Abstract
Infection by Staphylococcus aureus is on the rise, and there is need for a better understanding of host immune responses that combat S. aureus. Here we use DNA barcoding to enable deep sequencing of the paired heavy- and light-chain immunoglobulin genes expressed by individual plasmablasts derived from S. aureus-infected humans. Bioinformatic analysis of the antibody repertoires revealed clonal families of heavy-chain sequences and enabled rational selection of antibodies for recombinant expression. Of the ten recombinant antibodies produced, seven bound to S. aureus, of which four promoted opsonophagocytosis of S. aureus. Five of the antibodies bound to known S. aureus cell-surface antigens, including fibronectin binding protein A. Fibronectin binding protein A-specific antibodies were isolated from two independent S. aureus infected patients and mediated neutrophil killing of S. aureus in in vitro assays. Thus, our DNA barcoding approach enabled efficient identification of antibodies involved in protective host antibody responses against S. aureus.
1. Introduction
S. aureus, the second-most common bacterial pathogen isolated from humans, is estimated to cause over half a million cases of invasive infection and over 18,000 deaths annually in the United States [1] [2] [3]. The emergence of multidrug-resistant strains, such as methicillin-resistant Staphylococcus aureus (MRSA), has complicated treatment procedures, prompting research into vaccines and novel immunotherapies. Still, how S. aureus interacts with the human immune system is unclear, and no vaccines are currently approved for preventing or treating S. aureus infections [4]. Antibody titers against S. aureus proteins increase during acute infection [5], and patients with immunoglobulin deficiencies are more susceptible to S. aureus infections [6], suggesting that the host antibody response is important for containing S. aureus. However, the critical S. aureus antigens that elicit protective host immunity remain undefined, with little or no correlation observed between disease severity and levels of serological antibodies against known S. aureus virulence factors [7].
Antibodies circulating in the blood are produced by plasma cells and their transient B-cell precursors (termed plasmablasts). After successfully completing antigen-dependent affinity maturation, naïve and memory B cells differentiate into plasmablasts and proliferate rapidly in the blood before trafficking to infected tissues. Although plasmablast frequencies in peripheral blood are typically low before and after antigen stimulation of an immune response (~0.2% of circulating B cells), they can account for over 30% of circulating B cells during an infection [8, 9]. Thus, the expansion, specificity and accessibility of circulating plasmablasts provides a powerful approach to characterize the active B-cell response.
Accurate identification of the affinity matured and expanding B-cell lineages requires methods that can both approximate the frequency of B-cells expressing similar variable-region genes and recover the endogenous heavy- and light-chain variable-region pairings from individual B-cells. Previously published high-throughput approaches that sequence only the heavy-chain or heavy- and light-chain complementarity-determining-region 3 (CDR3) from bulk B-cells and/or do not implement any methods to normalize the relative abundance of sequencing reads to the number of cells expressing a given sequence [10–13], making it difficult to determine whether highly-represented sequences are the result of antigen-driven B-cell clonal proliferation or from sequence biases introduced during PCR amplification. Additionally, the inability to recover heavy- and light-chain pairs precludes the functional characterization of the endogenous antibodies.
To overcome these limitations, we use a novel DNA barcode system to sequence the paired heavy- and light-chain antibody genes expressed by individual plasmablasts, and in doing so to accurately determine the proportion of plasmablasts that express specific germline sequences and somatic mutations. Herein we sequence the endogenous antibodies expressed by individual plasmablasts circulating in the blood of individuals with S. aureus bacteremia, bioinformatically identify clonal families of antibodies sharing heavy and light chain VJ sequences, and recombinantly produce and characterize of the binding and functional properties of representative antibodies.
2. Material and methods
2.1. Single-cell sorting of peripheral blood plasmablasts
Blood was collected from individuals with culture-confirmed S. aureus bacteremia after obtaining informed consent and under human subject protocols approved by the Investigational Review Board at Stanford University. For all individuals, blood specimens were obtained within 24 hours after the initiation of standard-of-care antibiotic treatment. PBMCs were isolated using a Ficoll layer and stained with CD3-V450 (BD 560365), IgA-FITC (AbD Serotec STAR142F or Miltenyi #130-093-071), IgM-FITC (AbD Serotec STAR146F), IgM-APC (BD 551062) or IgM-PE (AbD Serotec STAR146PE), CD20-PerCP-Cy5.5 (BD 340955), CD38-PE-Cy7 (BD 335808), CD19-APC (BD 340437) or CD19-Brilliant Violet 421 (Biolegend 302233), and CD27-APC-H7 (BD 560222). IgG+ plasmablasts were gated on CD19+/intCD20-CD27++CD38++IgA-IgM- cells and individually sorted using a BD FACSAria III into a 96-well PCR plate containing a hypotonic lysis buffer (10mM Tris-HCl pH 7.6) containing 2 mM dNTPs (NEB), 5 µM oligo(dT)20VN, and 1 unit/µL of Ribolock (Fermentas), an RNase inhibitor.
2.2. Reverse transcription (RT) and Polymerase Chain Reaction (PCR) with well-ID and plate-ID adaptors
We added 6 mM MgCl2 with Ribolock, Superscript III (Life Technologies), and 1 µM final concentration of the appropriate well-ID oligonucleotide barcode to sorted plasmablast plates and performed RT at 42°C for 120 minutes. RT products from each plate were pooled, extracted with phenol-chloroform-isoamyl alcohol, and concentrated with Amicon Ultra-0.5 30 kDa (Millipore) units.
We used Phusion Hot Start II DNA polymerase (NEB/Fermentas) for both the first PCR (PCR1) and the second PCR (PCR2). The following PCR reaction conditions were used: 200 µM of dNTPs, 0.2 µM of primers, 0.2 U of Phusion polymerase, 4% DMSO, and 2 µL of RT product. PCR1 was performed with forward (FW) primers containing, at their 5’ end, a plate-ID barcode oligonucleotide, as well as a 454 Titanium adaptor, and with gene-specific reverse primers (GSP) for amplifying the gamma, kappa, and lambda chains. PCR2 was performed with FW primers containing a 454 Titanium adaptor at their 5’ end and reverse GSP containing a plate-ID barcode oligonucleotide and a 454 Titanium adaptor at their 3’ end.
We pooled the amplified DNA, gel purified them, and purified them with Ampure XP beads (Beckman Coulter). Amplicon concentrations were determined by using Picogreen DNA assay kits (Invitrogen), and amplicons were sent to Roche for 454 sequencing.
2.3. Bioinformatic analysis and generation of dendrograms
Sff output files from 454 sequencing, containing sequences and quality scores for each nucleotide, were read into Python by using the Biopython package, and sequences were grouped and parsed into separate sff files on the basis of their plate- and well-ID combinations. Consensus sequences corresponding to plasmablast heavy- and light-chain sequences were generated using Newbler 2.6. Where multiple assemblies occurred per well, an assembly was accepted if it represented >50% of all reads in the given well. Otherwise, we disregarded all reads from that well to preclude instances where a well contained more than one cell and to account for sequencing errors introduced during pyrosequencing.
