Abstract
Background
Common variation at the 11p11.2 locus, encompassing MADD, ACP2, NR1H3, MYBPC3 and SPI1, has been associated in genome-wide association studies with fasting glucose (FG) and insulin (FI). In the Cohorts for Heart and Aging Research in Genomic Epidemiology Targeted Sequencing Study, we sequenced five gene regions at 11p11.2 to identify rare, potentially functional variants influencing FG or FI levels.
Method & Results
Sequencing (mean depth 38×) across 16.1kb in 3,566 non-diabetic individuals identified 653 variants, 79.9% of which were rare (MAF <1%) and novel. We analyzed rare variants in five gene regions with FI or FG using the Sequence Kernel Association Test (SKAT). At NR1H3, 53 rare variants were jointly associated with FI (p=2.73 × 10−3); of these, seven were predicted to have regulatory function and showed association with FI (p=1.28 × 10−3). Conditioning on two previously associated variants at MADD (rs7944584, rs10838687) did not attenuate this association, suggesting that there are more than two independent signals at 11p11.2. One predicted regulatory variant, chr11:47227430 (hg18; MAF 0.00068), contributed 20.6% to the overall SKAT score at NR1H3, lies in intron 2 of NR1H3 and is a predicted binding site for FOXA1, a transcription factor associated with insulin regulation. In human HepG2 hepatoma cells, the rare chr11:47227430 A allele disrupted FOXA1 binding and reduced FOXA1-dependent transcriptional activity.
Conclusion
Sequencing at 11p11.2- NR1H3 identified rare variation associated with FI. One variant, chr11:47227430, appears to be functional, with the rare A allele reducing transcription factor FOXA1 binding and FOXA1-dependent transcriptional activity.
Keywords: fasting glucose, fasting insulin, chr11p11.2, target sequencing, next-generation sequencing
INTRODUCTION
High fasting blood levels of glucose (FG) and/or insulin (FI) are hallmarks of type 2 diabetes. Since 2007, genome-wide association studies (GWAS) of type 2 diabetes and diabetes-related quantitative traits have identified 53 common, consistently replicated single nucleotide variants (SNVs) associated with FG and FI. 1 One especially intriguing and complex region identified as influencing FG is the chromosome 11p11.2 locus. This region contains rs7944584, located in intron 25 of the MAP kinase-activating death domain (MADD) gene, and was associated with a 0.021 mg/dl per (A) allele increase in FG (p = 2.0 × 10−18) in a large GWAS of individuals of European ancestry.2 In a related GWAS of proinsulin, an additional signal at the 11p11.2 locus was identified for rs10838687 (r2 with rs7944584 = 0.074 in HapMap 2 CEU), which is intronic in MADD and associated with a 0.08 mg/dl difference in levels of fasting proinsulin (p = 1.1×10−88). 3 Thus, the 11p11.2-MADD locus appears to be consistently associated with insulin and glucose regulation, but the region encompasses additional genes plausibly linked to glycemic regulation. Follow-up studies to more precisely localize and characterize genes underlying GWAS-identified signals and to discover functional variants in genes or regulatory regions are a next step to further insights into the genetic pathways involved in glycemic regulation and type 2 diabetes risk. In this study we conducted high-throughput next-generation deep sequencing at the polygenic 11p11.2 locus to localize previously observed association signals and identify rare, potentially functional variants influencing FG and FI levels.
METHODS
The design of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Targeted Sequencing Study, including the study cohort sampling design, laboratory methods for targeted next generation deep sequencing, and rare and common variant statistical analyses, has been described in detail in Lumley et al. 4 and Lin et al. 5 These methods are described briefly here, with a focus on details specific to the analysis of FG and FI.
Study Cohort Sample
The CHARGE Targeted Sequencing Study is comprised of individuals of European ancestry from three cohorts that are part of the larger CHARGE consortium: the Atherosclerosis Risk in Communities (ARIC) study, Cardiovascular Health Study (CHS), Framingham Heart Study (FHS).6 The CHARGE Targeted Sequencing Study included a cohort random sample and selected case groups from a variety of related cardio-metabolic phenotypes, including a sample of approximately 200 participants (100 ARIC, 50 CHS, 50 FHS) from the high extremes of fasting (≥8-hour fast) insulin (FI) in individuals without diabetes, defined as either being diagnosed by a physician (ARIC), treated for diabetes or having a fasting glucose (FG) >7mmol/L (ARIC, FHS and CHS). FHS participants with type 1 diabetes were excluded from selection. Men and women were selected equally from each cohort, giving 3,566 individuals with successful sequencing and measured trait available for analysis of FG and FI as continuously distributed quantitative traits. All ARIC, FHS and CHS subjects provided written, informed consent to participate in research protocols that were approved by the University of North Carolina at Chapel Hill, Chapel Hill, NC (ARIC), Boston University, Boston, MA (FHS) and University of Washington, Seattle, WA (CHS) institutional review boards.
Quantitative Traits Measurement
FG and FI were measured from fasting plasma (FHS) or fasting serum (CHS, ARIC).7-9 In FHS, plasma was collected after a ≥8 hour overnight fast. FG was measured using a hexokinase assay (A-gent glucose test, Abbott, South Pasadena, California) and FI was measured on frozen specimen using the DPC Coat-A-Count RIA (total immunoreactive insulin) assay (assay sensitivity, 1.2 microU/mL). In CHS, FG (≥12-hour fast) was measured using a Kodak Ektachem 700 Analyzer assay and FI was measured using a competitive RIA (Diagnostic Products Corp., Malvern, PA). In ARIC (after a ≥8 hour fast), FG was measured using the hexokinase/glucose-6-phosphate dehydrogenase method and FI was measured by radioimmunoassay (125 Insulin kit; Cambridge Medical Diagnosis, Bilerica, MA)(assay sensitivity, 2μU/ml).
