Abstract
Significance: microRNAs (miRNA) have been characterized as master regulators of the genome. As such, miRNAs are responsible for regulating almost every cellular pathway, including the DNA damage response (DDR) after ionizing radiation (IR). IR is a therapeutic tool that is used for the treatment of several types of cancer, yet the mechanism behind radiation response is not fully understood. Recent Advances: It has been demonstrated that IR can alter miRNA expression profiles, varying greatly from one cell type to the next. It is possible that this variation contributes to the range of tumor cell responsiveness that is observed after radiotherapy, especially considering the extensive role for miRNAs in regulating the DDR. In addition, individual miRNAs or miRNA families have been shown to play a multifaceted role in the DDR, regulating multiple members in a single pathway. Critical Issues: In this review, we will discuss the effects of radiation on miRNA expression as well as explore the function of miRNAs in regulating the cellular response to radiation-induced damage. We will discuss the importance of miRNA regulation at each stage of the DDR, including signal transduction, DNA damage sensing, cell cycle checkpoint activation, DNA double-strand break repair, and apoptosis. We will focus on emphasizing the importance of a single miRNA targeting several mediators within a pathway. Future Directions: miRNAs will continue to emerge as critical regulators of the DDR. Understanding the role of miRNAs in the response to IR will provide insights for improving the current standard therapy. Antioxid. Redox Signal. 21, 293–312.
Introduction
Radiation therapy has been used for more than a century to combat tumor progression and metastasis. Ionizing radiation (IR) causes damage to many cellular structures, including DNA. IR induces DNA damage either by directly damaging the DNA or by indirectly generating reactive oxygen species (ROS), which, ultimately, alter the chemical structure of DNA. Radiation-induced damage typically results in double-strand breaks (DSBs), forcing tumor cells to halt cellular proliferation and initiate a DNA damage response (DDR). With extensive DNA damage, a cell usually undergoes programmed cell death. Massive cell death is the mechanism by which radiation functions therapeutically to reduce tumor burden. Unfortunately, tumors often possess or acquire radioprotective characteristics that enable them to escape radiation-induced cell death. Several factors can influence cellular response to radiation, including the tumor microenvironment and the cellular gene expression profile. It is important to understand how specific cells respond to IR in order to improve the effectiveness of cancer therapy.
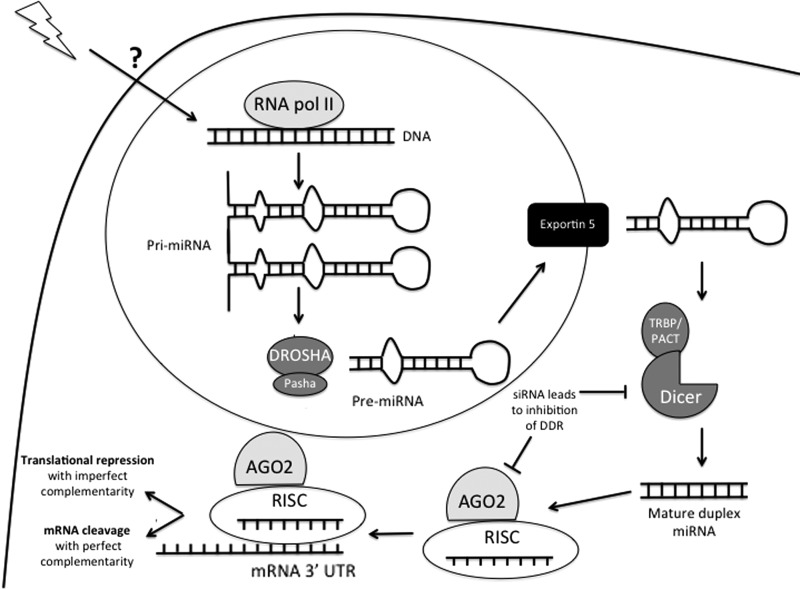
microRNAs (miRNAs) are a class of small non-coding RNA molecules that are involved in the regulation of gene expression. More than 1000 miRNAs have been identified in humans and are predicted to target at least 60% of all protein coding genes (40). miRNA biogenesis is a fairly well-understood process. After transcription and cleavage in the nucleus, an miRNA is exported into the cytoplasm as a hairpin. Further cleavage by an enzyme called Dicer generates duplex RNA. Mature miRNAs are loaded onto the RNA-induced silencing complex (RISC), where they can bind to a specific seed sequence in the 3′ untranslated region (UTR) of target genes (Fig. 1). If the 3′ UTR binding site is fully complementary to the miRNA, as is most commonly seen in plants, the mRNA is targeted for degradation. In animals, base pair mismatches are more common, triggering translational repression of the mRNA, and in some cases, promoting deadenylation of the polyA tail, accelerating the normal process of mRNA degradation (37, 151). Despite their fairly recent discovery, miRNAs have been implicated in a wide variety of diseases, including their ever-expanding role in cancer and cancer therapy.
FIG. 1.
miRNA biogenesis and radiation response. miRNAs are transcribed in the nucleus by RNA polymerase II (RNA pol II) generating a primary miRNA transcript (pri-miR). DROSHA and Pasha cleave the pri-miR into the precursor miRNA (pre-miRNA) hairpin. As a hairpin, Exportin 5 exports the pre-miRNA into the cytoplasm, where it is further cleaved by an enzyme called Dicer and its binding partner TRBP/PACT. This final cleavage step results in a duplex RNA structure. The mature miRNA is separated from the passenger strand and loaded into the RISC. Along with RISC, the miRNA binds to the 3′ UTR of a target gene and mediates translational repression, or in some cases, mRNA degradation. In response to IR, miRNA expression is dysregulated. Although it remains to be proved, it seems likely that IR will result in modification of the biogenesis pathway. Importantly, when AGO2 and Dicer are repressed, the DDR is inhibited (67). DDR, DNA damage response; IR, ionizing radiation; miRNA, microRNA; RISC, RNA-induced silencing complex; AGO2, Argonaute-2.
Understanding the regulation and function of miRNAs is essential to improving current cancer therapy. miRNA expression is tightly regulated under normal conditions. As such, miRNA dysregulation is often reported as a driver of tumorigenesis and metastasis. miRNA expression is controlled by a number of mechanisms, including transcription factor recruitment, genetic or epigenetic changes, and proper function of miRNA biogenesis pathway effectors (55). Changes to any of these mechanisms can result in altered miRNA expression. Exogenous stressors such as hypoxia or IR play a role in promoting miRNA regulatory changes. On exposure to a stressor such as IR, the cell should respond by attempting to repair the damage to its DNA. This response initiates a number of cellular changes, including the up- or down-regulation of genes that are directly involved in the DDR. Multiple examples of miRNA expression changes in response to IR have been reported. In this review, we will examine the most recent advances in understanding the role of IR in miRNA regulation as well as the role of miRNAs in cellular response to IR via the DDR. We will discuss the possibility of using miRNAs to improve current standard cancer therapies. It is important to note that miRNAs can have both direct interactions with targets involved in the DDR and indirect effects, such as targeting transcription factors that are necessary for the expression of a particular DDR protein. In the interest of being concise, we will focus primarily on the direct effect of miRNAs on the DDR.
IR Affects miRNA Expression
Although miRNA expression profiles differ from one cell type to the next, environmental stressors can influence miRNA expression in any cell type. IR induces a number of cellular changes, especially with regard to the DDR. One mechanism by which IR might affect the DDR is by altering the expression of translational regulators such as miRNAs. It is tempting to speculate that IR could affect miRNA biogenesis; however, little work has been done to study the effect of IR on miRNA biogenesis pathway members such as Dicer, Drosha, Exportin-5, or Argonaute-2 (AGO2) (Fig. 1).
One study, published this year, demonstrated that IR has little to no affect on Dicer1 or AGO2 expression in keratinocytes (63). In addition, Kraemer et al. used siRNA to Dicer and AGO2 to demonstrate an essential function for miRNAs in IR survival response in endothelial cells (67). In the absence of miRNA biogenesis, a decrease in cell survival was observed along with a reduction in cell cycle checkpoint activation and an increase in apoptosis, suggesting a global role for miRNAs in maintaining cellular homeostasis by mediating cell cycle checkpoints and apoptosis in response to IR (67). Taken together, these data suggest a critical role for miRNA biogenesis in response to IR.
Despite a lack of evidence for miRNA biogenesis changes, IR clearly plays a role in regulating miRNA expression as has been demonstrated several times in gene expression profiling studies post-IR. The earliest of these came from Sungkwan An's group in Korea looking at the miRNA expression profile in human B lymphoblast cells and human non-small cell lung cancer (NSCLC) cells (13, 124). Not surprisingly, a completely different panel of miRNAs emerged as having altered expression in response to IR in the two cell lines. Interestingly, in the lymphoblast cell line, a larger number of miRNAs were up-regulated after IR as opposed to the NSCLC cells, which largely showed down-regulation of miRNAs (13, 124). One explanation for this could be the dose of IR. In the lymphoblast line, a maximum dose of 10 Gy was used; while in the NSCLC cells, a maximum dose of 40 Gy was used (13, 124). It is possible that too much IR might result in global miRNA repression due to extensive DNA damage and a cell fate decision to undergo apoptosis rather than DNA repair. It would be interesting to study miRNA expression changes at a lower dose in NSCLC cells to compare with the changes seen in lymphoblast cells.
Other such gene expression profiling studies have been done across a wide range of cell types, including endothelial cells, human embryonic stem cells, squamous cell carcinoma cells, and primary glioma cells (16, 67, 102, 125, 139). In each study, a different expression pattern was observed after IR. A summary of some of these miRNA changes can be found in Figure 2. Some overlap in dysregulation exists between cell types, suggesting an important role for a few select miRNAs in mediating IR response. For example, at least one member of the miR-15/16 family (miR-15 a/b, miR-16, miR-195, miR-424, and miR-497) was often down-regulated across a number of different cell lines, including endothelial, NSCLC, and lymphoblast cells (13, 16, 67, 124). Interestingly, this family of miRNAs has been shown to regulate a number of cell cycle checkpoint genes (83, 86), further implicating the miR-15/16 family in contributing to IR response.
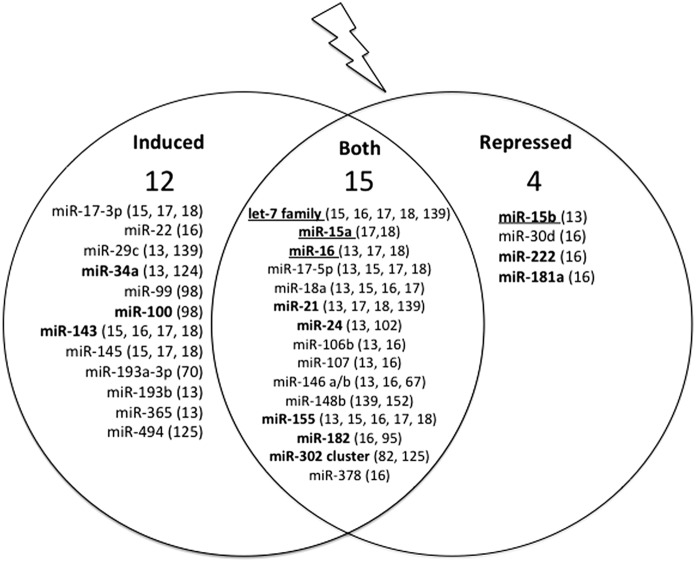
FIG. 2.
miRNAs are differentially regulated in response to IR. On exposure to IR, miRNA expression is often dysregulated. IR induces some miRNAs, while others are repressed, a decision likely dependent on the target genes involved. A summary of the miRNAs discussed in this review whose expression changes in response to IR can be found in this diagram. On the left and right are lists of miRNAs whose induction or repression, respectively, have been observed. In the center are miRNAs where both induction and repression have been observed in different cell types. Interestingly, the largest group of miRNAs exists in the center, illustrating how different the miRNA profile can be from one cell type to the next. Bold miRNAs function in multiple aspects of the DDR. Additional underlining identifies miRNA families that are involved in almost every stage of the DDR.
