Abstract
Cell division across members of the protozoan parasite phylum Apicomplexa displays a surprising diversity between different species as well as between different life stages of the same parasite. In most cases, infection of a host cell by a single parasite results in the formation of a polyploid cell from which individual daughters bud in a process dependent on a final round of mitosis. Unlike other apicomplexans, Toxoplasma gondii divides by a binary process consisting of internal budding that results in only two daughter cells per round of division. Since T. gondii is experimentally accessible and displays the simplest division mode, it has manifested itself as a model for apicomplexan daughter formation. Here we review newly emerging insights in the prominent role that assembly of the cortical cytoskeletal scaffold plays in the process of daughter parasite formation.
Keywords: Toxoplasma, cytoskeleton, IMC, ISP, Rab, intermediate filaments, centrin
1. Introduction
Toxoplasma gondii is the obligate intracellular apicomplexan parasite responsible for toxoplasmosis-associated encephalitis and birth defects (Montoya, Liesenfeld, 2004). Other medically significant members of the phylum Apicomplexa include the causative agent of malaria (Plasmodium spp.) (Haldar, Mohandas, 2009), opportunistic infections that cause acute gastroenteritis (Cryptosporidium spp.) (Tzipori, Ward, 2002), and several costly veterinary scourges (Eimeria, Theileria, and Babesia spp.) (Bishop et al., 2004; Bock et al., 2004; Shirley et al., 2005). The pathogenesis of Toxoplasma results from its rapid replication cycle and the ensuing immune response, which causes destructive tissue lesions. This replication cycle takes only about six to seven hours to complete (Gubbels et al., 2008b; Radke et al., 2001; Reilly et al., 2007) and is predicated on proper formation of the cytoskeleton (Agop-Nersesian et al., 2010; Agop-Nersesian et al., 2009; Anderson-White et al., 2011; Beck et al., 2010; Heaslip et al., 2010; Lorestani et al., 2010; Stokkermans et al., 1996; Tran et al., 2010). Due to the importance of the development of the cytoskeleton, the components of this structure make attractive potential therapeutic targets and have become the basis of a growing area of research.
The cell division process of Toxoplasma was first studied by electron microscopy in the late nineteen fifties and throughout the sixties (Goldman et al., 1958; Ogino, Yoneda, 1966; Sheffield, Melton, 1968). An internal budding process wherein two daughter parasites are assembled within the confinement of an intact mother cell was observed and dubbed endodyogeny (Fig. 1). The advent of genetic tools and fluorescence microscopy in the past 15 years has lead to the identification and characterization of a number of the molecules driving the budding process. Notably, the visualization of cytoskeletal components in Toxoplasma by fluorescence microscopy has revealed daughter budding to be a highly coordinated phenomenon. These observations suggest there are four general stages of cytoskeletal dynamics: initiation, early bud assembly, mid-budding, and late stage budding. During the initiation of budding and early bud assembly, foundations are laid for each layer and component of the cytoskeleton (Agop-Nersesian et al., 2010; Anderson-White et al., 2011; Hu et al., 2006). These fast progressing stages are followed by elongation of the cytoskeleton to the midpoint of budding. This is the widest point in the bud and is marked by the accumulation of the contractile ring components known as the basal complex on the leading edge of the developing parasites (Anderson-White et al., 2011; Gubbels et al., 2006; Hu, 2008). After reaching the midpoint, the growing daughters begin to taper toward the basal end, mediated by contraction of the basal complex, and move into the late stages of budding. The late stages of budding are indicated by the maturation of the daughter cytoskeletons, disassembly of the mother's cytoskeleton, and the incorporation of the mother's plasma membrane along with newly synthesized plasma membrane onto the daughter cells.
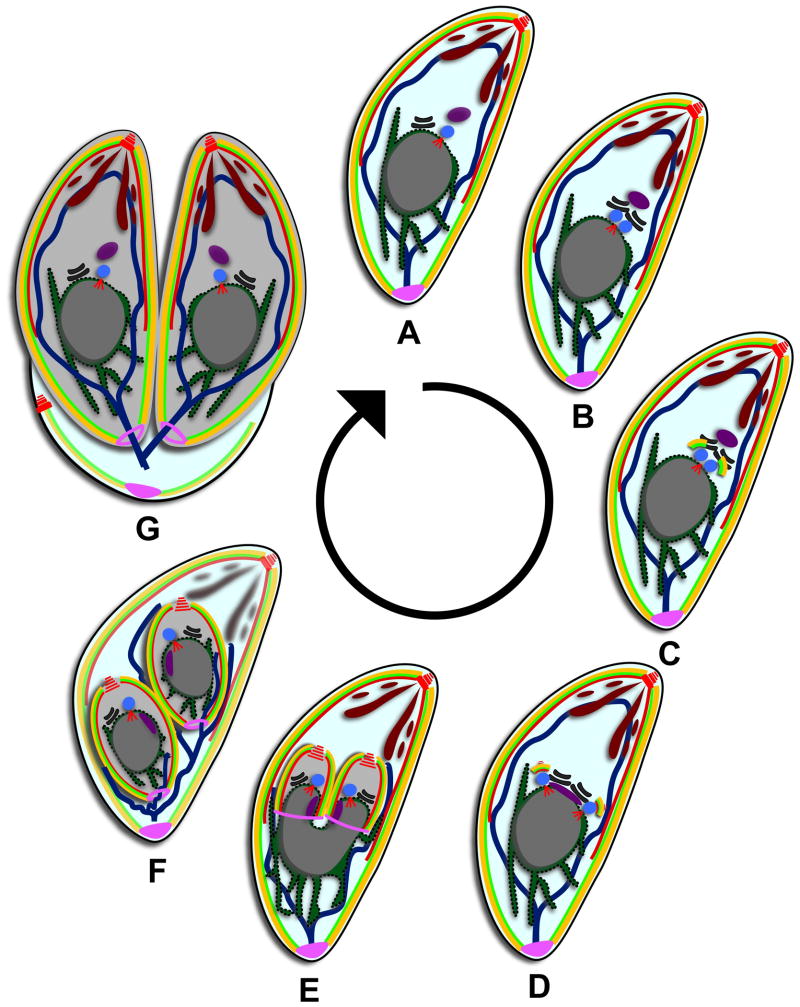
Figure 1.
