Abstract
Adipose tissue development is poorly understood. Here we use a lineage-tracing strategy optimal for adipocytes to provide evidence that Myf5 precursors are not the exclusive source of brown adipocytes and contribute more to the mature white and brite adipocyte populations than previously thought. Moreover, Myf5 lineage distribution in adipose tissue changes in response to modifiable and non-modifiable factors. We also find that the Pax3 lineage largely overlaps with the Myf5 lineage in brown fat and subcutaneous white fat, but exhibits gender-linked divergence in visceral white fat while the MyoD1 lineage does not give rise to any adipocytes. Finally, by deleting insulin receptor beta in the Myf5 lineage, we provide in vivo evidence that the insulin receptor is essential for adipogenesis and that adipocyte lineages have plasticity. These data establish a conceptual framework for adipose tissue development and could explain body fat patterning variations in healthy and lipodystrophic or obese humans.
Introduction
Adipose tissue development is not well understood. One view is that adipose tissue is a single organ that develops from a single lineage1, 2, 3, 4. However, lipodystrophies often manifest as lipid distribution disorders in which regionalized depot atrophy coexists with preservation and often expansion of other depots5. The basis for this regionalization is not understood. Adipose tissues exist in multiple discrete white (WAT) and brown (BAT) depots. WAT is typically generalized as being subcutaneous or visceral2, 6. Visceral WAT is considered unfavorable because its accumulation (e.g. in obesity) correlates with metabolic disease, while subcutaneous WAT is considered more metabolically favorable because its accumulation can be protective, although this generalization is likely oversimplified because metabolic properties vary between visceral depots and even between adipocytes within the same depot2, 6. Why this metabolic heterogeneity exists within and between depots is also not understood. One possible explanation is that adipocytes arise from diverse lineages and that this influences fat distribution and function. However, the ability to determine this is precluded by the fact that the developmental origin of adipocytes is largely unknown7.
We recently provided both genetic and lineage-tracing evidence that brown adipocytes and a subset of white adipocytes descend from progenitors that express Myf5 (i.e. myf5-cre), which may originate in the dermomyotome8, 9, and were previously thought to give rise to brown adipocytes and myocytes, but not to any white adipocyte10. However, due to inefficiencies associated with using cytoplasmic reporters [e.g. R26R-LacZ; R26R-YFP] in lipid-laden adipocytes, our previous ability to accurately quantify the Myf5-lineage contribution to each depot in vivo was limited to the precursor pool and the number of mature adipocytes traced with Myf5-Cre could not be determined8. Myf5 along with Pax3 and MyoD1 are transcription factors that function in a classic myogenic transcriptional cascade9. During embryonic muscle development, Myf5 is thought to function downstream or simultaneously with Pax3 and upstream of MyoD1, but whether Pax3 and/or MyoD1 expressing precursors also give rise to brown or white adipocytes in vivo is not known.
Here, by lineage tracing we quantify the Myf5 lineage contribution to the mature brown and white adipocyte population by combining a Myf5-Cre knock-in allele with a reporter optimal for fluorescently labeling mature adipocytes. By a similar strategy, we also trace the fate in adipose tissue of precursors that express Pax3-Cre and MyoD1-Cre. We find that the Myf5-Cre and Pax3-Cre adipose tissue lineages largely overlap, but also exhibit some significant differences, while MyoD1-Cre does not label any adipocytes. We additionally uncover evidence of lineage dynamics and plasticity. These results help clarify the Myf5-lineage contribution to adipose tissue and provide support for a model in which brown, white, and brite adipocytes have multiple developmental origins.
Results
Strategy for tracing adipocyte lineages
We recently discovered in a conditional gene-knockout study combined with lineage tracing that Myf5-Cre targets a subset of white adipocytes in addition to brown adipocytes8. However, at that time were using cytoplasmic reporters for lineage tracing (i.e. R26R-YFP and R26R-LacZ) on fixed tissues and because white adipocytes are filled with lipid and contain very little cytoplasm, we could not reliably determine how many mature adipocytes were actually traced with Myf5-Cre. To overcome this roadblock, we switched to the R26R-mTmG reporter for lineage tracing, which is a dual-color labeling system that utilizes membrane-targeted fluorescent reporters11. The use of membrane-targeted reporters alleviates the caveat of using cytoplasmic reporters in adipocytes and allows fluorescence detection at single cell resolution in whole mount preparations. In Cre-expressing mice that harbor the R26R-mTmG reporter, all Cre+ cells and their descendants are irreversibly labeled with membrane-targeted GFP (mGFP) while all Creneg lineages express membrane-targeted Tomato (mTFP). As a positive control we generated adiponectin-cre;mTmG mice in which only mature adipocytes are labeled mGFP+ (Supplementary Fig. 1a); as a negative control we analyzed R26R-mTmG mice in which all cells are mTFP+ (Supplementary Fig. 1b).
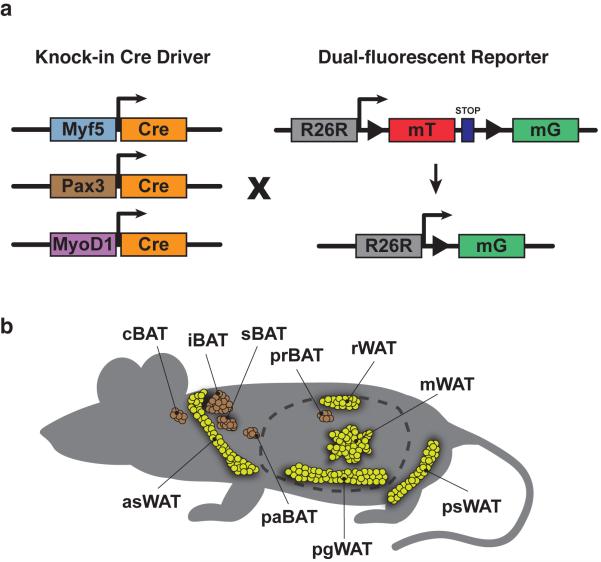
To compare the Myf5, Pax3, and MyoD lineage contributions globally to adipose tissue, we generated myf5-cre;mTmG, pax3-cre;mTmG, and MyoD1-cre;mTmG mice by combining the R26R-mTmG reporter with Myf5-Cre, Pax3-Cre, and MyoD1-Cre knock-in alleles12, 13, 14 in C57Bl/6J mice (Fig. 1a). Notably, while mGFP+ expression in the strictest sense only indicates that the endogenous promoter is or was previously active in the cell lineage, we refer here to labeled cells as being Myf5-lin+/neg, Pax3-lin+/neg, or MyoD-lin+/neg respectively. We determined that skeletal muscles (quadriceps, gastrocnemius, triceps and trapezius) are uniformly mGFP+ in the myf5-cre;mTmG, pax3-cre;mTmG, and myoD1-cre;mTmG mice, while the liver in each strain is mTFP+ (Supplementary Fig. 2a–b). Having validated the robustness of the reporter and functionality of each Cre driver, we proceeded to use the myf5-cre;mTmG, pax3-cre;mTmG, and MyoD1-cre;mTmG mice to perform lineage tracing of all major adipose tissue depots (Fig 1b).