Targeted Next Generation Deep Sequencing
Target selection in the CHARGE Targeted Sequencing Study included genomic regions shown to exhibit pleiotropy, or apparent association of a single locus on multiple traits, and included the 11p11.2 locus encompassing ACP2, NR1H3, MADD, MYBPC3 and SPI1. 5 We selected and sequenced regions with high regulatory potential for a total of 16.08kb, sequenced at a mean depth of 38× across the five gene regions (Supplemental Material Table 1). Sequence regions at each locus included key variants selected by GWAS trait associations or known gene expression (eQTL) associations, and HapMap CEU LD of r2>0.5 with other SNPs identified by GWAS or eQTL approaches, and regions displaying high sequence conservation across mammals and/or vertebrates (28-species PHAST conservation scores >500 10). As detailed in Lin et al. 5 sequencing was performed on the SOLiD™ next-generation sequencing platform, with cross-validation of sequence-identified genotypes by comparison with genotypes on the Affymetrix Gene Chip 500K Array Set & 50K Human Gene Focused Panel in 1,096 FHS samples. A total of 558 SNPs were shared between the two platforms. After excluding missing genotypes, 98.0% of genotypes were concordant between the two platforms, suggesting high accuracy of the sequenced genotypes.
Variant Classification and Annotation
We classified variants identified by sequencing across the 11p11.2 locus as common if the study-wide minor allele frequency (MAF) in the merged data sets (FHS, CHS and ARIC) was ≥1% and rare if the MAF was <1%. Variants were classified as novel if they were not described in dbSNP or the 1000 Genomes Project.11 Functional prediction annotations were obtained from the RegulomeDB database,12 which scores variants for regulatory potential, and includes data from ENCODE (ENCODE transcription factor (TF) ChIP-seq, histone ChIP-seq, FAIRE, and DNase I hypersensitive sites) 12 and other sources (including transcription factor ChIP-seq data available from the NCBI Sequence Read Archive 13-20 as well as eQTL,21-29 dsQTL,30 and ChIP-exo31 data) in addition to computational predictions and manual annotations to identify putative regulatory and functional variants. RegulomeDB prediction categories can be found in Supplemental Table 2. Based on RegulomeDB prediction scores, variants were grouped to determine which specific types of variants contributed most to the overall joint variant effect at a locus. For example, variants likely to influence local gene transcription have prediction scores 1-3 which includes transcription factor binding sites, transcription factor motifs, DNase Footprints and/or DNase peaks.
Follow-Up Genotyping of Rare Variants in FHS
To test whether chr11:47227430 had a discernable effect on FI levels we genotyped the variant in FHS subjects using TaqMan (ABI PRISM 7700 HT Sequence Detection System, Applied Biosystems, Foster City, California). Genotyping success rate was >98%. Genotyping quality was tested using duplicate samples in each 384-well assay along with synthetic oligonucleotide positive controls for the rare A/G variant. The average agreement rate comparing duplicates was greater than 99% and the agreement of positive controls with the expected genotype was 100%.
Electrophoretic Mobility Shift Assays
Allele-specific binding efficiency of the forkhead box A1 (FOXA1) transcription factor to the motif around chr11:47227430 site was evaluated using Electrophoretic Mobility Shift Assays (EMSA). FOXA1 protein was expressed using the TnT coupled Reticulocyte Lysate System (Promega, Madison, WI) programmed with a expression vector containing FOXA1 cDNA cloned into the KpnI and BamH1 sites of pcDNA3.1(+) (Invitrogen, Burlington, ON). Expression of FOXA1 protein in the reticulocyte lysate was confirmed by western blotting. DNA oligonucleotides were annealed and labelled using the DIG Gel Shift Labelling Kit (Roche Applied Science, Laval, QC). For EMSA, 5 μL of programmed reticulocyte lysate was incubated with 0.6 pmol of digoxigenin-labelled probe in EMSA binding buffer (DIG Gel Shift Labelling Kit, Roche Applied Science, Laval, QC) supplemented with 100 ng single-stranded non-specific oligonucleotide 32 and 50 ng denatured single-stranded DNA. Unlabelled competitor oligonucleotides were added at 5×-100× molar excess and the reaction was incubated for 20 minutes at room temperature. DNA-protein complexes were separated using a non-denaturing polyacrylamide gel (5% acrylamide / bisacrylamide (29:1); 0.5× Tris/Borate/EDTA), detected by chemiluminescence using an alkaline phosphatase-conjugated antibody (Anti-Digoxigenin-AP, Roche); and imaged using autoradiography. Sequences for these oligonucleotides are provided in Supplemental Table 5.
Transient transfection assays
Allele-specific effects of chr11:47227430 on FOXA1-mediated gene transcription was studied using dual luciferase assay experiments. Chromatin immunoprecipitation studies completed by the ENCODE project show that chr11:47227430 is located in a 280-bp region of active chromatin in HepG2 cells that binds FOXA1 33 This 280bp DNA fragment (containing the reference G allele), was amplified and cloned into the pGLuc Mini-TK expression vector (New England Biolabs, Whitby, ON), which contains a gaussia luciferase reporter gene controlled by a minimal promoter from the HSV thymidine kinase. Overlap extension PCR was used to create a second reporter containing the FI-associated A allele at the chr11:47227430 site. To isolate the effects of FOXA1 in this region, a second set of reporter plasmids was constructed that contained a 40bp genomic region centered on chr11:47227430 and flanked by approximately two helical turns of DNA on each side. As a positive control for FOXA1-mediated transcription, we tested expression of a reporter plasmid containing a single copy of the well characterized albumin gene enhancer FOXA1 motif or FB (FOXA1 Binding, 5′-TGTTTGTTCTT-3′)34. The control firefly luciferase expression vector (pGL3 Control Vector, Promega, Cat. # E1741) was used to correct for transfection efficiency. All the constructs were verified by Sanger DNA sequencing. Transfections were performed in quadruplicate using HepG2 cells grown in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies, Burlington, ON) supplemented with 10% fetal bovine serum (Sigma-Aldrich, Oakville, ON). The cells were seeded in 96-well plates; grown to 50% confluence; and co-transfected with 100 ng of plasmid DNA using Trans-LT1 transfection reagent (Mirus Bio, Madison WI). The transfection mix contained 86 ng pGluc Mini-TK reporter, 4 ng internal control (25:1 ratio) and 10 ng pcDNA3.1(+)-FOXA1 or pcDNA3.1(+) alone. The cells were lysed 48 hours following transfection and measured gaussia and firefly luciferase activities using the Dual Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instructions. Cells transfected with an empty pGluc Mini-TK backbone were used to adjust for background activity. The responsiveness of each construct to FOXA1 co-expression, was measured by comparing the transcriptional activity of each construct in cells transfected with pcDNA3.1-FOXA1 with cells transfected with empty pcDNA3.1.