Toxoplasma divides by internal daughter budding. (A) Mature parasite in G1. Red are MT (conoid, subpellicular, and spindle pole), yellow are the alveoli, bright green is the IMC meshwork, brown are secretory vesicles, dark blue line is the mitochondrion, purple is the apicoplast, blue is the centrosome, black is the Golgi apparatus, dark green is the ER, grey is the nucleus, and pink is the posterior cup or basal complex. (B) Mitosis is initiated at 1.2N with the duplication of the Golgi apparatus followed by the duplication of the centrosomes. (C) Budding is initiated with the appearance of early components of the cytoskeleton. (D) The spindle pole duplicates and the apicoplast moves below the centrosomes, elongating as the centrosomes separate. (E) The organelles begin to partition as the daughter buds elongate. The components of the basal complex accumulate at the leading edge of the bud. (F) The daughter buds contract and all the organelles are partitioned except for the mitochondrion. The secretory vesicles and cytoskeleton of the mother begin to degrade. (G) Daughter buds emerge and the plasma membrane from the mother is incorporated onto the daughters. The mother falls away as a residual body.
The experimental tractability of Toxoplasma in combination with its sequenced genome has made the rapid identification of entire cytoskeletal protein families possible. The advent of Δku80 knockout strains (Fox et al., 2009; Huynh, Carruthers, 2009) and conditional expression systems (Meissner et al., 2002) has eased the generation of cytoskeletal protein coding gene knockouts. In addition, the conditional overexpression system using a ligand-mediated (Shield1) destablilization domain has been used to study dominant negative alleles of genes (Agop-Nersesian et al., 2010; Agop-Nersesian et al., 2009; Breinich et al., 2009; Herm-Gotz et al., 2007). Moreover, a forward genetic strategy using random mutagenesis has been applied to identify genes with essential roles in Toxoplasma development throughout the lytic cycle (Gubbels et al., 2008a). These tools have led to the creation of a substantial amount of data on the constituents of the cytoskeleton in recent years, and, to a lesser extent, the regulatory controls of development. In this review we have gathered these findings together and created a spatial model of the organization of the cytoskeleton in accordance with the parasite's ultrastructure. Furthermore, we have attempted to organize the temporal dynamics of the cytoskeletal components in relation to each other during parasite development. Finally, we address the findings of current cytoskeletal knockouts and dominant negative alleles and offer questions that remain open in this research field.
2. Division by internal daughter budding
The fast replicating Toxoplasma tachyzoite divides asexually by a process of internal daughter budding known as endodyogeny within any nucleated cell type from a homoiothermic host (Hu et al., 2002a; Sheffield, Melton, 1968; Striepen et al., 2007; White et al., 2007) (Fig. 1). The process begins with the duplication of the Golgi apparatus late in G1 (Ogino, Yoneda, 1966; Pelletier et al., 2002; Sheffield, Melton, 1968). This is followed by the duplication of the centrosome early in S phase (Hartmann et al., 2006; Hu et al., 2002a; Nishi et al., 2008) (Fig. 1B). Budding is initiated late in S-phase before the onset of mitosis when the earliest components of the cytoskeleton begin to assemble apical to the recently duplicated centrosomes (Agop-Nersesian et al., 2010; Hu et al., 2006; Hu et al., 2002a; Mann, Beckers, 2001; Radke et al., 2001; Sheffield, Melton, 1968; Tilney, Tilney, 1996; White et al., 2005) (Fig.1C and D). Mitosis and cytokinesis progress concurrently as the cytoskeleton grows from the apical end toward the posterior end encapsulating first the divided Golgi (Nishi et al., 2008), then the apicoplast (Striepen et al., 2000) (Fig. 1E), the nucleus and endoplasmic reticulum (ER) (Hager et al., 1999; Hu et al., 2002a) (Fig. 1F), and finally the mitochondrion (Nishi et al., 2008) (Fig. 1G and A). The secretory organelles (micronemes, rhoptries, and dense granules) are created de novo (Nishi et al., 2008; Ogino, Yoneda, 1966; Sheffield, Melton, 1968) in a process requiring an alveolate specific dynamin related protein, DrpB, that appears to generate vesicles from the late Golgi compartment (Breinich et al., 2009) (Fig. 1G). As the daughter parasites reach maturity, the cytoskeleton of the mother breaks down and the plasma membrane of the mother is recycled onto the now emerging daughters (Sheffield, Melton, 1968) along with newly generated membrane (Morrissette, Sibley, 2002b; Tran et al., 2010) (Fig. 1F and G). Subsequent parasite division rounds continue until host cell resources are depleted and culminates in active egress and destruction of the infected host cell.
3. Composition of the cytoskeleton
The cortical Toxoplasma cytoskeleton (also known as the pellicle) is a complex, layered structure comprised of an outer plasma membrane and underlying inner membrane complex (IMC). The IMC is itself composed of a double membrane system (Fig. 2, yellow) with an undergirding protein meshwork (Fig. 2D bright green) that lines the cytoplasmic side of the IMC membranes (Mann, Beckers, 2001; Porchet, Torpier, 1977; Sheffield, Melton, 1968). The IMC rests on a final cytoskeletal layer composed of microtubules (MT) emanating from the apical end of the parasite. Specialized cytoskeletal structures are present at the extreme anterior and posterior ends of the parasite which are known as the apical and basal complex, respectively.
Figure 2.