To assign V(D)J families for consensus sequences, we analyzed heavy- and light-chain sequences with the IMGT HighV-QUEST15 database[14] to predict germline allele usage, germline sequence recombination, and shared mutations relative to the germline sequence.
To generate dendrograms, we first binned heavy- and light-chain sequences according to their V-gene usage and aligned them with Muscle28 [15]. They were clustered with PhyML maximum-likelihood clustering and rooted by their germline V gene [16]. Each V-gene phylogenetic tree was then arranged by heavy-chain V-gene families and drawn by using ETE30 [17].
2.4. Cloning and expression of recombinant antibodies
Antibodies representative of the heavy-chain families were selected for expression on the basis of (1) having a corresponding light-chain plate- and well-ID combination and (2) being the heavy- and light-chain pair most representative of the consensus sequence of the family. Gamma heavy chains were inserted into vector pEE6.4 (Lonza), and kappa and lambda light chains were inserted into vector pEE12.4 (Lonza). We used well-ID-specific FW primers and GSP reverse primers to selectively amplify cDNA for insertion into the expression vectors. Heavy- and light-chain sequences were cloned by first inserting the sequences of the antibody constant regions into the vector and inserting the leader and V(D)J sequences by using standard molecular biology techniques.
Expression of recombinant antibodies was performed by dual transfection of heavy-chain-containing pEE6.4 constructs and light-chain-containing pEE12.4 constructs in 293T cells, using Lipofectamine 2000 (Life Technologies). Antibodies were purified from culture supernatants by using Protein A Plus agarose beads (Pierce), eluted with IgG elution buffer (Pierce), and quantified by BCA Protein Assay (Thermo Scientific).
2.5. Bacterial culture and preparation
S. aureus Wood46 (ATCC 10832) were grown in LB to early-logarithmic (0.25 optical density units (ODU)), mid-logarithmic (0.5 ODU), or stationary (2.0 ODU) growth phases, as determined by spectrophotometry at 550 nm. We pelleted bacteria by spinning at 5000g for 10 minutes, and resuspended them in 0.1% sodium azide in PBS to halt bacterial metabolism and to capture snapshots of the protein expression.
For experiments in which S. aureus grown in the presence of fibroblasts, NIH/3T3 mouse embryonic fibroblasts (ATCC CRL-1658) were cultured in DMEM (ATCC 30-2002) supplemented with 10% FBS, 0.6 µg/ml L-glutamine, 0.2 U/ml penicillin, and 0.2 µg/ml streptomycin and stimulated with 300 pM recombinant mouse TGF-β1 (R&D Systems) in culture media minus antibiotics for 72 hours. S. aureus Wood46 were grown to stationary phase in LB resuspended in DMEM, and incubated with NIH/3T3 cells for 2 hours. NIH/3T3 cells were then lysed with hypotonic buffer and washed, and bacterial cells were resuspended in 0.1% sodium azide in PBS.
For experiments with E. coli, DH10B E. coli were grown in LB for 14 hours, pelleted, and resuspended in 0.1% sodium azide in PBS.
2.6. S. aureus lysate dot-blot analysis
To prepare S. aureus lysates for dot-blot, 0.25 g of growth phase-specific bacteria in sodium azide (dry weight) was incubated with B-PER lysis solution (Thermo Scientific), 1x Halt protease inhibitor, and 0.05 µg/ml freshly prepared lysostaphin (Sigma) for 35 minutes at 37°C, then for 15 minutes at ambient temperature with shaking. Samples were centrifuged at 15,000g for 15 minutes at 4°C, and the supernatants were collected and quantitated by BCA Assay.
Dot-blot was performed on 0.45 µm nitrocellulose transfer membrane with the Bio-Dot Microfiltration Apparatus (Bio-Rad) as per manufacturer’s instructions. 500 ng lysate in 50 µl TBS was spotted per well and blocked with 3% BSA-TBS. Recombinant test antibodies were diluted to 2 µg/ml in 0.1% TBST with 3% BSA and added to appropriate wells. 1:15,000 goat anti-human IgG-Fc HRP (Bethyl Laboratories) secondary antibody was used for detection, and dot-blots were visualized with West Femto Substrate (Thermo Scientific).
2.7. Flow cytometric analysis of antibody binding
We blocked sodium-azide-treated S. aureus Wood46 or DH10B E. coli cells with 1% BSA-PBS for 45 minutes at 4°C, then added 10 µg/ml of recombinant test antibodies in 1% BSA-PBS for 60 minutes at 4°C, washed three times with 1% BSA-PBS, and incubated for 20 minutes at 4°C in the dark with 1:5 mouse anti-human-IgG PE secondary antibody (BD Pharmingen) diluted 1:5. Cells were washed three times with PBS, and antibody surface binding was detected with an LSR Fortessa.
2.8. S. aureus antigen array analysis
A full-proteome microarray of the S. aureus USA300 strain—a strain prevalent in community-acquired S. aureus [18]— containing approximately 2,700 S. aureus proteins per array (Antigen Discovery Inc, Irvine, CA) was used to determine the specificity of recombinant antibodies derived from S. aureus infected humans.
2.9. S. aureus antigen ELISA
We used ELISA to determine whether the recombinant antibodies derived from bacteremic individuals bind to purified S. aureus antigens. We coated Nunc Maxisorp plates with 1.5 µg/ml lipoteichoic acid or peptidoglycan purified from S. aureus (Sigma Aldrich) in pH 7.4 PBS and blocked the plates overnight with PBS containing 1% BSA. Recombinant antibodies were added at 1 µg/ml and diluted serially 3x. After washing with PBST (PBS with 0.05% Tween20), antibody binding was detected by using an HRP-conjugated goat anti-human IgG-Fc antibody and TMB substrate. Absorbance was measured with a SpectroMax M5 spectrophotometer at 450 nm.
2.10. Flow cytometric analysis of S. aureus opsonophagocytosis
To label sodium-azide-treated S. aureus for detection of bacterial internalization, we washed and resuspended the bacteria in PBS, and then incubated them with 2 µl of CellTrace CFSE (Invitrogen) for 30 minutes at 37°C with constant shaking at 250 rpm. CFSE-staining was stopped by incubating the cells for 5 minutes with 500 µl ice-cold PBS and washing them three times in PBS.
We cultured THP-1 human monocyte-like cells (ATCC TIB-202) in RPMI-1640 supplemented with 10% FBS, 0.6 µg/ml L-glutamine, 0.2 U/ml penicillin, and 0.2 µg/ml streptomycin at a density below 5x105 cells/ml to minimize macrophage differentiation[19].