On the other hand, some miRNAs were up-regulated in certain cell lines, down-regulated in others, and/or had no change in expression. As an example, miR-148b is induced by IR in non-Hodgkin's lymphoma, but repressed by IR in endothelial cells (139, 152), demonstrating the variability in IR-induced expression changes. It is interesting to note that miRNA expression changes were not always consistent within a single cell line, specifically with regard to IR dose and recovery time post-IR. In fact, one study suggested that miRNAs may have different temporal expression patterns such that a single miRNA might be induced at 8 h post-IR, repressed at 12 h post-IR, and re-induced at 24 h (16). This likely indicates some type of feedback loop regulation for miRNA expression in response to IR.
IR activates p53, a DDR pathway member. It is important to note that p53 has been implicated in the regulation of expression of the miR-34 family (14, 47, 113). It has been shown that the miR-34 family of miRNAs contains p53 responsive elements within the miR-34a and miR-34b/c promoters (47). Activation of p53 by IR and subsequent induction of the miR-34 family can lead to increased levels of apoptosis. Although regulation of the miR-34 family is the most well-understood example of p53-dependent miRNA regulation, several reports have suggested that p53 plays a role in the regulation of several other miRNAs, including let-7 (116). Taken together, these data identify a role for p53 in regulating miRNA expression, especially in response to IR.
Another DDR pathway member, ataxia telangiectasia mutated (ATM), has also been identified as a regulator of miRNA expression. The KH-type splicing regulatory protein (KSRP) is involved in the cleavage events of miRNA processing and is associated with the induction of a specific subset of miRNAs (133). Recently, it was confirmed that ATM expression is necessary for this KSRP-driven induction of miRNAs in response to DNA DSBs (169). Interestingly, these KSRP-induced miRNAs included miR-16 and miR-21, both of which play a role in the DDR, which are discussed in detail later (169). This suggests that activation of ATM is necessary to promote the expression of specific miRNAs that are essential for the survival response to IR.
Obviously, IR plays a role in regulating miRNA expression, although the mechanism behind this regulation remains largely unknown. Looking forward, it will be crucial to understand how these miRNAs are regulated in response to IR as a potential radiation sensitization tool. If changes in miRNA expression promote cell survival in response to IR, it is possible that inhibiting these changes might sensitize tumor cells to IR therapy. The first step in moving toward this type of adjuvant therapy will be to elucidate the underlying regulation of these miRNAs in response to IR.
IR Induces PI3K/AKT and MAPK Signaling Pathways
Two main pro-survival signal transduction pathways are activated in response to IR: the PI3K/AKT pathway and the mitogen-activated protein kinase (MAPK) pathway [reviewed in Dent et al. (34)]. IR activates the EGFR/ERBB family of receptor tyrosine kinases, which, subsequently, initiates the PI3K/AKT or MAPK pathways (34).
PI3K/AKT pathway
Briefly, phosphoinositide 3-kinase (PI3K), through the conversion of phosphatidylinositol diphosphates to triphosphates, activates 3′-phosphoinositide-dependent protein kinase 1 (PDK1), which, in turn, activates AKT by phosphorylation. Phosphatase and tensin homolog (PTEN), a tumor suppressor, can inhibit this activation by suppressing PI3K function. Activation of AKT results in a pro-survival pathway, one that is frequently seen overactive in tumors (34). AKT acts through a number of different mediator proteins to inhibit apoptosis by blocking the function of several pro-apoptotic proteins, including the FOXO proteins, BIM, and BAX, or by promoting anti-apoptotic factors such as Bcl-2 (Fig. 3).
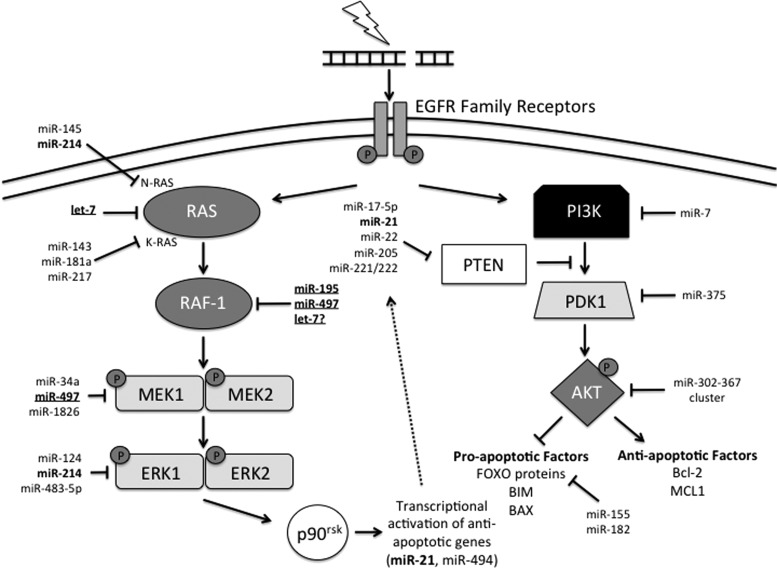
FIG. 3.
PI3K/AKT and MAPK signal transduction pathway activation in response to IR. IR activates the EGFR family of RTKs, which triggers both PI3K/AKT and MAPK signaling. On the left, activation of RAS by the RTK leads to activation of RAF-1. Active RAF-1 initiates a cascade of phosphorylation-dependent activation, first of MEK1/2 followed by ERK1/2. ERK1/2 activation results in the activation of p90rsk, which promotes the transcription of anti-apoptotic factors, including miRNAs such as miR-21. Importantly, miR-21 targets PTEN, which inhibits the PI3K/AKT pathway, promoting PI3K/AKT signaling. On the right, the RTK activates PI3K, which, in turn, phosphorylates AKT. Activated AKT promotes the transcription of anti-apoptotic factors and inhibits the transcription of pro-apoptotic factors. miRNA regulation of the FOXO proteins is shown. Other members of the apoptotic pathway will be discussed later in Figure 7. miRNA regulation is extensive in both these pathways. miRNAs that have been bolded indicate miRNAs which have multiple targets in the same pathway. Additional underlining identifies miRNAs that are involved in almost every pathway of the DDR. Dotted arrow indicates potential for a positive feedback loop. ERK1/2, extracellular signal-regulated kinase 1/2; MAPK, mitogen-activated protein kinase; MEK1/2, MAPK kinase 1/2; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homolog; RAS; RTK, receptor tyrosine kinase.
The PI3K/AKT pathway has several miRNA regulators acting at different levels in the pathway. miR-7 is the first and only miRNA that has been demonstrated to target PI3K with three independent binding sites in the 3′ UTR (146). In addition to PI3K, AKT is directly mediated by miRNA regulation, specifically the miR-302–367 cluster. Two groups simultaneously published evidence supporting this regulation, showing that miR-302 is responsible for repressing AKT expression with a single 3′ UTR binding site (9, 82). In addition, miR-302 is repressed in response to IR in breast cancer cells (82), perhaps as a mechanism to activate AKT and promote cell survival. Despite having several poorly conserved putative miRNA binding sites in its 3′ UTR, it is more likely that further AKT regulation occurs either directly or indirectly by other members of the PI3K/AKT pathway.
PTEN is a negative regulator of AKT expression and is subject to regulation by multiple miRNAs. One of the first miRNAs identified to target PTEN was miR-21 (92, 93). miR-21 is often induced after IR (4, 17, 81, 121, 139), facilitating the repression of PTEN and the activation of AKT to promote the DDR and cell survival. Several articles have demonstrated sensitization to IR by antagonizing miR-21, ultimately inhibiting the PI3K/AKT pathway (46, 51, 88). In addition to miR-21, several other miRNAs have also been shown to target PTEN, including miR-17-5p, miR-22, miR-205, and miR-221/222 (26, 77, 112, 129, 163). Similar to miR-21, repression of miR-205 or miR-221/222 confers radiosensitivity by inhibiting AKT pro-survival function (26, 112, 163). Interestingly, the passenger strand of miR-17, miR-17-3p, also targets a protein that is involved in the DDR, MDM2 (77). Although this will be discussed later, it illustrates the concept that a single miRNA might target multiple members of the DDR in order to regulate the response more efficiently. The considerable regulation of PTEN by miRNAs illustrates their necessity in maintaining genomic integrity by tightly mediating the PI3K/AKT pro-survival pathway.
Another member of the PI3K/AKT pathway, PDK1, is also under direct miRNA regulation. Multiple groups have demonstrated that miR-375 targets PDK1 at a single binding site in the 3′ UTR (36, 79, 135). miR-375 down-regulation has been observed in both gastric and esophageal cancers, presumably reducing levels of PDK1 (79, 135). As expected, driving expression of miR-375 induces apoptosis (79, 135), suggesting a mechanism by which these cancer cells evade apoptosis via the up-regulation of PDK1.
Several of the downstream pro- or anti-apoptotic effectors of AKT signaling are regulated by miRNAs. For the most part, each of the FOXO proteins have their own miRNA regulators, with FOXO1 and FOXO3a being the most widely studied. As an example, FOXO3a is targeted by miR-155 (66). miR-155 has been shown to confer radioresistance, potentially by down-regulating FOXO3a, promoting survival (5). FOXO1 has several miRNA regulators as well, including the well-studied miR-182 (45), which also targets BRCA1, discussed later (95). Other AKT effectors have miRNA regulators, which we will discuss later as mediators of apoptosis.
MAPK pathway
The second signal transduction pathway that is associated with the DDR after IR is the MAPK pathway. Again, activated by radiation-induced EGFR family receptors, the MAPK pathway begins with activation of RAS, leading to translocation of the RAF proteins to the plasma membrane. Notably, RAS can also activate the PI3K pathway. A series of phosphorylation and dephosphorylation steps results in active RAF, namely RAF-1. RAF-1 phosphorylates MAPK kinase 1/2 (MEK1/2), which subsequently phosphorylates extracellular signal-regulated kinase 1/2 (ERK1/2). ERK1/2 activate p90rsk, which phosphorylates several transcription factors that induce anti-apoptotic gene expression (Fig. 3) (136). As with the PI3K pathway, the MAPK pathway has a number of miRNA regulators.
The RAS family of proteins has several members, including the most clinically relevant H-RAS, N-RAS, and K-RAS (136). Each of these members has been shown to be direct targets of at least one miRNA. In fact, N-RAS and K-RAS have dozens of predicted miRNA binding sites in their respective 3′ UTRs. Interestingly, the let-7 family of miRNAs is capable of targeting all three RAS members (62). It is well established that the let-7 family is often suppressed in cancer, leading to an over-activation of RAS and a pro-survival phenotype in response to IR and other DNA damaging agents (62, 147). Importantly, let-7 family members have been shown to regulate other components of the DDR, including several cell cycle checkpoint proteins (61). In addition to let-7, many other miRNAs have been identified as RAS regulators such as miR-145 and miR-214 targeting N-RAS and miR-143, miR-181a, and miR-217 targeting K-RAS (22, 84, 123, 170, 174). All of these miRNAs are down-regulated in tumors when compared with normal tissue (22, 123, 170, 174), again suggesting a mechanism by which RAS is over-expressed, inhibiting apoptosis.
