Abstract
AIM: Recently, germ-line mutation in the base excision repair gene MYH has been identified to cause a novel autosomal recessive form of familial adenomatous polyposis (FAP). Interestingly, a striking evidence for MYH mutations within different ethnic groups has been demonstrated. In this study, we screened 30 patients with multiple adenomatous polyps for MYH mutations to assess its prevalence and ethnic specificity in Korea.
METHODS: Thirty patients (21 men and 9 women; mean age 62.3 years) with multiple adenomatous polyps were examined for MYH mutations. The mean number of adenomas per patient was 10.0. Sixteen exonic regions and their intronic sequences were amplified by PCR and subjected to SSCP and DNA sequencing analyses.
RESULTS: None of the patients was identified to carry any truncating or sequence alterations in MYH. Our screening for the mutational regions, which were recognized from Caucasian patients or affected Indian families, also failed to detect sequence substitutions.
CONCLUSION: Mutation in MYH may be rarely involved in the pathogenesis of multiple sporadic colorectal adenomas in Korean population, although a large-scale analysis will be required to clarify the presence of specific MYH variants in a subset of patients and their role in the predisposition of multiple colorectal adenomas in Korean population.
Keywords: MY, Multiple adenomatous polyps, Germ-line mutation, Familial adenomatous polyposis, Ethnic difference
INTRODUCTION
It has been well established that genetic factors play a pivotal role in up to 35% of all colorectal cancers (CRC)[1-3]. Familial adenomatous polyposis (FAP) is an autosomal dominant disorder with an increased predisposition to multiple colorectal adenomatous polyps, and thence to CRCs[4]. Classic FAP is caused by inherited mutations in the adenomatous polyposis coli (APC) gene, which encodes a protein that plays a critical role in the regulation of colonic cell growth[5,6]. Attenuated FAP (AFAP) is associated with smaller numbers of adenomas and is caused by mutations in the extreme of 5’ or 3’ ends of APC or in the alternatively spliced region of exon 9[4,5].
Somatic mutations of APC consist of the substitution of a thymine-adenine pair for a guanine-cytosine pair (G:C→T:A), which is a typical change caused by oxidative damage to DNA[7-11]. Oxidative DNA damage produces the stable 8-oxo-7, 8-dihydro2’deoxyguanosine (8-oxoG)[9]. It is highly mutagenic because it tends to mispair with adenine residues, leading to an increased frequency of spontaneous G:C→T:A transversion mutations in repair-deficient bacteria and yeast cells[10-13].
Recently, Al-Tassan et al[14] reported a novel autosomal recessive form of FAP. They found that three of seven siblings from a single British Caucasian family were all compound heterozygous for two non-conservative missense variants, Y165C and G382D, in the base excision repair gene MutY homologue (MYH). This finding first implicated a defect in the base excision repair mechanism in inherited predisposition to colorectal tumors in human beings.
Base excision repair (BER) is cell’s mechanism of protection against oxidative DNA damage[15]. The BER pathway repairs mutations caused by reactive oxygen species that are generated during aerobic metabolism[16]. Three main components of BER have been identified in human. MutT homologue (MTH) removes oxidized base from 8-oxoG:C pairs to reduce the chances of incorporation of 8-oxoG during DNA replication, and 8-oxoG glycosylase (OGG) initiates base-excision of 8-oxoG from 8-oxoG:C pairs[17]. Adenine-specific DNA glycosylase MYH removes adenine mispaired with 8-oxoG in 8-oxoG:A pairs. [18-20]
The prevalence of MYH mutations has been determined in patients with multiple colorectal polyps in Caucasian populations. However, to our knowledge, a few studies have been carried out regarding the prevalence of MYH mutations in Asian patients with multiple colorectal adenomatous polyps. In the present study, we therefore explored the possible implication of germ-line mutations of MYH in the development of multiple sporadic colorectal adenomatous polyps in Korean population.