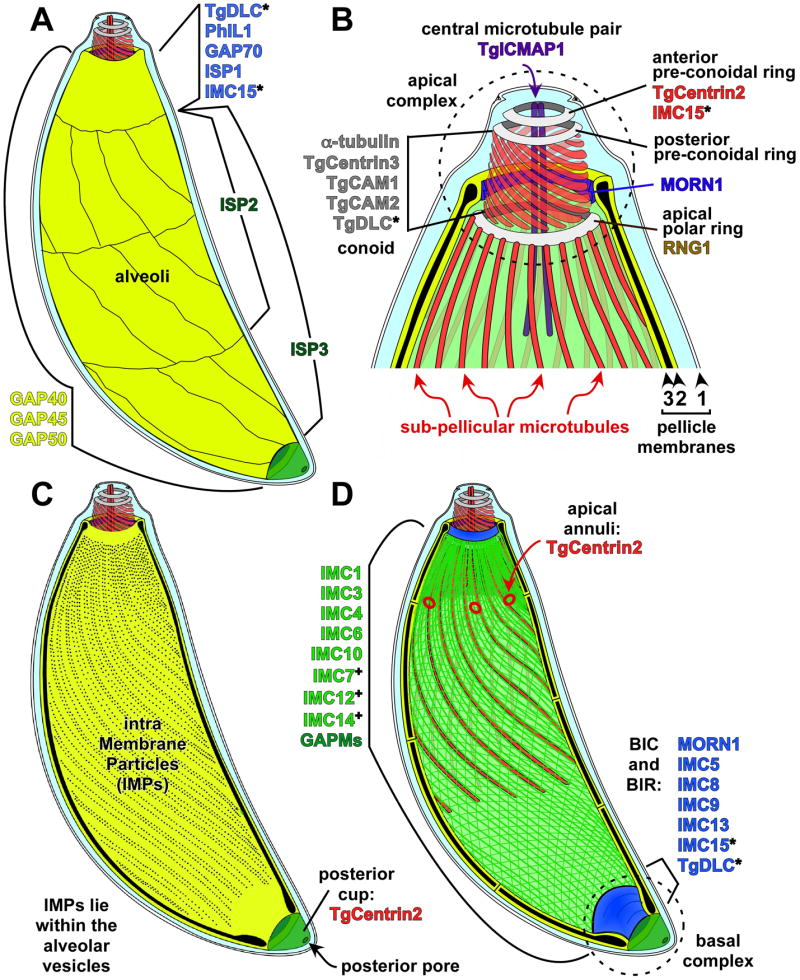
Schematic representation of the structures in the Toxoplasma cytoskeleton. (A) Directly under the plasma membrane lie the alveolar vesicles, shown in yellow. The most unique alveolar vesicle called the apical cap forms a cone around the parasite apex and three bands of rectangular, elongated vesicles fill in the remainder of the IMC below this cap. As marked, different proteins localize to different sections of the alveolar vesicles. (B) Representation of the most apical end of the cytoskeleton. The alveolar vesicles and IF-like protein filament meshwork have been removed to expose the 22 sub-pellicular microtubules and the conoid. A series of three cytoskeletal rings are located at the apex (grey). Components known to localize to these structures are indicated. Other structures are marked and named in the panel. (C) Representation of the Intra Membrane Particles (IMPs) lying within the flattened alveolar vesicles. Their molecular nature is unknown. At the basal end is the posterior cup (green), which contains TgCentrin2. (D) Representation wherein the alveolar vesicles have been removed from the top, exposing the IMC protein meshwork (green) containing the IF-filament IMC proteins. Other proteins localizing to the same region on either side of the alveoli are indicated here as well. Proteins localizing to this section of the parasite are shown in green (note that not all these proteins are part of the same structure). TgCentrin2 annuli are shown at the boundary between the most apical alveolar vesicle and the other vesicles (see panel A). The basal inner ring (BIR) and the basal inner complex (BIC) are located at the basal end of the alveoli. Proteins occupying several regions and appearing in different colors are indicated with an asterisk (*). Proteins that exhibit localization only in mature parasites are indicated with a plus (+). Figure inspired by (Nichols, Chiappino, 1987).
3.1. Tubulin, actin & company
There are 22 subpellicular MT that spiral two-thirds of the length of the parasite (Fig. 2B,D). The minus ends of the MT are anchored in a microtubule organizing center (MTOC) called the apical polar ring (Fig. 2B,D). The MT grow from the apical end of the parasite toward the posterior end, with the (+)ends remaining free in the mature parasite in a state of stalled depolymerization (Cyrklaff et al., 2007; Nichols, Chiappino, 1987; Russell, Burns, 1984). Unlike other eukaryotes, single MT in apicomplexans are extremely stable (Morrissette et al., 1997). Some of this stability could be attributed to the microtubule-associated proteins (MAPs) that are suspected to connect the MT to the pellicle (Morrissette et al., 1997; Morrissette, Sibley, 2002a) such as the newly described subpellicular microtubule protein 1 (SPM1) (Tran et al., 2011). An additional atypical MT structure known as the conoid is present at the apical end of the parasite. The conoid is a basket of spiraling MT filaments composed exclusively of α-tubulin fitted with a pair of preconoidal rings at its anterior face (Fig. 2B). These preconoidal rings are connected to the apical polar ring by the spiraling α-tubulin polymers of the conoid (Hu et al., 2002b; Morrissette, Sibley, 2002a; Nichols, Chiappino, 1987; Scholtyseck, 1970; Sheffield, Melton, 1968; Swedlow et al., 2002) (Fig. 2B). Finally, two short intraconoidal MT reside within the conoid and may serve in the release of secretory proteins from the apical end of the parasite during invasion (Carruthers, Sibley, 1997; Nichols, Chiappino, 1987) (Fig. 2B). A protein marker for these MT, intra-conoid microtubule associated protein1 (ICMAP1), was recently identified but how this protein functions in the MT organization has not been determined (Heaslip et al., 2009).