Figure 1. Lineage Tracing Strategy.
(a) Cre drivers and fluorescent reporter system used in this study. Each Cre driver is a knock-in allele meaning Cre activity is indicative of the activity of the endogenous promoter.
(b) Anatomical disposition of BAT and WAT depots examined in this study. Interscapular BAT (iBAT, subsapular BAT (sBAT), cervical BAT (cBAT), periaortic BAT (prBAT), perirenal BAT (prBAT), anterior-subcutaneous WAT (asWAT), posterior-subcutaneous WAT (psWAT), retroperitoneal WAT (rWAT), perigonadal WT (pgWAT) and mesenteric WAT (mWAT) depots are indicated.
Myf5-Cre and Pax3-Cre mark similar adipocyte populations
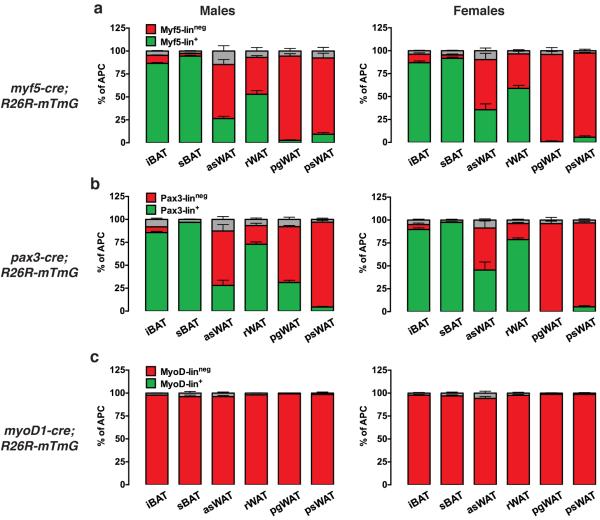
First we asked whether Pax3-Cre and/or MyoD1-Cre trace adipocyte precursor cells (APCs) in patterns similar to Myf5-Cre. Importantly, the percentages of Myf5-lin+ (i.e. mGFP+) APCs in each depot match our previous data obtained with the cytoplasmic R26R-YFP reporter (Fig. 2a)8 confirming the R26R-mTmG reporter's accuracy. Pax3-Cre lineage tracing recapitulates a nearly identical pattern to that of the Myf5-Cre in the APC populations except for one major depot difference: 31.2% of the APCs in male pgWAT are Pax3-lin+ compared to only 2.6% being Myf5-lin+ (Fig. 2a–b). In contrast, less than 1% of the female pgWAT APCs are Pax3-lin+ hinting at a gender-linked difference in Pax3-lineage distribution (Fig. 2b). The MyoD1-Cre does not label APCs in any depot we examined (Fig. 2c) suggesting that developmental lineages expressing MyoD1 do not typically give rise to adipocytes. We conclude that Myf5-Cre and Pax3-Cre label similar pools of APCs in most depots with the one exception being male pgWAT, in which Pax3-Cre but not Myf5-Cre labels a significant number of APCs.
Figure 2. Lineage tracing of adipocyte precursor cells (APCs).
(a–c) Number of mTFP+ and mGFP+ APCs isolated from each of the indicated depots from 6 week old myf5-cre,R26R-mTmG (a), pax3-cre,R26R-mTmG (b) and myod1-cre,R26R-mTmG mice (c). Data from males and females are shown separately (n=3–5; mean+S.E.M). A small number of cells in each case that could not be distinguished are indicated in grey
Brown adipocytes have multiple origins
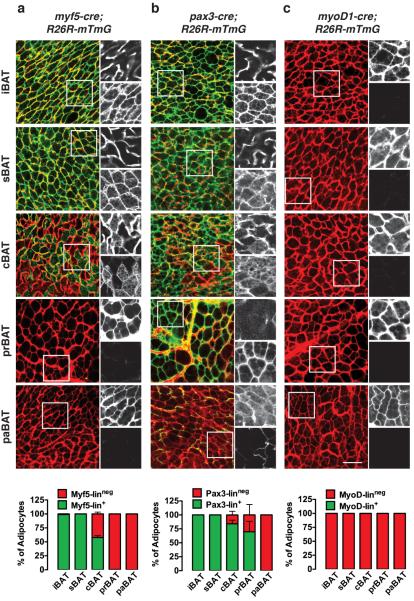
Next we asked whether all mature brown adipocytes arise from Myf5-Cre expressing precursors as one model posits15. In the interscapular and subscapular depots (iBAT & sBAT) we find that 99% and 100% of the brown adipocytes respectively are indeed Myf5-lin+ in both males and females (Fig. 3a & Supplementary Fig. 3a). However, in cervical BAT (cBAT) only 58% of male and 66% of female brown adipocytes are traced with Myf5-Cre exhibiting a mosaic distribution pattern (Fig. 3a & Supplementary Fig. 3a). In peri-renal (prBAT) and peri-aortic (paBAT) brown fat, no brown adipocytes are Myf5-lin+ (Fig. 3a & Supplementary Fig. 3a). We confirmed the brown fat identity of each individual depot by showing high levels of ucp1, prdm16, and cidea mRNA expression compared to WATs, by Hematoxylin & Eosin staining showing multilocular lipid droplets, and by UCP1 immunohistochemistry (Supplementary Fig. 4). These results indicate that only a subset of the total brown adipocyte population is traced with Myf5-Cre.
Figure 3. The Myf5 and Pax3 lineages are heterogeneously distributed in BAT.
(a) Representative images of whole-mount preparations of the indicated BATs of 6 week old myf5-cre;R26R-mTmG male mice. The mTFP and mGFP channels within the boxed inset are shown enlarged to the right of each panel (top = mTFP; bottom = mGFP). The number of mTFP+ and mGFP+ mature brown adipocytes in each depot is indicated in a graph at the bottom of each column (n=3–8 mice; mean+S.E.M).
(b) Same as (a) except using pax3;R26R-mTmG male mice. (n=3–8; mean+S.E.M).
(c) Same as (a) & (b) except using myod1;R26R-mTmG male mice. (n=3–4; mean+S.E.M). Scale bar= 50 μm.
We also find that Pax3-Cre traces a similar population of brown adipocytes. For example, the Pax3-Cre trace mirrors the Myf5-Cre lineage-tracing pattern in iBAT (100% and 99% Pax3-lin+ in males and females respectively) and sBAT (100% in both genders)(Fig. 3b & Supplementary Fig. 3b). However, the Pax3-lineage contributes more than the Myf5-lineage to brown adipocytes in cBAT (84% in males; 91% in females), prBAT (68% in males; 52% in females), and female paBAT (73%)(Fig. 3b & Supplementary Fig. 3b). We were unable to detect any Pax3-lin+ brown adipocytes in male paBAT (Fig. 3b). On the other hand, MyoD1-Cre did not label any brown adipocytes confirming that brown adipocytes do not typically arise from MyoD1 expressing precursors (Fig. 3c). These data argue that the Pax3-lineage largely overlaps with the Myf5-lineage in iBAT and sBAT, but is more broadly represented in the global brown adipocyte population.