MATERIALS AND METHODS
Specimens
Thirty patients (21 men and 9 women; mean age, 62.3 years) with multiple sporadic colorectal adenomatous polyps were recruited from Kyung Hee University, Medical Center, Seoul, Korea. All patients were Korean and none had a family history of vertical transmission of colorectal cancer or adenomatous polyps. Five individuals who did not have any recognizable diseases were selected as healthy controls. Informed consent was taken from all patients. Ten milliliters of blood were obtained from all the patients and healthy controls for extraction of genomic DNA. The characteristics of patients are summarized in Table 1.
Table 1.
Characteristics of patients
| Characteristics | Patient with CRC and adenomatous polyp (n = 10) | Patient with adenomatous polyp only (n = 20) |
| Sex | 3 males : 1 females | 15 males : 5 females |
| Age (yr) | Mean 45.9 (range: 21 ~ 78) | Mean 51.8 (range: 44 ~ 69) |
| Number of adenomatous polyps | Mean 9 (range: 1 ~ 10) | Mean 7 (range: 3 ~ 100) |
| Family history of adenomatous polyps | 0 | 0 |
CRC: colorectal cancer.
PCR amplification of MYH gene
Genomic DNA was prepared from venous blood samples using QIAGEN Genomic-tip system (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of extracted DNA was determined by a spectrophotometric measurement (Schimadzu Scientific Instruments, Inc., Concord, CA). Exons 1-16 of MYH and their flanking intronic sequences were amplified by PCR with 14 primer sets which cover the entire coding region of MYH. Primer sequences used for PCR amplification are shown in Table 2. PCR was performed with 200 ng genomic DNA as template for 38 cycles at 95°C (1 min), 58-62°C (30 s), and 72°C (30 s) in 1.5 mmol/L MgCl2-containing reaction buffer (PCR buffer II, Perkin Elmer-Cetus, Concord, CA). Ten microliters of PCR products were electrophoresed on 20 g/L agarose gel containing ethidium bromide, visualized under ultraviolet light, and photographed.
Table 2.
Primer sequences of oligonucleotide primers used for PCR-SSCP analysis of MYH
| Exon | Primers | Sequences (5’ to 3’) | Length (bp) |
| 1 | M1S | CAGAGCGCAGAGGCTTTGAACA | 240 |
| M1AS | CTGAACGGAAGTTCGACCCATC | ||
| 2 | M2S | AATTTGGCCTCATTGTGACTGA | 221 |
| M2AS | AATCTGCCTTTCATGGCCAATG | ||
| 3 | M3S | CACAGGCTGCTGTGTCCCAAGA | 259 |
| M3AS | CCCACCCACTGTCCCTGCTCCT | ||
| 4+5 | M45S | AACTCCTCATCTGGGGTTGCAT | 296 |
| M45AS | GGTCTGACCCATGACCCTTCCC | ||
| 6+7 | M67S | ACCACCTTCACCCTTGACCTTG | 268 |
| M67AS | ACCCAAGACTCCTGGGTTCCTA | ||
| 8 | M8S | GGAACCCAGGAGTCTTGGGTGT | 222 |
| M8AS | AAGGAGGCTGGGCACGCACAAA | ||
| 9 | M9S | TTTGTGCGTGCCCAGCCTGGTT | 223 |
| M9AS | TGCTGTGAAGCAGAGCTCCAAA | ||
| 10 | M10S | AAAGGAGCTCTGCTTCACAGCA | 227 |
| M10AS | CACTCCTTAGGACTTCTCACTG | ||
| 11 | M11S | GTAAGCCTACTGGGGAAGGGG | 222 |
| M11AS | GCAGAATCTTACTCAGGTTAG | ||
| 12 | M12S | GCCCTCTTGGCTTGAGTAGGGT | 278 |
| M12AS | TCTCTTGTTACTCATGCCACTG | ||
| 13 | M13S | AGGGAATCGGCAGCTGAGGCCT | 226 |
| M13AS | AAAAGCCAACATCCTTGGCTAT | ||
| 14 | M14S | TATATCCACAGGCCTATTTGAA | 256 |
| M14AS | ATATTCATGTAGAACATGTAGG | ||
| 15 | M15S | GACATGAAGTTAAGGGCAGAAC | 212 |