RAF-1 is directly downstream of RAS. Although it does not have quite as many predicted miRNA mediators as the RAS proteins, it is still amenable to miRNA regulation. Family members, miR-195 and miR-497 have been shown to target RAF-1 through a single 3′ UTR binding site (76). Similar to RAS-regulating miRNAs, miR-195 and miR-497 are also down-regulated in cancer (76), presumably to enhance RAF-1 expression and promote cell survival. It is interesting to note that let-7 has two putative binding sites in the 3′ UTR of RAF-1, although they have yet to be validated. Let-7 targeting both RAS and RAF-1 would suggest a multifaceted role for let-7 in the DDR to more efficiently regulate signaling.
RAF-1 kinase is responsible for the activation of MEK1/2. MEK2 has no known miRNA regulators, which is consistent with an absence of conserved putative miRNA binding sites in its 3′ UTR. Conversely, MEK1 is targeted by miR-34a and miR-1826 with one or two binding sites, respectively, in its 3′ UTR (48, 54). Interestingly, miR-497, which targets RAF-1, mentioned earlier, also targets MEK1 via at least one 3′ UTR binding site (171), providing yet another example of a single miRNA acting on multiple members within the same pathway.
ERK1/2 are activated by MEK1/2. Although some miRNA regulation of ERK1/2 exists, it is the regulation of miRNA expression by ERK1/2 that is arguably more important with regard to IR response. miR-124, miR-214, and miR-483-5p have been demonstrated to target ERK1 and are down-regulated in cancers, presumably to enable the over-expression of ERK1 and promote cell survival (142, 157). One mechanism by which ERK1/2 regulates survival is by activating the expression of anti-apoptotic genes, including miRNAs. Both miR-21 and miR-494 have been identified as targets of ERK1/2 activation (52, 114). It is tempting to speculate that IR induces the MAPK pathway, activating ERK1/2, which induces the expression of miR-21. miR-21 subsequently targets PTEN (93), as discussed earlier, and CDC25A (143), mentioned next, repressing their expression. The repression of PTEN promotes the activation of the PI3K/AKT pathway, while the repression of CDC25A triggers cell cycle arrest and enables the cell sufficient time to repair IR-induced damage.
Considering the amount of evidence supporting miRNA regulation of the PI3K/AKT and MAPK pathways, it is fair to say that miRNAs are essential for the proper function of these pathways. More specifically, miRNAs are necessary to regulate the radiation response and promote cell survival, especially in the case of therapy-resistant tumor cells. It is plausible that targeting miRNAs involved in these pathways may be a clinical tool in the near future to improve radiation therapy by sensitizing tumor cells and promoting apoptosis.
miRNAs Target Many Members of the DDR
miRNAs are responsible for regulating many cellular pathways, including those involved in the DDR. Here, we will discuss specific miRNAs that have been implicated in the DDR with a focus on cell cycle checkpoint activation, DSB repair, and apoptosis.
DNA damage sensing and cell cycle checkpoint activation
IR induces several different types of DNA damage, including single-strand break (SSB) and DSB. Initial steps after this damage include DNA damage sensing and cell cycle checkpoint activation. DNA damage sensors and signal transducers include poly [ADP-ribose] polymerase-1 (PARP1), the MRN complex, ATM, ataxia telangiectasia and Rad3 related (ATR), and H2AX. SSB sensing is often performed by PARP1. Currently, there are no validated miRNAs that target PARP1; however, the PARP1 3′ UTR contains a number of putative miRNA binding sites, including one for miR-7.
The MRN complex is composed of MRE11, RAD50, and NBS1 and senses DSBs, the more common form of radiation-induced DNA damage. Once a DSB has been identified, the MRN complex recruits ATM, which phosphorylates a number of downstream mediators and effectors. There are no experimentally validated miRNAs targeting the MRN complex, but prediction websites suggest that the 3′ UTRs of each component of the MRN complex contain one or more putative miRNA-binding sites.
Two of the major players in DNA damage sensing and cell cycle checkpoint activation are ATM and ATR, serine-threonine kinases responsible for initiating a signaling cascade in the presence of DNA damage. ATR typically responds to SSBs and stalled replication forks. ATM is the main activator of the DDR after radiation-induced DSBs. As such, its expression is tightly regulated.
The first indication for miRNA-mediated regulation of ATM was published in 2010 when Hu et al. linked miR-421 to ATM expression (49). It was suggested that miR-421 induction by the oncogene N-MYC resulted in a repression of ATM and increased sensitivity to IR (49). Since then, several other miRNAs have been identified as mediators of ATM expression, including miR-18a, miR-100, and miR-101. All three of these miRNAs target ATM, resulting in increased radiosensitivity (100, 126, 149, 158). Interestingly, miR-18a is down-regulated after IR (139), likely to enable an induction of ATM expression and initiation of the DDR.
ATM is responsible for the phosphorylation of several downstream mediators and effectors, including histone variant H2AX, a mediator of DSB repair that is responsible for recruitment of DNA repair proteins to the site of damage. This phosphorylation step is crucial to the recruitment of several important DDR factors, including BRCA1 and 53BP1. Two miRNAs, miR-24 and miR-138 have been shown to target H2AX (72, 145). miR-24 targets H2AX, promoting chromosomal instability and sensitization to DNA damage in terminally differentiated blood cells (72). miR-138 sensitizes cells to IR by targeting H2AX and preventing downstream DSB repair (145). Importantly, both miRNAs have been shown to be up-regulated by IR in squamous cell carcinoma and glioma cell lines (102), identifying one possible mechanism by which these cells evade cell cycle checkpoint activation and promote tumor progression.
Apart from DNA damage sensors and transducers, there are many effectors of the DDR, including those involved in cell cycle checkpoint activation. The cell has two main checkpoints to maintain genomic integrity throughout the cell cycle; one at the G1/S interphase (Fig. 4) and one at the G2/M interphase (Fig. 5). An additional checkpoint exists during the S phase, but we will not discuss that in detail here. Cyclins and cyclin-dependent kinase (CDK) proteins are essential for the progression of the cell through the cell cycle. In response to DNA damage, these cyclins and CDKs should be inhibited at the checkpoint to arrest cell proliferation and allow time for repair (33). Each of these checkpoints has a different set of key players that orchestrate the progression from one phase of the cell cycle to the next.
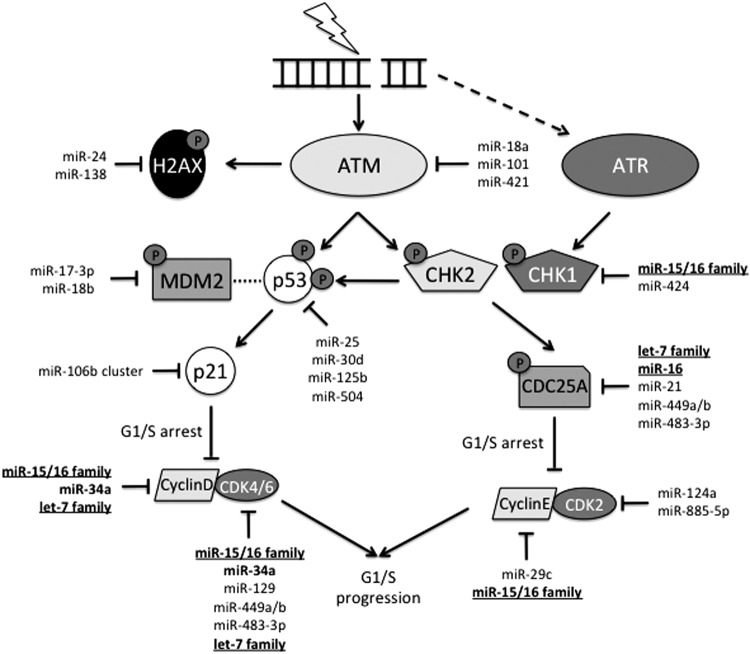
FIG. 4.
G1/S cell cycle checkpoint activation in response to radiation-induced DNA damage. IR induces DNA DSBs. ATM and ATR are recruited to sites of DNA damage, but ATM is specific for DSBs. ATM activates several down-stream mediators by phosphorylation, including H2AX, p53, and the CHK proteins, typically CHK2. Activated CHK2 phosphorylates CDC25A, targeting it for degradation. In the absence of CDC25A, Cyclin E and CDK2 are impaired, resulting in cell cycle arrest. CHK2 is also capable of activating p53 by phosphorylation. Concurrently, activated p53 is unable to bind to MDM2 (indicated by dotted line), leading to its accumulation. Accumulation of p53 stimulates p21 expression. p21 binds to both cyclin/CDK complexes and inhibits them from promoting progression through the cell cycle. miRNAs regulate almost every member of this checkpoint. Bold miRNAs indicate multiple targets within the pathway. Underlined miRNA families are seen throughout the DDR. Dotted arrow indicates limited secondary activation of ATR in response to DSBs. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; CDK, cyclin-dependent kinase.
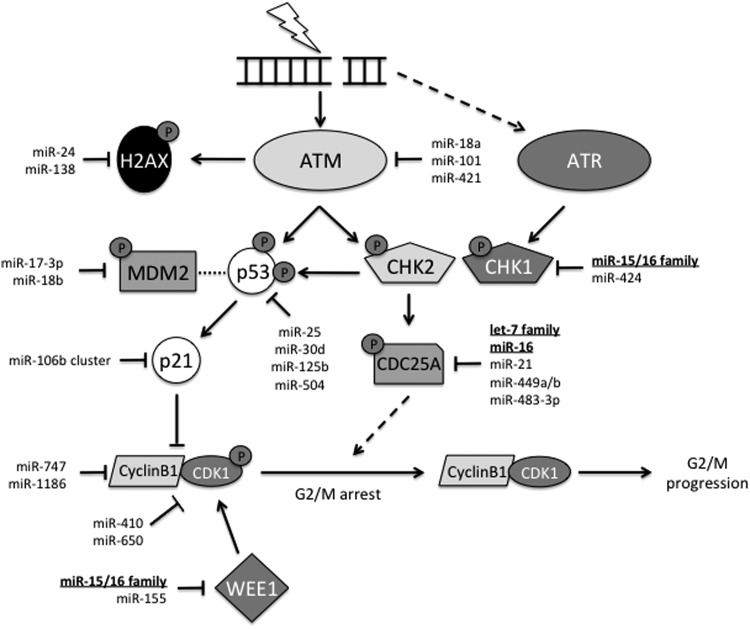
FIG. 5.
G2/M cell cycle checkpoint activation in response to radiation-induced DNA damage. Similar to the G1/S checkpoint, IR induces DNA DSBs that are recognized by the MRN complex (not pictured), which activates ATM and ATR. ATM phosphorylates CHK2, which targets CDC25A for degradation. CDC25A is necessary for the removal of an inhibitory phosphate on CDK1. WEE1 kinase is responsible for this phosphorylation of CDK1. Degradation of CDC25A inhibits progression through the cell cycle. In addition, ATM activates p53, preventing its association with MDM2. p53 accumulation promotes p21 expression. p21 inhibits cyclin B1/CDK1, inducing cell cycle arrest. As with G1/S, miRNA regulation is observed in almost every member of the G2/M checkpoint. Bold miRNAs are multifaceted within the pathway. Underlined miRNAs play a role in almost every aspect of the DDR from signal transduction to apoptosis.