To assess opsonophagocytosis, we incubated 2x106 CFSE-labeled S. aureus for 60 minutes with 10 µg/ml antibody in 1% BSA-PBS, washed them three times , and resuspended them in pre-warmed 25 µl RPMI-1640 with 1% FBS. 100 µl THP1 cells were then added to the CFSE-labeled S. aureus and incubated for 40 minutes at 37°C with shaking at 200 rpm. Phagocytosis was stopped by adding 1 ml ice-cold PBS to the cell mixture and centrifuging it at 350g for 3 minutes at 4°C. Cell-surface and extracellular fluorescence was quenched by resuspending the cell pellet with 200 µl ice-cold Trypan Blue (2 mg/ml in PBS) in 2% paraformaldehyde [20]. >10,000 THP1 cellular events were counted with an LSR Fortessa.
2.11. Neutrophil bactericidal assay
HL-60 cells were cultured with RPMI-1640 supplemented with 10% FBS, 0.6 µg/ml L-glutamine, 0.2 U/ml penicillin, 0.2 µg/ml streptomycin, and 50 µM β-mercaptoethanol. HL-60 cells were differentiated for 96 hours in culture media containing 1.3% DMSO and 2.5 µM all-trans retinoic acid as previously described [21] and resuspended in HBSS supplemented with 10% human AB serum at 106 cells/ml.
S. aureus Wood46 were grown in LB overnight at 37°C, resuspended in HBSS with 10% human AB serum at 106 bacteria/ml, and 10 µg/ml recombinant test antibody was added to the bacteria. HL-60 cells and bacteria were then added at a 1:1 volume ratio and incubated for 1 hour at 37°C with shaking. Three 1:10 serial dilutions of the HL-60:bacteria mixture were performed in PBS. Bacteria were plated on TSA plates (BD), grown at 37°C for 16 hours, and bacterial colony-forming units were counted.
3. Results
3.1. Plasmablasts are elevated during S. aureus acute bacteremia
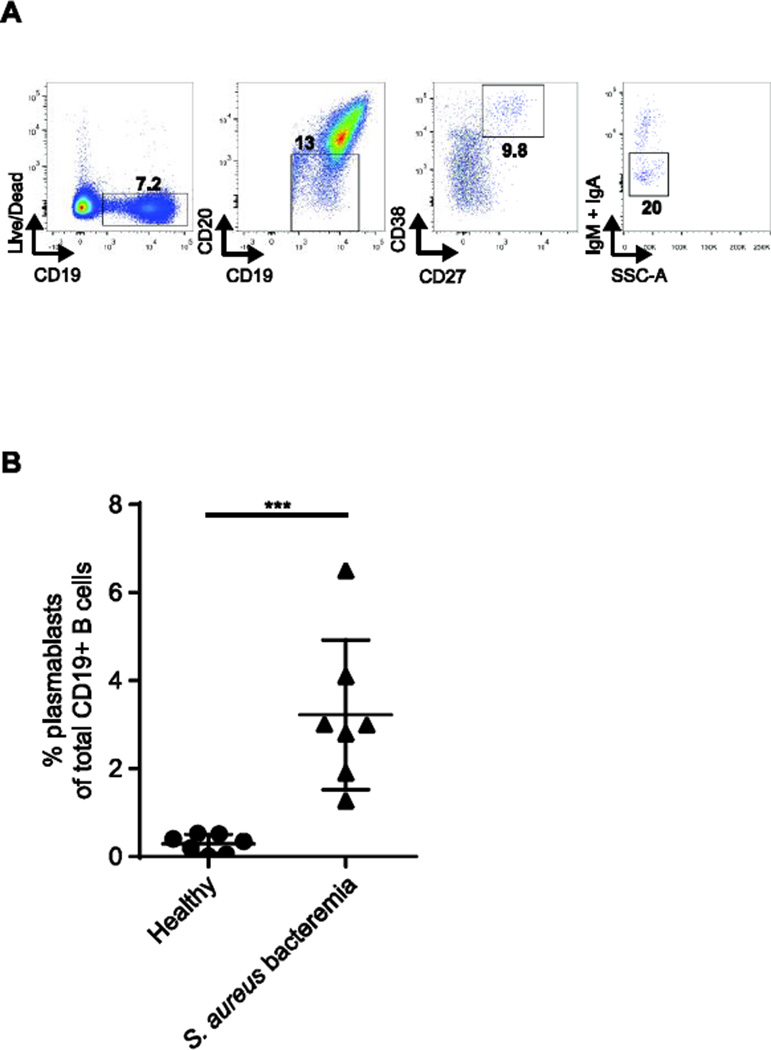
Previous studies detected transient elevations of peripheral blood plasmablasts that peak approximately 7 days following an immune challenge [9]. We investigated whether patients with community-acquired S. aureus bacteremia and acute clinical manifestations exhibit elevated plasmablasts (CD19+/int/CD20-/CD27++/CD38++) in peripheral blood. All individuals with S. aureus bacteremia had greater than 1% plasmablasts per total peripheral blood B cells, with a mean of 3.2% (Fig. 1A and 1B).
Figure 1.
Plasmablasts are elevated during acute S. aureus bacteremia. (A) Fluorescenceactivated gating strategy for single-cell sorting IgG-secreting plasmablasts from PBMCs of S. aureus-infected individuals. Plasmablasts were gated on CD19+/intCD20−CD27++CD38++IgA−IgM− live B cells. (B)
3.2. IgG plasmablast single-cell sequencing reveals expression of antibody families with shared gene segments
To obtain paired heavy and light chain antibody sequences from individual single-cell sorted IgG+ plasmablasts, we utilized a DNA barcoding method that our laboratory developed . This method utilizes the dual function of Superscript III reverse transcriptase to synthesize cDNA from the mRNA of single cells, and to thereby enable tagging of the newly generated cDNA with well-specific (and thus cell-specific) oligonucleotide barcodes at the 5' end of mRNA via template-switching [23]. Primers containing plate-specific barcodes were then added and hybridized to a universal linker sequence flanking the 5’ end of the well-IDs. The resulting double-tagged amplicons were sequenced by 454 GS FLX+, generating ~800,000 reads per run with a mean read length of 600 base-pairs (bp) for heavy chains and 480 bp for light chains. These lengths were sufficient for the recovery of the entire V(D)J region of heavy- and light-chains spanning the Framework 1 and Framework 4 conserved regions.
After discarding any incomplete VH and VL sequence assemblies, we generated dendrograms of each patient’s plasmablast heavy-chain (VH dendrograms) and paired heavy- and light-chain (VH:VL dendrograms) repertoire to visualize the number of antibodies using an identical germline IGHV genes, which we define as an IGHV families (Fig. 2 and Supplemental Fig. S1). Because the VH dendrograms contained more sequences than the paired-chain dendrograms, we focused on plasmablast heavy-chain dendrograms in determining which immunoglobulin rearrangements were preferentially selected for in response to S. aureus infection.
Figure 2.