| M15AS | TGTTCACCCAGACATTCGTTAG | ||
| 16 | M16S | AACTACAAGGCCTCCCTCCTTCCA | 270 |
| M16AS | AACAACAGGATTCTCAGGGAATG |
Single strand conformation polymorphism analysis
To identify sequence alterations in the MYH gene, we performed nonisotopic PCR-SSCP analysis as described previously[14]. Briefly, 20 μL of the PCR products was mixed with 5 μL of 0.5 mol/L NaOH, 10 mmol/L EDTA and 10 μL of denaturing loading buffer (950 mL/L formamide, 20 mmol/L EDTA, 0.5 g/L bromophenol blue, and 0.5 g/L xylene cyanol). After heating at 95°C for 5 min, the samples were rapidly loaded in wells pre-cooled to 4°C and run simultaneously on two 80 g/L non-denaturing polyacrylamide gels with or without 100 g/L glycerol. These two gels were run at 18-20°C and then repeated at 6-10°C in a buffer-jacketed gel apparatus (DGGE-II; Aladin Enterprises, Inc., San Francisco, CA). Following an 8 h run at 400 volts, the gels were stained with ethidium bromide and photographed under ultraviolet light.
Automated DNA sequencing
PCR amplification products were purified using the PCR purification kit (Quiagen). DNA sequencing was carried out using ABI PRISM 377 automated DNA sequencer (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Sequencing was carried out in both directions to confirm the findings.
RESULTS
To explore the presence of germ-line alterations of MYH, we, using quantitative genomic PCR analysis, initially evaluated genomic status of MYH. Blood DNA samples from 30 patients with multiple sporadic colorectal adenomatous polyps and 5 healthy individuals were subjected to PCR amplification of MYH. All of the 16 exons and flanking intronic sequences of the gene were amplified as 14 fragments using intron-specific primer sets, and their levels in the patients were compared with those in healthy controls. As shown in Figure 1, genomic levels of MYH, which were evaluated for 14 separate gene regions, showed no detectable difference between patients and healty individuals, and none of the patients was found to have structural alteration within these protein-encoding regions of the gene. Although systemic analysis for the genomic status was not carried out in this study, the results suggested that our patients did not harbor germ-line deletion or structural abnormalities of the gene, which could result in loss or significant reduction of MYH protein function.
Figure 1.
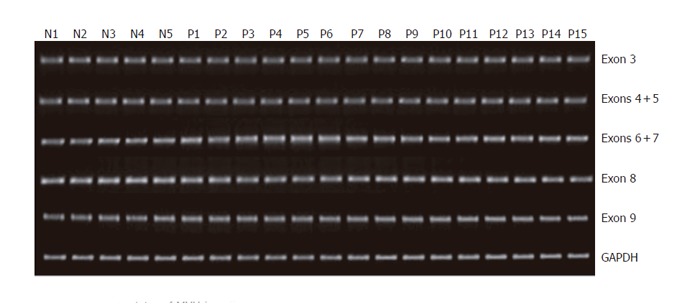
Genomic status of MYH in patients with multiple colorectal adenomas determined by quantitative DNA-PCR analysis. Representative examples for exons 3-9 amplification are shown. Two hundred nanograms of DNA obtained from 30 patients and 5 healthy controls were used as templates for PCR amplification of 16 exonic regions using 14 intronic primer sets. GAPDH was used as an internal control for PCR. Ten microliters of PCR product was resolved on 20 g/L agarose gel and visualized by ethidium bromide staining. No significant reduction of gene levels was identified in samples from patients as compared with normal controls. Lanes N1-N5: Normal controls; Lanes P1-P15: Patients.