Some of the few overlapping effectors involved in all three checkpoints are the CHK proteins, CHK1 and CHK2. Typically, CHK2 is activated by ATM in response to DSBs, while CHK1 is activated by ATR after SSBs; however, it is believed that there is some cross-talk between the two transducers (104). While there are no reports of miRNA regulation of CHK2, CHK1 has at least a few miRNA regulators, including the miR-15/16 family. Several members of this family have been experimentally validated, including miR-15 and miR-424 (110, 155). Pouliot et al. showed that multiple members of the miR-15/16 family could down-regulate protein expression of CHK1, including miR-15a, miR-15b, miR-16, miR-16-1*, and miR-424* (110). More conclusively, Xu et al. demonstrated that miR-424 targets CHK1 by multiple molecular assays, including a 3′ UTR luciferase assay (155). Notably, it has been shown that miR-424 is repressed in cervical cancer tissue compared with normal tissue (155), suggesting a mechanism by which cancer cells can bypass cell cycle checkpoints and promote tumor progression. In addition, as mentioned earlier, miR-15/16 family members are often down-regulated in response to IR to promote DNA repair and cell survival (13, 16, 67, 124). Taken together, it is tempting to speculate that the regulation of expression of the miR-15/16 family might be crucial to the DDR and tumor cell survival.
Another checkpoint effector that plays a role in all three checkpoints is the phosphatase, CDC25A. In response to DSBs, CDC25A is targeted for proteasomal degradation by the CHK proteins. Loss of CDC25A prevents proper function of the Cyclin E/Cdk2 complex and slows progression of the cell into the S phase. Several miRNAs are predicted to target CDC25A, a few of which have been experimentally validated, namely let-7, miR-16, miR-21, miR-449a/b, and miR-483-3p (6, 32, 61, 81, 109, 143, 159). Of these, miR-21 is the most well studied in its relationship to CDC25A. miR-21 has a single binding site in the 3′ UTR of CDC25A (143). Importantly, miR-21 has been shown to be up-regulated in a number of different cancer types (4, 43, 122), and its expression is induced upon exposure to IR (16, 17, 121, 139). It has also been suggested that miR-21 expression is induced by hypoxia (32), a common tumor microenvironment characteristic, and that its over-expression is responsible for radiation resistance by activating cell cycle checkpoints and preventing cell cycle progression, specifically at the G2/M checkpoint (4). Together, these data demonstrate a crucial role for miR-21 targeting CDC25A in cell cycle checkpoint activation in response to IR. miR-21 likely prevents apoptosis after IR by enabling DSB repair at the cell cycle checkpoint. In addition, miR-21 has been shown to target multiple members of the DDR, including the PTEN mentioned earlier, suggesting a more global role for miR-21 in regulating the response to radiation-induced DNA damage.
At the G1/S checkpoint, there is evidence for miRNA regulation of cyclin and CDK proteins as well. Both miR-124a and miR-885-5p have been identified as miRNAs that target CDK2 (2, 65); however, only miR-124a has a predicted binding site in the 3′ UTR of CDK2. Interestingly, there are several putative miRNA binding sites in the CDK2 3′ UTR, suggesting that miRNA regulation might be important for the function of CDK2, but will need further validation. Cyclin E also has a number of validated miRNA regulators, including miR-29c and the miR-15/16 family (35, 86). Notably, the miR-15/16 family is also involved in the regulation of other cyclin and CDK proteins, which will be discussed later.
Cell cycle arrest at G1/S driven by CDC25A is only temporary. The secondary and more long-term pathway of G1/S arrest involves p53. If DNA damage is present in G1, ATM activation leads to CHK2 phosphorylation and subsequent phosphorylation of p53. It is this phosphorylation event that prevents p53 from interacting with MDM2, enabling p53 to accumulate. p53 stimulates expression of p21, which will bind to CDK-Cyclin complexes, cyclin D/CDK4/6, and cyclin E/CDK2, and arrest the cell in G1 until the damage is repaired (Fig. 4) (33). Not surprisingly, there are a number of miRNAs that regulate MDM2, p53, and p21 expression.
It has been shown that MDM2 is targeted by both miR-17-3p and miR-18b (31, 77). MDM2 harbors 3 binding sites for miR-17-3p in the 3′ UTR and a single binding site for miR-18b (31, 77). There is evidence which suggests that miR-17, both 5p and 3p, are up-regulated after IR (15, 18), which, subsequently, down-regulates MDM2, enabling p53 accumulation and G1 arrest in response to radiation-induced damage. Conversely, it has been suggested that miR-18b is down-regulated in certain cancers, including melanoma (31), which would enable over-expression of the oncogene MDM2 and inhibition of the tumor suppressor, p53. Interestingly, the seed sequence of miR-18b is identical to that of miR-18a, which targets ATM (126, 149). Obviously a finite level of MDM2 should be maintained, at least in part by these two miRNAs, to preserve cellular homeostasis without promoting tumorigenesis.
p53, a well-established tumor suppressor, is also regulated by several miRNAs. The first miRNA that directly regulates p53 was identified as miR-125b in 2009. Le et al. demonstrated that IR can repress expression of miR-125b in zebrafish and that this repression contributes to the well-known radiation-induced expression of p53 (73). miR-125b has only one binding site in the 3′ UTR of p53 (73); while miR-504, another p53-targeting miRNA, has two 3′ UTR binding sites (50). miR-504 over-expression significantly reduces G1 arrest due to the repression of p53 (50). Recently, two more p53-targeting miRNAs were discovered through a screen for miRNAs having a direct interaction with the 3′ UTR of p53. Both miR-25 and miR-30d were identified, each with a single predicted binding site in the 3′ UTR of p53 (69). Both of these miRNAs were able to independently reduce the number of G1 arrested cells (69). Extensive miRNA regulation such as this suggests a critical role for miRNAs in the expression of p53 to maintain genomic integrity through cell cycle checkpoint activation.
p21 expression is also miRNA-mediated during the G1 checkpoint. Recent evidence suggests that multiple members of the miR-106b cluster are responsible for targeting the 3′ UTR of p21 (56). Specifically, miR-106b was shown to reduce p21 expression and to promote cell cycle progression despite the presence of DNA damage (56). In addition, the miR-106b family is known to be up-regulated in several cancer types (3, 80, 106), likely promoting tumorigenesis by repressing cell cycle checkpoint activation.
Cyclin D and CDK 4/6 are targets of miRNA regulation as well. The miR-15/16 family is responsible for regulating cyclin D1, D3, and CDK6 with two, two, and one 3′ UTR binding sites, respectively (86). miR-34a has been shown to target both cyclin D1 and CDK6 (128). It is important to note that cyclin D proteins have dozens of predicted miRNA binding sites, indicating that miRNAs may play a significant role in regulating expression of the cyclin proteins, especially in response to IR. In fact, many miRNAs have been experimentally validated as regulators of cyclin D expression (60, 117, 128, 161). In addition to the miR-15/16 family and miR-34a, CDK6 is also targeted by miR-129 in at least three complementary binding sites in its 3′ UTR (150). miR-129 repression has been observed across a number of different cancers (150), suggesting that over-expression of CDK6 might drive progression through the cell cycle even in the presence of DNA damage, although this has yet to be demonstrated. Interestingly, CDK6 is targeted by miR-449a/b (159) and CDK4 is targeted by miR-483-3p (6), both of which also target CDC25A, mentioned earlier. Finally, let-7 family members target CDK6, cyclin D2, and CDC25A (61). This illustrates the idea that some miRNAs might be regulating pathways by targeting multiple members of a particular pathway. This concept will be discussed later in greater detail.
The second main cell cycle checkpoint occurs during the G2 to M phase transition. Many of the same players from the G1/S checkpoint are also associated with G2/M, but a different set of cyclin/CDK complexes are involved, specifically cyclinB1 and CDK1. Again, ATM responds to a DSB by phosphorylating CHK2, which, in turn, phosphorylates CDC25A and p53, resulting in two independent pathways for arresting G2/M. Similar to G1/S, p53 accumulates and drives expression of p21, a CDK inhibitor, which prevents the function of the cyclin B1/CDK1 complex. On the other hand, phosphorylated CDC25A is degraded, which prevents the dephosphorylation of CDK1 (usually phosphorylated by WEE1), keeping the cyclin B1/CDK1 complex inactive (Fig. 5). Further phosphorylation of CDC25A occurs via PLK1, which also phosphorylates WEEl, targeting it for degradation (33). In addition to the miRNA regulation already discussed earlier for ATM, p53, p21, and CDC25, other members of the G2/M checkpoint are also subject to miRNA repression.
Similar to the cyclin D proteins, the 3′ UTR of cyclin B1 contains several putative miRNA binding sites; however, none have been validated. It is interesting to note that one article, published in 2012, has suggested a role for three miRNAs in activating expression of cyclin B1 (53). miRNA activation of gene expression is a highly controversial concept, although evidence demonstrating its existence continues to accumulate [see these reviews for more information (75, 107)]. In addition, it has been reported that CDK1 is targeted by miR-410 and miR-650 (25, 97), but further studies are necessary to elucidate their role with regard to the DDR.
On the other hand, another G2/M player, WEE1 has several validated miRNA regulators. Inhibition of WEE1 sensitizes cells to IR (108). In the absence of WEE1, CDK1 does not get phosphorylated and a cell with excess DNA damage from IR proceeds through the cell cycle without repair, ultimately facing apoptosis. Although no one has shown convincing evidence for miR-155 interacting with the WEE1 3′ UTR, several studies have suggested its role in targeting WEE1 (7, 8, 110). Moreover, prediction websites suggest that the WEE1 3′ UTR harbors at least one miR-155 putative binding site. A recurring theme, the miR-15/16 family has also been shown to target WEE1 with two binding sites identified in the 3′ UTR (110). Multiple members of this family have been validated, including miR-195 and miR-497 (7, 132). It has been suggested that miR-195 targets WEE1 in malignant melanoma. Interestingly, miR-195 was up-regulated, and, consequently, WEE1 was down-regulated in metastases compared with primary melanoma (7). This contradicts the notion that inhibition of WEE1 sensitizes cells to IR. Bhattacharya et al. rationalized this by suggesting a finite regulation of WEE1 that is necessary to facilitate metastasis by forcing a damaged cell through the cell cycle without sensitizing the cell to therapy-driven apoptosis (7). In line with WEE1 inhibition sensitizing cells to IR, it has been shown that high miR-497 expression, corresponding with an increase in apoptosis, correlates with a better prognosis in neuroblastoma patients (29). Obviously, miRNAs play an important role in regulating G2/M transition and might serve as targets to improve current cancer therapies such as IR.
DNA DSB repair
Once the cell cycle has been halted by checkpoint activation, the cell should initiate DNA repair pathways. There are two main types of DNA repair, SSB repair and DSB repair. We will focus on DSB repair, as this is the major type of DNA damage that is caused by IR. DSB repair occurs via one of two pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR) (Fig. 6). As the names would suggest, NHEJ does not require a homologous template, and, as such, is the more common repair pathway, occurring throughout the cell cycle. The lack of template, however, makes NHEJ much more error prone. Conversely, HR is almost exclusively performed during S phase with some activity in G2, when a homologous template is available via replication (33). As with cell cycle checkpoint activation, DNA repair has several components that are susceptible to miRNA regulation.