Most dorsal-anterior adipocytes are Pax3/Myf5-lin+
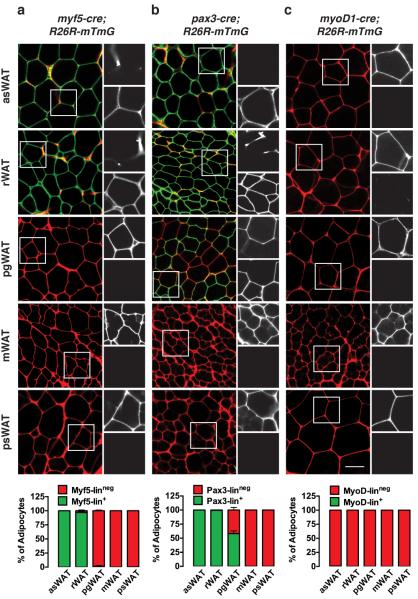
Next we quantified the number of mature white adipocytes traced with Myf5-Cre. Because the APC pool in the asWAT contains a mixture of Myf5-lin+ and Myf5-linneg precursors (Fig. 2a), we speculated that the mature adipocyte population in this depot would also be a heterogeneous mix. Interestingly however, we find that almost all of the adipocytes in the asWAT are traced with Myf5-Cre (Fig. 4a & Supplementary 5a) and the only Myf5-linneg adipocytes detectable lie in the ventral tip of the depot arranged in a gradient pattern such that the extreme ventral tip is completely Myf5-linneg (Supplementary Fig. 5c). Myf5-Cre also traces 97% and 98% of the mature rWAT adipocytes in males and females respectively (Fig. 4a & Supplementary Fig. 5a). This again is more than we predicted for this depot based on the Myf5-Cre trace of the APC pool (Fig. 2a), which indicates a mixture of Myf5-lin+ and Myf5-linneg APCs (roughly 60:40) that can differentiate into adipocytes in vitro with equal efficiency8. In contrast Myf5-Cre does not label any mature pgWAT adipocytes in either sex (Fig. 4a & Supplementary Fig. 5a) consistent with pgWAT APCs being mostly Myf5-linneg (Fig. 2a). Myf5-Cre also does not label any mWAT adipocytes (we did not examine the mWAT APC pool)(Fig. 4a & Supplementary Fig. 5a) or psWAT adipocytes (Fig. 4a & Supplementary Fig. 5a), which is consistent with most psWAT APCs being Myf5-linneg (Fig. 2a). These data indicate that most of the mature white adipocytes in the dorsal-anterior fat depots in 6-week old mice are derived from Myf5-lin+ precursors.
Figure 4. Most dorsal-anterior white adipocytes trace with Myf5-Cre and Pax3-Cre.
(a) Representative images of whole-mount preparations of the indicated WATs of 6 week old myf5;R26R-mTmG male mice. The mTFP and mGFP channels within the boxed inset are shown enlarged to the right of each panel (top = mTFP; bottom = mGFP). The number of mTFP+ and mGFP+ mature brown adipocytes in each depot is indicated in a graph at the bottom of each column (n=3–5 mice; mean+S.E.M).
(b) Same as (a) except using pax3;R26R-mTmG male mice. (n=3–8 mice; mean+S.E.M).
(c) Same as (a) & (b) except using myod1;R26R-mTmG male mice. (n=4 mice; mean+S.E.M). Scale bar= 50 μm.
Next we examined the Pax3-lineage contribution to the mature white adipocyte population. We find that the Pax3-Cre trace in WAT largely overlaps with the Myf5-Cre pattern in both male and female asWAT and rWAT (Fig. 4b & Supplementary Fig. 5b & 5d). Also similar to the Myf5-Cre pattern, no mature adipocytes in mWAT or psWAT trace with Pax3-Cre (Fig. 4b & Supplementary Fig. 5b). However, 58% of the male pgWAT adipocytes trace with Pax3-Cre and they are distributed in a variegated pattern throughout the depot (Fig. 4b). This is a sharp divergence from the Myf5-Cre trace, which did not mark any male pgWAT adipocytes (Fig. 4a), but is consistent with the Pax3-Cre trace to the male pgWAT APC pool (Fig. 2b). No adipocytes in female pgWAT trace with Pax3-Cre (Supplementary Fig. 5b) confirming a gender-linked difference in lineage distribution. Notably, the Pax3-lin+ male pgWAT adipocytes are smaller than the Pax3-linneg adipocytes cohabitating the same depot (Supplementary Fig. 5e) indicating lineage heterogeneity correlates with size heterogeneity. MyoD1-Cre does not label any white adipocytes (Fig 4c) further confirming that white adipocytes do not typically arise from MyoD1 expressing precursors.
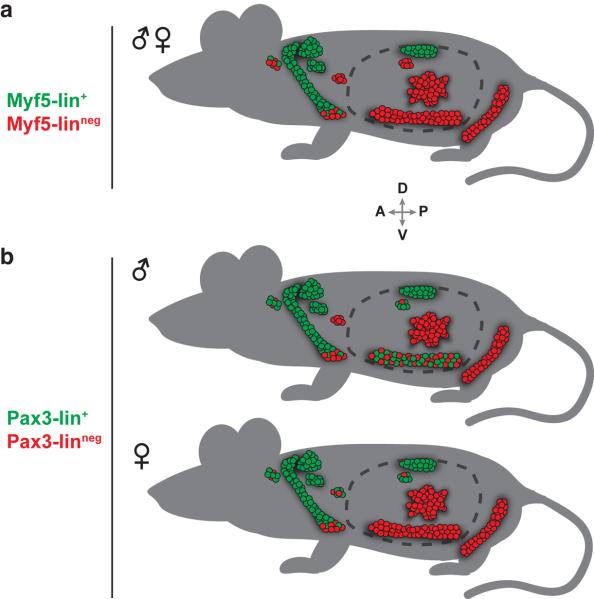
Collectively, these data argue that the Pax3-lineage largely overlaps with the Myf5-lineage in asWAT and rWAT, but exhibits a striking gender-linked difference in male pgWAT only. A comparative summary of the lineage tracing patterns to both BAT and WAT that are described above is depicted in Figure 5a (for Myf5-Cre) and Figure 5b (for Pax3-Cre).
Figure 5. Model depicting the Myf5-Cre and Pax3-Cre lineage tracing patterns.
(a) Anatomical distribution of Myf5-Cre traced adipocytes in BAT and WAT (refer to Figure 1b for depot identity). The distribution is not significantly different between males and females. Anterior (A), posterior (P), dorsal (D), and ventral (V) are indicated.