The parasite has an atypical actin cytsokeleton wherein the majority of actin is present in the globular form (Dobrowolski et al., 1997). Short filaments only form transiently and act during gliding and host cell invasion (Dobrowolski, Sibley, 1996; Shaw, Tilney, 1999) as part of a motility apparatus known as the glideosome, which contains Myosin A (MyoA), Myosin Light Chain 1 (MLC1), and the membrane anchoring proteins gliding associated protein 45 (GAP45) and GAP50 (Keeley, Soldati, 2004). Inhibition of or interference with actin polymerization significantly decreases motility and invasion (Dobrowolski, Sibley, 1996; Poupel, Tardieux, 1999). In contrast, actin appears to play only a minor role during the early stages of endodyogeny, since interference with actin polymerization does not result in an early block of cell division. Obvious defects can only be observed during the final stages of endodyogeny, culminating in enlarged residual bodies (Shaw et al., 2000), implicating a role for actin during the final stages of daughter cell assembly and cytokinesis. To date, studies on the eleven Toxoplasma myosins have focused on class XIVa/b motors and their molecular interaction partners, since they are implicated in gliding motility (Delbac et al., 2001; Foth et al., 2006; Frenal et al., 2010; Gaskins et al., 2004; Herm-Gotz et al., 2006; Johnson et al., 2007; Meissner et al., 2002). Interestingly a phylogenetic analysis and the presence of different domains in the tail domains of apicomplexan myosins suggest different roles for these unconventional motor proteins during the life cycle of the parasite. For example, different subclasses of myosins with tail domains, likely to be involved in signaling cascades, tubulin dynamics, or chromatin regulation, have been described (Foth et al., 2006). Furthermore, Toxoplasma myosins B (MyoB) and C localize to the basal end of the parasite and overexpression of MyoB results in enlarged residual bodies (Delbac et al., 2001). A detailed analysis of myosins will help to understand the role of these motors and actin during endodyogeny in more detail since currently their contribution appears to be underestimated.
3.2. The pellicle and inner membrane complex
The Inner Membrane Complex (IMC) is composed of a double membrane system (Fig. 2, yellow) with an undergirding protein mesh (Fig. 2D bright green). This protein mesh is composed of 8-10 nm wide filaments, which contain intermediate filament-like (IF-like) proteins that line the cytoplasmic side of the alveoli and overlay the subpellicular MT (Mann, Beckers, 2001; Porchet, Torpier, 1977; Sheffield, Melton, 1968). The IMC runs the entire length of the parasite with openings at the apical and posterior ends (Bommer et al., 1968; Gonzalez Del Carmen et al., 2009; Mondragon, Frixione, 1996; Nichols, Chiappino, 1987; Scholtyseck, 1973). The membrane portion of the IMC is a patchwork of flattened membranous sacs called alveoli that are assembled from vesicles trafficked through the Golgi apparatus in a process mediated by the small GTPase Rab11B (Agop-Nersesian et al., 2010; Sheffield, Melton, 1968; Vivier, Petitprez, 1969). The alveoli are rectangular and arranged in three rows encircling the parasite with a single cone-shaped vesicle at the anterior end known as the apical cap (Dubremetz, Elsner, 1979; Porchet, Torpier, 1977) (Fig. 2A).
Cortical alveoli are the defining feature of the Alveolata, a superphylum consisting of ciliates, dinoflagellates, and apicomplexans (Keeling et al., 2005). The combination of alveoli and underlying intermediate filamentous meshwork are believed to serve diverse functions in the alveolates including structural supports, cellulose-reinforced armor (Lau et al., 2007), and calcium storage (Plattner, Klauke, 2001; Stelly et al., 1991). In the Apicomplexa, the alveoli and their associated cytoskeletal elements (together termed the IMC) give structure to the cell, form a scaffold for daughter parasites assembly, and serve as a support for the glideosome mediated motility (Gaskins et al., 2004; Mann, Beckers, 2001). In support of the taxonomic grouping of the Alveolata, many conserved proteins are found in all three phyla where they associate with the alveoli-supporting meshwork (Gould et al., 2011; Gould et al., 2008). The first such protein group to be identified was the aveolins, articulin-like proteins that possess a characteristic “alveolin” repeat motif. The aveolins were discovered in Toxoplasma where they are known as IMC proteins and constitute a fourteen-member family of IF-like proteins with several different cytoskeletal localizations and timing of appearance during endodyogeny, suggesting distinct roles in cell division (Anderson-White et al., 2011; Mann, Beckers, 2001). In addition to forming the IMC meshwork, these IMC proteins are likely anchored in the alveolar membrane sacs by palmitoylation and may also bind to a family of multi-membrane spanning glideosome associated membrane proteins (GAPMs) embedded in the membrane on the cytoplasmic side of the alveoli (Bullen et al., 2009) (Fig. 2D).
Within the double membranes of the alveoli there are double rows and single rows of intramembranous particles (IMPs) organized with a 32 nm periodicity, reflecting the periodicity of the subpellicular MT (Fig. 2C) (Dubremetz, Elsner, 1979; Morrissette et al., 1997; Porchet, Torpier, 1977). It has been hypothesized that the double rows of IMPs anchor the MAPs that interact with the MT to further stabilize the cytoskeleton (Morrissette et al., 1997; Morrissette, Sibley, 2002a); but the rows of IMPs run the entire length of the parasite suggesting they may instead interact with the IMC IF-like proteins (Dubremetz, Torpier, 1978; Morrissette et al., 1997), possibly mediated by the GAPMs (Bullen et al., 2009).
3.3. IMC sub-compartments
Studies of the cell biology of Toxoplasma reveal a more compartmentalized cytoskeleton than initially suggested by the ultrastructure and a growing complexity in the number of protein components. The anterior cone-shaped alveolus called the apical cap is delimited by five to six annuli containing TgCentrin2 at its base (Fig. 2A, B, D) (Hu et al., 2006). Over the past ∼5 years or so an increasing number of proteins localizing to the apical cap have been described. For instance, the (-)end-directed MT motor dynein light chain (TgDLC) localizes to the apical cap of the parasite and may transport cargo along the subpellicular MT (Hu et al., 2006). The meshwork component IMC15 is also enriched in the apical cap (Anderson-White et al., 2011). Another protein, which is not essential, localizing predominantly to the apical cap is Photosensitized INA-labeled protein 1 (PhIL1) (Barkhuff et al., 2011; Gilk et al., 2006). Finally, a component of the glideosome, gliding-associated protein 70 (GAP70), localizes specifically to the apical cap (Fig. 2A). This protein is closely related to GAP45, which recruits the members of the glideosome to the IMC (Frenal et al., 2010; Gaskins et al., 2004). Because GAP70 is slightly longer than GAP45, the space between the plasma membrane and the IMC outer membrane is a slightly wider (Frenal et al., 2010). The big mystery is why is the structure of the apical cap different from the rest of the IMC, and what is the function of the proteins localizing specifically to the cap, and in particular, what is the function of the TgCentrin2 annuli?