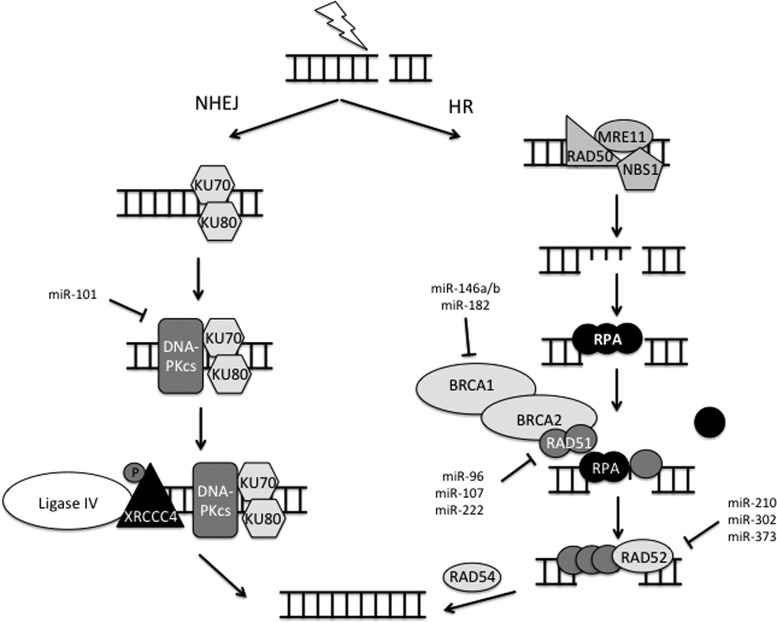
FIG. 6.
miRNA regulation of radiation-induced DNA DSB repair via NHEJ or HR. After cell cycle checkpoint activation, radiation-induced DSBs are repaired by one of two pathways; NHEJ or HR. NHEJ occurs throughout the cell cycle, while HR occurs only during S and early G2 phase when a homologous template is available. NHEJ begins with recognition of the DSB by KU70 and KU80 (XRCC6 and XRCC5 in humans). DNA-PK, including its catalytic subunit DNA-PKcs, is recruited to the site of damage, and phosphorylates the DNA Ligase IV cofactor XRCC4. After end-processing by a yet-unidentified nuclease, DNA Ligase IV ligates the two ends together, often resulting in deletions of DNA. On the other hand, HR is less error prone, as it utilizes a homologous template to repair the break. The MRN complex binds the DSB and facilitates resection of the DNA, resulting in single-strand overhangs. RPA binds to the single-stranded DNA and protects it from nuclease activity. RAD51, with the help of RAD52 and BRCA1/2, displaces RPA to coat the single-stranded DNA, forming a nucleoprotein filament to initiate the homology search. RAD54 facilitates the removal of RAD51 once a homologous template has been located to enable DNA synthesis and ligation to repair the gap. miRNAs target key members of these pathways, namely DNA-PKcs, BRCA1/2, RAD51, and RAD52. HR, homologous recombination; NHEJ, non-homologous end-joining.
NHEJ is the simpler of the two pathways. KU70/KU80 (XRCC6/XRCC5 in humans) acts as a heterodimer to recognize a DSB and functions as a scaffold for NHEJ machinery. DNA-PK is recruited to the site of damage and phosphorylates XRCC4, the cofactor necessary for DNA Ligase IV function. Since radiation-induced DSBs are often not clean enough to ligate directly, DNA processing should occur, frequently resulting in large deletions. The mechanism of this step is not well understood. Nucleases are required, but their identity remains elusive. Some have suggested that MRE11 and/or Artemis play a role in this processing (140), but further evaluation is necessary. Once the DSB ends have been properly resected, DNA Ligase IV and its cofactor XRCC4 seal the gap (140).
Currently, the only member of this pathway with validated miRNA regulation is DNA-PK, specifically the catalytic subunit, DNA-PKcs. miR-101 has been shown to target the 3′ UTR of DNA-PKcs with a single binding site. It has also been shown that over-expression of miR-101 is capable of sensitizing cancer cells to IR both in vitro and in vivo (21, 158). Presumably, the over-expression of miR-101 down-regulates both ATM (previously mentioned) and DNA-PKcs, inhibiting the DDR and promoting cell death.
It is possible that indirect miRNA regulation occurs in the NHEJ pathway. One example would be the role of miR-7 in regulating DNA-PKcs. It has been shown that miR-7 targets EGFR with at least two binding sites and PI3K with at least 3 binding sites in their respective 3′ UTRs (39, 146). On over-expression of miR-7, this regulation of the EGFR/PI3K/AKT pathway reduces active DNA-PKcs, resulting in radiosensitization (74). This highlights the possibility of other indirect miRNA regulation effects on the NHEJ pathway that remain to be described.
On the other hand, HR has a number of pathway members under miRNA regulation, namely the RAD and BRCA proteins. Unlike NHEJ, HR requires a homologous template to repair the DSB. A homologous template is only present during the S and early G2 phase, which explains why HR is the more accurate, but less commonly used repair pathway. Briefly, the DSB is identified by the localization of the MRN complex, γH2AX, and BRCA1. The DSB is processed, leaving a 3′ SS DNA overhang. It is believed that the MRN complex is partly responsible for this nuclease activity. RPA binds to the SS DNA, protecting the SS DNA from nuclease activity and preventing secondary DNA structures from forming. RAD51, with the help of several mediator proteins, including RAD52, RAD54, and BRCA2, displaces RPA to load onto the SS DNA, forming a nucleoprotein filament and initiating the homology search for the template of interest. Once a template is found, RAD54 facilitates the removal of RAD51 from the SS 3′ end and enables DNA synthesis and ligation (Fig. 6) (94).
miRNAs are responsible for regulating the RAD proteins. Both RAD51 and RAD52 have validated miRNA regulators, although most of the RAD proteins harbor putative miRNA binding sites in their 3′ UTR. Recently, it was shown that both miR-107 and miR-222 could target RAD51 with one and two 3′ UTR binding sites, respectively (99). It is important to note that these sites do not contain perfect complementarity to the seed region of either miRNA (99). Surprisingly, with a binding site in the coding region, miR-96 has also been shown to target RAD51, leading to an inhibition of HR (144). Along with the fact that the 3′ UTR of RAD51 contains dozens of other putative sites similar to these, it is possible that abundant RAD51 miRNA regulation exists, but has yet to be described.
Despite its somewhat mysterious function in HR in human cells, RAD52 knock-down has been shown to be conditionally lethal with BRCA2, indicating an independent function, but similar to that of BRCA2. In fact, it has been hypothesized that RAD52 plays a role in loading RAD51 onto the SS DNA in the absence of BRCA2, suggesting the possibility of redundant function (89). miRNA regulation of RAD52 has been demonstrated by miR-210, miR-302, and miR-373 (30, 82). Crosby et al. has shown that miR-210 and miR-373 are induced in hypoxia (30). Hypoxia is a characteristic of the tumor microenvironment which is defined by oxygen deprivation that facilitates the resistance of tumor cells to IR. Although much of this resistance is due to the inherent lack of oxygen that is necessary to generate oxygen radicals, some resistance can be attributed to other molecular changes, including miRNA expression. An over-expression of miR-210 and miR-373 in hypoxia forces the repression of RAD52, possibly reducing the efficiency of HR and promoting genomic instability in these hypoxic cells. On the other hand, over-expression of miR-302 is sufficient to sensitize cells to IR, presumably by down-regulating RAD52 and HR and promoting cell death (82). Interestingly, miR-302 is down-regulated by IR (82, 102, 125), while there is no evidence supporting expression changes for miR-210 or miR-373 in response to IR. It is possible that miR-302 is more acutely involved in response to IR, as its repression enables over-expression of RAD52, promoting DNA repair and progression through the cell cycle, while the regulation by miR-210 and miR-373 remains unchanged in response to IR. Conversely, hypoxic induction of miR-210 and miR-373 may serve to decrease the repair capacity and decrease the radiation survival of chronically hypoxic cells, but this remains to be determined.
In addition to the RAD proteins, the BRCA proteins are also subject to miRNA regulation. BRCA1 is a ubiquitously important protein that is associated with every step of the DDR [reviewed in Roy et al. (115)]. BRCA1 plays a role in both cell cycle checkpoint activation and SSB and DSB repair. Arguably, its most prominent function is its role in HR, such that in the absence of BRCA1, HR is significantly impaired. Multiple miRNAs have been identified that target BRCA1, including miR-146a/b and miR-182. miR-182 targets BRCA1 in at least three confirmed 3′ UTR binding sites, suppressing HR and leading to an increase in DSBs as measured by neutral comet assay (95). In response to IR, miR-182 is down-regulated (95), presumably to induce BRCA1 expression and promote DSB repair via HR. In addition, the over-expression of miR-182 confers radioresistance and has been observed in several cancer types, including cervical, breast, and ovarian (68, 87, 95, 130). miR-146a/b target BRCA1 and are predicted to target BRCA2 as well (41, 118). Interestingly, a single-nucleotide polymorphism in the seed region of miR-146a has been associated with an earlier age of onset in familial breast and ovarian cancers lacking a BRCA1 or BRCA2 mutation (105, 118); however, data supporting this association are highly controversial (11, 42). miR-146a/b target BRCA1 at a single 3′ UTR binding site. Over-expression of miR-146a/b has been observed in triple-negative breast cancer when compared with estrogen receptor and/or progesterone receptor-positive breast cancer and is associated with a higher grade, more aggressive tumor. The over-expression of miR-146a/b inhibits HR, likely due to the repression of BRCA1 (41). As would be expected, miRNAs that target BRCA1 are often over-expressed in cancer to promote tumorigenesis and genomic instability by inhibiting BRCA1 function in HR and other pathways.
Of note, another significant example of indirect miRNA regulation comes from the suppression of BRCA1 function by miR-99 targeting the chromatin-remodeling factor, SNF2H (98). SNF2H functions to recruit BRCA1 to sites of DSBs independent of γH2AX. The miR-99 family, specifically miR-99a and miR-100, is up-regulated by IR in cancer cells, conferring radiation resistance by indirectly inhibiting BRCA1 function. In addition, the repression of SNF2H by the miR-99 family results in an increase in DSBs and a decrease in HR (98). Obviously, it is necessary to consider indirect miRNA regulation as well as direct regulation when defining the role of miRNAs in the DDR.
Although not all members of the DSB repair pathway have experimentally validated miRNA mediators, it is important to note that several key players experience extensive miRNA regulation. For that reason, it is possible that targeting these miRNAs may prove to be a useful adjuvant therapy to enhance the efficacy of IR treatment.
Apoptosis
In the presence of excess DNA damage or in the absence of functional DNA repair after IR, a cell will often undergo radiation-induced cell death. Several mechanisms of cell death can occur after IR, including mitotic catastrophe, senescence, and apoptosis. In this review, we will specifically focus on apoptotic cell death. Not surprisingly, miRNA regulation plays a role in maintaining this pathway as well. Apoptosis is a relatively complex process. In the interest of being concise, we will only discuss a few major players involved in apoptosis, but it is important to note that many of the factors involved in this process are likely regulated by miRNA repression. All of the pro- and anti-apoptotic factors we will cover are in some way mediated by the signaling pathways previously discussed.