(b) Anatomical distribution of Pax3-Cre traced adipocytes in BAT and WAT. Male and female distribution patterns are shown separately because of the gender-linked variation in paBAT and pgWAT.
Myf5 lineage distribution is dynamic
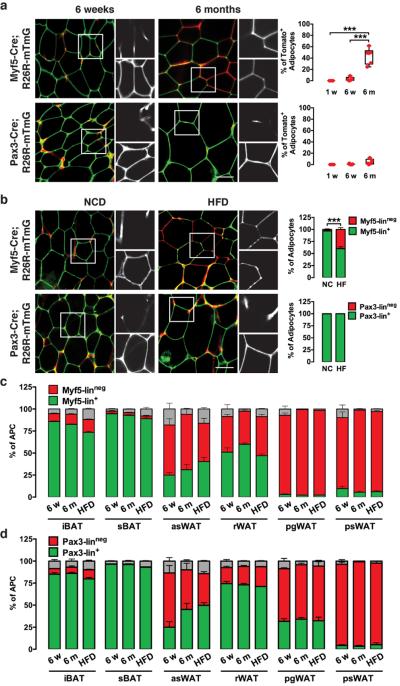
Next, we wondered whether age or diet influence adipocyte lineage distribution. Interestingly, we find that Myf5-linneg adipocytes appear in the rWAT of myf5-cre;mTmG mice with age, increasing in prevalence from 0% at one week old to around 3% at 6 weeks and up to 44% at 6 months (Fig. 6a). In contrast, age has no influence on the Pax3 lineage distribution in rWAT, which remains almost exclusively mGFP+ at 6 months (Fig. 6a). Short-term high fat diet (HFD; 2 weeks, 45% calories from fat) has similar effects, increasing the percentage of Myf5-linneg adipocytes in rWAT to around 40% even though these mice are young (6 weeks) at the time of analysis (Fig. 6b). HFD (similar to age) has no effect on the Pax3-lineage distribution (Fig. 6b). Importantly, aging and HFD does not significantly alter the Pax3 or Myf5 lineage contribution to the APC pools in any depot we examined (Fig. 6c–d) indicating that the change in Myf5-lineage distribution in the mature adipocyte population is not due to a change in the lineage composition of the precursor pool. We do not observe any differences in the lineage composition of brown fat with age or HFD (Fig. 6c–d & not shown). In summary, the Myf5-lineage distribution in fat changes in response to both non-modifiable (e.g. age) and modifiable (e.g. diet) factors while the Pax3-lineage distribution appears to be more static.
Figure 6. The Myf5-lineage distribution is dynamic.
(a) Representative images of whole-mount rWAT from myf5-cre;R26R-mTmG and pax3-cre;R26R-mTmG males at 6-weeks and 6-months of age. Quantification of mTFP+ (Myf5-Linneg) adipocytes at 1-week, 6-weeks and 6-months of age is shown at the right. Each dot represents one mouse (n=3–6; mean+S.E.M; ***, p<0.001; ANOVA test).
(b) Four weeks old myf5-cre;R26R-mTmG and pax3-cre;R26R-mTmG males were fed a HFD (45% of calories from fat) for 2 weeks and whole-mount preparations of rWAT were imaged at 6-weeks of age. Bar graphs (right) are the quantifications of mTFP+ and mGFP+ mature rWAT adipocytes in mice fed the normal chow diet (NC) or high fat diet (HF) (n=5–10 mice for myf5-cre; n=3–8 mice for pax3-cre. mean+S.E.M; ***, p<0.001; t-test).
(c) Number of mTFP+ and mGFP+ APCs isolated from each of the indicated depots from myf5-cre;R26R-mTmG at 6-weeks of age (6w), 6-months of age (6m) and from 6 weeks of age after 2 weeks high fat diet (HFD). A small number of cells in each case that could not be distinguished are indicated in grey (n=3–4; mean+S.E.M).
(d) Same as (c) except using pax3-cre;R26R-mTmG mice (n=3–6; mean+S.E.M). Scale bar= 50 μm.
Depot differences in brite adipocyte origins
Chronic cold temperature or treatment with a β3-adrenergic receptor agonist such as CL316,243 increases the abundance of Ucp1+ multilocular adipocytes in white fat that are called brite (brown in white) adipocytes16. We previously proposed that brite adipocytes are derived from both Myf5-lin+ or Myf5-linneg precursors depending upon the depot8. To confirm this, we treated myf5-cre;mTmG mice with CL316,243 and asked whether any multilocular adipocytes are mGFP+. We find that all multilocular adipocytes induced by CL316,243 appearing in the asWAT are mGFP+ while all multilocular adipocytes appearing in the psWAT are mTFP+ (Fig. 7a). Interestingly, CL316,243 also induces the formation of nascent mTFP+ adipocytes in rWAT, which represent around 40% of the total adipocytes after 1 week of treatment (Fig. 7b) thereby changing the Myf5-lineage composition of rWAT in a manner similar to the effects of aging and HFD. As with age and HFD, CL316,243 does not alter the distribution of Myf5-lin+ APCs in any depot17. This supports the notion that some Myf5-lin+ adipocytes may transform into brite adipocytes while others may arise de novo from Myf5-linneg precursors3, 8, 18, 19. To verify that the multilocular adipocytes induced by CL316,243 are indeed brite adipocytes, we analyzed whether they express UCP1. By immunohistochemistry we find that all multilocular cells induced by CL316,243 in psWAT and rWAT strongly stain positive with a UCP1 antibody (Supplementary Fig. 6a) consistent with the increase in ucp1 mRNA (Supplementary Fig. 6b) and protein expression (Supplementary Fig. 4b) in these depots after CL316,243 treatment. This suggests that brite adipocytes can be Myf5-lin+ or Myf5-linneg depending upon the depot.
Figure 7. Brite adipocytes are Myf5/Pax3-lineage positive or negative depending upon the depot.
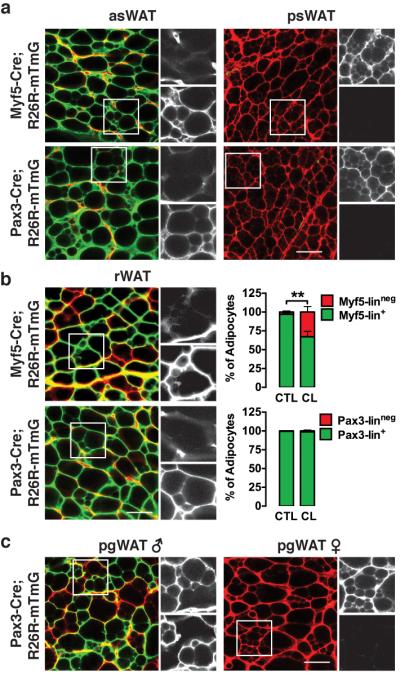
(a) Representative images of whole-mount preparations of asWAT and psWAT of 6-week old male myf5-cre;R26R-mTmG (top) and pax3-cre;R26R-mTmG (bottom) mice that were treated with CL316,243 for one week.