Heavy-chain dendrograms of the plasmablast antibody repertoires of S. aureus-bacteremic individuals and shared HC V-J analysis. Each peripheral node depicts a sequenced VH region with a unique plate and well-ID barcode combination. Plasmablast antibody repertoires of two individuals with S. aureus bacteremia: (A) patient 1 and (B) patient 2. Red lines indicates the IGHV families with greater than 5 recovered full-length VH sequences, from which analysis of shared IGHJ was performed. Pie charts of the IGHJ gene usage for each IGHV family are displayed adjacent to the branch representing each IGHV. For presentation, red bolded lines indicate the IGHV families where a prominent IGHJ gene was used. S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10 denote the recombinant antibodies that were selected for recombinant expression. Circles denote antibodies that bind S. aureus, and squares denote antibodies that do not. Recombinant antibodies from shared gene families are denoted in blue, and antibodies not sharing gene rearrangements are denoted in green.
For patients 1 and 2, who mounted robust plasmablast responses and successfully cleared their clinical infections (and from whom we obtained 120 and 125 full-length VH sequences, respectively), we analyzed the extent to which IGHV families containing more than five VH sequence assemblies also used the same IGHJ genes (Fig. 2). In patient 1, nine IGHV families contained more than five recovered VH sequences, and five of these IGHV families used the same IGHJ gene in at least 50% of the sequences (Fig. 2A). Of the nine IGHV families, IGHV3-7 had the highest percentage of shared germline IGHV and IGHJ usage (64% IGHJ4). Notably, five of the seven VH sequences in the IGHV5-a family in patient 1 used IGHJ6 and had highly similar somatic mutations in both the heavy- and light-chain variable regions, suggesting that the five plasmablasts containing these VH sequences may have arisen from the same B-cell lineage (Supplemental Fig. S2). Patient 2 displayed greater heterogeneity in the combination of IGHV and IGHJ genes used in the rearranged sequences. Six IGHV families contained more than five VH sequences and family IGHV3-30 had the highest percentage of shared germline IGHJ usage (40% IGHJ4) (Fig. 2B).
3.3. Rational selection of highly represented antibody sequences for recombinant antibody expression
Because we observed that certain IGHV and IGHJ alleles were combinatorially selected for during the response to S. aureus infection and thus appeared more frequently in the antibody repertoire, we asked whether antibodies encoded by these sequences exhibited anti-S. aureus binding. We selected representative antibodies from IGHV families with highly shared IGHJ for recombinant human monoclonal IgG expression (S3, S4, S5, S8, S10) (Supplemental Fig. S1 and Table 1). For comparison, we also expressed five VH:VL pairs that did not contain shared intra-patient IGHV and IGHJ usage (S1, S2, S6, S7, S9), which we term singletons.
3.4. Recombinant antibodies from families with shared IGHV-IGHJ usage are enriched for anti-S. aureus binding
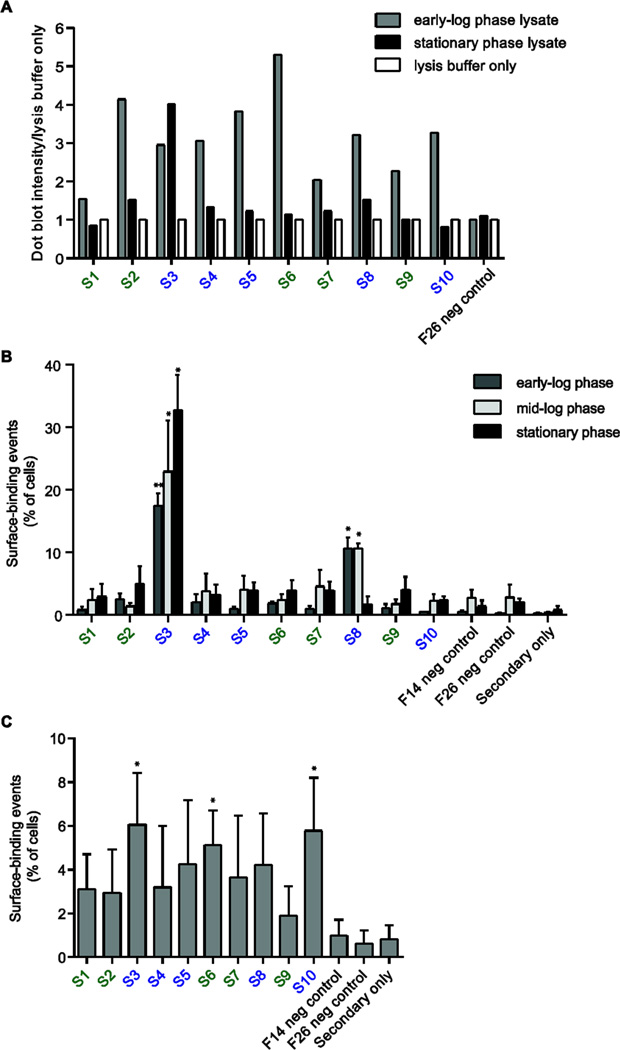
To determine whether the recombinant antibodies we expressed could bind to S. aureus, we first performed dot-blots using crude lysates of the Wood46 strain of S. aureus. The Wood46 strain of S. aureus contains only low levels of protein A, thereby minimizing non-specific binding to immunoglobulin. Because bacterial gene expression differs significantly between early logarithmic (early-log) and stationary growth phases [24], we assessed antibody binding to lysates from bacteria at both growth phases. Quantitative dot-blot analysis revealed that the antibodies S2, S4, S5, S6, S8, and S10 bound to S. aureus in the early-log growth phase with at least three-fold greater intensity as compared to the lysis buffer alone, whereas only S3 bound strongly to S. aureus in the stationary phase (Fig. 3A).
Figure 3.
Screening the binding activity of recombinant antibodies generated from the plasmablasts of S. aureus-infected individuals. (A) Dot-blot analysis of antibody binding to lysates prepared from S. aureus Wood46 at early-log and stationary growth phases. Data are dot-blot densitometry values shown as a fold increase relative to control (lysis buffer only). (B,C) Flow cytometric detection of antibody binding to the surface of S. aureus Wood46 at different growth phases in LB (B) or in the presence of NIH/3T3 fibroblasts (C). Recombinant antibodies from shared gene families are denoted in blue, and antibodies not sharing gene rearrangements are denoted in green. Influenza-specific antibodies F14 and F26 were used as isotype controls. Data in (B) and (C) are shown as the mean ± s.e.m. of triplicates and are representative of three or more independent experiments *P < 0.05 by two-tailed unpaired t-test, comparing binding of S. aureus recombinant antibodies to that of the isotype control antibodies.
We next used flow cytometry to determine whether any of the recombinant antibodies could bind to the surface of the S. aureus Wood46 strain at three different growth phases. We found that antibody S3 bound to bacteria at all growth phases, whereas S8 bound to bacteria at early-log and mid-log phases, but not at the stationary phase (Supplemental Fig. S3). Antibodies S2, S4, S5, S6 and S10, which bound S. aureus in the dot-blot assay, did not bind S. aureus in any of the growth phases using medium alone (Fig. 3B).