Apoptosis signaling begins in one of two ways: Either activation of a death-receptor, such as the Fas receptor (FAS-R), initiates a caspase activation cascade or the Bcl-2-like family of proteins initiates a pathway through the mitochondria. IR typically induces the Bcl-2 specific pathway, although both pathways can be active. The extensive activation of p53 in the presence of massive DNA damage initiates apoptosis by transcriptionally activating several pro-apoptotic factors, including PUMA, FAS-R, NOXA, and BAX and inhibiting anti-apoptotic factors, Bcl-2 and MCL1. The Bcl-2 family includes pro-apoptotic factors, BAX, BAK, PUMA, and BIM, and anti-apoptotic factors, Bcl-2, Bcl-xL, and MCL1. BAX and BAK are activated by homo-oligomerization, ultimately releasing cytochrome c into the cytoplasm. Cytochrome c forms a complex with caspase 9 and apoptotic protease-activating factor −1 (APAF-1), which activates caspases 3 and 7, facilitating apoptosis. Apoptosis is negatively regulated by Bcl-2 and MCL1, which sequester the pro-apoptotic factors and inhibit progression of apoptosis (Fig. 7) (91). Again, many of these factors involved in apoptosis are subject to miRNA post-transcriptional regulation.
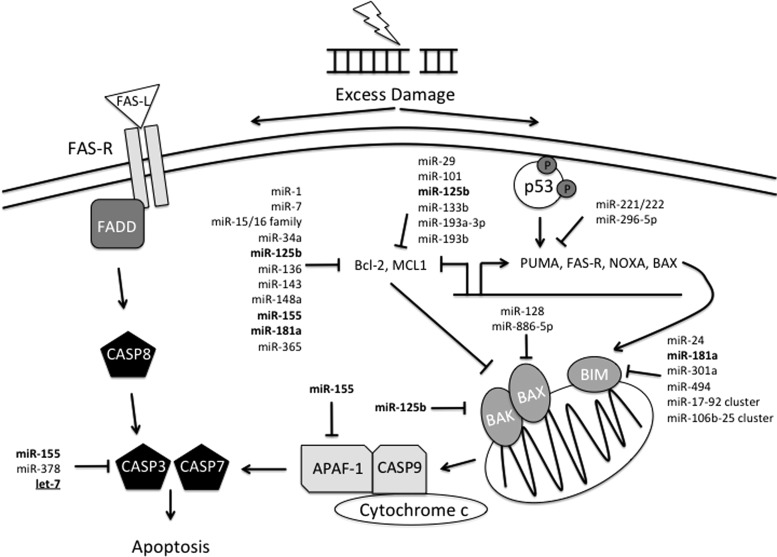
FIG. 7.
miRNA regulation of radiation-induced apoptosis. Excessive radiation-induced DNA damage leads to activation of apoptosis. Here, we depict two mechanisms by which apoptosis is initiated in response to IR. Activated p53 transcriptionally activates pro-apoptotic factors, including PUMA, FAS-R, and BAX, while inhibiting transcription of anti-apoptotic factors Bcl-2 and MCL1. Activation of pro-apoptotic factors BAX, BAK, and BIM in the mitochondria, ultimately, results in cytochrome c release into the cytoplasm. Cytochrome c forms a complex with APAF-1 and caspase 9, which activates downstream caspases 3 and 7. Activation of caspase 3 and 7 can also be accomplished via death-receptor signaling. FAS-R, a death-receptor, is activated by its ligand, FAS-L, resulting in caspase 8 activation and subsequent activation of caspases 3 and 7. miRNA regulation is significantly involved in the regulation of apoptosis signaling, especially with regard to the Bcl-2-like family of pro- and anti-apoptotic factors. miRNAs in bold regulate multiple targets within the abridged pathway depicted here. Underlined miRNAs are involved throughout DDR. FAS-R, Fas receptor.
Despite having several putative miRNA binding sites in its 3′ UTR, PUMA has only a few validated miRNA regulators, namely miR-221/222 and miR-296-5p (12, 164, 165). Interestingly, miR-221/222 is more commonly over-expressed in higher-grade glioblastomas, likely repressing PUMA expression and reducing apoptosis (165). As previously mentioned, miR-221/222 also targets PTEN (26), suggesting a multifaceted role for this miRNA in the DDR.
Other members of the apoptosis pathway are also regulated by miRNAs, including several of the BCL-2 family members. BAX is regulated by at least two different miRNAs, miR-128 and miR-886-5p (1, 59, 78). Surprisingly, miR-128 is repressed in a number of cancer types, including glioblastoma and breast cancer (27, 173), likely resulting in an up-regulation of BAX and an increase in apoptosis. Interestingly, Adlakha and Saini have demonstrated that over-expression of miR-128 leads to a down-regulation of BAX and an unanticipated increase in apoptosis (1). This could be explained by a redundant role for BAX and BAK, but further work is necessary to support this hypothesis. BAK has a single known miRNA regulator, miR-125b, despite having a few putative binding sites for miRNAs in its 3′ UTR. miR-125b is overexpressed in a number of different cancer types, including prostate cancer, leading to a down-regulation of BAK (120). Repression of BAK likely inhibits apoptosis, promoting tumorigenesis.
Another member of the Bcl-2 family, BIM, is a target of miRNA regulation with dozens of predicted binding sites in its 3′ UTR. Several of these miRNAs have been experimentally validated, including the miR-17–92 cluster, miR-24, the miR-106b-25 cluster, specifically miR-25, miR-181a, miR-301a, and miR-494 (24, 64, 90, 103, 111, 114, 137, 168). The majority of these miRNAs mentioned earlier also target other players in the DDR. In addition to BIM, miR-181a targets K-RAS, as well as two other members of the Bcl-2 family, Bcl-2 itself and MCL1 (103, 123). Notably, BIM is a pro-apoptotic protein, while both Bcl-2 and MCL1 are anti-apoptotic. As is likely the case with many such miRNA-mRNA interactions, the regulation by other miRNAs as well as the basal expression level in a particular tissue might help determine which proteins will be targeted (103). Importantly, miR-181a is down-regulated in glioblastoma and confers radioresistance to these cells (19, 119), suggesting a potential therapeutic advantage by inducing miR-181a expression to sensitize tumor cells to IR.
Several other miRNAs have been shown to target anti-apoptotic factors, MCL1 and Bcl-2. Not surprisingly, many of the miRNAs that are capable of targeting MCL1 and/or Bcl-2 have already been discussed as regulators of other DDR pathway members. In addition to miR-181a, mentioned earlier, miR-7, miR-34a, miR-125b, miR-143, miR-155, and members of the miR-15/16 family, including miR-15b, miR-16, miR-195, and miR-497, target Bcl-2 as well as other DDR proteins (57, 58, 85, 148, 153, 154, 156, 166, 172). In fact, one key report found that a frequent deletion of chromosome 13q14 seen in chronic lymphocytic leukemia (CLL) contained the miR-15 and miR-16 loci, resulting in repression of miR-15 and miR-16 expression (10). This would suggest an over-expression of Bcl-2 in CLL. Similarly, miR-29, miR-101, and miR-125b target MCL1 and at least one other member of the DDR (44, 96, 127). MCL1 is also regulated by miR-133b, miR-193a-3p, and miR-193b, while Bcl-2 is regulated by miR-1, miR-136, miR-148a, and miR-365 (20, 28, 70, 101, 131, 160, 167). Strikingly, miR-136 and miR-181a are down-regulated in glioblastoma (27, 160), and miR-7, miR-34a, miR-148a, miR-195, miR-365, and miR-497 are repressed in either tumor tissue or cancer cells when compared with normal tissue or cells (23, 57, 76, 85, 101, 154). This suggests a frequent over-expression of Bcl-2 in tumors, resulting in a decrease in radiation-induced apoptosis. Likewise, miR-125b, miR-133b, miR-101, and miR-193b are down-regulated in tumor tissue compared with normal tissue (20, 28, 44, 127), suggesting a similar up-regulation of MCL1 and subsequent decrease in apoptosis. In addition, expression of miR-193a-3p is induced in response to IR (70), presumably to enable an increase in radiation-induced apoptosis. Taken together, this suggests a vital role for miRNAs in regulating the Bcl-2-like apoptotic pathway, especially in response to IR.
Finally, the caspase proteins, essential to the apoptotic response, are also subject to direct miRNA regulation, albeit less than the Bcl-2-like family. Briefly, caspase 8 is activated by FAS-R in response to excess radiation-induced damage. Caspase 8 can directly activate caspase 3 and 7. Caspase 3 and 7 can also be activated by the release of mitochondrial cytochrome c into the cytosol and subsequent complex formation (Fig. 7). miR-155 has been shown to target both APAF-1 and caspase 3 (141, 162). miR-155 is up-regulated in several tumor types, including lung, breast, and pancreatic cancer (138, 162) and, as mentioned earlier, has been shown to promote radiation resistance (5). Over-expression of miR-155 may result in a repression of APAF-1 and caspase 3, leading to an inhibition of FAS-R-mediated apoptosis. In addition to miR-155, miR-378 and let-7a have also been shown to target caspase 3 (38, 134). Let-7a over-expression confers radioresistance to cancer cells (134), which is consistent with the fact that let-7a targets caspase 3, inhibiting radiation-induced apoptosis. The let-7 family represents yet another example of a miRNA family with a multidimensional role in the DDR.
Innovation
As key regulators of gene expression, miRNAs are intricately involved in the ionizing radiation (IR)-induced DNA damage response (DDR). Recently, it has become evident that individual miRNAs or miRNA families are responsible for regulating a number of different genes within a particular pathway. In addition, specific miRNAs are often up- or down-regulated in response to a stressor such as IR. It is tempting to speculate that a single miRNA or miRNA family will emerge as a potential target for adjuvant therapy to reduce tumor cell survival after IR by inhibiting the DDR. A better understanding of miRNA regulation and function will be critical to enhancing IR therapy.
Concluding Remarks
Radiation therapy is a critical modality of treatment for cancer patients; however, the complete mechanism behind both its successes and limitations remains largely unknown. As has been discussed, it has been well established that miRNAs play a crucial role in the cellular response to IR. We have summarized the most recent advances in miRNA regulation and function in response to IR and shed light on the potential for the use of these small non-coding RNA molecules as therapeutic adjuvants to improve radiation treatment.
As mentioned several times, it is important to emphasize the role of miRNAs at a global level in response to IR. A number of the radiation-responsive miRNAs discussed actually play a multifaceted role in the DDR such that a single miRNA or miRNA family is responsible for the regulation of multiple DDR proteins. Examples of this include, but are not limited to, the miR-15/16 family, the let-7 family, miR-21, and miR-155, each regulating at least two essential players in the DDR. It is conceivable that the concept of a single miRNA targeting multiple members of a particular pathway will become increasingly more abundant in the next decade of miRNA research. Several articles have already begun to identify this phenomenon, including Krishnan et al. and Liu et al. (68, 86), both discussed in this review. Finally, an even more abstract possibility is the expansion of potential miRNA targets, as suggested by Lal et al., proposing that miRNAs do not require seed sequence complementarity to target a particular mRNA (71). In fact, Lal et al. demonstrated that miR-24 is capable of targeting several DDR members through highly complementary, yet “seedless” binding sites (71). Although this remains the first evidence of its kind, it is not implausible to imagine that miRNAs might play an even more complex and vital role in pathway regulation than we currently understand, especially considering the youth of this field. Improving our understanding of miRNA regulation in response to radiation-induced DNA damage will, ultimately, improve the efficacy of current cancer therapy.