(b) Representative images of whole-mount preparations of rWAT from male myf5-cre;R26R-mTmG (top) and pax3-cre;R26R-mTmG (bottom) mice treated with CL316,243 for one week starting at 5 weeks of age. (Right) Quantification of mTFP+ and mGFP+ mature adipocytes in the rWAT of control (CTL) and treated (CL) mice is shown (n=4–5 mice for myf5-cre; n=4–8 mice for pax3-cre. mean+S.E.M; **, p<0.01; t-test).
(c) Representative images of whole-mount preparations of male and female pgWAT of 6-week old pax3-cre;R26R-mTmG mice that were treated with CL316,243 for one week. Scale bar= 50 μm.
We next asked whether CL316,243 induces Pax3-lin+ brite adipocytes in the pax3-cre;mTmG mice. Similar to the case with the Myf5-lineage nearly all of the multilocular adipocytes induced by CL316,243 in the asWAT and rWAT are mGFP+ (Fig. 7a). However, unlike in the myf5-cre;mTmG mice, no nascent mTFP+ brite adipocytes are detectable in the rWAT following CL316,243 treatment (Fig. 7b) further suggesting that the Pax3-lineage is more static. In male pgWAT, CL316,243 also induces the formation of both mGFP+ and mTFP+ brite adipocytes without affecting Pax3-lineage distribution (Fig. 7c). CL316,243 also does not affect the Pax3-lineage distribution in any APC population examined (Supplementary Fig. 6c, d). In sum, these data obtained from a survey of all major depots argue that although most brite adipocytes reside in the psWAT, these cells cannot be generally classified as being Myf5-lineage positive or negative (or Pax3-lineage positive or negative) because brite adipocytes residing in other depots can also be traced with Myf5-Cre or Pax3-Cre.
Adipocyte lineages have plasticity
We previously discovered that deleting PTEN (a negative regulator of insulin/PI3K signaling) with Myf5-Cre results in a body fat distribution phenotype in which iBAT, sBAT, asWAT, and rWAT (the Myf5-lin+ depots) expand while the psWAT, mWAT, and pgWAT (the Myf5-linneg depots) disappear8. Furthermore, by lineage tracing with the R26R-YFP reporter we found that deleting PTEN with Myf5-Cre selectively expands the Myf5-lin+ APC population at the expense of Myf5-linneg APCs. We therefore predicted that all mature adipocytes in these mice are likely Myf5-lin+. To test this, we generated PTENmyf5cKO mice that additionally carry the R26R-mTmG reporter (myf5-cre;PTENfl/fl;mTmG). Lineage tracing with the R26R-mTmG reporter confirms that there is an expansion of the Myf5-lin+ APCs in the PTENmyf5cKO mice (Supplementary Fig. 7a) and that all mature adipocytes in these mice are indeed traced with Myf5-Cre (Supplementary Fig. 7b). Thus, differences in insulin/PI3K signaling between lineages can alter the lineage composition of adipose tissue.
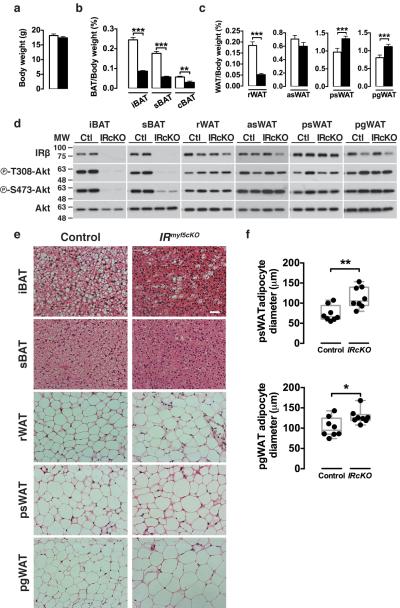
To further investigate the role of insulin signaling in Myf5 lineage adipocytes, we generated myf5-cre;IRβfl/fl conditional knockout (IRmyf5cKO) mice. Six-week old IRmyf5cKO mice show no significant difference in total body weight compared to controls (Fig. 8a) and no overall difference in individual lean tissue mass (Supplementary Fig. 8a). However, iBAT, sBAT, cBAT and rWAT weigh over 3 fold less in the IRmyf5cKO mice (Fig. 8b–c) while the mass of psWAT and pgWAT is increased (Fig. 8c). Despite altered fat distribution, the IRmyf5cKO mice have normal insulin, glucose, and cold tolerance (Supplementary Fig. 8b–d). Thus, IRmyf5cKO mice have a body fat distribution phenotype characteristically opposite to the PTENmyf5cKO mice.
Figure 8. IRmyf5cKO mice have altered body fat distribution.
(a) Total body weight of IRmyf5cKO and littermate control mice at 6-weeks of age (n=14 mice).
(b) BAT mass relative to body weight for the indicated depots at 6-weeks of age (n=14 mice; mean+S.E.M; **, p<0.01***, p<0.001; t-test).
(c) WAT mass relative to body weight for the indicated depots at 6-weeks of age (n=14 mice; mean+S.E.M; **, p<0.01***, p<0.001; t-test).
(d) Western blots of the indicated tissues from IRmyf5cKO and littermate control mice at 6-weeks of age.
(e) H&E images of the indicated BATs and WATs from IRmyf5cKO and control mice at 6-weeks of age.
(f) Cell size quantification of mature adipocytes from psWAT and pgWAT of IRmyf5cKO and littermate control mice at 6-weeks of age (n=8 mice; boxplot indicating maximum, minimum, media and all data points; *, p<0.05; **, p<0.01; t-test).
Consistent with our lineage tracing experiments, IRβ levels and AKT phosphorylation are reduced in the IRmyf5cKO iBAT and sBAT (Fig. 8d). Also consistent, IRβ levels and AKT phosphorylation are normal in the Myf5-linneg psWAT and pgWAT (Fig. 8d). In contrast to our lineage-tracing results, IRβ levels and AKT phosphorylation are also similar to controls in the rWAT and asWAT (Fig. 8d). This was unexpected because most of the mature adipocytes in these depots trace with Myf5-Cre.
Histological analysis indicates that most of the IRβ-deficient brown adipocytes are smaller because they have reduced lipid content (Fig. 8e) consistent with a role for insulin in driving lipogenesis genes in BAT including srebp1c (sterol regulatory biding protein 1c), acl (ATP-citrate lyase), acc (acetyl-CoA carboxilase) and fas (fatty acid synthase) (Supplementary Fig. 8e). Notably however, a few interspersed brown adipocytes containing normal lipid droplet content are detectable in the mutant BATs (Fig. 8e) suggesting that a small number of brown adipocytes might also be IRβ-positive. The adipocytes in the IRmyf5cKO rWAT, asWAT, psWAT, and pgWAT exhibit the typical white adipocyte appearance of having a single large lipid droplet (Fig. 8e & not shown). Cell size analysis also reveals that the mature adipocytes in the psWAT and pgWAT of the IRmyf5cKO mice are larger (hypertrophic) than the corresponding adipocytes in controls (Fig. 8e–f) likely explaining at least in part why these depots weigh more in the mutant.
