Abstract
Effective tracking of biospecimens within a biobank requires that each biospecimen has a unique identifier (ID). This ID can be found on the sample container as well as in the biospecimen management system. In the latter, the biospecimen ID is the key to annotation data such as location, quality, and sample processing. Guidelines such as the Best Practices from the International Society of Biological and Environmental Repositories only state that a unique identifier should be issued for each sample. However, to our knowledge, all guidelines lack a specific description of how to actually generate such an ID and how this can be supported by an IT system. Here, we provide a guide for biobankers on how to generate a biospecimen ID for your biobank. We also provide an example of how to apply this guide using a longitudinal multi-center research project (and its biobank). Starting with a description of the biobank's purpose and workflows through to collecting requirements from stakeholders and relevant documents (i.e., guidelines or data protection concepts), and existing IT-systems, we describe in detail how a concept to develop an ID system can be developed from this information. The concept contains two parts: one is the generation of the biospecimen ID according to the requirements of stakeholders, existing documentation such as guidelines or data protection concepts, and existing IT-infrastructures, and the second is the implementation of the biospecimen IDs and related functionalities covering the handling of individual biospecimens within an existing biospecimen management system. From describing the concept, the article moves on to how the new concept supports both existing or planned biobank workflows. Finally, the implementation and validation step is outlined to the reader and practical hints are provided for each step.
Introduction
The role of high quality biospecimens has gained importance in biomedical research as it has become recognized that poor or unknown quality might prevent reproducibility and generate incorrect research results.1,2 Increasing numbers of biospecimens are required to answer research/scientific questions, and samples are therefore collected from multiple sites.3 This increases the importance of labeling sample containers and entering specimen-related information into a biospecimen management system in a consistent and informed manner.
The landscape of labeling is very heterogeneous. Multiple labeling methods exist, as there is no agreement upon a gold standard.4–6 According to the 2012 International Society of Biological and Environmental Repositories (ISBER) Informatics Working Group survey (more than one option possible), more than half of the respondents (51%) already use printed 2D barcode labels, 47% used text labels, and 40% used linear barcode labels.7 From 2010 to the 2012 survey, the percentage of handwritten labels decreased from 39% to 24%.7 In our opinion, these inconsistencies are mainly influenced by such factors as inadequate funding to cover adequate Information Technology (IT) infrastructure, lack of personnel with biospecimen management IT-expertise associated within the institution or biobank, and the availability of professional software.
For multi-site recruiting centers, the existence of this heterogeneity causes huge challenges if not addressed at the outset of the study. As a solution we need a process comparable to a citation in an article, which reflects the process of managing biospecimens in many ways. Both processes consist of two components: a short unique identifier (ID) at the point of interest (sample container or a reference after distinct idea or finding coming from someone else in an article) and more details at some other location, which can be tracked down using the unique ID (biospecimen management system with detailed information or reference list at the end of an article).
Labeling a sample with a unique ID can be done in different ways. The easiest, but most error prone way of identifying biological samples, is handwriting text and/or numbers onto the collection tubes. Machine-written labels or even barcode (1D or 2D) stickers are a more sophisticated method but both may fall off at low temperatures.8 For the past few years, companies have offered alternative solutions such as tubes with laser-etched barcodes, avoiding the loss of identification. Storing information within DNA is another alternative for identifying the sample.9,10 Radio-frequency identification (RFID) chips may be the best current alternative, but are not yet common as the costs are still too high.11 The tools for management of biospecimens and their identification data also range from very simple manual options to complex automated solutions. At one end of the identification scale is a handwritten entry into a paper-based laboratory notebook. This manual entry approach progresses to Excel sheets and to state-of-the-art options, such as professional biobanking software products.12,13 An overview of the latter is available on the ISBER marketplace.
Despite the commercial availability of solutions in the fields of labeling and management of biospecimen identification data, many researchers still struggle when deciding what information their biospecimen ID should include and how to implement the generation of suitable labels or identification systems associated with the biospecimen and representation of this data within their biospecimen management systems. Here, the best practices as developed by the Biorepositories and Biospecimen Research Branch of National Cancer Institute (NCI) in the US14 or the ISBER151 only provide statements but no how-to-guide or enough practical details to help individuals accomplish this critical operational task.