The recent discovery of a family of three membrane-tethered proteins known as IMC Sub-compartment Proteins (ISPs) visualized further sub-compartmentalization within the alveoli (Beck et al., 2010). These closely related proteins contain no recognizable domains and are distributed into 3 distinct alveolar compartments (Fig. 2A). ISP1 occupies the apical cap while ISP2 localizes to a central region beginning at the basal end of the apical cap and extending about two-thirds the length of the cell body. ISP3 also targets to this central region but additionally localizes all the way to the posterior end of the IMC membranes. The ISPs are not part of the IMC cytoskeletal meshwork as they are easily extracted in mild detergent conditions; rather, they are anchored in the alveoli membranes through coordinated myristoylation and palmitoylation at the extreme N-terminus of each protein. Fusions of the extreme N-termini of these proteins to YFP are generally properly trafficked to their respective sub-compartments, suggesting a model wherein this compartmentalization is determined by palmitoylation activity within the IMC membranes. While both myristoylation and palmitoylation are essential for targeting of ISP1/2/3 to the IMC membranes, other IMC proteins only contain predicted palmitoylation signals that are likely to mediate membrane association (Anderson-White et al., 2011). Consistent with this, a fourth ISP family member has recently been identified whose IMC localization is dependent solely on palmitoylation (P. Bradley, unpublished data). Protein palmitoylation is catalyzed by a family of multi-pass transmembrane enzymes known as palmitoyl acyltransferases (PATs), 18 of which are encoded within the Toxoplasma genome (Beck et al., 2010). While no Toxoplasma PATs have yet been studied, future identification and characterization of PATs resident within the alveoli will allow validation of this sub-compartmentalization model and provide greater understanding of the role of this lipid modification in organization of the IMC.
3.4. Apical complex
The EF-hand containing calcium binding protein TgCentrin2 localizes to the anterior preconoidal ring at the extreme apical end of the parasite (Fig. 2B). TgCentrin2 additionally localizes to a series of annuli at the posterior edge of the apical cap and is also detected in the basal complex (Fig. 2D). Given the contractile functions ascribed to many centrin proteins, it has been hypothesized that TgCentrin2 plays a role in constriction of the basal complex during endodyogeny (Hu, 2008). The function of TgCentrin2 in the apical portions of the parasite is not known but may similarly use its contractile activity for roles in organizing the IMC or in parasite division. Similar to TgCentrin2, IMC15 is found at the extreme apical end of the parasite and also in the basal complex. IMC15 is a member of the alveolin protein family that makes up the meshwork of the IMC (Anderson-White et al., 2011). IMC15 is of particular interest as it is the earliest known cytoskeletal protein to appear at the onset of daughter assembly, suggesting that it plays a role in the organization of early parasite development. Posterior to TgCentrin2 at the apex of the IMC, a protein known as Ring 1 (RNG1) localizes to the apical polar ring (Fig. 2B). The function of RNG1 is unknown but it is likely essential and appears in daughters just before disassembly of the mother parasite (Tran et al., 2010). Another ring-shaped structure at the apical end is defined by the membrane occupation and recognition nexus 1 (MORN1) protein, which localizes to the apical extreme of the alveoli (Gubbels et al., 2006; Hu, 2008) (Fig. 2B). Next to the modest presence of MORN1 in the apical complex, more MORN1 is found in the spindle pole as well as the basal complex. TgCentrin3, a paralog of TgCentrin1 and TgCentrin2, is found faintly in the conoid as well, though its main localization is in the centrosome (Hu et al., 2006). In addition, two proteins containing an EF-hand, calcium-binding domain, TgCAM1 and TgCAM2, are found in the conoid (Fig. 2B). The latter two proteins may play a role in conoid extrusion in response to calcium (Hu et al., 2006).
3.5. Basal complex
At the opening in the IMC at the apical end lies the conoid, whereas at the basal/posterior end a structure is present known as the posterior cup or basal complex (Mann, Beckers, 2001) (Fig. 2C, D). At the ultrastructure level, the basal complex consists of two electron dense structures dubbed the basal inner ring (BIR) and the basal inner complex (BIC) and several unit membranes (UM) (Anderson-White et al., 2011). The function of the basal complex in the mature parasite is unknown, but it could function in resisting mechanical stress during the host cell invasion process. MORN1 and TgCentrin2 are both present in this structure, suggesting a role for these components in constriction of the parasite during cytokinesis (Gubbels et al., 2006; Hu, 2008; Hu et al., 2006). In addition, the alveolin-repeat containing IMC proteins 5, 8, 9, 13, and 15 are found in the basal complex (Anderson-White et al., 2011), highlighting the complexity of this structure and suggesting a key role in the proper formation of daughter parasites. TgDLC localizes to the basal end as well; however its abundance is quite low (Hu et al., 2006) (Fig. 2D). It should be noted that neither the conoid nor the posterior cup are features conserved across different apicomplexan parasites and/or life stages.