During infection, S. aureus often adhere to connective tissues and extracellular matrix proteins [25]. Fibroblasts are the most abundant cell type in soft connective tissue and secrete many of the factors that comprise the extracellular matrix [26]. Therefore, we sought to determine whether culturing S. aureus in the presence of fibroblasts more closely resembles the environment of S. aureus colonization and thereby induces differential regulation of S. aureus surface antigens. Flow cytometric analysis of S. aureus Wood46 grown in the presence of TGF-β-stimulated NIH/3T3 fibroblasts revealed that not only the S3, but also the S6 and S10 recombinant antibodies, can bind robustly to the bacterial surface (Fig. 3C and Supplemental Fig. S3). None of these antibodies bind to E. coli (Supplemental Fig. S3). Importantly, in all surface-binding experiments, two recombinant antibodies that recognize the trivalent inactivated flu vaccine did not bind to S. aureus, indicating the specificity of the antibody binding.
Collectively, our antibody-binding analyses show that all five antibodies from clonal families with shared intra-patient IGHV and IGHJ usage (i.e., S3, S4, S5, S8, and S10) bind to S. aureus, whereas only two of the five antibodies from families lacking common IGHV and IGHJ usage (i.e., S2 and S6) bind to S. aureus (Table 1). Further, our results demonstrate differential binding of individual antibodies to S. aureus at different growth phases.
Table 1.
Summary of recombinant antibodies derived from S. aureus bacteremic patients.
| Monoclonal Antibody a |
Selection Rationale |
HC+LC (HC) V-J mutation count b |
Binds S. aureus? |
Antigen(s) c | Enhances phagocytosis / killing |
|---|---|---|---|---|---|
| S3 | Shared HC V-J | 29 (21) | Yes | LTA | Yes |
| S4 | Shared HC V-J | 33 (26) | Yes | SSL5 | No |
| S5 | Shared HC V-J | 45 (34) | Yes | Unknown | - |
| S8 | Shared HC V-J | 60 (38) | Yes | Unknown | Yes |
| S10 | Shared HC V-J | 21 (16) | Yes | FnBPA | Yes |
| S1 | Singleton | 28 (22) | No | - | - |
| S2 | Singleton | 60 (32) | Yes | PGN | No |
| S6 | Singleton | 28 (18) | Yes | FnBPA, PGN | Yes |
| S7 | Singleton | 34 (19) | No | - | - |
| S9 | Singleton | 54 (35) | No | - | - |
Recombinant monoclonal IgG antibodies were generated from two patients with S. aureus infections. S2, S3, S8, S9, and S10 were derived from patient 1. S1, S4, S5, S6, and S7 were derived from patient 2.
Mutations of the heavy- and light-chain V and J segments were obtained from IMGT.
LTA = lipoteichoic acid; SSL5 = superantigen-like protein 5; FnBPA = fibronectin-binding protein A; PGN = peptidoglycan
3.5. Recombinant antibodies derived from peripheral blood plasmablasts bind to S. aureus virulence factors and surface antigens
We used a full-proteome microarray of the S. aureus USA300 strain (Antigen Discovery, Inc.)—a strain prevalent in community-acquired S. aureus outbreaks [18]—to identify the targets of these S. aureus-specific recombinant antibodies. Antibodies S6 and S10 bound to SAUSA300_2441 and SAUSA300_2441-s1 with four-fold intensity over background (Fig. 4A). These two hits correspond to different segments of the same protein, fibronectin-binding protein A (FnBPA). FnBPA is a MSCRAMM (microbial surface components recognizing adhesive matrix molecules) expressed on the bacterial membrane that allows the bacteria to attach to the extracellular-matrix proteins fibronectin and fibrinogen and thereby promotes invasion of host tissues [27–29]. Subsequent binding analyses showed that S6 and S10 bind to full-length FnBPA and to an N-terminal fragment of FnBPA but not a C-terminal fragment of FnBPA (Fig. 4B-C). Furthermore, antibody S4 bound moderately to superantigen-like protein 5 (SSL5), a secreted virulence factor that inhibits neutrophil migration (Fig. 4A) [30] [31].
Figure 4.
Identification of protein antigens targeted by plasmablast-derived recombinant antibodies. (A) Screen of recombinant antibody binding to a S. aureus USA300 proteome microarray. Recombinant antibodies from shared gene families are denoted in blue, and antibodies not sharing gene rearrangements are denoted in green. Measurement of antibody binding ranged from 5000 units (low-affinity) to 20,000 units (high-affinity). Mean fluorescence intensity is displayed and the scale bar represents the range of fluorescence units measured. Only antigens with a binding intensity greater than two standard deviations of signal above background are shown. (B) Schematic representation of the recombinant full-length and truncated N-terminal and C-terminal fragments of FnBPA with domains annotated; numbers indicate the amino-acid residue position. (C) Antibody-binding assay plotting S6 and S10 binding against recombinant full-length, truncated N-terminal, and truncated C-terminal fragments of FnBPA.
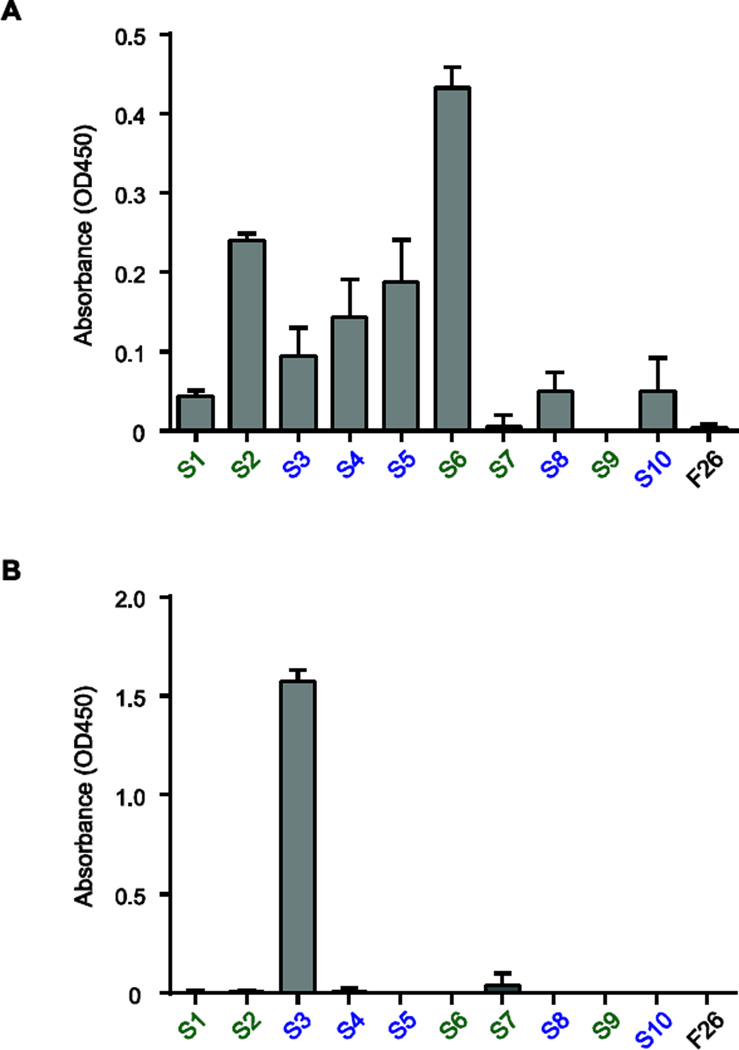
A large majority of healthy and infected individuals contain serum reactive to S. aureus peptidoglycan (PGN) and lipoteichoic acid (LTA) [7] [32]. ELISAs using purified S. aureus PGN and LTA revealed that S6 bound robustly to PGN while antibodies S2 and S5 bound weakly (Fig. 5A), and S3 bound strongly to LTA (Fig. 5B).