For example, the NCI Best Practices in Section B 6.1.2 limits its recommendation to “linking the labels on the physical biospecimen container […] to other information regarding that biospecimen in the system,”14 and in Section B 6.2.2 notes that a “method to have global unique identification of biospecimen”14 should be employed.
In Section I2.000 the ISBER Best Practices advises biobankers that “The [biospecimen management] system should have the capacity to assign a unique ID to each specimen entered in the database”.15 Section I3.000 mentions “Labels should contain an ID linking to a database containing details about the specimen collection and processing information”.15 I3.100 Labels for Specimens recommends that there should be a unique ID per specimen not containing any identifiable information about the donor. Lastly, (I3.200) “each aliquot or container should be labeled with a unique barcode/number”.15 However, details of how to act on these guidelines are not described.
In this article, we provide a guide for you as biobankers on how-to design a biospecimen ID for your biobank and fully integrate IDs and ID-related functions into your biospecimen management system, based on our experience with a major biomedical research project. This guide takes the statements on this topic found in Best Practices guidelines and gives you a step-by-step guide of how to do it. In addition, we provide an example of what you can end up with following the guide, if you want to improve the already existing ID generation solution in your biospecimen management system to satisfy your users. Our example solution allows high quality annotations of biospecimens while minimizing the documentation workload for the researchers.
How-To-Guide
In order to design biospecimen IDs for your biobank and fully integrate them into your biospecimen management system with related functions that are necessary for a smooth workflow, the steps below have to be applied in a sequential order.
To make this how-to-guide more practical, we provide an example of how we designed a biospecimen ID and fully integrated this and the accompanying functions into the biospecimen management software of an existing biobank (see Supplementary “biobank ID implementation example” document; supplementary material is available online at www.liebertonline.com/bio). This biobank is a large (>1000 cases), multi-site (∼20 collaborating centers from northern Germany will participate in recruiting donors), longitudinal (samples will be collected at four time points over 18 months from each participant but stored at one central site in Göttingen) mono-user biobank. Mono-user biobank is the design that is intended to serve a single research project only.16 This biobank became operational in April 2012, as of the end of May 2014 approximately 560 participants had provided>18000 biospecimen samples in 1725 kits as documented in the biospecimen management system.
From planning the ID concept up to the point where the programming by the vendor could be done took approximately 6 months. From then until the biomedical research project, the KFO 241, was able to use its IT-infrastructure,12 which consists of more IT components than just the biospecimen management system, took another 10 months. This also included setting up and developing further IT-components, programming of interfaces between the components and buying and installing dedicated server hardware. As this study was the pilot for this IT-infrastructure, the development period was relatively long. For further projects, it will now only take a few weeks depending on the complexity of the study design. Approximately 2.5 full-time equivalents (FTE) were involved over more than a year to set up the complete IT-infrastructure.
Description of the biobank
A plan for the biobank, for which the biospecimen IDs need to be designed must exist. This plan should include the expected size and type of biobank16 (e.g., how many cases are expected, will there be a long-term follow-up of the participants, will the biobank serve only one project or many, how many sites will contribute biospecimens). The current or planned biospecimen management software should also be described as well as the party (internal vs. external vendor) who would be able to implement it.
Analysis of the biobank's workflows
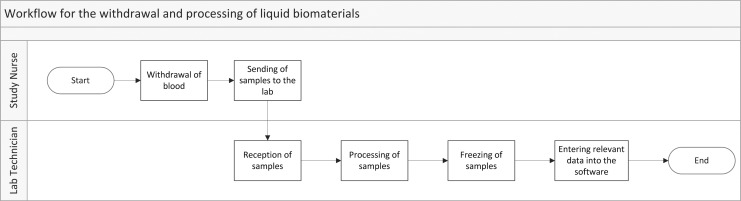
In order to design a biospecimen ID and fully integrate related functionalities into a biospecimen management system, it is important to first analyze and fully understand the workflows of a biobank. These are typically all workflows connected to biospecimen handling starting from biospecimen withdrawal and ending with either the withdrawal of consent or the feedback of data after distribution of the biospecimen to one or more requesting parties. This process of workflow analysis must take place in close collaboration with all biobank stakeholders and if applicable, with the personnel involved from the respective research project for which the specimens are collected. This might require several iterative cycles and we recommend visualizing the workflows in an understandable manner for all sites, such as flow charts or swim-diagrams17 as depicted in Figure 1. A more detailed outline of the processing of samples can be seen in the Supplementary Material available in the online article at www.liebertpub.com/bio.