4. Coordinated development and dynamics of the cytoskeleton
New components of the cytoskeleton are being described continuously, illustrating ever more elaborate and sophisticated structures. This creates additional challenges in determining timing of the assembly of these structures into the cytoskeleton during the budding process. Questions that arise include; how many steps are present, what are the requirements to establish certain elements or triggers certain transitions, and how are these steps coordinated? To start answering these questions, the timing and sequence of cytoskeletal element incorporation into the daughter buds has been determined individually based on comparisons to developmental markers such as MORN1 for early bud formation (Gubbels et al., 2006; Hu, 2008), GAP45 for late stage budding (Gaskins et al., 2004), and IMC1 for everything in between (Hu et al., 2002a; Mann et al., 2002). However, these studies cannot easily be compared due to the usage of different reference markers, which is especially true for early budding when a large number of proteins are converging on a small subcellular area in a short time span (about 30 minutes) (Nishi et al., 2008). We recently established a detailed timeline of the various steps in the assembly process of the intermediate filament cytoskeleton (Anderson-White et al., 2011). This timeline revealed several previously not appreciated steps in the budding process. Here we made an effort to map the appearance of various other cytoskeletal proteins onto this time line by using a combination of fluorescent protein fusion or epitope fusion reporters as well as specific antisera. Although protein abundance and the sensitivity of the various reagents can result in some variability in evaluating the timing of markers relative to one another, this provides a global overview of the sequence of events and is more inclusive than previous efforts. Four different periods with cytoskeletal changes are currently appreciated and are discussed per stage below and summarized in Figure 8.
Figure 8.
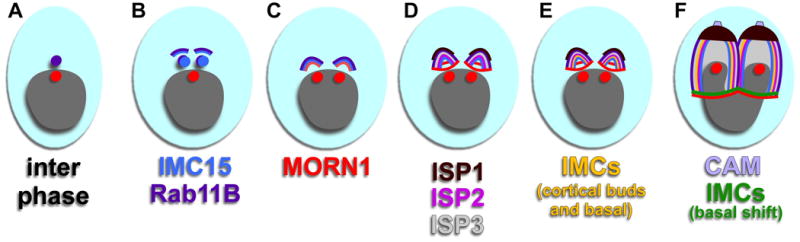
Timeline of early budding activity. Time progresses from panel A through F. (A) Inter phase represents G1 without any budding. Bud components correspond to the text colors below the panels. Included components are those whose timing has been verified by comparative IFA.
4.1. Initiation of budding
After division of the Golgi apparatus, at a DNA content of about 1.2N, the centrosome duplicates (Hartmann et al., 2006; Hu et al., 2002a; Nishi et al., 2008). The centrosome appears to coordinate the mitotic cycle with the cytokinetic cycle and at the same time provides the spatial cue for daughter cytoskeleton assembly (Gubbels et al., 2008b). The dynamics of the centrosomes can be monitored using the three identified centrin proteins in Toxoplasma, TgCentrin1, 2, and 3 (Hu et al., 2006; Hu et al., 2002a). At the point of centrosome duplication IMC15 and Rab11B colocalize apical of the centrosomes marking the first outlines of the daughter parasites (Fig. 3A and B). IMC15 is the earliest member of the IMC meshwork to appear in the initial bud after it transitions from the duplicated centrosomes on which it first accumulates (Anderson-White et al., 2011). Rab11B traffics the vesicles of the alveoli to the budding daughters (Agop-Nersesian et al., 2010) and it is reasonable that the protein meshwork and the membranous alveoli components of the IMC would develop in tandem. Consistent with this model is that many of the IMC proteins contain predicted palmitoylation sites that likely anchor them into the alveoli suggesting the IMC meshwork cannot be assembled in the absence of alveolar membrane (Section 5). In addition to Rab11B, the actin-like protein 1 (ALP1) may assist in the development of the IMC membranes. It has been shown that ALP1 appears at the bud before other members of the IMC family, such as IMC1, suggestive of a role in early daughter formation (Gordon et al., 2008). The end of bud initiation is marked by the appearance of MORN1 on the daughter buds.
Figure 3.
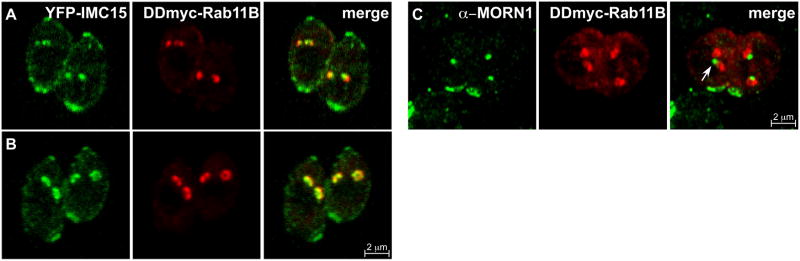
IMC15 and Rab11B precede MORN1 into the initial daughter bud. (A-B) Parasites expressing YFP-IMC15 (green) and DDmyc-Rab11B (red) in the presence of Shield1 colocalize at the centrosomes (A) and then expand into the forming daughter buds (B). (C) DDmyc-Rab11B (red) co-stained with anti-MORN1 (green). Arrow indicates an unduplicated spindle pole. Both constructs are driven by the ptub promoter.
The MT structures of the cytoskeleton alos begin to form at the initial budding stage. The subpellicular MT and conoid both begin to assemble shortly after the duplication of the centrosome (Agop-Nersesian et al., 2010; Hu et al., 2006). The MT binding protein associating with the two intraconoid MT, TgICMAP1, appears concurrently with this event as well (Heaslip et al., 2009); however, its exact timing relative to other events has yet to be determined. Together, the IMC and underlying MT filaments form the foundation for the forming daughter buds.