Figure 5.
Identification of S. aureus cell wall components targeted by plasmablast-derived recombinant antibodies. (A) ELISA of recombinant antibody binding to S. aureus peptidoglycan. (B) ELISA of recombinant antibody binding to S. aureus lipoteichoic acid. Recombinant antibodies from shared gene families are denoted in blue, and antibodies not sharing gene rearrangements are denoted in green. Influenza-specific antibody F26 was used as an isotype control. ELISAs were performed in triplicate and are presented as mean values of all replicates ± s.e.m.
3.6. Antibodies that bind to S. aureus differentially promote opsonization and killing of S. aureus
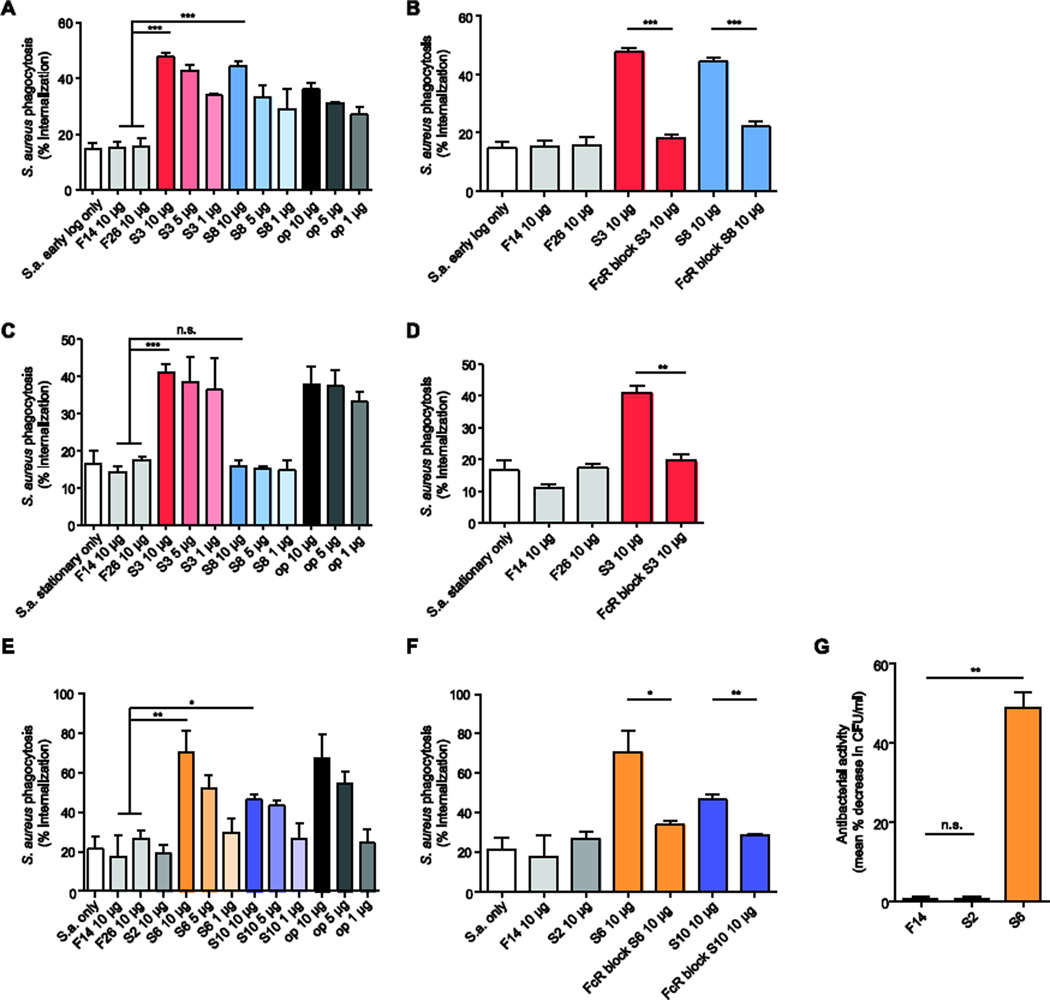
A major way in which antibodies protect against pathogens is through opsonization, a process in which antibodies coat the pathogen and thus mark it for ingestion and destruction by phagocytes. We used undifferentiated THP1 (human monocyte-like) cells and CFSE-labeled inactivated S. aureus to test whether the recombinant antibodies that bound to surface S. aureus proteins could function as opsonins. Using positive and negative control antibodies and halting the assay at 30-minute intervals allowed us to optimize the duration of the assay (Supplemental Fig. S4A). We found that S3 and S8 promoted phagocytosis of S. aureus at the early-log growth phase (Fig. 6A), whereas only S3 promoted phagocytosis of S. aureus at the stationary phase (Fig. 6C). Antibodies S6 and S10 promoted phagocytosis of S. aureus grown in the presence of fibroblasts (Fig. 6E and Supplemental Fig. S4B). Blocking FcγRs with FcR Block before co-incubation of monocytes and bacteria abrogated the antibody-induced increase in phagocytosis, confirming that S3, S6, S8, and S10 function as opsonins (Fig. 6B, Fig. 6D, Fig. 6F, Supplemental Fig. S4B). Notably, antibody S2, which bound to early-log phase bacterial lysate with the second highest intensity in the dot-blot screen, did not enhance internalization of S. aureus cells (Fig. 6E).
Figure 6.
Opsonophagocytic and bactericidal capability of recombinant antibodies generated from S. aureus-infected individuals. (A,C,E) Flow cytometric analysis of antibody-mediated phagocytosis. CFSE-labeled S. aureus Wood46 fixed in early-log growth phase (A), fixed in stationary growth phase (C), or grown with NIH/3T3 fibroblasts (E), were incubated with undifferentiated THP1 monocyte-like cells and 1 µg/ml, 5 µg/ml, or 10 µg/ml of recombinant antibody. (B,D,F) Flow cytometric analysis of FcR dependence of antibody-mediated phagocytosis. Early-log phase (B), stationary phase (D), and NIH/3T3-co-cultured (F), antibodyopsonized, CFSE-labeled S. aureus Wood46 were incubated with undifferentiated THP1 cells pre-treated with FcR-blocking human IgG. (G) Evaluation of the antibodies’ antibacterial activity. Live S. aureus Wood46 were incubated with antibodies and activated HL-60 neutrophils. Bacteria were plated following HL-60 lysis, and mean percent decrease in CFU was measured. Flu-specific antibodies F14 and F26 were used as isotype controls. op = anti-S. aureus polyclonal rabbit IgG antibodies. Data are from three experiments performed on separate days and are presented as mean values of all replicates ± s.e.m. Statistical significance was determined by using two-tailed unpaired t-tests; n.s., not significant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001.