Statistical Analyses
All analyses were adjusted for age, sex and study design variables (i.e. clinic site for CHS and ARIC and recruitment cohort for FHS). FG was analyzed as per mg/dL. FI, in pmol/L, was naturally log transformed to improve normality and was additionally adjusted for body mass index (BMI), measured using standard methods as previously described.35 The analytic strategy for sequence-based variants, outlined in brief below, followed the approach described in detail in Lumley et al 4; and Lin et al. 5 We used two-sample t-tests and chi-square tests for analysis of functional assay results and of trait values by genotype category for follow-up genotyping in FHS.
Rare Variant Analyses
Rare variants within each of the five 11p11.2 gene regions (ACP2, NR1H3, MADD, MYBPC3 and SPI1) were jointly tested for association with FG or BMI-adjusted ln(FI) using the Sequence Kernel Association Test (SKAT).36 FHS used a SKAT test that accounted for family structure.37 SKAT tests were conducted within each of three cohorts and meta-analysed using a weighted sum of squares of z-statistics from single-variant score tests. These variant scores were squared, weighted by a function of MAF, and summed to create a Q statistic. The significance of the Q statistics was determined using an asymptotic distribution, as described in Wu et al.36 The Q statistic, calculated by taking the weighted squared z-score for each variant and dividing by the total Q statistic, can be used to identify variants contributing most to the signal.
Primary rare variant analyses included all rare variants in each gene region. Secondary analyses included only variants that were considered likely to be functional based on annotation predictions (specifically, variants with RegulomeDB prediction scores 1 – 3, see Supplemental Table 2) in both non-coding and coding regions. To control type 1 error a p-value < 0.05/20 = 0.0025 (corrected for 20 tests: 2 traits × 5 gene regions × 2 analyses) was used to define statistical significance for rare variant tests, where significance indicates that in aggregate the tested rare variants at the locus are associated with variation in FG and/or FI levels to a greater degree than expected by chance.
Common Variant Analyses
We tested common variants identified by deep sequencing for association with FG or BMI-adjusted ln(FI) using standard additive genetic linear regression models within each cohort. Mixed effects models were used in FHS to account for familial correlation. We meta-analyzed summary statistics from each cohort using standard fixed-effect inverse-variance weighted meta-analysis. 38 P-values were obtained from unweighted regression models. Analyses weighted by the inverse of the sampling probability were used to obtain unbiased estimates of effect size. 4 To control type 1 error the significance threshold for common variant analyses was set at p < 2.16 × 10−4 (0.05 corrected for 232 tests: 2 traits × 116 variants).
Conditional Analyses
The MADD locus harbors two common variants (r2=0.07 in HapMap 2 CEU) associated with diabetes-related quantitative traits, rs794584 (associated with FG; Dupuis et al.2) and rs10838687 (fasting proinsulin levels; Strawbridge et al.39). A second reported signal at MADD, rs10501320, was in high linkage disequilibrium with rs794584 (r2 = 0.924 in HapMap 2 CEU) and thus was not included in conditional rare variant analyses. The conditional analysis tested the hypothesis that these two known common variants explain (at least in part) the observed rare variant association at any of the five genes at 11p11.2. The conditional analysis was conducted by including the two common variants along with the rare variants at each of five gene regions in SKAT. In CHS and FHS we used rs794584 and rs10838687 and in ARIC rs4752979 was used as a proxy for rs10838987 (r2 = 1.00 in HapMap CEU). Conditioned SKAT tests were conducted within each of the three cohorts, then meta-analysed, with results compared with the unconditioned meta-analysed SKAT p-values. If unconditioned and conditioned p-values are similar, this suggests that known common and novel rare variation represent different signals at the locus.
All sequence variant-trait association analyses were conducted using custom scripts implemented in R 2.9.2. Adjustment for familial correlation (FHS) was done using the R kinship package. Functional assays were conducted using eight replicates and differences between groups in reporter assay results were tested using two sample t-tests.
RESULTS
Participant Characteristics
About half of the 3,566 non-diabetic study participants were women, the mean age ranged from 52 to 72 years, the mean BMI was in the “overweight” range and mean FG was in the high normal range (Table 1). FI values varied widely across studies, as have been observed previously. 2
Table 1.