FIG. 1.
Workflow for the withdrawal and processing of liquid biospecimens.
Requirements analysis
Requirements for the biospecimen labeling and IT system can be divided into three categories: a) stakeholders, b) (existing) documentation, and c) (existing) IT systems.18 All stakeholders should be interviewed regarding their requirements related to the biospecimen ID and the integration into the biospecimen management system. Existing documentation, with which the biobank should strive to be in compliance, include Best Practice guidelines or data protection guidelines, laws, or concepts. Lastly, some requirements (e.g., limitations) might come from the IT-system, which you plan to use for the management of your biospecimen.
Once you have collected the requirements from all three sources, you want to put them together and, as described by Schwanke et al.,19 you should group similar requirements and break down and re-arrange complex ones into simpler ones. Ideally, you want the stakeholders to prioritize their requirements as essential or desirable.20 A second round of discussion with all stakeholders together might be useful to get further input.
Tables 1–3 depict organizational, ID-related, and administration-related requirements grouped into the respective tables as identified in our example. See the Supplementary Material available in the online article at www.liebertpub.com/bio.
Table 1.
Requirements Related to the Organization of the Research Project
| No. | Requirements related to the organization of the research project |
|---|---|
| O.1 | All participating centers should use the same labeling system to standardize and simplify the biospecimen management. |
| O.2 | The sets of tubes should be packable in advance for later use or shipping to increase flexibility for research nurses as they can quickly take any the pre-packed set of tubes. |
| O.3 | Each set of tubes should be usable for any study participant to provide flexibility for study nurses. |
Table 2.
Requirements Related to the Identification of Biospecimen
| No. | Requirements related to the biospecimen ID |
|---|---|
| I.1 | The ID on the biospecimen container should include the name or abbreviation of the research project for intellectual property rights. |
| I.2 | The biospecimen ID should contain a unique arbitrary number not related to IDs in other IT-components of the same IT-infrastructure (e.g., study participant ID in the study participant management system) |
| I.3 | It should be visible from the biospecimen ID which tubes belong to the same study participant from the same visit (to improve tube handling in the lab). |
| I.4 | The IDs should be unique for each tube to allow management on a single tube level and to avoid mistaken identity. |
| I.5 | The biospecimen ID should include the type of biospecimen that is in the tube to allow for easy handling of samples in the lab and in the biobank. |
| I.6 | The biospecimen ID should not contain any identifying information of the participant.14 |
| I.7 | In terms of quality management and assurance: The biospecimen ID should be human (text) and machine readable (barcodes) to reduce manual errors and allow automatic processing of samples.14 |
Table 3.
Requirements Related to the Administration of Biospecimen in a Biospecimen Management System
| No. | Requirements for the administration of biospecimen in a biospecimen management software |
|---|---|
| M.1 | Data which belongs to all biospecimens from one participant of one visit should only need to be documented once to minimize the documentation effort, e.g. date of phlebotomy. |
| M.2 | It should be possible to add further tubes (having the same ID-root; print new sticker and attach to spare tube) to an already existing set of tubes if needed. |
| M.3 | It should be traceable in the database where aliquots or processed samples (children samples) originate from (parental sample). |
| M.4 | The generation of unique IDs should be fully integrated into the software used for the administration of the biomaterial samples to avoid duplicates. |
| M.5 | The amount of already packed and not used sets of tubes should be monitorable (inventory control) in the database. |
| M.6 | Each biospecimen has to be assigned to a participant's ID19,20 to be in accordance with data protection regulations. |
| M.7 | Relevant pre-analytic variables should be documented to be able to distinguish between the quality of samples.21,22 |
| M.8 | Adaptation of the biospecimen management software should not interfere with other components of the IT-infrastructure of this research project. |
Developing the concept for biospecimen IDs and full integration into a biospecimen management software
Concept for biospecimen IDs
The results of the requirements analysis, together with the precise documentation of the workflows, should lead to a concept for biospecimen IDs and their full integration into biospecimen management software, respectively. Full integration means that the generation of IDs, the management of related biospecimen data, and further requirements are tightly connected to the generation of biospecimen IDs and are all in one IT system (supporting the identified workflows). From our experience, several iteration cycles including discussions with all stakeholders and refinement of the concept are necessary to come up with a final concept agreed upon by all stakeholders.
