Abstract
AIM: To detect aneusomic changes with respect to chromosome 11 copy number in esophageal precancers and cancers wherein the generation of cancer-specific phenotypes is believed to be associated with specific chromosomal aneuploidies.
METHODS: We performed fluorescence in situ hybridization (FISH) on esophageal tissue paraffin sections to analyze changes in chromosome 11 copy number using apotome-generated images by optical sectioning microscopy. Sections were prepared from esophageal tumor tissue, tissues showing preneoplastic changes and histologically normal tissues (control) obtained from patients referred to the clinic for endoscopic evaluation.
RESULTS: Our results demonstrated that aneusomy was seen in all the cancers and preneoplastic tissues, while none of the controls showed aneusomic cells. There was no increase in aneusomy from precancers to cancers.
CONCLUSION: Our results suggest that evaluation of chromosome 11 aneusomy in esophageal tissue using FISH with an appropriate signal capture-analysis system, can be used as an ancillary molecular marker predictive of early neoplastic changes. Future studies can be directed towards the genes on chromosome 11, which may play a role in the neoplastic transformation of esophageal precancerous lesions to cancers.
Keywords: Esophageal cancer, Aneusomy, Chromosome 11, Fluorescence in situ hybridization, Early detection
INTRODUCTION
More than 30% of the adult population exhibits upper gastrointestinal tract disorders associated with symptoms such as regurgitation, heartburn and dysphagia, warranting an endoscopic evaluation. Some of these esophageal pathologies require medication while others can be managed by altering life-style and dietary habits. A certain percentage of these, however, progress into esophageal malignancies. In India, esophageal cancer is the second leading cancer in men and fourth leading cancer in women[1].
Epithelial tumors of the esophagus [squamous cell carcinoma (SCC) and adenocarcinoma (ADC)] are responsible for more than 95% of all esophageal carcinomas. This malignancy presents generally as a locally advanced disease, hence leading to poor prognosis with an average 5-year survival of < 12% in India[2]. Abnormal proliferation of the esophageal epithelial cells with hyperplasia and dysplasia in the normal squamous lining are regarded as premalignant lesions[3,4]. Another common premalignant condition is Barrett’s esophagus (BE), where patients have a forty-fold increased risk for developing adenocarcinoma as compared to normal individuals. Although significant advances have been made in the diagnosis and treatment of esophageal carcinomas, not many studies have evaluated markers in the target tissue, in association with increased risk for malignant transformation. Hence, there is a need to identify the genetic and molecular factors responsible for the progression of these esophageal lesions into malignancy.
Neoplastic progression is a complex multistep process associated with gross chromosomal alterations and mutations in regulatory genes, culminating in tumorigenesis. Genomic instability is a prominent feature of most cancers, wherein aneuploidy, a change in chromosomal number caused by unequal partitioning of chromosomes during cell division, occurs frequently in many solid tumors[5,6]. Aneuploidy then generates specific aneusomies autocatalytically, due to errors in chromosome segregation and repair processes[7]. Aneusomies have been reported in non-cancerous conditions such as Down’s syndrome, wherein the extra chromosome 21 is considered to be associated with an increased risk for leukemias. The study of human cancers shows evidence for cancer-specific aneusomies, despite a plethora of unspecific aneuploidies[8,9]. Recently, it has been demonstrated cytogenetically that chromosome 11 may be important in the etiology of SCC of the esophagus[10,11].
The present study was a hospital-based, unmatched, case-control study in 25 cases referred to our clinic from different hospitals in Hyderabad, South India. Chromosome 11 aneusomy was investigated in esophageal biopsies taken from controls, premalignant and malignant lesions, using fluorescence in situ hybridisation (FISH). Our results demonstrated that aneusomy was seen in all the cancers and preneoplastic tissues, while none of the controls showed aneusomic cells. There was no increase in aneusomy from precancers to cancers. Evaluation of chromosome 11 aneusomy in esophageal tissue can be used as an ancillary molecular marker predictive of early neoplastic changes.