Abbreviations Used
- AGO2
Argonaute-2
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3 related
- CDK
cyclin-dependent kinase
- CLL
chronic lymphocytic leukemia
- DDR
DNA damage response
- DSB
double-strand break
- ERK1/2
extracellular signal-regulated kinase ½
- FAS-R
Fas receptor
- Gy
gray
- HR
homologous recombination
- IR
ionizing radiation
- KSRP
KH-type splicing regulatory protein
- MAPK
mitogen-activated protein kinase
- MEK1/2
MAPK kinase 1/2
- miRNA, miR
microRNA
- MRN
MRE11-RAD50-NBS1 complex
- NHEJ
non-homologous end-joining
- NSCLC
non-small cell lung cancer
- PARP1
poly [ADP-ribose] polymerase-1
- PDK1
3′-phosphoinositide-dependent protein kinase 1
- PI3K
phosphoinositide 3-kinase
- PTEN
phosphatase and tensin homolog
- RISC
RNA-induced silencing complex
- ROS
reactive oxygen species
- RTK
receptor tyrosine kinase
- SSB
single-strand break
- UTR
untranslated region
References
- 1.Adlakha YK. and Saini N. MicroRNA-128 downregulates Bax and induces apoptosis in human embryonic kidney cells. Cell Mol Life Sci 68: 1415–1428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, Theissen J, Fischer M, Zapatka M, Brors B, Savelyeva L, Sagulenko V, Speleman F, Schwab M, and Westermann F. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ 18: 974–984, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, and Croce CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 68: 6162–6170, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anastasov N, Hofig I, Vasconcellos IG, Rappl K, Braselmann H, Ludyga N, Auer G, Aubele M, and Atkinson MJ. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol 7: 206, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, and Slack FJ. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther 12: 908–914, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertero T, Gastaldi C, Bourget-Ponzio I, Mari B, Meneguzzi G, Barbry P, Ponzio G, and Rezzonico R. CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell Death Differ 20: 800–811, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya A, Schmitz U, Wolkenhauer O, Schonherr M, Raatz Y, and Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 32: 3175–3183, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Butz H, Liko I, Czirjak S, Igaz P, Khan MM, Zivkovic V, Balint K, Korbonits M, Racz K, and Patocs A. Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas. J Clin Endocrinol Metab 95: E181–E191, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Cai N, Wang YD, and Zheng PS. The microRNA-302–367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA 19: 85–95, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, and Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524–15529, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K, Sutter C, Wappenschmidt B, Dick M, Arnold N, Bugert P, Niederacher D, Meindl A, Schmutzler RK, Bartram CC, Ficarazzi F, Tizzoni L, Zaffaroni D, Manoukian S, Barile M, Pierotti MA, Radice P, Burwinkel B, and Peterlongo P. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat 31: E1052–E1057, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Masuoka HC, Charlton MR, and Gores GJ. A role for miR-296 in the regulation of lipoapoptosis by targeting PUMA. J Lipid Res 52: 1517–1525, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha HJ, Shin S, Yoo H, Lee EM, Bae S, Yang KH, Lee SJ, Park IC, Jin YW, and An S. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol 34: 1661–1668, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, and Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhry MA. and Omaruddin RA. Differential regulation of microRNA expression in irradiated and bystander cells. Mol Biol (Mosk) 46: 634–643, 2012 [PubMed] [Google Scholar]
- 16.Chaudhry MA, Omaruddin RA, Brumbaugh CD, Tariq MA, and Pourmand N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J Radiat Res 54: 808–822, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhry MA, Omaruddin RA, Kreger B, de Toledo SM, and Azzam EI. Micro RNA responses to chronic or acute exposures to low dose ionizing radiation. Mol Biol Rep 39: 7549–7558, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhry MA, Sachdeva H, and Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol 29: 553–561, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, and Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep 23: 997–1003, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Zhang X, Lentz C, Abi-Daoud M, Pare GC, Yang X, Feilotter HE, and Tron VA. miR-193b Regulates Mcl-1 in Melanoma. Am J Pathol 179: 2162–2168, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Wang H, Ng WL, Curran WJ, and Wang Y. Radiosensitizing effects of ectopic miR-101 on non-small-cell lung cancer cells depend on the endogenous miR-101 level. Int J Radiat Oncol Biol Phys 81: 1524–1529, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, and Zhang CY. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28: 1385–1392, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, and Liu Z. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg 14: 1170–1179, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Chen LY, Dai HY, Wang P, Gao S, and Wang K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting Bim expression. J Cell Biochem 113: 3229–3235, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Chien WW, Domenech C, Catallo R, Kaddar T, Magaud JP, Salles G, and Ffrench M. Cyclin-dependent kinase 1 expression is inhibited by p16(INK4a) at the post-transcriptional level through the microRNA pathway. Oncogene 30: 1880–1891, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, and Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10: 367, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, and Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351–1358, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Crawford M, Batte K, Yu L, Wu X, Nuovo GJ, Marsh CB, Otterson GA, and Nana-Sinkam SP. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun 388: 483–489, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creevey L, Ryan J, Harvey H, Bray IM, Meehan M, Khan AR, and Stallings RL. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol Cancer 12: 23, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crosby M, Kulshreshtha R, Ivan M, and Glazer P. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69: 1221–1229, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, Sagebiel R, Miller JR, 3rd, and Kashani-Sabet M. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst 105: 433–442, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Oliveira PE, Zhang L, Wang Z, and Lazo JS. Hypoxia-mediated regulation of Cdc25A phosphatase by p21 and miR-21. Cell Cycle 8: 3157–3164, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Deckbar D, Jeggo PA, and Lobrich M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit Rev Biochem Mol Biol 46: 271–283, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dent P, Yacoub A, Contessa J, Caron R, Amorino G, Valerie K, Hagan MP, Grant S, and Schmidt-Ullrich R. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res 159: 283–300, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F, Shao K, Li N, Qiu B, and He J. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 32: 1025–1032, 2011 [DOI] [PubMed] [Google Scholar]
- 36.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, and van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 57: 2708–2717, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, and Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA 15: 21–32, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, Ren AJ, Yuan WJ, and Lin L. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 17: 410–423, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Fang Y, Xue JL, Shen Q, Chen J, and Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 55: 1852–1862, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Friedman RC, Farh KK, Burge CB, and Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaelian I, and Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med 3: 279–290, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia AI, Cox DG, Barjhoux L, Verny-Pierre C, Barnes D, Antoniou AC, Stoppa-Lyonnet D, Sinilnikova OM, and Mazoyer S. The rs2910164:G >C SNP in the MIR146A gene is not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum Mutat 32: 1004–1007, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, and Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res 70: 4528–4538, 2010 [DOI] [PubMed] [Google Scholar]
- 44.Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, and Zhuang SM. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 32: 3071–3079, 2012 [DOI] [PubMed] [Google Scholar]
- 45.Guttilla IK. and White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 284: 23204–23216, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, Kim JH, Yin J, Yoo H, Lee SH, and Park JB. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One 7: e47449, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, and Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirata H, Hinoda Y, Ueno K, Shahryari V, Tabatabai ZL, and Dahiya R. MicroRNA-1826 targets VEGFC, beta-catenin (CTNNB1) and MEK1 (MAP2K1) in human bladder cancer. Carcinogenesis 33: 41–48, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H, Du L, Nagabayashi G, Seeger RC, and Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A 107: 1506–1511, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, and Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell 38: 689–699, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang S, Li XQ, Chen X, Che SM, Chen W, and Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus 26: 823–831, 2012 [DOI] [PubMed] [Google Scholar]
- 52.Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, Horiuchi D, Lebbink RJ, Mo YY, Goga A, and McManus MT. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem 284: 18515–18524, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, and Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40: 1695–1707, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ichimura A, Ruike Y, Terasawa K, Shimizu K, and Tsujimoto G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol Pharmacol 77: 1016–1024, 2010 [DOI] [PubMed] [Google Scholar]
- 55.Iorio MV. and Croce CM. Causes and consequences of microRNA dysregulation. Cancer J 18: 215–222, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, and Cleary MA. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, and Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 8: 266, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, and Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One 4: e6816, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji S, Shao G, Lv X, Liu Y, Fan Y, Wu A, and Hu H. Downregulation of miRNA-128 sensitises breast cancer cell to chemodrugs by targeting Bax. Cell Biol Int 37: 653–658, 2013 [DOI] [PubMed] [Google Scholar]
- 60.Jiang Q, Feng MG, and Mo YY. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer 9: 194, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, and Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–7722, 2007 [DOI] [PubMed] [Google Scholar]
- 62.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, and Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647, 2005 [DOI] [PubMed] [Google Scholar]
- 63.Joly-Tonetti N, Vinuelas J, Gandrillon O, and Lamartine J. Differential miRNA expression profiles in proliferating or differentiated keratinocytes in response to gamma irradiation. BMC Genomics 14: 184, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, Yang J, Abraham JM, Mori Y, and Meltzer SJ. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 136: 1689–1700, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawano S. and Nakamachi Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann Rheum Dis 70Suppl 1: i88–i91, 2011 [DOI] [PubMed] [Google Scholar]
- 66.Kong W, He L, Coppola M, Guo J, Esposito N, Coppola D, and Cheng J. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 285: 17869–17879, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Kraemer A, Anastasov N, Angermeier M, Winkler K, Atkinson MJ, and Moertl S. MicroRNA-mediated processes are essential for the cellular radiation response. Radiat Res 176: 575–586, 2011 [DOI] [PubMed] [Google Scholar]
- 68.Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, and Grimmond SM. MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 19: 230–242, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, Young KH, and Li Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 30: 843–853, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon JE, Kim BY, Kwak SY, Bae IH, and Han YH. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis 18: 896–909, 2013 [DOI] [PubMed] [Google Scholar]
- 71.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, and Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell 35: 610–625, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, and Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 16: 492–498, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, and Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 23: 862–876, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee KM, Choi EJ, and Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol 101: 171–176, 2011 [DOI] [PubMed] [Google Scholar]
- 75.Lee S. and Vasudevan S. Post-transcriptional stimulation of gene expression by microRNAs. Adv Exp Med Biol 768: 97–126, 2013 [DOI] [PubMed] [Google Scholar]
- 76.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, Zhao D, Sun Q, Zeng Z, Dress A, Lin MC, Kung HF, Rui H, Liu LZ, Mao F, Jiang BH, and Lai L. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res 17: 1722–1730, 2011 [DOI] [PubMed] [Google Scholar]
- 77.Li H. and Yang BB. Stress response of glioblastoma cells mediated by miR-17-5p targeting PTEN and the passenger strand miR-17-3p targeting MDM2. Oncotarget 3: 1653–1668, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li JH, Xiao X, Zhang YN, Wang YM, Feng LM, Wu YM, and Zhang YX. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol Oncol 120: 145–151, 2011 [DOI] [PubMed] [Google Scholar]
- 79.Li X, Lin R, and Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci 56: 2849–2856, 2011 [DOI] [PubMed] [Google Scholar]
- 80.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, and Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci 100: 1234–1242, 2009 [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Zhao S, Zhen Y, Li Q, Teng L, Asai A, and Kawamoto K. A miR-21 inhibitor enhances apoptosis and reduces G(2)-M accumulation induced by ionizing radiation in human glioblastoma U251 cells. Brain Tumor Pathol 28: 209–214, 2011 [DOI] [PubMed] [Google Scholar]
- 82.Liang Z, Ahn J, Guo D, Votaw JR, and Shim H. MicroRNA-302 replacement therapy sensitizes breast cancer cells to ionizing radiation. Pharm Res 30: 1008–1016, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, and Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Luo XJ, Xiong AW, Zhang ZD, Yue S, Zhu MS, and Cheng SY. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J Biol Chem 285: 26599–26607, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu L, Chen L, Xu Y, Li R, and Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun 400: 236–240, 2010 [DOI] [PubMed] [Google Scholar]