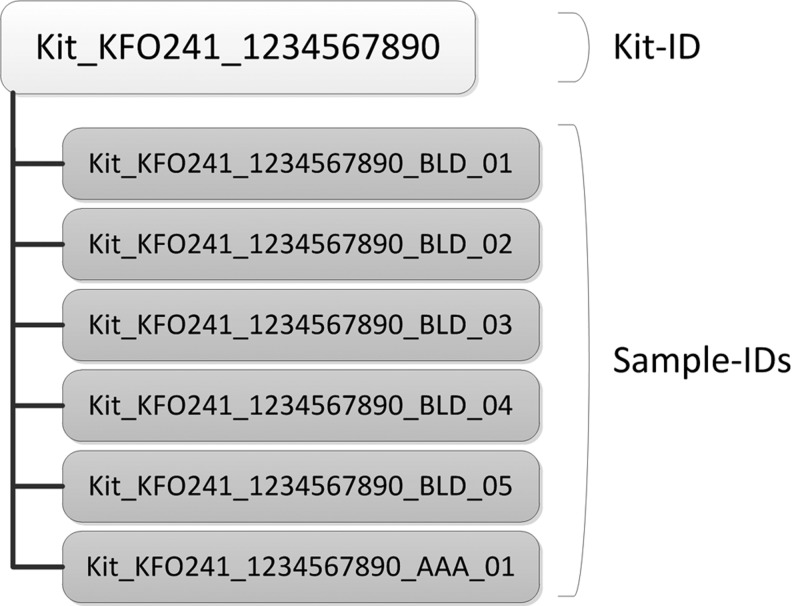
Figure 2 depicts one solution of what sample-IDs could look like. A kit-ID is the basis for all biospecimens taken from one study participant at one research visit. Within a kit, each sample is uniquely identifiable by the addition of a material code (e.g., BLD for blood based on the SPREC21,22) and an ascending number. This is in compliance with ISBER Best Practices15 Section I3.100, where it is stated that the biospecimen label should not contain any information about the donor or its health status. The material code has advantages for handling the sample in the lab and does not increase the risk of donor re-identification. Further details on how we designed our biospecimen IDs are provided in the Supplementary Material available in the online article at www.liebertpub.com/bio.
FIG. 2.
Biospecimen IDs as contained in one kit with the kit-ID on the top.
Description of how the developed concept will support the biospecimen workflow from an IT-perspective
Once the concept is agreed upon, a document should be written to explain in detail how the developed concept supports the biospecimen workflow from an IT-perspective. This document will be very helpful for the understanding of the context for the programmer and may serve as a basis for the contract between you and the programmer. A detailed description of the functions that need to be implemented in the biospecimen management software should be included as well. See the Supplementary Material available in the online article at www.liebertpub.com/bio
Implementation and validation of the concept
Depending on the stage of development of your biobank (planning or in operation), you either select the software product that is most similar to your concept, implement the concept with the assistance of the vendor or have in-house IT-personnel integrate it into your existing biospecimen management system.