Characteristics of 3,566 non-diabetic participants in three cohorts of European in the CHARGE Targeted Sequencing Study
| Framingham Heart Study | Cardiovascular Health Study | Atherosclerosis Risk in Communities | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Fasting Insulin | Fasting glucose | Fasting Insulin | Fasting glucose | Fasting Insulin | Fasting glucose | |
| N | 811 | 838 | 967 | 1,761 | ||
| Sex (% Women) | 51.7 | 55.3 | 50.3 | |||
| Age [years]a | 54.1 (10.7) | 72.5 (5.4) | 54.7(5.7) | |||
| Body Mass Index [kg/m2]* | 27.9 (6.5) | 26.4 (5.0) | 27.2(5.7) | |||
| Fasting Insulin [pmol / l]* | 32.6 (21.3) | 103.1 (63.9) | 83.1(73.2) | |||
| Fasting Glucose [mg/dL]* | 94.7 (9.4) | 100.4 (9.7) | 99.2(9.6) | |||
mean (standard deviation)
Variants Identified by Targeted Deep Sequencing
Counts and characteristics of the rare and common variants identified by deep sequencing at five genes at the chromosome 11p11.2 locus are shown in Table 2, Supplemental Table 1 and Supplemental Table 2. Deep (mean coverage 38×) sequencing across 16.1 kb produced 653 variants, 79.9% (N = 522) of which were rare and novel, and 116 were common. Annotation information for coding and non-coding variation was available for 485 (74.3%) of the variants. These included 61 rare variants with RegulomeDB Prediction scores 1-3, suggesting potential effects on gene or chromatin regulation (Supplemental Table 2).
Table 2.
Counts and characteristics of 653 single nucleotide variants identified by deep sequencing at five genes at the chromosome 11p11.2 locus in the CHARGE Targeted Sequencing Study
| ACP2 | NR1H3 | MADD | MYBPC3 | SPI1 | Total | |
|---|---|---|---|---|---|---|
|
| ||||||
| Total | 42 (9) | 53 (13) | 226 (45) | 57 (10) | 159 (39) | 537 (116) |
| Coding | 3 (0) | 5 (0) | 17 (4) | 4 (1) | 0 (0) | 29 (5) |
| Novel | 41 (4) | 53 (5) | 220 (10) | 53 (0) | 155 (12) | 522 (31) |
|
Predicted Function
(RegulomeDB Prediction scores 1-3) |
11 (4) | 7 (3) | 26 (19) | 0 (9) | 17 (19) | 61 (54) |
Counts above include common (MAF ≥ 1%) variants in parentheses and rare (MAF < 1%) variants.
Rare Variant Associations with FI and FG
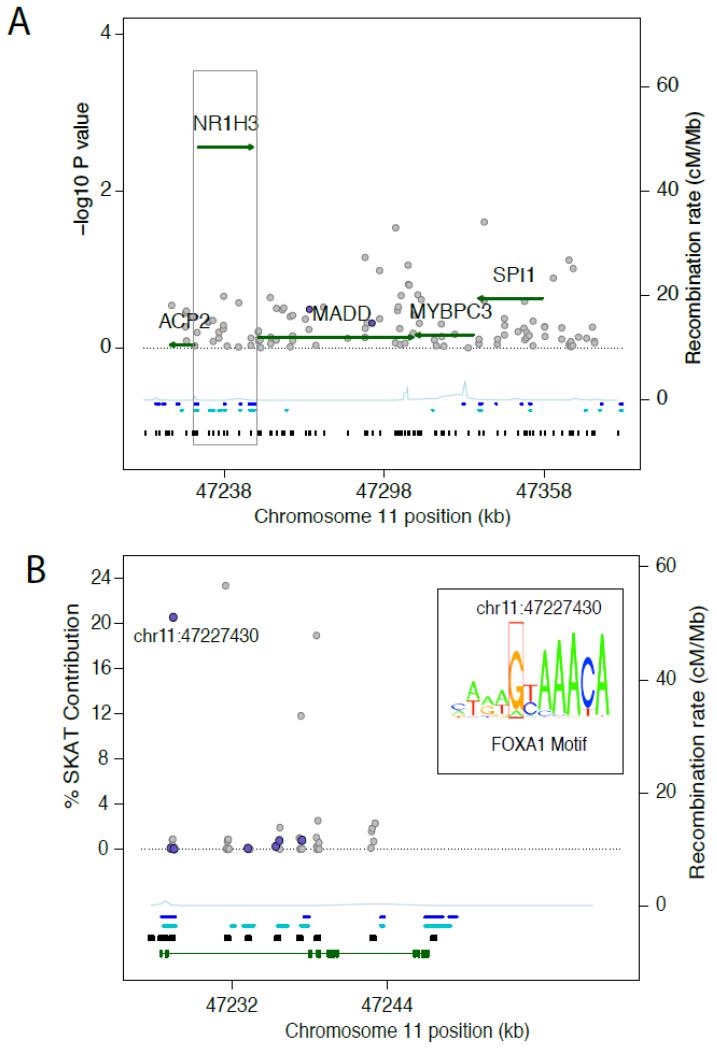
Meta-analyzed SKAT results (Table 3 and Figure 1), showed that 53 rare variants at the NR1H3 (nuclear receptor subfamily 1 group H member 3) locus were significantly associated with FI (pmeta =2.73 ×10−3). A subset of variants with RegulomeDB prediction scores 1-3 (N = 7) was also significantly associated (pmeta =1.28 ×10−3) with FI. Four rare NR1H3 variants accounted for 74.6% of the overall SKAT Q statistic (Figure 1 Panel B and Supplemental Table 3). One of these variants, chr11:47227430 (MAF = 0.00068; 6 individuals in the source cohorts with the risk allele) accounted for 20.54% of the overall SKAT Q statistic. Chr11:47227430 was located in intron 2 of the longer isoform of NR1H3 in a predicted binding site for the forkhead box A1 (FOXA1) transcription factor, with the A to G substitution altering the fifth position of the FOXA1 consensus motif (Figure 1 Panel B inset). FOXA1 is a key transcriptional regulator of hepatic and pancreatic development as well as insulin, glucagon, glucose and adipocyte metabolism.40, 41 Three individuals without diabetes carrying the A allele for this variant (N=1, N=1, N=1 in each of ARIC, CHS and FHS) had increased FI levels (ARIC: 172.2 pmol / l, CHS: 194.5 pmol / l, FHS: 68 pmol / l) compared to their corresponding population mean (Table 1), but in FHS 7 individuals with the AG genotype (all were non-diabetic and no one had a AA genotype) had a similar FI level (72.0 pmol / l) compared with 2,458 with the GG genotype (84.9 pmol / l, p = 0.045). No additional rare variant associations were observed for FG or FI.