To test the antibacterial activity of the recombinant antibodies, we incubated them with live S. aureus and differentiated HL-60 (human neutrophil-like) cells and evaluated any antibody-associated decreases in formation of bacterial colonies, as a measure of the antibodies’ antibacterial activity. The S. aureus-binding antibody S6 reduced colony formation, whereas the S. aureus-binding antibody S2 and a flu-specific control antibody did not (Fig. 6G).
4. Discussion
In this present study, we use a novel barcoding methodology that we developed to sequence the full-length sequencing of the paired VH and VL antibody sequences expressed by individual plasmablasts. This method, through cell-specific barcoding of mRNA, further enables us to bioinformatically index the reads recovered from high-throughput sequencing to a given cell to accurately determine the proportion of B cells with shared V(D)J sequences and somatic mutations, thereby correcting for biases in amplification that may occur during library amplification, emulsion PCR or sequencing. This accurate determination of frequently used antibody rearrangements and similar somatic mutations enables accurate identification of clonal plasmablast populations as well as highly selected somatic mutations. Secondly, template-switch addition of well-ID barcodes and the hybridization of plate-ID barcodes onto universal adapter oligonucleotide at the 5’ end of the well-ID barcodes avoids the need for degenerate V-region primers which may fail to amplify certain allelic variants or highly mutated V-regions, and thereby provide unbiased hybridization of PCR primers.
Barcode-enabled pairing of entire VH and VL sequences from single cells allows us not only to profile the expansion of plasmablast clonal families, but also to recombinantly produce relevant monoclonal antibodies secreted by plasmablasts to characterize the binding and functional properties of endogenous antibodies generated in human immune responses to S. aureus infection. Previous high-throughput methods for evaluating antibody repertoires have largely focused on the sequence diversity of VH or heavy-chain CDR3 regions [10–12], precluding characterization of the binding and functional properties of the specific individual antibodies generated by each cell. Other methods that sequence paired VH and VL [12, 33] do not sequence through the full-length V-region, and as a result necessitate surrogate, non-native V-region sequences for recombinant antibody expression. Our method, through barcoding of all the cDNA generated by each individual B cell, enables identification and matching of the cognate heavy- and light-chain antibody sequences expressed by the same cell, and thereby the recombinant production of the bona fide antibody expressed by that cell. Recombinant antibody production enables binding and functional analyses of the expressed antibodies in the absence of other blood antibodies, an advantage over approaches based on analysis of serum antibodies [34].
We utilized this approach to identify the antibodies expressed by plasmablasts generated in S. aureus bacteremic individuals, and to use the recombinant antibody products to determine whether they bind and/or kill to S. aureus. Seven of the ten (70%) of the recombinant antibodies derived from two S. aureus-infected individuals bound to S. aureus. This percentage is similar to the findings of previous studies that investigated plasmablast specificity against vaccine protein antigens and viral peptides [9] [8]. The high percentage of S. aureus-specific antibodies from families with shared IGHV and IGHJ usage suggests that the analysis of antibody families can facilitate identification of candidate immunoglobulin rearrangements and somatic hypermutations preferentially selected for during infection. This also suggests that characterization of antibodies expressed by plasmablasts, the population of activated B cells present in an immune response, may provide a strategy to identify antibodies targeting the critical antigenic determinants in an anti-microbial immune response. Further studies with additional S. aureus-infected patients or other infectious diseases will be necessary to elucidate whether antibodies produced by rapidly expanding B-cell populations are most relevant to the containment of infection.
Of the seven antibodies that bound to S. aureus, four enhanced S. aureus opsonization. Pro-opsonophagocytic antibodies S6 and S10, which were derived from separate patients, both bound to an extracellular region of fibronecting binding protein A (FnBPA) away from its fibronectin-binding domains and C-terminal membrane-spanning domain [35], suggesting that these antibodies can recognize FnBPA and mediate opsonization even when it is bound to fibronectin. These antibodies also bind away from the FnBPA fibronectin-binding domain repeats, which are known to exhibit inter-strain variance [36]. The finding that two antibodies containing unique germline gene segments from different patients both bound to the FnBPA N-terminus suggests that the FnBPA N-terminus may be an immunodominant epitope during S. aureus bacteremia. Further analysis is necessary to determine whether the epitopes recognized by these antibodies are conserved across multiple bacterial strains. Antigens targeted robustly by multiple patients with favorable clinical outcomes may be useful for vaccine design.
In contrast, antibody S2, which bound S. aureus, didn’t promote opsonophagocytosis or bacterial killing. Emerging evidence reveals that antibodies that bind to S. aureus antigens can fail to promote opsonophagocytic killing independent of staphylococcal evasion mechanisms [37] [38], which may be due an antibody binding orientation that masks the accessibility of the Fc domain. This underscores the utility and need for recombinant antibody-enabled functional analyses.
Multiple recombinant antibodies from plasmablasts bound to non-protein S. aureus antigens PGN and LTA. The majority of plasmablasts in peripheral blood are believed to differentiate from B-cell precursors that have undergone conventional CD4+ T-cell-dependent activation and somatic hypermutation in B-cell follicles, and are therefore specific for protein antigens and zwitterionic polysaccharides. Marginal zone and B1 B-cells are important in the immune response against bloodborne pathogens and can circulate in the blood as plasmablasts; however, their immunoglobulin genes are believed to undergo limited somatic hypermutation and antibodies secreted by these B-cell subsets show limited evidence of affinity maturation. Interestingly, the sequence analysis revealed that these antibodies had high mutation numbers (Table 1), suggesting that the plasmablasts expressing these antibodies were activated by uncharacterized mechanisms of T-cell-dependent activation or that some extrafollicular B cells may undergo considerable affinity maturation that has not previously been appreciated.
As anticipated, all recombinant antibodies bound to IgG-binding proteins A and Sbi in the antigen microarrays (data not shown), which are believed to have a role in bacterial evasion of the host immune system by disrupting opsonization and complement fixation [35, 39]. Protein Sbi has also been implicated in immune evasion through the degradation of C3 [40]. Further tests with Fab fragments are required to determine whether the antibodies bind specifically to protein Sbi. That S2, S5, and S8 did not bind to any proteins on the microarray could be because of inter-strain variation. Also, the cell-free Rapid Translation System used for expressing the S. aureus proteins for the microarray may have unexpectedly altered the conformation of certain S. aureus proteins, masking epitopes normally recognized by these antibodies.