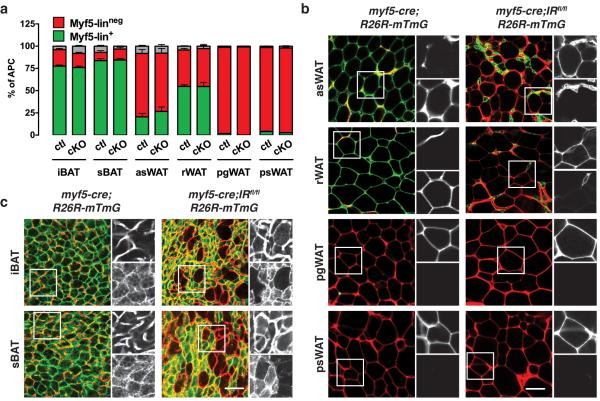
Why is IRβ levels normal in the rWAT and asWAT of IRmyf5cKO mice when these depots are normally traced with Myf5-Cre? We hypothesized that the apparent contradiction could reflect lineage plasticity. To test this, we generated myf5-cre;IRβfl/fl;mTmG mice to ask whether Myf5-linneg (mTFP+) adipocytes replace Myf5-lin+ (mGFP+) adipocytes in the mutant rWAT and asWAT. This is indeed the case; while deleting IRβ with Myf5-cre does not significantly affect the Myf5-lineage composition of the APC pool in either depot (Fig. 9a) it completely changes the lineage composition of the mature adipocyte population (Fig. 9b). For example, the mature adipocytes in the mutant asWAT and rWAT, which are normally Myf5-lin+, are entirely Myf5-linneg in the IRβmyf5cKO mice (Fig. 9b). Notably, the Myf5-lin+ (mGFP+) IRβ knockout cells are detectable in the mutant rWAT and asWAT, but they are much smaller and interspersed among the larger lipid-filled Myf5-linneg adipocytes (Fig. 9b). Thus, IRβ is not essential in the Myf5-lineage for establishing APCs but it is essential for adipogenesis.
Figure 9. Adipocyte lineages have plasticity.
(a) Number of mTFP+ and mGFP+ APCs isolated from each of the indicated depots from 6 week old male IRmyf5cKO-R26R-mTmG and control mice (n=10 males; mean+S.E.M).
(b–c) Representative images of whole-mount preparations of the indicated WATs (b) and BATs (c) of 6 week old IRmyf5cKO-R26R-mTmG mice. Scale bar= 50 μm.
In the mutant iBAT and sBAT the more abundant smaller brown adipocytes are all mGFP+ consistent with the robust decrease in IRβ and phospho-AKT levels in these depots (Fig. 8d). However, the few large adipocytes detectable in these depots are Myf5-linneg (mTFP+)(Fig. 9c) indicating that some expansion of Myf5-linneg (IRβ+) brown adipocytes also occurs in IRβmyf5cKO mice. However, there is no compensatory “browning” in psWAT (i.e. increased multilocularity and ucp1 induction) as would be expected if BAT were completely dysfunctional (Fig. 8e and Supplementary Fig. 8f)20. Collectively, these findings indicate that adipocyte lineages have plasticity.
Discussion
In this study, we compare the lineage tracing patterns of Myf5-Cre, Pax3-Cre, and MyoD1-Cre in adipose tissue using a reporter optimal for labeling mature adipocytes. We provide evidence that only a subset of brown adipocytes arise from Myf5-Cre expressing precursors. Furthermore, we find that most dorsal-anterior white adipocytes also derive from Myf5-Cre expressing precursors, and that there is a large degree of overlap between the Pax3-Cre and Myf5-Cre lineages in both brown and white adipocytes. In contrast, no adipocytes trace to MyoD1-Cre expressing precursors. Finally, we find that adipocyte lineages are not necessarily fixed, but can vary with gender, and in response to modifiable and non-modifiable factors. Based on these findings, we propose a revised model of the Myf5-lineage contribution to adipose tissue (Fig. 10) and the following concepts in adipose tissue development: First, adipocytes originate from multiple developmental lineages that are distributed heterogeneously in depot-specific patterns; Second, adipocyte lineage contribution to the mature adipocyte pool varies with age; Third, adipocyte lineages may selectively expand in response to certain factors; Fourth, adipocyte lineages can compensate for each other indicating lineage plasticity exists. Another study found that cephalic adipocytes arise from neural crest precursors further supporting model in which adipocytes have multiple developmental origins 21. Although we do not yet know the origin of the Pax3/Myf5-lineage negative adipocytes in our study, we hypothesize that they may arise from multiple distinct lineages that might be dynamically distributed in patterns similar to those described here. Thus, our work provides a conceptual framework for future investigations.
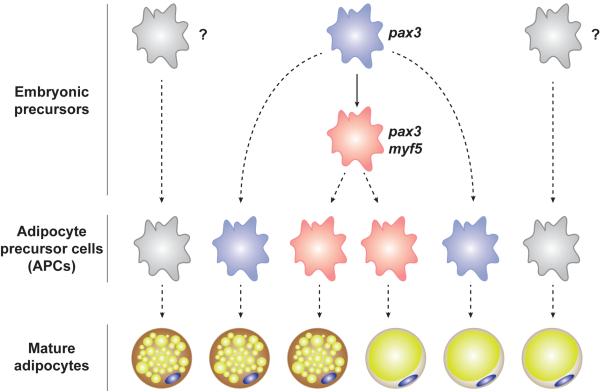
Figure 10. A revised model of adipocyte origins based on Pax3-Cre and Myf5-Cre lineage tracing to mature adipocytes.
Embryonic precursors expressing Pax3-Cre and Myf5-Cre give rise to a subset of APCs and mature brown and white adipocytes. A subset of brown and white adipocytes also arises from Pax3-Cre+ precursors that do not express Myf5-Cre. The origin of Pax3-Cre and Myf5-Cre negative adipocytes is not yet clear. Dotted lines are used to indicate lineages because the intermediate stages are poorly understood. Note that the knock-in Cre drivers used in this study are indicative of the activity of the endogenous promoters.