Table 3.
Fasting insulin and glucose association meta-analyses of multivariate rare variants SKAT tests at five genes at the chromosome 11p11.2 locus in the CHARGE Targeted Sequencing Study
| Fasting Insulin | Fasting Glucose | |||
|---|---|---|---|---|
|
| ||||
| All | Predictions 1-3 | All | Predictions 1-3 | |
| ACP2 | 0.91 | 0.93 | 0.10 | 0.57 |
| NR1H3 | 2.73 × 10−3 | 1.28 × 10−3 | 0.73 | 0.08 |
|
NR1H3 Conditioned on
GWAS variants * |
0.08 | 1.71 × 10−3 | ||
| MADD | 0.73 | 0.83 | 0.73 | 0.95 |
| MYBPC3 | 0.69 | - | 0.26 | - |
| SPI1 | 0.23 | 0.51 | 0.7 | 0.24 |
P-values from meta-analysis of multivariate SKAT analyses for fasting insulin and fasting glucose from three cohort samples. Included are results from all rare variants and the subset of variants with prediction scores between 1 and 3 obtained from RegulomeDB.12 Variants with these prediction scores were predicted to be any (or a combination) of the following: transcription factor binding site, transcription factor motif, DNase Footprint and/or DNase peak.
NR1H3 Fasting Insulin analyses were conditioning on GWAS variants rs7944584 and rs10838687 to access if known common variants explain (at least in part) the observed rare variant association.
Significance level: α = < 0.05/20 = 0.0025 (2.5 × 10−3)
Figure 1.
Regional plots for association of fasting insulin with common and rare variants at the chromosome 11p11.2 locus.
(A) Regional association at chromosome 11p11.2. The −log10P value of variant associations with fasting insulin is plotted on the left y-axis versus chromosomal location on the x-axis (NCBI Genome Build 36). Regions targeted for sequencing are marked with black boxes along the x-axis. Common variant associations are shown as grey circles, and rare variant associations as green arrows that mark the −log10 P from meta-analysis of SKAT and the genomic position of the gene region containing the aggregate rare variants tested. DNAse I hypersensitivity sites in pancreatic (blue dots) and hepatic cells (teal dots) are marked by their chromosomal position along the x-axis. The regional recombination rate is plotted in light blue with values on the right y-axis. The grey box indicates the NR1H3 region (meta-analysis P-value = 2.73 × 10−3) that is expanded and shown in Panel B.
(B) Regional plot of 53 rare variants at NR1H3 and their individual percent contribution to the overall regional meta-analytic SKAT Q test statistic. Variants with functional prediction 1-3 are shown as purple circles and variants with other functional prediction or without annotation by grey circles. The x-axis shows the chromosomal position of the variants with the regions sequenced in black boxes. Additionally, DNAse I hypersensitivity sites in pancreatic (blue dots) and hepatic cells (teal dots) and an ideogram for the NR1H3 gene body (green line) are marked. The left y-axis shows the per-cent contribution of each variant to the overall meta-analysis SKAT Q statistic for the multi-variant test at NR1H3. The marked variant chr11:47227430 is predicted to be functional and has a large contribution to the SKAT Q statistic. The regional recombination rate is plotted in light blue with values on the right y-axis. The inset shows the consensus logo for the FOXA1 binding motif where the chr11:47227430 A-G substitution produces an adenine instead of a guanine in the fifth position, marked in the red box. The relative conservation of each nucleotide in the motif is displayed by its height. 12
Common Variant Association Tests
None of the 116 common variants was significantly associated with FG or FI (see Supplemental Figure 1 and Figure 1 Panel A, respectively), including known MADD region GWAS SNPs rs7944584 (MAF= 0.278, pmeta = 0.48) and rs10838687 (MAF = 0.194, pmeta = 0.32) (Supplemental Material Table 4).
Conditional Analyses of Known Common Variants on Rare Variants Tests
SKAT of all 53 rare variants at NR1H3 conditioned on rs7944584 and rs10838687 eliminated the significance of their association with FI (pconditioned = 0.08), but conditioned SKAT of the 7 highly functional variants remained significantly associated with FI (pconditioned = 1.71 × 10−3)(Table 3).