If you decide not to implement the developed concept using existing IT personnel (e.g., a lack of expertise within the biobank) or the software product chosen requires vendor modifications to implement it, you will need to approach the producer of the software product. You will describe the deficiencies of the software from your perspective (requirements),20 and based on this the vendor will describe their solutions to address these issues. The vendor will provide a cost and time estimate to implement their solutions and you will have a chance to adjust your original order. It is important that you will have the chance to test and validate the delivered product thoroughly and claim bug fixes, in case the revised software does not perform as expected. Finally, before going live with the customized system, users should be offered a training session and a manual on how to use the new features should be written and delivered.
If you decide to implement the developed concept yourself or use someone with IT-expertise from within your institution, you will still require a document describing the deficiencies of the software regarding your requirements as collected from stakeholders. The person implementing the requirements should at a minimum draft a concept for internal discussion and properly document all programming changes within the project files. Given that the programmer is from within the same institution, you should use this opportunity to let the programmer regularly interact with the stakeholders. This close collaboration will be very helpful to implement new functions in a user-friendly way. But you should also be aware of at least three issues that can occur. First, you will absolutely need comprehensive documentation of the source code so it is understandable and self-explaining, even without the person that produced the code. This should be done for sustainability and risk management in case the developer leaves the institute. If you choose to implement functions by yourself, the second issue is that you need software that allows these changes. If you rely on a biospecimen management system with closed source code, you will not be able to implement your own features. The third issue is that you need to establish a procedure for validating your code, especially if you are planning to use the system for clinical trials or drug efficacy studies to conform with, for example, US Food and Drug Administration (FDA) regulations.
Note that advantages of having the vendor do the implementation are that the vendor is responsible for all changes while ensuring the system still works and programming is likely to be done faster. On the other hand, self-implementation allows you more flexibility and may be cheaper.
Practical Hints and Recommendations
In general, before addressing the issue of generating biospecimen IDs, it is important that the scope of the biobank is well defined and accepted by all stakeholders. A detailed project plan should be made transparent, and risk management processes should be established and followed throughout setting up and operating the biobank.
Description of the biobank
Describing the biobank during the proposal writing process or at an early stage of a research project can be a difficult undertaking. The description must be in line with the study protocol and the ethics approval document. This step is important as it will help you to understand the challenges your biobank encounters biospecimen ID-wise.
Analysis of the biobank's workflows
Analysis of the biobank workflows in the early stages of a research project can be a long process, especially when the workflows have not been finalized. The point of the exercise is that all stakeholders understand the content and that you have a common base to discuss and understand the workflows. The comprehension is increased if you have visualized the workflows, as well as described them in written text.
Sometimes it is difficult with more complex biobanks to fit all workflows into one big figure as described above. As long as it is understandable and clear to everyone, it does not matter if you go for several workflow figures instead of only one. All information relevant for understanding the workflows should be included, such as who is doing what and where.
Requirements analysis
Collecting requirements should be an iterative process with all stakeholders involved in biospecimen handling. Here it is especially important to listen to the needs of the lab technicians and not only to the principal investigators. To address this issue, the different stakeholders should be interviewed separately to ensure all personnel were comfortable expressing their opinions. IT-personnel should know the details on relevant documents such as data protection concepts and the requirements coming from the IT-system. In a final discussion with all stakeholders, it should be ensured that everyone understands all requirements. This effort requires a large investment in time that usually turns out to be well worth it as all users will then be very satisfied with the final product.
Prioritizing the requirements is a good thing to do, especially if you only have limited funds for adapting the biospecimen management system. This method allows categorizing the requirements into different classes (i.e., essential and nice to have). Thus, less important requirements might be postponed to a later stage for implementation in the software or not implemented at all. If many requirements need to be implemented, a prioritization might also indicate a timely order for implementation.
Developing the concept for biospecimen IDs and full integration into a biospecimen management software
Concept for biospecimen IDs
Several iterative cycles should be conducted alternating conceptualizing and discussion of the concept until everyone is satisfied and accepts the result. Only then should the document for the adapting party be designed.
Double check that all of the requirements collected are addressed in the concept and thus are specified in the document you send to the vendor or give to the IT-person and in the document you will receive from the developing company of the biospecimen management software. A good way of answering all open questions and to make sure that there are no misunderstandings is a face-to-face workshop with the programmers.