- 86.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, Hernando E, and Wei JJ. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol 228: 204–215, 2012 [DOI] [PubMed] [Google Scholar]
- 88.Liu ZL, Wang H, Liu J, and Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem 372: 35–45, 2013 [DOI] [PubMed] [Google Scholar]
- 89.Lok BH. and Powell SN. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res 18: 6400–6406, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lwin T, Lin J, Choi YS, Zhang X, Moscinski LC, Wright KL, Sotomayor EM, Dalton WS, and Tao J. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood 116: 5228–5236, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marquez RT, Bryan W.Tsao , Nicolas F.Faust , and Liang Xu. Drug resistance and molecular cancer therapy: apoptosis versus autophagy. In: Apoptosis, edited by Rudner J. ISBN: 978-953-51-1133-7, InTech, DOI: 10.5772/55415 Available from: www.intechopen.com/books/apoptosis/drug-resistance-and-molecular-cancer-therapy-apoptosis-versus-autophagyInTech;2013 [DOI]
- 92.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, and Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130: 2113–2129, 2006 [DOI] [PubMed] [Google Scholar]
- 93.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, and Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mladenov E, Magin S, Soni A, and Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol 3: 113, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, and Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41: 210–220, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mott JL, Kobayashi S, Bronk SF, and Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26: 6133–6140, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S, Borsky M, Verner J, Doubek M, Brychtova Y, Trbusek M, Hampl A, Mayer J, and Pospisilova S. MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood 119: 2110–2113, 2012 [DOI] [PubMed] [Google Scholar]
- 98.Mueller AC, Sun D, and Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 32: 1164–1172, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neijenhuis S, Bajrami I, Miller R, Lord CJ, and Ashworth A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair (Amst) 12: 394–402, 2013 [DOI] [PubMed] [Google Scholar]
- 100.Ng WL, Yan D, Zhang X, Mo YY, and Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 9: 1170–1175, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 101.Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y, Du X, and Han W. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis 33: 220–225, 2012 [DOI] [PubMed] [Google Scholar]
- 102.Niemoeller OM, Niyazi M, Corradini S, Zehentmayr F, Li M, Lauber K, and Belka C. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol 6: 29, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ouyang YB, Lu Y, Yue S, and Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 12: 213–219, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Palii SS, Cui Y, Innes CL, and Paules RS. Dissecting cellular responses to irradiation via targeted disruptions of the ATM-CHK1-PP2A circuit. Cell Cycle 12: 1105–1118, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pastrello C, Polesel J, Della Puppa L, Viel A, and Maestro R. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis 31: 2124–2126, 2010 [DOI] [PubMed] [Google Scholar]
- 106.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, Munker R, Volinia S, Boccadoro M, Garzon R, Palumbo A, Aqeilan RI, and Croce CM. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A 105: 12885–12890, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Portnoy V, Huang V, Place RF, and Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA 2: 748–760, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.PosthumaDeBoer J, Wurdinger T, Graat HC, van Beusechem VW, Helder MN, van Royen BJ, and Kaspers GJ. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 11: 156, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, and Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J 28: 2090–2099, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pouliot LM, Chen YC, Bai J, Guha R, Martin SE, Gottesman MM, and Hall MD. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res 72: 5945–5955, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, and Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med 208: 549–560, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qu C, Liang Z, Huang J, Zhao R, Su C, Wang S, Wang X, Zhang R, Lee MH, and Yang H. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 11: 785–96, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, and Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743, 2007 [DOI] [PubMed] [Google Scholar]
- 114.Romano G, Acunzo M, Garofalo M, Di Leva G, Cascione L, Zanca C, Bolon B, Condorelli G, and Croce CM. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci U S A 109: 16570–16575, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roy R, Chun J, and Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12: 68–78, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saleh AD, Savage JE, Cao L, Soule BP, Ly D, DeGraff W, Harris CC, Mitchell JB, and Simone NL. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One 6: e24429, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schultz J, Lorenz P, Gross G, Ibrahim S, and Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18: 549–557, 2008 [DOI] [PubMed] [Google Scholar]
- 118.Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, and Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29: 1963–1966, 2008 [DOI] [PubMed] [Google Scholar]
- 119.Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, and You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res 1236: 185–193, 2008 [DOI] [PubMed] [Google Scholar]
- 120.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, and deVere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A 104: 19983–19988, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi Y, Zhang X, Tang X, Wang P, Wang H, and Wang Y. MiR-21 is continually elevated long-term in the brain after exposure to ionizing radiation. Radiat Res 177: 124–128, 2012 [DOI] [PubMed] [Google Scholar]
- 122.Shibuya H, Iinuma H, Shimada R, Horiuchi A, and Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 79: 313–320, 2010 [DOI] [PubMed] [Google Scholar]
- 123.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, and Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun 404: 896–902, 2011 [DOI] [PubMed] [Google Scholar]
- 124.Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, and An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol 35: 81–86, 2009 [PubMed] [Google Scholar]
- 125.Sokolov MV, Panyutin IV, and Neumann RD. Unraveling the global microRNAome responses to ionizing radiation in human embryonic stem cells. PLoS One 7: e31028, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, Li M, and Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One 6: e25454, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, and Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 69: 1135–1142, 2009 [DOI] [PubMed] [Google Scholar]
- 128.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582: 1564–1568, 2008 [DOI] [PubMed] [Google Scholar]
- 129.Tan G, Shi Y, and Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun 417: 546–551, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, and Choy KW. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol 129: 199–208, 2013 [DOI] [PubMed] [Google Scholar]
- 131.Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, and Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J 50: 377–387, 2009 [DOI] [PubMed] [Google Scholar]
- 132.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, and Croce CM. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A 108: 4908–4913, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, and Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsang WP. and Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 13: 1215–1222, 2008 [DOI] [PubMed] [Google Scholar]
- 135.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, and Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res 70: 2339–2349, 2010 [DOI] [PubMed] [Google Scholar]
- 136.Valerie K, Yacoub A, Hagan MP, Curiel DT, Fisher PB, Grant S, and Dent P. Radiation-induced cell signaling: inside-out and outside-in. Mol Cancer Ther 6: 789–801, 2007 [DOI] [PubMed] [Google Scholar]
- 137.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, and Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, and Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103: 2257–2261, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, and Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol 5: 25, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang C. and Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining. Int J Radiat Oncol Biol Phys 86: 440–449, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, and Luo ZJ. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol 225: 232–242, 2011 [DOI] [PubMed] [Google Scholar]
- 142.Wang L, Shi M, Hou S, Ding B, Liu L, Ji X, Zhang J, and Deng Y. MiR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett 586: 1312–1317, 2012 [DOI] [PubMed] [Google Scholar]
- 143.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr., Lazo JS, Wang Z, Zhang L, and Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 69: 8157–8165, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Huang JW, Calses P, Kemp CJ, and Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res 72: 4037–4046, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, and Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res 9: 1100–1111, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, and Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731–5741, 2009 [DOI] [PubMed] [Google Scholar]
- 147.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, and Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 67: 11111–11116, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Willimott S. and Wagner SD. miR-125b and miR-155 contribute to BCL2 repression and proliferation in response to CD40 ligand (CD154) in human leukemic B-cells. J Biol Chem 287: 2608–2617, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu CW, Dong YJ, Liang QY, He XQ, Ng SS, Chan FK, Sung JJ, and Yu J. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One 8: e57036, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C, Spira A, Cardoso WV, and Lu J. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle 9: 1809–1818, 2010 [DOI] [PubMed] [Google Scholar]
- 151.Wu L, Fan J, and Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 103: 4034–4039, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu Y, Liu GL, Liu SH, Wang CX, Xu YL, Ying Y, and Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin's Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res 53: 516–525, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, and Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer 123: 372–379, 2008 [DOI] [PubMed] [Google Scholar]
- 154.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, and Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci 7: 805–814, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, and Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 32: 976–987, 2013 [DOI] [PubMed] [Google Scholar]
- 156.Yadav S, Pandey A, Shukla A, Talwelkar SS, Kumar A, Pant AB, and Parmar D. miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem 286: 37347–37357, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yamane K, Jinnin M, Etoh T, Kobayashi Y, Shimozono N, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara T, Aoi J, Oike Y, and Ihn H. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med (Berl) 91: 69–81, 2013 [DOI] [PubMed] [Google Scholar]
- 158.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, and Wang Y. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One 5: e11397, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, and Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev 23: 2388–2393, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang Y, Wu J, Guan H, Cai J, Fang L, Li J, and Li M. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett 586: 3608–3612, 2012 [DOI] [PubMed] [Google Scholar]
- 161.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, McCue P, and Pestell RG. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 182: 509–517, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zang YS, Zhong YF, Fang Z, Li B, and An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther 19: 773–778, 2012 [DOI] [PubMed] [Google Scholar]
- 163.Zhang C, Kang C, Wang P, Cao Y, Lv Z, Yu S, Wang G, Zhang A, Jia Z, Han L, Yang C, Ishiyama H, Teh BS, Xu B, and Pu P. MicroRNA-221 and -222 regulate radiation sensitivity by targeting the PTEN pathway. Int J Radiat Oncol Biol Phys 80: 240–248, 2011 [DOI] [PubMed] [Google Scholar]
- 164.Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, and Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol 37: 1621–1626, 2010 [DOI] [PubMed] [Google Scholar]
- 165.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, and Kang CS. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9: 229, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang H, Cai X, Wang Y, Tang H, Tong D, and Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep 24: 1363–1369, 2010 [DOI] [PubMed] [Google Scholar]
- 167.Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng H, and Lai M. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ 18: 1702–1710, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang H, Zuo Z, Lu X, Wang L, Wang H, and Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep 27: 594–598, 2012 [DOI] [PubMed] [Google Scholar]
- 169.Zhang X, Wan G, Berger FG, He X, and Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41: 371–383, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, and Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis 31: 1726–1733, 2010 [DOI] [PubMed] [Google Scholar]
- 171.Zheng D, Radziszewska A, and Woo P. MicroRNA 497 modulates interleukin 1 signalling via the MAPK/ERK pathway. FEBS Lett 586: 4165–4172, 2012 [DOI] [PubMed] [Google Scholar]
- 172.Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, and Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol 29: 384–391, 2012 [DOI] [PubMed] [Google Scholar]
- 173.Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao HR, Gong C, Chen JN, Su FX, Zhang Y, and Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res 17: 7105–7115, 2011 [DOI] [PubMed] [Google Scholar]
- 174.Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, Jiang Y, Chen X, Qi Y, Zhang X, Wang X, Sun Q, Kung HF, Lin MC, Dress A, Wardle F, Jiang BH, and Lai L. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle 11: 2137–2145, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]