MATERIALS AND METHODS
Tissue specimens
Esophageal biopsy specimens were endoscopically resected from patients referred for histopathological evaluation by a qualified gastroenterologist. The control samples were taken from those patients undergoing an endoscopy, which showed normal tissue histology. All samples were included in the study after informed consent was obtained from the patients. The study was approved by our Institutional Ethical Committee. Formalin-fixed, paraffin-embedded tissue sections from 25 selected cases were subjected to FISH analysis subsequent to confirmation by histology.
Histopathological analysis
Paraffin-embedded tissue sections (4 μm thick) were first subject to deparaffinization. Slides were placed in xylene (3 min × 3 min), 100% alcohol (3 min × 3 min), rinsed in running water and stained with haematoxylin (Harries, Merck) for 5-10 min. They were then placed in running water, dipped in 1% hydrochloric acid and subsequently transferred to Eosin yellow staining (Merck) for 30 s. After that the slides were passed through graded alcohol series for dehydration, placed in xylene and mounted in DPX (Ref: Histopathology Laboratory, Armed Forces Institute of Pathology, Washington DC, 20 305, USA). Suitable images of the required areas from representative tissue sections were taken using a CCD camera (Figure 1).
Figure 1.
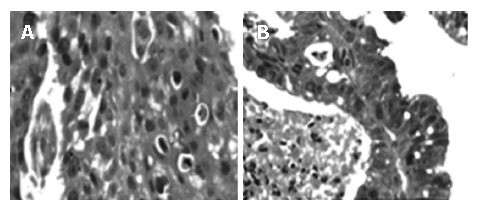
Representative HE stained sections at × 400 magnification selected for FISH analysis. A: Neoplastic streaks with eosin stained keratin pearls indicating a well-differentiated squamous cell carcinoma; B: Adenocarcinomatous tissue showing columnar epithelial replacement of the normal squamous lining.
Based on the endoscopic and histopathological evaluation, 25 tissue biopsies were selected for FISH analysis using a Spectrum Green labeled, centromere enumeration probe (CEP) for chromosome 11 [Vysis India Ltd] (This was used instead of the LSI probes that would help evaluate gene amplification).
FISH on esophageal tissue sections
Paraffin sections of 4 μm thick were deparaffinized in an oven at 95°C for 20 min, then immediately placed in xylene (3 min × 3 min) and transferred into 100% ethanol (3 min × 5 min). Dried slides were incubated in 2 × SSC solution at 75°C for 10 min followed by treatment with proteinase K solution (2 mg/mL) at 37°C for 15 min. The slides were rinsed in 2 × SSC solution at room temperature. They were then immersed in a 75°C denaturant bath (70% formamide/2 × SSC) for 5 min and dehydrated in gradient ethanol. Dry slides were placed on a 45-50°C slide warmer. Probe mixture was simultaneously prepared at room temperature (7 μL of hybridization buffer + 1 μL Spectrum Green labeled CEP 11 DNA probe + 2 μL purified double distilled H2O) and then denatured at 95°C. Ten microliters of the probe mix were applied to the slide, and a coverslip was placed on it immediately. The slides were hybridized in a pre-warmed humidified chamber overnight (12-16 h) at 37°C. Post-hybridization washes were done with freshly prepared 0.4 × SSC/0.3% NP-40 solution at 55°C followed by 2 × SSC/0.1% NP-40 at room temperature. The slides were then air-dried in the dark. Ten microliter DAPI counterstain was applied to the target area and a coverslip was placed on it carefully to avoid formation of air bubbles.
The slides were viewed under a fluorescence microscope (Olympus, Optical sectioning microscope attached to an Axioplan imaging Apotome apparatus, Zeiss, Germany) using a suitable filter set (DAPI Exc: 367nm; Emi: 452 nm and Spectrum Green Exc: 509 nm; Emi: 538 nm). The optical sectioning microscope provided a high-quality image enhancement required for proper signal visualization. The sections were visualized through Optical sectioning mode using the Apotome for the best probe signals in a 3D mode. Ten to fifteen areas per slide were taken for analysis based on the density of the nuclei. Each area allowed optical sectioning of about 8-10 consecutive sections using the apotome. The images were captured at 1500 × magnification with oil immersion. Analysis was done using the 3D analysis of the Axiovision Apotome 1 imaging and Adobe Photoshop 7.0 version softwares.