Full integration of biospecimen-ID related features into the biospecimen management system: How the developed concept will support the biospecimen workflow from an IT-perspective
One issue you will hear regularly when interviewing stakeholders for their workflows is “How would it work best for the IT?” IT has to follow the workflow of biospecimen, not the other way round. IT should support the operations of the project/biobank. Thus, it is important for a successful implementation of a biobank to involve IT-personnel early on during this process. On the one hand, their priorities might be at odds with those of the biobank as they usually have to support several projects throughout an institution and want to minimize their own effort, but on the other hand, IT personnel therefore can be extremely powerful in any biobank's oversight organizational structure. If the concept is developed in close collaboration with all stakeholders involved in handling the biospecimen at different stages, an optimized workflow support will be reached.
Implementation and validation of the concept
Depending on the scope of the changes and the capability and availability of any programmers (internal or external), the implementation of changes might take up a few weeks or even longer. The general rule should be followed that the person validating the system should not be the one, who developed it.
Achieving a very high user satisfaction and acceptance of the adapted biospecimen management system can result when users are deeply engaged right from the beginning. The users therefore see it as their product as well.
Note that this how-to-guide's example only considered designing biospecimen IDs for internal use. It is common practice, although not clearly stated in best practice guidelines,15 that upon giving any sample away to an external researcher, its ID should be changed to a different release ID. This release-ID is also often varied if aliquots of the same sample are given to two different requesting researchers. Nevertheless, the how-to guide could also be used for that purpose by simply adding the requirement of a different release ID.
Discussion
This How-to-Guide was developed for biobankers who are planning to or already running a biobank and do not have access to a sophisticated biospecimen management system that could generate their biospecimen-IDs. For smaller projects with limited budget and tight time-frame, independent researchers, or PhD students, the kit-example provided here is probably too ambitious. However, following the How-To-Guide recommendations for collecting requirements could be done on a smaller scale and followed by a decision on what the biospecimen-IDs should look like. Therefore, some of the box tool will be useful for smaller projects.
Conclusion
When a system is developed by users for users and these stakeholders are involved in all decision making processes, their acceptance of the end-product is very high. This is in accordance with other findings23 that involving end-users in decisions creates a sense of ownership and may extend the development time but the outcome will be an easier implementation and acceptance.
As described in the introduction, best practices provide general guidelines to be followed but lack a detailed description of how to implement these guidelines. This article is a first step in generating a whole set of how-to guides based on the best practices in biobanking.14,15 By providing a real-life practical example of designing a biospecimen ID and fully integrating this into a biospecimen management system, this how-to guide could be called an evidence-based guideline.24 Evidence-based guidelines for biobanks were demanded recently by several researchers25,26 and How To`s to assist biobanks to operationalize ‘best practices’ should be created with input and endorsement from organizations that have already generated or endorsed best practice documents (e.g., ISBER or ESBB).
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through the Clinical Research Group 241 ‘Genotype-phenotype relationships and neurobiology of the longitudinal course of psychosis’ (http://www.kfo241.de; grant number SCHU 1603/5-1) as well as the University Medical Center Göttingen Biobank. SY Nussbeck was supported by a fellowship within the Postdoc-Program of the German Academic Exchange Service (DAAD) with the personal ID 91510522 while writing the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Silberman S. Libraries of Flesh: The Sorry State of Human Tissue Storage. Wired Magazine. 2010; 157 Available from: http://www.wired.com/magazine/2010/05/ff_biobanks/ Last accessed: 2013November26