Using the same Myf5-Cre knock-in allele employed in this study, a previous study concluded that Myf5+ precursors give rise to all brown adipocytes and myocytes, but not to any white adipocyte10. Why Seale et al. arrived at a different conclusion is unclear but could reflect the use of a lineage-tracing reporter that is suboptimal for labeling mature adipocytes or limiting the analysis to a few depots. Seale et al also concluded that all brite adipocytes (sometimes called beige adipocytes) arise from a Myf5neg lineage10. However, our results suggest that brite adipocytes can descend from precursors that are either Myf5-Cre (or Pax3-Cre) positive or negative depending upon the depot. Most studies of brite/beige adipocytes focus on psWAT likely explaining why this was previously missed. Notably, the Cre-drivers used in this study are knock-in alleles meaning that Cre expression reflects the activity of the endogenous promoter. While this is ideal for lineage tracing, whether Myf5 and Pax3 proteins co-express or have a functional role in adipocyte development cannot be inferred from studies using these drivers. It also cannot be determined whether the Myf5-Cre and Pax3-Cre drivers express in precursor cells that are common to both myocytes and adipocytes or in separate co-existing lineages. Nevertheless, our results favor of a model in which brown and white adipocytes have multiple developmental origins, a subset of which can be defined by Myf5 and/or Pax3 promoter expression.
The in vivo regulation of adipose tissue development is poorly understood in part because, without knowing the developmental origins of adipocytes, tools to genetically target adipocyte lineages have been lacking. Previous in vivo genetic studies of adipocyte regulation have largely relied on Cre drivers that express in mature adipocytes22, 23. Taking advantage of the fact that Myf5-Cre targets precursors to both brown and white adipocytes, we investigated the function of the insulin receptor in adipocyte development. Consistent with the hypothesis that preadipocytes become insulin-dependent during differentiation24, we found that IRβ is required in the Myf5-lineage for adipogenesis but nor for establishing APCs, and that Myf5-linneg adipocytes can increase in abundance and size to compensate. Such plasticity has been reported previously in skeletal muscle in which Myf5 and MyoD define different skeletal muscle lineages that can compensate for each other during development25, 26. However, we also observed that while adipocyte lineages have plasticity, the degree of plasticity varies depending upon the depot. For example, while Myf5-linneg adipocytes fully compensate for IRβ knockout lineages in asWAT (indicated by the normal size of this depot in the IRβmyf5cKO mice mice), the rWAT is much smaller in the mutant suggesting Myf5-linneg adipocytes can only partially compensate in this depot. Notably, the degree to which Myf5-linneg adipocytes compensate is roughly proportional to the number of Myf5-linneg APCs present in each depot (Fig. 2a). Despite this depot-dependent variation in lineage plasticity however, the IRβmyf5cKO mice maintain similar total body mass as controls by additionally increasing the mass (by increasing individual cell size) of the psWAT and pgWAT depots. This combined presence of lipoatrophy and lipohypertrophy is often observed in human partial lipodystrophy syndromes5.
While lineage tracing in mammals currently can only be performed in mice, we hypothesize that the concepts outlined here may help explain in humans some of the heterogeneity observed within and between adipose tissue depots, body fat patterning variations in the population, and/or the pathology of certain lipodystrophies. Further elucidating other developmental origins of adipocytes, the signals regulating their distribution, and their functional differences will be critical to understanding the pathogenesis of obesity and obesity-associated complications.
Methods
Materials
Antibodies to IRβ, P-T308-Akt, P-S473-Akt and Pan-Akt (all used at 1/1000 dilution) were from Cell Signaling. UCP1 antibody (#10983) was from AbCam (used at 1/100 for IHC and at 1/1000 for western blot). Live/dead Blue (Molecular Probes, L23105), CD31-PE-CY7 (eBioscience, 25-0311)(used at 1/1000), CD45-PE-CY7 (eBioscience, 25-0451) (used at 1/1000), CD29-AlexaFluor700 (Biolegend, 102218)(used at 1/400), CD34-AlexaFluor647 (Biolegend, 119314)(used at 1/200), Sca1-Pacific Blue (BD Bioscience, 560653)(used at 1/1000), CD45-FITC (eBioscience, 11-0451) (used at 1/200) were used for flow cytometry. CL316,243 was from Tocris.
Mice
myf5-cre (stock 007893), R26R-mTmG (stock 007676), pax3-cre (stock 005549), myoD1-cre (stock 01440), adiponectin-cre (stock 010803) and IRβ (stock 006955) mice were from Jackson Laboratory. Mice were housed in the Animal Medicine facilities of the UMMS in a room set at 22°C and 45% humidity under a daily 12h light/dark cycles. All animal experiments were approved by the University of Massachusetts Medical School animal care and use committee.
In vivo experiments
For high-fat-diet experiments, 4-week-old male and female mice were fed high-fat diet of 45% calories from fat (Research Diets) for 2 weeks. CL316,243 (1mg/Kg of body weight) and vehicle control were administered in PBS via i.p. in 5-week-old male and female mice for one week. Both, high-fat diet and CL316,243 treated mice were analyzed at 6 weeks of age. Overnight 6-week-old fasted male and females were subjected to glucose tolerance test by injecting glucose at 2g/Kg of body weight by i.p. injection. Insulin tolerance tests were performed in 6 hours fasted animals. Insulin (HumulinR) was administrated at 0.75 U/Kg of body weight by i.p. injection. Blood glucose was measured by tail bleeding using a commercial blood glucose tester. For acute cold challenge 6-week-old males and female mice were transferred to pre-acclimated cages in a cold room and rectal temperature was measured every hour using the Microtherma 2 system (Braintree Scientific, INC.).
Gene expression and western blot
Total RNA was isolated from cells or tissues using Qiazol (Invitrogen) and an RNeasy kit (Invitrogen). Equal amounts of RNA were retro-transcribed to cDNA using a High Capacity cDNA reverse transcription kit (#4368813, Applied Biosystems). qRT-PCR was run in 10 μL reactions in a StepOnePlus real-time PCR machine from Applied Biosystems using SYBR Green PCR master mix (#4309156, Applied Biosystems) according to manufacturers instructions. Standard and melting curves were run in every plate for every gene to ensure efficiency and specificity of the reaction. Primer sequences: TBP Fw:5'- GAAGCTGCGGTACAATTCCAG, TBP Rv: 5'- CCCCTTGTACCCTTCACCAAT, SREBP1c Fw: 5'- AAGCAAATCACTGAAGGACCTGG, SREBP1c Rv: 5'- AAAGACAAGCTACTCTGGGAG, ACL Fw: 5'- CTCACACGGAAGCTCCATAA, ACL Rv: 5'- ACGCCCTCATAGACACCATC, ACC Fw: 5'- GGAGATGTACGCTGACCGAGAA, ACC Rv: 5'- ACCCGACGCATGGTTTTCA, FAS Fw: 5'- GCTGCGGAAACTTCAGGAAAT, FAS Rv: 5'- AGAGACGTGTCACTCCTGGACTT, UCP1 Fw: 5'- CTGCCAGGACAGTACCCAAG, UCP1 Rv: 5'- TCAGCTGTTCAAAGCACACA, PRDM16 Fw: 5'- GACATTCCAATCCCACCAGA, PRDM16 Rv: 5'- CACCTCTGTATCCGTCAGCA, cidea Fw: 5'- ATCACAACTGGCCTGGTTACG, cidea Rv: 5'- TACTACCCGGTGTCCATTTCT, tdTomato Fw: 5'- GCTGAAGGGCGAGATCCA, tdTomato Rv: 5'- GTGGGAGGTGATGTCCAGCTT, eGFP Fw: 5'- CATGCCCGACTACGT, eGFP Rv: 5'- GCTTGTGCCCCAGGATGTT.