Criteria for evaluating FISH signals
Data were scored from areas showing uniform fluorescence intensity. We screened a large number of nuclei per sample in order that we did not miss any aneusomic cells, especially with respect to precancer tissues. In each case, clear, distinct FISH signals were evaluated by counting 250 non-overlapping nuclei. Only those samples in which the artifactual nullisomy (negative nuclei) did not exceed the prescribed 25% were chosen for evaluation. A baseline frequency of monosomic population was established to control ‘truncation’ artifacts resulting from cut nuclei during microtomy.
RESULTS
From the patients with upper GI tract disorders referred for endoscopic evaluation, 25 cases were selected into the FISH study based on endoscopic and histopathological categorization. Of these 25 cases, 20 gave analyzable results; they included five normal tissues, four precancers (two squamous dysplasias and two BE with high-grade dysplasia) and eleven esophageal cancers as shown in Table 1.
FISH was performed on a few metaphase cells and, in conjunction with G-banded chromosome analysis, we confirmed the probe hybridization. Subsequently the method was applied to esophageal tissue sections obtained from paraffin blocks. Optical sectioning generated 3D images were evaluated and screened for chromosome 11 probe signals (Figure 2).
Figure 2.
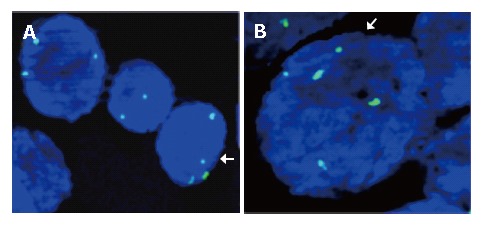
Images showing aneusomy with regard to chromosome 11 in esophageal cancer cells. A: Trisomy and tetrasomy chromosome 11 in the same area; B: Pentasomy chromosome 11 with normal disomy.
The results in each sample were tabulated as nullisomy (absence of signals), monosomy (single signal), normal disomic condition (two signals), trisomy (three signals), tetrasomy (four signals) and pentasomy (five signals). Esophageal tumor cells with trisomic, tetrasomic and pentasomic signals indicating the presence of extra chromosome 11 are shown in Figure 3.
Figure 3.
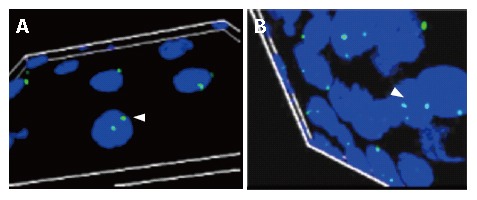
Axiovision Apotome software generated 3D-image from overlay of 10 consecutive images captured by the Optical sectioning microscopy. A: A portion of the esophageal tissue section from the control showing normal disomy of chromosome 11 (indicated by an arrow); B: Tissues from patients with well-differentiated squamous cell carcinoma (arrow indicating cell with trisomy for chromosome 11).
Artifactual nullisomy was found to be less than 15% in all 20 cases studied. Monosomy was seen in < 35% of the cells in all three groups of controls, precancers and cancers; this was not included in the analysis as it was considered as a technical artifact. The controls showed normal disomy in all the remaining cells analyzed. Premalignant tissues showed an increase in the copy number of chromosome 11; all the four precancer cases showed trisomy of chromosome 11 (4%, 10%, 15.2%, 5.6%); of these, one case with severe squamous dysplasia exhibited tetrasomy (0.4%), and a case of BE with high grade dysplasia showed tetrasomy and pentasomy of chromosome 11 (2.4% and 0.8%), respectively (Table 1).
Table 1.