- 2.Ransohoff DF, Gourlay ML. Sources of bias in specimens for research about molecular markers for cancer. J Clin Oncol 2010;28:698–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF. The psychiatric GWAS consortium: Big science comes to psychiatry. Neuron 2010;68:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kay AB, Estrada DK, Mareninov S, et al. Considerations for uniform and accurate biospecimen labelling in a biorepository and research environment. J Clin Pathol 2011;64:634–636 [DOI] [PubMed] [Google Scholar]
- 5.Ratnasingham S, Hebert PDN. Bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes 2007;7:355–364 [DOI] [PMC free article] [PubMed]
- 6.Stoeckle MY, Hebert PDN. Barcode of life. Sci Am 2008;299:82–86, 88 [PubMed] [Google Scholar]
- 7.Fearn P, Michels C, Meagher K, Cada M. 2012 International Society for Biological and Environmental Repositories Informatics Working Group: Survey Results and Conclusions. Biopreserv Biobanking 2013;11:64–66 [DOI] [PubMed] [Google Scholar]
- 8.De Souza YG, Greenspan JS. Biobanking past, present and future: Responsibilities and benefits. AIDS 2013;27:303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman N, Bertone P, Chen S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 2013; 23:advance online publication. Available from: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11875.html Last accessed: 2013August22 [DOI] [PMC free article] [PubMed]
- 10.Davis JC, Shon J, Wong DT, et al. A DNA-based biological sample tracking method. Cell Preserv Technol 2005;3:54–60 [Google Scholar]
- 11.Bettendorf E, Malenfant C, Chabannon C. RFID technology and electronic tags to identify cryopreserved materials. Cell Preserv Technol 2005;3:112–115 [Google Scholar]
- 12.Demiroglu SY, Skrowny D, Quade M, et al. Managing sensitive phenotypic data and biomaterial in large-scale collaborative psychiatric genetic research projects: Practical considerations. Mol Psychiatry 2012;17:1180–1185 [DOI] [PubMed] [Google Scholar]
- 13.Prokosch HU, Beck A, Ganslandt T, et al. IT infrastructure components for biobanking. Appl Clin Inform 2010;1:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Office of Biorepositories and Biospecimen Research NCI. NCI Best Practices for Biospecimen Resources. U.S. Department of Health and Human Services; 2011. Available from: http://biospecimens.cancer.gov/bestpractices/2011-NCIBestPractices.pdf Last accessed: July17, 2013
- 15.International Society for Biological and Environmental Repositories. 2012 Best Practices for Repositories–Collection, storage, retrieval, and distribution of biological materials for research. Biopreserv Biobanking 2012;10:79–161 [DOI] [PubMed] [Google Scholar]
- 16.Watson PH, Barnes RO. A proposed schema for classifying human research biobanks. Biopreserv Biobanking 2011;9:327–333 [DOI] [PubMed] [Google Scholar]
- 17.Rummler GA, Brache AP. Improving Performance: How to Manage the White Space on the Organization Chart. San Francisco: Jossey-Bass Publishers; 1990 [Google Scholar]
- 18.Pohl K. Requirements Engineering: Fundamentals, Principles, and Techniques. Heidelberg; New York: Springer; 2010 [Google Scholar]
- 19.Schwanke J, Rienhoff O, Schulze TG, et al. Suitability of customer relationship management systems for the management of study participants in biomedical research. Methods Inf Med 2013;52:340–350 [DOI] [PubMed] [Google Scholar]
- 20.IEEE Computer Society. IEEE Std 830-1998: IEEE Recommended Practice for Software Requirements Specifications. IEEE; 1998 [Google Scholar]
- 21.Betsou F, Lehmann S, Ashton G, et al. Standard preanalytical coding for biospecimens: Defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prev 2010;19:1004–1011 [DOI] [PubMed] [Google Scholar]
- 22.Lehmann S, Guadagni F, Moore H, et al. Standard preanalytical coding for biospecimens: Review and implementation of the sample PREanalytical Code (SPREC). Biopreserv Biobanking 2012;10:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinsons MG. Comparing the Decision Styles of American, Chinese and Japanese Business Leaders. Rochester, NY: Social Science Research Network; Report No.: ID 952292. Available from: http://papers.ssrn.com/abstract=952292 Last accessed: November25, 2013 [Google Scholar]
- 24.Moore HM, Compton CC, Alper J, et al. International approaches to advancing biospecimen science. Cancer Epidemiol Biomarkers Prev 2011;20:729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson E, Betson F, Fuller BJ, et al. Translating cryobiology principles into trans-disciplinary storage guidelines for biorepositories and biobanks: A concept paper. Cryo Lett 2013;34:277–312 [PubMed] [Google Scholar]
- 26.Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet Med 2007;9:141–149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.