Functional characterization of the FI-associated variant at chr11:47227430
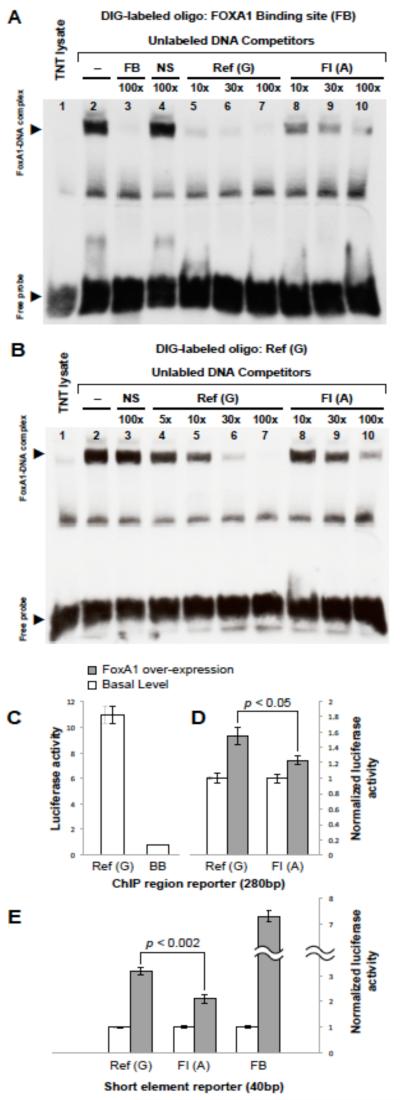
To assess the effects of the chr11:47227430 variant on FOXA1-mediated transactivation, we first used EMSA to examine allele-specific disruption of FOXA1-DNA complexes using a well-characterized FOXA1 binding site located in the albumin gene enhancer (FB).34 We found that the reference G allele at chr11:47227430 specifically and nearly completely displaced FOXA1 binding at its FB site at 10-fold excess, whereas a non-specific competitor oligonucleotide (NS) did not displace FOXA1 binding at 100-fold excess (Figure 2A). In addition, the reference G allele displaces binding of the FB oligonucleotide oligonucleotide at much lower concentrations than the FI-associated A allele, suggesting that the G allele bound with greater stability. We confirmed these findings by showing that the reference G allele was bound directly by FOXA1 and that the FI-associated allele could not completely displace the reference G allele even at 100× molar excess, highlighting the higher affinity of the reference G allele towards FOXA1 (Figure 2B). The functional consequences of these differences in FOXA1 binding were assessed in transient transfection assays. We showed that the 280-bp region surrounding chr11:47227430 exhibited enhancer activity in HepG2 cells (Figure 2C), which was significantly elevated after over-expression of FOXA1 (Figure 2D), supporting a role of FOXA1 in regulation of gene expression by binding to chr11:47227430. Most interestingly, the FI-associated A allele showed significantly reduced FOXA1-dependent transactivation (Figure 2D, E), suggesting that this variant reduces enhancer activity by attenuating FOXA1 binding.
Figure 2.
The FI-associated A allele disrupts FOXA1 binding and transactivation potential at chr11:47227430.
Panels A and B. Gel electrophoretic mobility shift assays (EMSA) show that FOXA1 binds to the chr11:47227430 site, with the reference G allele having higher affinity for FOXA1 compared to the FI-associated A allele.
(A) A DIG-labeled oligonucleotide containing well-characterized FOXA1 binding site (FB) in the albumin gene enhancer was incubated with unprogrammed reticulocyte lysate (lane 1) or reticulocyte lysate programmed to express FOXA1 (Lanes 2-10). A retarded complex (upper arrowhead) is seen after nondenaturing acrylamide gel electrophoresis (lane 2), which is competed by 100-fold molar excess unlabeled oligonucleotide (lane 3), but not by an equivalent amount of nonspecific competitor oligonucleotide (NS, lane 4). While a synthetic oligonucleotide surrounding the reference G variant can efficiently compete with the DIG-labeled synthetic oligonucleotide carrying the FB site (lanes 5 to 7), the FI-associated A variant cannot displace the labeled probe even when used up to 100-fold molar excess (lanes 8 to 10), suggesting that G->A substitution at the chr11:47227430 site disrupts FOXA1 binding.
(B) Binding and competition assays reciprocal to those in panel A confirm specific binding of FOXA1 to the reference G allele at chr11:47227430. Here, the synthetic oligonucleotide surrounding the reference G allele was labeled with DIG, and is shown to form a specific complex with FOXA1 (lane 2, upper arrowhead) that cannot be efficiently displaced by unlabeled NS oligonucleotide (lane 3). Increasing amounts of an unlabeled oligonucleotide containing the ref-G allele displaces the bound DIG-labeled ref-G oligonucleotide at lower molar fold excess than an unlabeled oligonucleotide carrying the FI-associated A variant (compare lanes 4-7 to lanes 8-10).
Panels C-E. The reference G allele at chr11:47227430 has significantly transactivation potential compared to the FI-associated A allele in luciferase reporter assays.
(C) Dual luciferase assays show that a reporter plasmid containing the 280bp region surrounding the chr11:47227430 site exhibits enhancer activity in HepG2 cells [Ref(G)] compared to the minimal HSV thymidine kinase (TK) promoter in the pGluc Mini-TK backbone (BB).
(D) The 280bp region surrounding chr11:47227430 shows FOXA1-dependent enhancer activity in HepG2 cells. The FI-associated A variant [Ref(A)] is significantly less responsive to FOXA1 over-expression compared to the reference G allele [Ref(G)]. For each construct, luciferase measurements are normalized by the corresponding reporter activity measured in the absence of FOXA1 co-expression.
(E) Luciferase activity was measured for reporter constructs containing 40-mer oligonucleotides centered on the chr11:47227430 reference [Ref(G)] or risk [FI(A)] alleles or a well characterized albumin enhancer FOXA1 binding site (FB). For each construct, luciferase measurements are normalized by the corresponding reporter activity measured in the absence of FOXA1 co-expression. All experiments were conducted using eight replicates.
DISCUSSION
We performed deep resequencing at the multi-gene locus chromosome 11p11.2 in 3,566 individuals of European ancestry from three cohort studies to identify rare, potentially functional variants associated with FG and FI, two key quantitative traits associated with type 2 diabetes physiology and risk. In the NR1H3 region seven rare variants predicted to be especially functional were multiple-testing-corrected significantly associated with levels of FI, even after conditioning on two known glycemia-associated variants in MADD, suggesting that there are more than two independent signals at 11p11.2, and that the signal at NH1R3 is new. One of these seven variants, chr11:47227430, appeared to account for a substantial portion of the gene-based signal. Chr11:47227430 lies in a binding motif for FOXA1, a transcription factor implicated in glucose metabolism and insulin secretion. 40, 41 We showed in functional studies that the rare A allele of chr11:47227430 has a lower affinity to bind to FOXA1 compared with the reference G allele and disrupts FOXA1-mediated luciferase assay activity at the locus. Thus, we show that targeted deep resequencing in regulatory regions can identify new rare variants with previously unidentified regulatory activity. In the three subjects from the sequenced sample, carriers of the A allele appeared to have higher FI levels than their reference populations, but additional genotyping of the variant in FHS did not confirm that A allele carriers have a discernable phenotype. The chr11:47227430 variant is very rare; additional genotyping in large numbers of additional samples is required to definitively determine if variation at this base pair is associated with a discernable glucoregulatory phenotype. Finally, we found that no rare variants around any of the five genes were associated with FG, and that no common variants at 11p11.2 were associated with FG or FI.