Using PCR-SSCP and DNA sequencing analyses, we next screened for mutations and sequence variants across the entire coding region of the MYH gene. To detect the MYH variants, such as Y165C, G382D, Y90X and E466X, which are commonly observed in Caucasian patients or affected Indian families, we performed comprehensive SSCP assay under four different running conditions[14]. However, we could not observe any truncating MYH mutations in patient or control samples. Neither did we identify any likely pathogenic mutations and sequence variants in the patient samples. Representative examples of SSCP analysis are shown in Figure 2.
Figure 2.
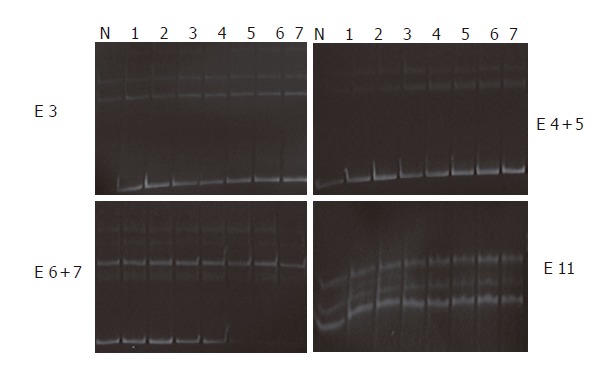
Non-isotopic PCR-SSCP analysis of MYH. For detection of sequence alterations, all of the 16 exons and flanking intronic sequences were amplified by PCR as 14 separate fragments, and 20 μL of PCR product was subjected to non-isotopic SSCP analysis. Genomic DNA isolated from the healthy controls was used as normal controls. None of patient samples showed abnormal migration shift of single strand DNA molecules. Lane N: Normal control; Lanes 1-7: Patients; Lane E: Exons.
DISCUSSION
Germ-line mutation of the MYH gene has been known to predispose persons in a variety of European populations to recessive inheritance of multiple colorectal adenomatous polyposis and classic FAP[4,22]. In addition, all patients with bialleic MYH mutations have an increased predisposition to CRCs.
MYH mutation presents with a distinguished genetic pathway in developing colorectal tumors. Many questions regarding its role in pathogenesis of multiple colorectal adenomatous polyps and CRCs still remain open. For example, it has not been identified why germ-line MYH mutations are associated with tumors of gastrointestinal tract, or why MYH polyposis differs in its phenotype and inheritance from classical FAP[21].
It is difficult to distinguish between patients with APC mutations and those with MYH germ-line mutation on the basis of clinical features or pathological findings. MYH mutations seem to be a more common cause of the multiple adenoma phenotypes than are APC mutations[23-26]. They are, however, a less common cause of classic colorectal adenomatous polyposis.
Jones et al[4] could not identify any pathogenic variants in BER genes OGG1 or MTH1 in cases with colorectal tumors. Thus, there is a possibility that these genes are less frequently mutated than MYH. It is also possible that those mutations do not predispose to tumors in humans due to unknown mechanism. However, further studies are required to determine whether OGG1 or MTH1 are involved in CRC predisposition.
On the basis of previous studies, genetic analysis of MYH mutation should be considered for patients with a phenotype resembling FAP or AFAP, when no clear evidence of vertical transmission is noted. However, whether it is worthwhile to perform genetic testing in the family of patients with MYH mutations is a question that still remains unclear.
In the present study, we failed to detect any truncating or sequence alterations of the MYH gene in Korean patients with multiple colorectal adenomas. Although patient numbers enrolled in this study are too small to exclude the association of MYH mutations with development of multiple colonic adenoma in Korean population, our data suggests the presence of ethnic difference in contribution of MYH mutations to disease development between the Korean patients and the white patients. Previous studies identified four British families that were either homozygous for Y165C or compound heterozygous for Y165C/G382D, three Indian families that were homozygous for E466X, and a single Pakistani family that was homozygous for Y90X[4]. Specific MYH mutations appear to be identified in different ethnic groups. A question still remains as to how frequently MYH mutations contribute to the phenotype of apparently sporadic AFAP/FAP. Further studies of patients from distinct geographical and ethnic groups will help to define the possible ethnic differences in actual mutations.