Details of cases analyzed for chromosome 11 aneusomy using FISH in different esophageal pathologies and controls
| Case, No. | Case type | Details |
Chromosome 11 % aneusomy |
||
| Trisomy (%) | Tetrasomy (%) | Pentasomy (%) | |||
| 1 | Controls | Normal | - | - | - |
| 2 | Normal | - | - | - | |
| 3 | Normal | - | - | - | |
| 4 | Normal | - | - | - | |
| 5 | Normal | - | - | - | |
| 6 | Precancers | Sq. dysplasia (High grade) | 4.0 | - | - |
| 7 | Sq. dysplasia (High grade) | 10.0 | 0.4 | - | |
| 8 | Barrett's dysplasia (High grade) | 15.2 | 2.4 | 0.8 | |
| 9 | Barrett's dysplasia (High grade) | 5.6 | - | - | |
| 10 | Cancers | ADC | 12.8 | 2.0 | - |
| 11 | ADC | 10.0 | 2.0 | - | |
| 12 | WDSCC | 11.2 | - | - | |
| 13 | WDSCC | 12.8 | 0.4 | - | |
| 14 | WDSCC | 8.8 | 2.4 | - | |
| 15 | MDSCC | 10.0 | 0.8 | - | |
| 16 | MDSCC | 12.8 | 0.4 | 0.4 | |
| 17 | MDSCC | 5.2 | 0.8 | - | |
| 18 | PDSCC | 1.6 | - | - | |
| 19 | PDSCC | 6.0 | - | - | |
| 20 | PDSCC | 3.2 | - | - | |
Percentage of aneusomy in different cases from the first 250 cells scored. (Monosomies and normal disomies are not shown in the table). ADC: Adenocarcinoma; WDSCC: Well differentiated SCC; MDSCC: Moderately differentiated SCC; PDSCC: Poorly differentiated SCC; Sq. : Squamous.
All eleven cancer cases showed varying degrees of aneusomy of chromosome 11 in the affected tissues. Both cases of adenocarcinoma showed trisomy and tetrasomy. Of the remaining SCC cases, 4/9 showed only trisomy, 5/9 also showed tetrasomy and 1/9 also exhibited pentasomy of chromosome 11 (Table 1).
DISCUSSION
Chromosomal instability leading to aneuploidy and subsequently, specific aneusomies is characteristic of cancers[7,12]. In spite of the somatic gene-mutation hypothesis supporting the role of oncogenes and tumor suppressor genes in carcinogenesis, it has not been possible to disprove the century-old aneuploidy hypothesis even today. Cancer independent evidence suggests that specific aneusomies encode the phenotypes of irreversible precancerous lesions and are sufficient to alter the phenotype of eukaryotic cells to trigger cellular transformation[13-16]. Given this, it follows that chromosome number mutation, not gene mutation alone, is a probable cause of many dominant cancer cell phenotypes. Aneusomy can alter the dosage and thus the relevant activities of thousands of genes on the chromosome involved. Dividing aneuploid cells become increasingly unstable and most of these cells die eventually; rarely, these generate a specific aneusomy which then promotes the cell toward neoplastic transformation.
Specific chromosomal aneuploidies are characteristic of solid tumors unlike the random aneuploidies exhibited in hematological malignancies[9,17]. Chromosome 11 has a number of oncogenes and tumor suppressors including Cyclin D1, which is overexpressed in a wide variety of human neoplasms[18,19]. Reports suggest that overexpression of cyclin D1 is not due to gene amplification but due to chromosome number changes[5]. Other studies show the partial segmental aneusomy involving chromosome 11 in esophageal cancers, indicating its probable role in the disease etiology[10,11]. In the present work, FISH was used to assess change in chromosome 11 copy number in esophageal tumors using centromeric DNA probes.