Our results suggest that there is additional allelic heterogeneity associated with glucose and insulin regulation at the 11p11.2 – MADD locus. Common polymorphisms have been have been identified through several GWA studies at MADD and here we identify at the locus new rare variants with predicted regulatory function that appear to be independent of known common variation. Our findings complement Huyghe et al.’s recent exome array analysis, drawn from a Finnish cohort of 8,229 men without diabetes. 42 They found a mutation, rs35233100, encoding a stop codon Arg766 in MADD and associated with lower proinsulin levels. In this sample, rs35233100 was in modest LD with rs7944584 and independent of rs1051006, pointing to MADD as a causal gene accounting for prior GWAS signals at the locus. However, proinsulin associations in their analysis were not completely eliminated by conditional analysis suggesting that additional variant(s) associated with insulin regulation at 11p11.2 remained to be identified. Our results from analysis of large numbers of deep sequences targeted at non-coding as well as coding variation suggests that rare regulatory variation at NH1R3 contributes to genetic glucose and insulin regulation and metabolism pathways encoded on chromosome 11p11.2. Notably, the 11p11.2 locus does not appear to be associated with risk of T2D in individuals of European ancestry, although may perhaps be weakly associated with T2D risk in Han Chinese. 43, 44 The locus seems to belong to the class of loci where variants with relatively large effect sizes for glycemic traits (including FI and FG) show little risk for T2D (and vice versa). 43 Thus, our findings appear to bear more on physiological homeostatic regulation of glucose and insulin than on physiological pathways involved in the development of T2D.
The functional variant we detected lies in intron 2 of the longer of two isoforms of NR1H3, a plausible biological candidate gene. NR1H3, a nuclear hormone receptor, belongs to a family of ligand-activated transcription factors activated by cholesterol-derived compounds. NH1H3, also known as liver X receptor (LXR), plays a role in beta cell proliferation and mass. LXR mRNA levels are elevated in the pancreatic islets of animal models of type 2 diabetes and in humans, LXR elevation is associated with pancreatic beta cell dysfunction in T2D. 45, 46 The variant chr11:47227430 is predicted in RegulomeDB to alter a binding motif for FOXA1. We showed that the rare allele at chr11:47227430 had sequence-specific lower binding affinity for FOXA1, resulting in reduced FOXA1-mediated transactivation activity. FOXA1 (also known as hepatic nuclear factor 3α, HNF3α) belongs to the family of FOXA transcription factor genes critical for diverse biological processes relevant to glucose and insulin metabolism. Studies in mice lacking one or both FOXA1 alleles show impaired insulin secretion and glucose intolerance, and mice lacking FOXA1 expression develop a complex metabolic syndrome characterized by neonatal persistent hypoglycemia, pancreatic alpha- and beta-cell dysfunction and increased expression of FOXA1 target genes. 47 Despite this supporting evidence, our data do not indicate that it is NH1R3, or in fact which gene at all, is the target influenced by variation at this transcript-specific intronic transcription factor binding site. Further functional work and correlation analyses using eQTL data in large cohorts where the rare variant might be represented are needed to elucidate how and which target gene(s) are influenced by the chr11:47227430 variant.
The Cohorts for Heart and Aging Research in Genomic Epidemiology Targeted Sequencing Study represents a new generation of high-throughput deep sequencing studies focused on coding and non-coding variation underlying common cardiovascular disease and metabolic risk factors and diseases. Strengths of the study include a creative case-cohort design to assemble well-characterized cohorts of European descent. To analyse these data a variety of new statistical methods and bioinformatics resources were developed, combined and applied in a rigorous framework intended to limit type 1 error in the setting where no similar data exist for replication. For instance, the very high average sequence depth combined with rigorous QC applied uniformly across three cohorts increased confidence that even the rarest observed variation is likely to be true variation and not technical artefact. Further, we repeat-genotyped the one variant of interest identified by the sequencing approach. Type 2 error remains a limitation of the analysis, however, where a sample of 3,566 individuals has very limited power to detect common variant associations and for rare variants, the sample size gave just 3 individuals with the interesting, potentially functional mutation at chr11:47227430. While high sequence depth is a strength, somewhat limited coverage breadth is a limitation. The genomic segments selected for sequencing favored exons, but did include generous intronic flanking regions around exons and within and beyond regulatory regions flanking genes, thereby including far more non-coding variation than with strictly exome-based sequencing approaches. Unbiased whole-genome sequence in large numbers will be needed to define the true allelic spectrum of functional variation at 11p11.2. Of note is that mean BMI was in the overweight range in all three populations. However, effect sizes in known variants associated with glycemic traits do not differ between BMI strata. 35 In addition, notable heterogeneity was seen in FI levels across the three study populations, which might likely be a reflection of limited standardization of insulin assays. We have addressed these issues by adjusting for BMI, which improves ability to detect insulin resistance signals, 35 and further, assay heterogeneity has not precluded the ability to detect novel variants in prior FI GWAS.2, 35 Finally, our results include only people of European ancestry, so our analysis was further constrained by limited ancestral spectrum of human allelic variation at the 11p11.2 – MADD locus.