For western blot, tissues were homogenized using a TissueLyser (Quiagen) system in a buffer containing 50mM Hepes, pH 7.4, 40mM NaCl, 2mM EDTA, 1.5mM NaVO4, 50mM NaF, 10mM sodium pyrophosphate, 10mM sodium β-glycerophosphate, 0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100. An equal amount of total protein was loaded into acrylamide/bis-acrylamide gels and transferred to PVDF membranes for detection with the indicated antibodies. All antibodies for Western blot were used at dilution 1/1000.
UCP1 IHC
Fat sections were subjected to UCP1 IHC according to27. Briefly, fat sections were re-hydrate and antigen retrieval was done by incubating the sections in citrate buffer at 90–95°C water for 20 min. After blocking, primary antibody was applied overnight at 4°C. Next day, SuperPicture 3rd Gen IHC Detection Kit (Novex) was used for detection.
Tissue harvest
Adipose tissues used were carefully dissected to avoid surrounding tissue contamination. Adipose tissue notation used here was described previously8, 28. psWAT is equivalent to inguinal WAT and paBAT is equivalent to mediastinic BAT in28.
Adipocyte precursor cell (APC) isolation and FACS analysis
Adipose progenitor cells (APC) were isolated as follows8, 29: Stromal-vascular fraction (SVF) was prepared from each fat pad by collagenase treatment and SVF pellets were resuspended in erythrocyte lyses buffer (0.15 M ClNH4, 0.01 M HCO3K in water). Cells were then pelleted by centrifugation, resuspended in staining media (HBSS + 2% FBS), and labeled with appropriate antibodies. After staining, cells were filtered through a 35-μm cell-strainer capped tube to ensure single cell suspension and stained with live/dead Blue. Live single cells were gated according to the expression of surface markers (CD31−CD45−CD29+CD34+Sca1+) in a BD LSRII analyser. Data was analyzed with FlowJo.
Whole-mount confocal microscopy
Fats were dissected and cut in approximately 5mm × 5mm cubes. Fragments were mounted with Fluoromount-G (Southern Biotech). Mounted samples were imaged the same day on a LSM 5 Pascal (Zeiss) point scanner confocal system. To acquire the images for quantification, a general survey of the entire tissue was first performed using a 10X objective to determine the uniformity of labeling throughout the tissue. Then, the tissue was subdivided into quadrants and a 40X oil immersion objective was used to obtain random images from each quadrant. Background fluorescence was offset using tissues from animals not carrying the R26R-mTmG reporter. tdTomato and eGFP settings were set using fat tissues from mice carrying the R26R-mTmG reporter and from adiponectin-Cre;R26R-mTmG mice, respectively. eGFP was excited at 488 nm and detected from 515 to 565 nm. tdTomato was excited at 543 nm and detected from 575 to 640 nm.
Histology and image analysis
For muscle histology, fresh dissected muscles were flash-frozen in OCT using dry-ice cold isopentane. Frozen transversal sections (10 μm) were obtained and mounted in Prolong Gold. Images were taken on the same day on a LSM 5 Pascal (Zeiss) point scanner confocal system. ImageJ (NIH) was used to calculate adipocyte diameter in formalin fixed tissues that were sectioned at 5 μm thickness. Embedding and sectioning was done by the UMass Morphology Core. ImajeJ was used for tdTomato and eGFP channel isolation. Cell counter plug-in of ImageJ was used to count by hand the tdTomato/eGFP adipocytes. More than 100 adipocytes per tissue were counted.
Statistical analysis
Data is presented as mean+S.E.M, unless stated otherwise. Student t-test or ANOVA, as appropriate, were used to determine statistical significance (P<0.05). No pre-test was used to choose sample size. No data points were excluded. Statistical analysis was done using GraphPad Prism v6.0c. The number of mice used per experiment is stated in each figure legend
Supplementary Material
Acknowledgements
We thank the UMMS flow core. We thank Dr. Marc R. Freeman and colleagues for access to their microscope and imaging assistance. We thank the Guertin lab for valuable discussions and especially to Huawei Li for assisting with animal colony management. D.A.G is supported by grants from the National Institutes of Health (R01DK094004), the Pew Charitable Trusts, and the American Diabetes Association. J.S.-G. was supported by the Beatriu de Pinós program from the Generalitat de Catalunya.
Footnotes
Author contribution J.S.-G. and D.A.G designed and conducted experiments, analyzed and interpreted data, and wrote the manuscript.
Competing financial interests The authors declared no competing financial interest.
References
- 1.Cinti S. The adipose organ. Prostaglandins, leukotrienes, and essential fatty acids. 2005;73:9–15. doi: 10.1016/j.plefa.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Cinti S. The adipose organ at a glance. Disease models & mechanisms. 2012;5:588–594. doi: 10.1242/dmm.009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst KL. Rare adipose disorders (RADs) masquerading as obesity. Acta pharmacologica Sinica. 2012;33:155–172. doi: 10.1038/aps.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahima RS, Lazar MA. Physiology. The health risk of obesity--better metrics imperative. Science. 2013;341:856–858. doi: 10.1126/science.1241244. [DOI] [PubMed] [Google Scholar]
- 7.Berry DC, Stenesen D, Zeve D, Graff JM. The developmental origins of adipose tissue. Development. 2013;140:3939–3949. doi: 10.1242/dev.080549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Gurmaches J, Hung CM, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell metabolism. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature reviews Molecular cell biology. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 10.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 12.Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 13.Lang D, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto M, et al. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon B, Nedergaard J. Cell biology: Neither brown nor white. Nature. 2012;488:286–287. doi: 10.1038/488286a. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Gurmaches J, Hung C-M, Sparks CA, Tang Y, Li H, Guertin DA. PTEN Loss in the Myf5 Lineage Redistributes Body Fat and Reveals Subsets of White Adipocytes that Arise from Myf5 Precursors. Cell Metabolism. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 19.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell metabolism. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billon N, et al. The generation of adipocytes by the neural crest. Development. 2007;134:2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bluher M, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Developmental cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 23.Boucher J, et al. Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nature communications. 2012;3:902. doi: 10.1038/ncomms1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith PJ, Wise LS, Berkowitz R, Wan C, Rubin CS. Insulin-like growth factor-I is an essential regulator of the differentiation of 3T3-L1 adipocytes. The Journal of biological chemistry. 1988;263:9402–9408. [PubMed] [Google Scholar]
- 25.Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- 26.Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited versus nonrecruited molecular signatures of brown, “brite” and white adipose tissues. Am J Physiol Endocrinol Metab. 2011 doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 29.Rodeheffer MS, Birsoy K, Friedman JM. Identification of White Adipocyte Progenitor Cells In Vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.