Interestingly, our results showed that 100% of the cancerous lesions of the esophagus were aneusomic for chromosome 11. These ranged from trisomies to pentasomies in the same sample, indicating heterogeneity in the tumor tissue (Table 1). The baseline monosomy established (due to experimental artifacts) was similar in all controls, precancers and cancers. This is the first study to report that chromosome 11 copy number is altered in both esophageal SCC and ADC tumors. Because the number of ADC cases was small, there was no significant difference in the levels of aneusomy between SCC and ADC tumors. None of the controls showed any aneusomy suggesting that this chromosomal alteration is associated only with the neoplastic changes.
In addition, we also made a detailed investigation regarding the esophageal pathologies in each sub-group (though small in number) and the aneusomy in them.
There have been very few reports of specific aneusomies in esophageal premalignant lesions; using FISH, some studies showed hyperdiploidy involving chromosomes 4 and 8 in BE, aneuploidy in chromosome 11 in esophageal tumors and Barrett’s dysplasias[20-23]. In the present work, all the precancerous lesions including BE with high-grade dysplasia exhibited aneusomy of chromosome 11. Our data indicate that levels of aneusomy of chromosome 11 seemed to occur increasingly in preneoplastic tissues suggesting that there may be genes on chromosome 11 that have a role in the initiation and neoplastic transformation of esophageal lesions.
Cytogenetic analysis by conventional chromosomal banding is labor-intensive and time-consuming. Besides, it is difficult to analyze some of the complex karyotypes characteristic of many human tumors. On the other hand, FISH is a relatively more convenient and quick method for evaluating tissue cell chromosomal changes[24,25]. Moreover, we have observed that by using adequate fluorescent signal capture (optical sectioning microscopic method) and analysis systems, an accurate estimate of the changes in chromosome copy number could be obtained from tumor tissues, based on which we could make satisfactory interpretations. Our study supports the finding that solid tumor cancers are often aneuploid for specific chromosomes and this may be used as an ancillary marker for detecting early changes associated with cancer.
Epigenetic changes like DNA methylation may lead to chromosomal instability, activation of endogenous parasitic sequences and mutations[26]. Cancer cells undergo methylation changes resulting in overexpression/silencing of several genes that are responsible for maintaining chromosomal integrity. Our study also revealed that 4/11 cancers and 2/5 precancers with increased copies of chromosome 11 had a hypermethylated hMLH1 repair gene promoter known to be responsible for altered repair efficiency (data not shown).
We did not perform any studies to assess whether this aneusomy correlates with the DNA content aneuploidy (i.e. gross chromosomal instability) of the tissues. The sample number (with a mix of ADCs and SCCs, which might have different etiologies), though, was not sufficient to make conclusions about predictive values, the results showed the differences in the frequencies of aneusomy in the two types of tumor. The present work clearly indicates that aneusomy is observed in early esophageal lesions and may be involved in neoplastic transformation. Aneusomy of other chromosomes cannot be ruled out, however here, chromosome 11 is clearly demonstrated in all the esophageal pathologies studied. This marker merits investigation in a larger number of cases to determine its potential as a predictive molecular marker for an increased risk for malignant transformation.
ACKNOWLEDGMENTS
We acknowledge help from Gastroenterology Units of Bhagwan Mahavir Medical Research Center, AC Guards, Hyderabad, Osmania General Hospital, Afzalgunj, Hyderabad and Kamineni Hospitals, LB Nagar, Hyderabad. We thank Mr. Srinivas and Mr.Mallesh, technicians of the Department of Pathology for their assistance with histopathology.
COMMENTS
Background
More than 30% of the adult population exhibits esophageal tract disorders, where a certain proportion of chronic cases even develop malignancy. Markers for early detection, which will help improve survival and treatment response are still lacking. This study investigates specific aneusomies which are believed to be associated with solid cancer phenotypes.
Research frontiers
Current biopsy surveillance programs are based on histopathological assessment of the tissue. However, molecular changes precede visible histopathological changes in cancer. FISH-based assay using brush cytology specimens (more easily accessible than biopsy) are increasingly used for investigation of chromosomal alterations and after validation these molecular markers can be used for routine surveillance in order to aid in early detection.