In conclusion, our observations suggest that MYH mutations are not common in patients with multiple colorectal adenomas in Korean population. Considering that number and histology of colorectal polyps are the cornerstones of detection of many CRC predisposition conditions, further larger-scale evaluation including the histologic analyses of the polyps, is required to determine whether MYH polymorphisms play a role in predisposition of a subset of multiple sporadic colorectal adenomas in Korean population.
Footnotes
S- Editor Kumar M and Guo SY L- Editor Elsevier HK E- Editor Bi L
References
- 1.Cannon-Albright LA, Skolnick MH, Bishop DT, Lee RG, Burt RW. Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med. 1988;319:533–537. doi: 10.1056/NEJM198809013190902. [DOI] [PubMed] [Google Scholar]
- 2.Houlston RS, Collins A, Slack J, Morton NE. Dominant genes for colorectal cancer are not rare. Ann Hum Genet. 1992;56(Pt 2):99–103. doi: 10.1111/j.1469-1809.1992.tb01136.x. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G: C-->T: A mutations. Hum Mol Genet. 2002;23:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 5.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 6.Fearnhead NS. Familial adenomatous polyposis and MYH. Lancet. 2003;362:5–6. doi: 10.1016/S0140-6736(03)13844-5. [DOI] [PubMed] [Google Scholar]
- 7.Halliwell B. Mechanisms involved in the generation of free radicals. Pathol Biol (Paris) 1996;44:6–13. [PubMed] [Google Scholar]
- 8.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 9.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 10.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proc Natl Acad Sci U S A. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G: C--> T: a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol Gen Genet. 1993;239:72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 13.Thomas D, Scot AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of G . C--> T . A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol Gen Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- 14.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, et al. Inherited variants of MYH associated with somatic G: C--> T: A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 16.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 17.Roldán-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol Cell Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine: 8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipton L, Halford SE, Johnson V, Novelli MR, Jones A, Cummings C, Barclay E, Sieber O, Sadat A, Bisgaard ML, et al. Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 2003;63:7595–7599. [PubMed] [Google Scholar]
- 22.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 23.Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H, Nohmi T, Kasai H, Yokota J. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–3225. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- 24.Wikman H, Risch A, Klimek F, Schmezer P, Spiegelhalder B, Dienemann H, Kayser K, Schulz V, Drings P, Bartsch H. hOGG1 polymorphism and loss of heterozygosity (LOH): significance for lung cancer susceptibility in a caucasian population. Int J Cancer. 2000;88:932–937. doi: 10.1002/1097-0215(20001215)88:6<932::aid-ijc15>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 25.Hanaoka T, Sugimura H, Nagura K, Ihara M, Li XJ, Hamada GS, Nishimoto I, Kowalski LP, Yokota J, Tsugane S. hOGG1 exon7 polymorphism and gastric cancer in case-control studies of Japanese Brazilians and non-Japanese Brazilians. Cancer Lett. 2001;170:53–61. doi: 10.1016/s0304-3835(01)00565-1. [DOI] [PubMed] [Google Scholar]
- 26.Lamlum H, Al Tassan N, Jaeger E, Frayling I, Sieber O, Reza FB, Eckert M, Rowan A, Barclay E, Atkin W, et al. Germline APC variants in patients with multiple colorectal adenomas, with evidence for the particular importance of E1317Q. Hum Mol Genet. 2000;9:2215–2221. doi: 10.1093/oxfordjournals.hmg.a018912. [DOI] [PubMed] [Google Scholar]


