serum immunglobulin E. Allergy. 2006;61:1351–1357. doi: 10.1111/j.1398-9995.2006.01192.x. [DOI] [PubMed] [Google Scholar]
- 27.Green DR. Fas Bim boom! Immunity. 2008;28:141–143. doi: 10.1016/j.immuni.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Whitham M, Fortes MB. Heat shock protein 72: release and biological significance during exercise. Front Biosci. 2008;13:1328–1339. doi: 10.2741/2765. [DOI] [PubMed] [Google Scholar]
- 29.Elst EF, Klein M, de Jager W, Kamphuis S, Wedderburn LR, van der Zee R, Albani S, Kuis W, Prakken BJ. Hsp60 in inflamed muscle tissue is the target of regulatory autoreactive T cells in patients with juvenile dermatomyositis. Arthritis Rheum. 2008;58:547–555. doi: 10.1002/art.23202. [DOI] [PubMed] [Google Scholar]
- 30.Prakken B, Kuis W, van Eden W, Albani S. Heat shock proteins in juvenile idiopathic arthritis: keys for understanding remitting arthritis and candidate antigens for immune therapy. Curr Rheumatol Rep. 2002;4:466–473. doi: 10.1007/s11926-002-0052-7. [DOI] [PubMed] [Google Scholar]
- 31.Hauet-Broere F, Wieten L, Guichelaar T, Berlo S, van der Zee R, van Eden W. Heat shock proteins induce T cell regulation of chronic inflammation. Ann Rheum Dis. 2006;65:iii65–iii68. doi: 10.1136/ard.2006.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthoin S, Allender H, Baquet G, Dupont G, Matran R, Pelayo P, Robin H. Plasma lactate and plasma volume recovery in adults and children following high-intensity exercises. Acta Paediatr. 2003;92:283–290. doi: 10.1080/08035250310009185. [DOI] [PubMed] [Google Scholar]
- 33.Kaczor JJ, Ziolkowski W, Popinigis J, Tarnopolsky MA. Anaerobic and Aerobic Enzyme Activities in Human Skeletal Muscle from Children and Adults. Pediatr Res. 2005;57:331–335. doi: 10.1203/01.PDR.0000150799.77094.DE. [DOI] [PubMed] [Google Scholar]
- 34.Zanconato S, Buchthal S, Barstow TJ, Cooper DM. 31P-magnetic resonance spectroscopy of leg muscle metabolism during exercise in children and adults. J Appl Physiol. 1993;74:2214–2218. doi: 10.1152/jappl.1993.74.5.2214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