Innovations and breakthroughs
FISH, when compared to conventional cytogenetic analysis, is a more convenient and quick method for evaluating tissue cell chromosomal changes. An apotome-attached optical sectioning microscope to capture fluorescent probe signals has shown maximum efficiency for signal analysis in tissue FISH. This will aid in satisfactory data collection and interpretation.
Applications
Chromosome 11 aneusomy has been demonstrated in all the cases of cancer and precancer studied by us indicating its potential as an early marker for neoplastic transformation. Our results call for evaluating this marker in a larger cohort of patients.
Terminology
Regurgitation: bringing back undigested food from the stomach; Dysphagia: difficulty in swallowing; Dysplasia: Nuclear atypia, loss of normal cell polarity, and abnormal tissue maturation.
Footnotes
S- Editor Liu Y L- Editor Zhu LH E- Editor Liu WF
References
- 1.Gajalakshmi V, Swaminathan R, Shanta V. An Independent Survey to Assess Completeness of Registration: Population Based Cancer Registry, Chennai, India. Asian Pac J Cancer Prev. 2001;2:179–183. [PubMed] [Google Scholar]
- 2.Gupta NM, Jindal R, Prakash O, Gupta R, Bhasin DK. Comparison of the clinical profile and outcome for squamous cell carcinoma and adenocarcinoma of the distal esophagus and cardia in India. Surg Today. 2001;31:400–404. doi: 10.1007/s005950170129. [DOI] [PubMed] [Google Scholar]
- 3.Kagawa Y, Yoshida K, Hirai T, Toge T, Yokozaki H, Yasui W, Tahara E. Microsatellite instability in squamous cell carcinomas and dysplasias of the esophagus. Anticancer Res. 2000;20:213–217. [PubMed] [Google Scholar]
- 4.Lehrbach DM, Nita ME, Cecconello I. Molecular aspects of esophageal squamous cell carcinoma carcinogenesis. Arq Gastroenterol. 2003;40:256–261. doi: 10.1590/s0004-28032003000400011. [DOI] [PubMed] [Google Scholar]
- 5.Lung JC, Chu JS, Yu JC, Yue CT, Lo YL, Shen CY, Wu CW. Aberrant expression of cell-cycle regulator cyclin D1 in breast cancer is related to chromosomal genomic instability. Genes Chromosomes Cancer. 2002;34:276–284. doi: 10.1002/gcc.10072. [DOI] [PubMed] [Google Scholar]
- 6.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 7.Fabarius A, Willer A, Yerganian G, Hehlmann R, Duesberg P. Specific aneusomies in Chinese hamster cells at different stages of neoplastic transformation, initiated by nitrosomethylurea. Proc Natl Acad Sci USA. 2002;99:6778–6783. doi: 10.1073/pnas.251670699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf NG, Abdul-Karim FW, Schork NJ, Schwartz S. Origins of heterogeneous ovarian carcinomas. A molecular cytogenetic analysis of histologically benign, low malignant potential, and fully malignant components. Am J Pathol. 1996;149:511–520. [PMC free article] [PubMed] [Google Scholar]
- 9.Alcaraz A, Takahashi S, Brown JA, Herath JF, Bergstralh EJ, Larson-Keller JJ, Lieber MM, Jenkins RB. Aneuploidy and aneusomy of chromosome 7 detected by fluorescence in situ hybridization are markers of poor prognosis in prostate cancer. Cancer Res. 1994;54:3998–4002. [PubMed] [Google Scholar]
- 10.Yen CC, Chen YJ, Lu KH, Hsia JY, Chen JT, Hu CP, Chen PM, Liu JH, Chiou TJ, Wang WS, et al. Genotypic analysis of esophageal squamous cell carcinoma by molecular cytogenetics and real-time quantitative polymerase chain reaction. Int J Oncol. 2003;23:871–881. [PubMed] [Google Scholar]