4.2. Early budding
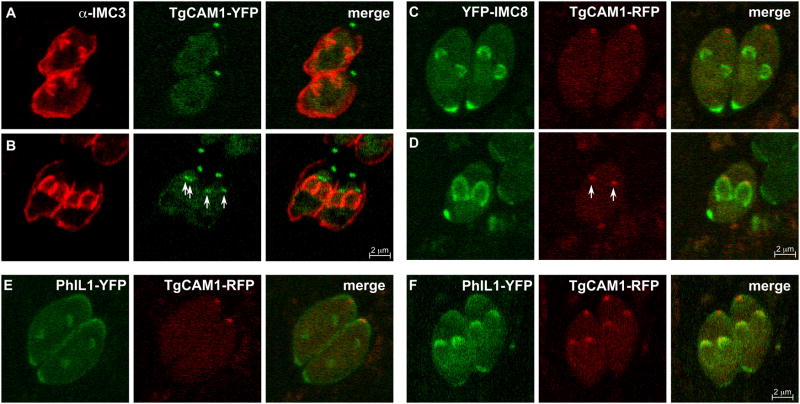
After recruitment of MORN1, the next components to enter the new buds are the ISP1-3 proteins (Fig. 4). About 30 minutes after centrosome duplication, at a DNA content of about 1.8N, IMC3 and IMC1 follow the ISP proteins into the daughters (Hu et al., 2002a; Nishi et al., 2008). Often there are faint accumulations of IMC3 near early ISP proteins, but IMC3 does not begin to form recognizable buds until the intensity of the ISP signals increase (Fig. 4A-C). It is currently assumed that the other IMC proteins that localize cortically in budding parasites, IMC4, 6, and 10, enter the daughters with the same timing as IMC1 and 3. One IMC protein pair that has been tested together is YFP-IMC8 (one of the IMC proteins that transitions halfway through budding from the cortex to the basal complex of the daughter) and cherry-IMC3. This permitted us to determine that the IMCs of the basal complex, IMC5, 8, 9, and 13, join the early bud concurrently with IMC3 (Fig. 5). The components of the glideosome begin to appear during this early stage of budding as well. GAP50 and GAP40 are the earliest glideosome elements (Frenal et al., 2010; Gaskins et al., 2004) but their time of arrival to the daughters compared to the ISP or IMC proteins has yet to be determined.
Figure 4.
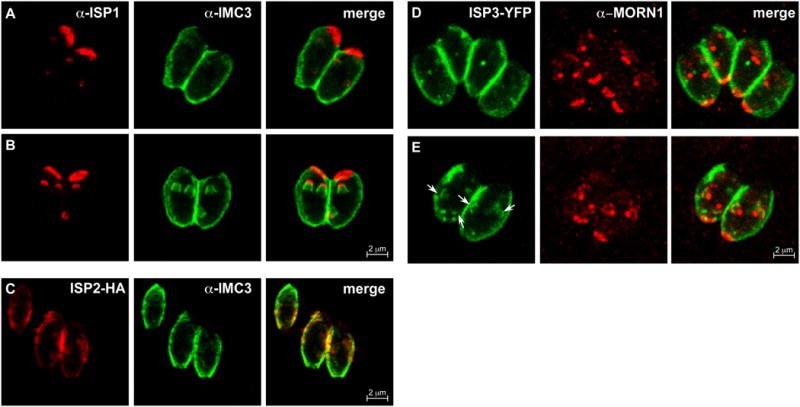
Timing of the recruitment of ISP proteins 1-3 relative to other assembly markers. (A) IMC3 is present in amorphous accumulations near the buds clearly indicated by ISP1. IMC3 does not fully associate with the buds until after ISP1 arrives at the buds; both proteins are clearly established in the daughters at an early stage (B). (C) ISP2-HA precedes IMC3 into the daughter buds as well. ISP2-HA is under the control of its native promoter. (D,E) Parasites expressing ISP3-YFP (green) are co-stained with anti-MORN1 (red) showing no ISP3 at the recently divided spindle poles (D) and then colocalization of ISP3 with the early MORN1 rings (E). ISP3-YFP parasites present with numerous inclusion bodies; therefore, the arrows indicate the bud-associated ISP3.
Figure 5.
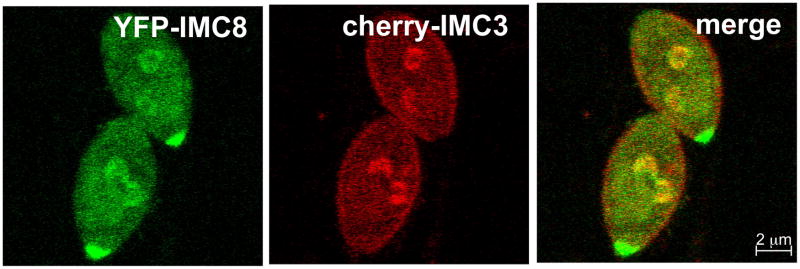
The basal and cortical IMCs associate with the buds concurrently. Parasites expressing YFP-IMC8 (green) and cherry-IMC3 (red) show concurrent localization to the daughter buds at an early stage. Both constructs are driven by the ptub promoter.
With all of the earliest components in place, the forming daughter cytoskeletons begin to elongate. A microtubule-based process likely powers this extension process, although the IMC filaments are able to autonomously assemble to some extent (Morrissette, Sibley, 2002b; Vaishnava et al., 2005). A ring of MORN1 protein marks the growing posterior end of the cytoskeleton from an early stage. MORN1 also localizes to the early conoid marking the apical end of the forming daughters (Gubbels et al., 2006; Hu et al., 2006). Once the advancing cytoskeleton reaches the edge of the forming apical cap, the annuli of TgCentrin2 form (Hu, 2008; Hu et al., 2006). As the cytoskeleton continues to grow past the cap region toward the budding midpoint, ISP1 remains behind in the cap (Beck et al., 2010). This suggests that the cytoskeleton scaffold first forms the apical end and grows in the posterior direction, and that many specific elements marking the sub-compartments are incorporated at the time the sub-compartment is formed.