There were several technical limitations to the approach used in this study. Selection of antibodies for recombinant expression based on shared gene segments did not include antibodies with different sequences sharing CDR protein structures and potentially similar binding properties. In addition, the limited sequencing depth per patient may have prevented detection of sequences representing small clonal families. Future studies sequencing additional patients and plasmablasts per patient will be necessary to elucidate clonal family frequencies and identify clonal families shared between multiple patients.
The efficient recovery of anti-S. aureus reactive antibodies from peripheral blood plamablasts derived from S. aureus bacteremic humans indicates that this approach can efficiently and effectively reveal antigens targeted by the anti-microbial B cell response. This strategy has the potential to be leveraged to identify and isolate valuable diagnostic and therapeutic antibodies, to monitor the status of the humoral immune response over the course of infection, and to determine critical epitopes for vaccine design and monitoring.
Supplementary Material
Highlights.
Deep-sequencing of plasmablast antibody repertoires in S. aureus infection.
Single-cell barcoding enables recovery of natively paired heavy- and light-chains.
Identification of clonal families of antibodies with shared rearrangements.
-
Antibodies from clonal populations are enriched for S. aureus binding.
Patient-derived S. aureus-specific antibodies differentially enhance phagocytosis.
Acknowledgments
Funding
This research was supported by National Institutes of Health (NIH) RC1 AR058713 and NIH NHLBI Proteomics Center N01-HV-00242 to W.H.R.; and an A*STAR NSS (PhD) fellowship to Y-C.T.
Abbreviations
- VH
Heavy chain variable region domain (comprised of the recombined immunoglobulin heavy chain V gene segment, D gene segment and J gene segment)
- VL
Light chain variable region domain (comprised of the recombined light chain V gene segment and J gene segment)
- S. aureus
Staphylococcus aureus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
Y-C.T. is an employee of and owns equity in Atreca, Inc.; J.S. and W.H.R. are consultants to and own equity in Atreca, Inc. All other authors have no disclosures.
Contributors
D.R.L. designed and performed experiments, analyzed results, interpreted results, and wrote the manuscript. Y-C.T. designed and performed experiments, analyzed results, interpreted results, and wrote the manuscript. S.K. performed bioinformatic analysis of antibody sequencing experiments. X.C. cloned and expressed antibodies. E.A.S. designed experiments and interpreted results. T.M.L. contributed to interpretation of results, and wrote and edited the manuscript. J.S. contributed to experimental design, data analysis and interpretation, and obtained blood samples. W.H.R. designed experiments, analyzed results, interpreted results, and wrote the manuscript.
REFERENCES
- 1.Talan DA, et al. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients 2004 and 2008. Clin Infect Dis. 2011;53(2):144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 2.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillinresistant Staphylococcus aureus, United States 1999–2005. Emerg Infect Dis. 2007;13(12):1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Proctor RA. Challenges for a universal Staphylococcus aureus vaccine. Clin Infect Dis. 2012;54(8):1179–1186. doi: 10.1093/cid/cis033. [DOI] [PubMed] [Google Scholar]
- 5.Holtfreter S, Kolata J, Broker BM. Towards the immune proteome of Staphylococcus aureus - The anti-S. aureus antibody response. Int J Med Microbiol. 2010;300(2–3):176–192. doi: 10.1016/j.ijmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Wood P, et al. Recognition, clinical diagnosis and management of patients with primary antibody deficiencies: a systematic review. Clin Exp Immunol. 2007;149(3):410–423. doi: 10.1111/j.1365-2249.2007.03432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dryla A, et al. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol. 2005;12(3):387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrammert J, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol. 2012;86(6):2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5(171):171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31(2):166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30(5):447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 14.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 17.Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother. 2009;64(3):441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 19.Ataide R, et al. Using an improved phagocytosis assay to evaluate the effect of HIV on specific antibodies to pregnancy-associated malaria. PLoS One. 2010;5(5):e10807. doi: 10.1371/journal.pone.0010807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuutila J, Lilius EM. Flow cytometric quantitative determination of ingestion by phagocytes needs the distinguishing of overlapping populations of binding and ingesting cells. Cytometry A. 2005;65(2):93–102. doi: 10.1002/cyto.a.20139. [DOI] [PubMed] [Google Scholar]
- 21.Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci U S A. 1980;77(5):2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan YC, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clinical Immunology. 2014;151(1):55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu YY, et al. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30(4):892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 24.Resch A, et al. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71(5):2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett AH, Hulten KG. Staphylococcus aureus pathogenesis: secretion systems, adhesins, and invasins. Pediatr Infect Dis J. 2010;29(9):860–861. doi: 10.1097/INF.0b013e3181ef2477. [DOI] [PubMed] [Google Scholar]
- 26.Ross R. The fibroblast and wound repair. Biol Rev Camb Philos Soc. 1968;43(1):51–96. doi: 10.1111/j.1469-185x.1968.tb01109.x. [DOI] [PubMed] [Google Scholar]
- 27.Jonsson K, et al. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202(3):1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz-Linek U, Hook M, Potts JR. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol Microbiol. 2004;52(3):631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 29.Ponnuraj K, et al. A "dock, lock, and latch" structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115(2):217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 30.Bestebroer J, et al. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectinmediated neutrophil rolling. Blood. 2007;109(7):2936–2943. doi: 10.1182/blood-2006-06-015461. [DOI] [PubMed] [Google Scholar]
- 31.Itoh S, et al. Staphylococcal superantigen-like protein 5 inhibits matrix metalloproteinase 9 from human neutrophils. Infect Immun. 2010;78(7):3298–3305. doi: 10.1128/IAI.00178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wergeland H, et al. Antibodies to Staphylococcus aureus peptidoglycan and lipoteichoic acid in sera from blood donors and patients with staphylococcal infections. Acta Pathol Microbiol Immunol Scand B. 1984;92(5):265–269. doi: 10.1111/j.1699-0463.1984.tb02832.x. [DOI] [PubMed] [Google Scholar]
- 33.Busse CE, et al. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur J Immunol. 2014;44(2):597–603. doi: 10.1002/eji.201343917. [DOI] [PubMed] [Google Scholar]
- 34.Sato S, et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat Biotechnol. 2012;30(11):1039–1043. doi: 10.1038/nbt.2406. [DOI] [PubMed] [Google Scholar]
- 35.Signas C, et al. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci U S A. 1989;86(2):699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice K, et al. Variance in fibronectin binding and fnb locus polymorphisms in Staphylococcus aureus: identification of antigenic variation in a fibronectin binding protein adhesin of the epidemic CMRSA-1 strain of methicillin-resistant S. aureus. Infect Immun. 2001;69(6):3791–3799. doi: 10.1128/IAI.69.6.3791-3799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HK, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28(38):6382–6392. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skurnik D, et al. Natural antibodies in normal human serum inhibit Staphylococcus aureus capsular polysaccharide vaccine efficacy. Clin Infect Dis. 2012;55(9):1188–1197. doi: 10.1093/cid/cis624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3(12):948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 40.Smith EJ, et al. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun. 2011;79(9):3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.