- 11.Jin Y, Jin C, Law S, Chu KM, Zhang H, Strombeck B, Yuen AP, Kwong YL. Cytogenetic and fluorescence in situ hybridization characterization of clonal chromosomal aberrations and CCND1 amplification in esophageal carcinomas. Cancer Genet Cytogenet. 2004;148:21–28. doi: 10.1016/s0165-4608(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 12.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 13.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 14.Duesberg P, Li R, Rasnick D, Rausch C, Willer A, Kraemer A, Yerganian G, Hehlmann R. Aneuploidy precedes and segregates with chemical carcinogenesis. Cancer Genet Cytogenet. 2000;119:83–93. doi: 10.1016/s0165-4608(99)00236-8. [DOI] [PubMed] [Google Scholar]
- 15.Lindsley DL, Sandler L, Baker BS, Carpenter AT, Denell RE, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethmann RC, et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb T. When theories collide: experts develop different models for carcinogenesis. J Natl Cancer Inst. 2001;93:92–94. doi: 10.1093/jnci/93.2.92. [DOI] [PubMed] [Google Scholar]
- 17.Mitelman F. Catalogue of chromosome aberrations in cancer. Cytogenet Cell Genet. 1983;36:1–515. doi: 10.1159/000131930. [DOI] [PubMed] [Google Scholar]
- 18.Hui AB, Or YY, Takano H, Tsang RK, To KF, Guan XY, Sham JS, Hung KW, Lam CN, van Hasselt CA, et al. Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res. 2005;65:8125–8133. doi: 10.1158/0008-5472.CAN-05-0648. [DOI] [PubMed] [Google Scholar]
- 19.Segas JV, Lazaris AC, Nikolopoulos TP, Kavantzas NG, Lendari IE, Tzagkaroulakis AM, Patsouris ES, Ferekidis EA. Cyclin D1 protein tissue detection in laryngeal cancer. ORL J Otorhinolaryngol Relat Spec. 2005;67:319–325. doi: 10.1159/000090041. [DOI] [PubMed] [Google Scholar]
- 20.Fahmy M, Skacel M, Gramlich TL, Brainard JA, Rice TW, Goldblum JR, Connor JT, Casey G, Legator MS, Tubbs RR, et al. Chromosomal gains and genomic loss of p53 and p16 genes in Barrett's esophagus detected by fluorescence in situ hybridization of cytology specimens. Mod Pathol. 2004;17:588–596. doi: 10.1038/modpathol.3800088. [DOI] [PubMed] [Google Scholar]
- 21.Doak SH, Saidely D, Jenkins GJ, Parry EM, Griffiths AP, Baxter JN, Parry JM. Generation of locus-specific probes for interphase fluorescence in situ hybridisation--application in Barrett's esophagus. Exp Mol Pathol. 2004;77:26–33. doi: 10.1016/j.yexmp.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Manoel-Caetano Fda S, Borim AA, Caetano A, Cury PM, Silva AE. Cytogenetic alterations in chagasic achalasia compared to esophageal carcinoma. Cancer Genet Cytogenet. 2004;149:17–22. doi: 10.1016/s0165-4608(03)00274-7. [DOI] [PubMed] [Google Scholar]
- 23.Finley JC, Reid BJ, Odze RD, Sanchez CA, Galipeau P, Li X, Self SG, Gollahon KA, Blount PL, Rabinovitch PS. Chromosomal instability in Barrett's esophagus is related to telomere shortening. Cancer Epidemiol Biomarkers Prev. 2006;15:1451–1457. doi: 10.1158/1055-9965.EPI-05-0837. [DOI] [PubMed] [Google Scholar]
- 24.Cortés-Gutiérrez EI, Dávila-Rodríguez MI, Muraira-Rodríguez M, Said-Fernández S, Cerda-Flores RM. Association between the stages of cervical cancer and chromosome 1 aneusomy. Cancer Genet Cytogenet. 2005;159:44–47. doi: 10.1016/j.cancergencyto.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Watters AD, Going JJ, Cooke TG, Bartlett JM. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003;77:109–114. doi: 10.1023/a:1021399923825. [DOI] [PubMed] [Google Scholar]
- 26.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]