In conclusion, deep sequencing at 11p11.2 in 3,566 individuals from three European cohorts identified rare functional variation in an intron NR1H3 associated with FI. Conditional analysis suggests that known common variants near MADD and rare functional variants at NR1H3 appear to represent independent signals. Functional analysis showed the rare A allele at the chr11:47227430 reduced FOXA1 binding in vitro, resulting in reduced FOXA1-mediated transcriptional activity in luciferase assays. Other genes localized to the MADD region – ACP2, MYBPC3, MADD, SPI1 – showed no significant association of rare variants with either FI or FG levels. Our results suggest that in addition to previously demonstrated associations with common variation in MADD, rare variation at NH1R3 contributes to genetic glucose and insulin regulation and metabolism pathways encoded on chromosome 11p11.2.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Christine Mendonca BS and Alessandro Doria MD PhD at the Joslin Diabetes Center Genetics & Epidemiology Research Core for conducting the FHS follow-up genotyping and Bianca Porneala at the MGH General Medicine Division for assistance with statistical analysis.
FUNDING SOURCES
Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium” was provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. Support was also received from the Baylor Genome Center (U54 HG003273)
The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C, and HHSN2682011000012C).
The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (Contract No. N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (Contract No. N02-HL-6-4278), for quality control by Framingham Heart Study investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Also supported by R01DK078616, K24 DK080140 and an American Diabetes Association Mentored Post Doctoral Fellowship Award (Dr. Meigs).
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
Footnotes
DISCLOSURES
James B. Meigs serves as an academic advisor to Quest Diagnostics and a consultant to LipoScience Inc. Bruce M. Psaty serves on the DSMB of a clinical trial of a device funded by ZollLifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Medtronic. Jose C. Florez has received consulting honoraria from Lilly and Pfizer. The authors have no other disclosures to report.
REFERENCES
- 1.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lumley T, Dupuis J, Rice KM, Barbalic M, Bis JC, Cupples LA, et al. Two-phase subsampling designs for genomic resequencing studies. submitted. [Google Scholar]
- 5.Lin H, Wang M, Brody JA, Bis JC, Dupuis J, Lumley T, et al. Strategies to design and analyze targeted sequencing data: The cohorts for heart and aging research in genomic epidemiology (charge) targeted sequencing study. doi: 10.1161/CIRCGENETICS.113.000350. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cushman M, Cornell ES, Howard PR, Bovill EG, Tracy RP. Laboratory methods and quality assurance in the cardiovascular health study. Clin Chem. 1995;41:264–270. [PubMed] [Google Scholar]
- 8.Meigs JB, Nathan DM, Wilson PW, Cupples LA, Singer DE. Metabolic risk factors worsen continuously across the spectrum of nondiabetic glucose tolerance. The framingham offspring study. Ann Intern Med. 1998;128:524–533. doi: 10.7326/0003-4819-128-7-199804010-00002. [DOI] [PubMed] [Google Scholar]
- 9.Smith NL, Barzilay JI, Shaffer D, Savage PJ, Heckbert SR, Kuller LH, et al. Fasting and 2-hour postchallenge serum glucose measures and risk of incident cardiovascular events in the elderly: The Cardiovascular Health Study. Arch Intern Med. 2002;162:209–216. doi: 10.1001/archinte.162.2.209. [DOI] [PubMed] [Google Scholar]
- 10.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, et al. Multiplexed massively parallel selex for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor cdx2. Dev Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ets-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo KA, Bauchmann MK, Baumann AP, Donahue CJ, Thiede MA, Hayes LS, et al. Genome-wide profiling of h3k56 acetylation and transcription factor binding sites in human adipocytes. PLoS One. 2011;6:e19778. doi: 10.1371/journal.pone.0019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, et al. Differential genomic targeting of the transcription factor tal1 in alternate haematopoietic lineages. EMBO J. 2011;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S, Cui K, Jothi R, Zhao DM, Jing X, Zhao K, et al. Gabp controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood. 2011;117:2166–2178. doi: 10.1182/blood-2010-09-306563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 22.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-qtls yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, et al. Transcriptome genetics using second generation sequencing in a caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with rna sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, et al. Dnase i sensitivity qtls are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Ahmad SM, Aboukhalil A, Busser BW, Kim Y, Tansey TR, et al. Differential regulation of mesodermal gene expression by drosophila cell type-specific forkhead transcription factors. Development. 2012;139:1457–1466. doi: 10.1242/dev.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from encode data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cirillo LA, Zaret KS. Specific interactions of the wing domains of foxa1 transcription factor with DNA. J Mol Biol. 2007;366:720–724. doi: 10.1016/j.jmb.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genetic Epidemiology. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by brg1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaestner KH. The foxa factors in organogenesis and differentiation. Curr Opin Genet Dev. 2010;20:527–532. doi: 10.1016/j.gde.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao N, Le Lay J, Qin W, Doliba N, Schug J, Fox AJ, et al. Foxa1 and foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol Endocrinol. 2010;24:1594–1604. doi: 10.1210/me.2009-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu C, Zhang R, Wang C, Wang J, Ma X, Hou X, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the chinese. PLoS One. 2010;5:e15542. doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krasowski MD, Ni A, Hagey LR, Ekins S. Evolution of promiscuous nuclear hormone receptors: Lxr, fxr, vdr, pxr, and car. Mol Cell Endocrinol. 2011;334:39–48. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng ZX, Nie J, Ling JJ, Sun JX, Zhu YX, Gao L, et al. Activation of liver x receptors inhibits pancreatic islet beta cell proliferation through cell cycle arrest. Diabetologia. 2009;52:125–135. doi: 10.1007/s00125-008-1174-x. [DOI] [PubMed] [Google Scholar]
- 47.Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci U S A. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.