4.3. Mid budding
In this stage the extending daughter cytoskeletons start to taper toward the basal end. In addition, TgCAM1, TgCAM2, and TgDLC localize to the MT region of the conoid, which occurs about 1.5 hr after centrosome duplication (Hu et al., 2006; Nishi et al., 2008). This corresponds to about the midpoint of budding based on comparison of TgCAM1 with IMC3 and IMC8 (Fig. 6A-D). The midpoint of budding coincides with redistribution of the posterior IMC proteins, IMC5, 8, 9, and 13, from the periphery of the daughter buds to the growing basal ends where MORN1 is located (Anderson-White et al., 2011). This transition marks the widest part of the future mature daughters and the growing buds begin to taper as they elongate from this point onward. Orthologous cell division related contractions in other systems are driven by an actinomyosin based process. However, support for a role for F-actin is lacking in Toxoplasma since basal complex contraction proceeds normally in the presence of actin depolymerizing compounds such as cytochalasin D and latrunculin A (Gubbels et al unpubished; (Shaw et al., 2000)). On the other hand, manipulation of the MyoB and C present in the basal complex alters, but does not prevent the progression of basal complex constriction (Delbac et al., 2001). Therefore, there appears to be some contribution of an actinomyosin based system, but it is not well understood. As for alternative mechanisms powering basal complex constriction, MT are absent from the basal complex making it unlikely that dyneins or kinesins power contraction. However, the Ca2+-dependent filament forming and contractile protein TgCentrin2 has been put forward as a candidate to drive constriction of the basal complex (Hu, 2008). TgCentrin2 starts to assemble on the basal complex simultaneously with the translocation of IMC5, 8, 9, and 13 to the this structure (Anderson-White et al., 2011; Hu, 2008). Soon after the basal complex begins to contract, heat shock protein 20 (Hsp20) localizes in a discontinuous striped pattern to the outer membrane of the IMC (de Miguel et al., 2008). A recent study of Hsp20 in Plasmodium suggests the protein is not essential for cell division, but controls different gliding motility modes of sporozoites (Montagna et al., 2012). PhIL1 was originally reported to localize to the apical cap in these later stages of daughter development (Gilk et al., 2006). However, when PhIL1 is compared to TgCAM1, PhIL1 localizes to the forming daughter buds much earlier than TgCAM1, suggesting that PhIL1 is incorporated into the daughters earlier then previously thought (Fig. 6E, F). Since the exact function of PhIL1 and many of the other components is unknown, the biological meaning of this particular sequence of events is unknown.
Figure 6.
TgCAM1 enters the conoid near the midpoint of budding just after recruitment of PhIL1. (A-B) Parasites expressing TgCAM1-YFP (green) co-stained with anti-IMC3 (red) show an absence of TgCAM1 in the early bud (A). TgCAM1 enters the conoid around the midpoint of budding as indicated by the arrows (B). (C-D) Parasites expressing YFP-IMC8 (green) support the entrance of TgCAM1 to the conoid at the midpoint since TgCAM1 is again absent from the early bud as indicated by YFP-IMC8 (C), but appears as YFP-IMC8 transitions to the growing edge of the midbud (D). Arrows indicate localization of TgCAM1 to the conoid. TgCAM1-YFP, TgCAM1-RFP, and YFP-IMC8 are driven by the ptub promoter. (E-F) PhIL1 enters the daughter buds prior to the midpoint. Parasites expressing PhIL1-YFP (green) and TgCAM1-RFP (red) show PhIL1-YFP in the early bud prior to the appearance of TgCAM1-RFP (E). At the midpoint of budding, as indicated by TgCAM1-RFP, PhIL1-YFP is well established in the daughter buds (F). PhIL1-YFP, YFP-IMC8, and TgCAM1-RFP are driven by the ptub promoter.
4.4. Late budding
At this stage the cytoskeletons of the daughter parasites mature and the mother parasite's cytoskeleton is broken down. The beginning of this stage is marked by the appearance of RNG1 at the apical polar ring just before the mother cytoskeleton starts to disassemble (Tran et al., 2010). The mother cytoskeleton then begins to break down, starting from the apical end, and the plasma membrane of the mother is incorporated into the pellicles of the new daughters in a Rab11A-dependent process (Agop-Nersesian et al., 2009; Sheffield, Melton, 1968). The glideosome assembles between the forming plasma membrane and the IMC as GAP45 is recruited to GAP50, bringing along MLC1 and MyoA. GAP45 anchors the plasma membrane on the outer alveolar membrane of the IMC for the length of the parasite except for the apical cap region (Frenal et al., 2010; Gaskins et al., 2004). At the apical cap, GAP70 bridges the space between the plasma membrane and the IMC (Frenal et al., 2010). With the completion of basal complex contraction, the mature basal complex is formed. Upon final emergence of the daughters a small residual body containing the remnants of the mother is left behind.
The cytoskeleton of the mother appears to be disassembled in a well-organized pattern following an apical to basal direction and occurs only at the point where mother and daughter cytoskeleton are closely apposed to the plasma membrane. The nature of or mechanism driving disassembly is not known, but it appears that the IF-like filaments and alveoli are dissembled on the spot, whereas the conoid migrates in an apical to basal direction and ends up in the residual body, from which it quickly disappears thereafter (Morrissette, Sibley, 2002b; Tran et al., 2010).
Maturation of the daughter cytoskeleton coincides with proteolytical cleavage of the C-terminus of IMC1, which is concurrent with cross-linking of the IMC proteins into a non-ionic detergent-resistant meshwork (Mann et al., 2002). Currently it is unknown whether other IMC proteins undergo similar processing. Based on fluorescence recovery after photobleaching (FRAP) experiments IMC1 is generated de novo in the growing daughters and not recycled from the mother parasite (Hu et al., 2002a). On the other hand, FRAP experiments with IMC4 suggest some of this IMC protein may be salvaged from the mother (Hu et al., 2006). These results support the idea that multiple and complex mechanisms operate even within the same family of proteins to construct the daughter buds.
4.5. Mature parasites in G1
The newly emerged daughter parasites are now fully mature and the remnants of the mother parasite have been left behind as a residual body. For poorly understood reasons, three more IMC proteins are incorporated into the cytoskeleton during G1: IMC14 in the first third of G1 and IMC7 and IMC12 at about the midway point of G1 (Anderson-White et al., 2011). One hypothesis is that all or some of these IMC proteins differentially mark the mother from the daughters because only the mother cytoskeleton needs to be dissembled late in the budding process while the daughters should continue to mature.
Throughout G1 IMC1 is continually added to the mature cytoskeleton but at a slower rate than during budding (Hu et al., 2002a). It is unknown if there is active turnover and replacement of the other cytoskeletal proteins after budding is complete. It is reasonable to speculate that this is true for proteins like IMC1 and IMC4 that maintain their level of intensity in IFA between daughter development and G1 phase (Anderson-White et al., 2011). Proteins like IMC3, 6, and 10 that exhibit significantly weakened signals during G1 are probably less dynamic during G1, being degraded but not being replaced (Anderson-White et al., 2011).
5. Mechanistic insights from disruption of cytoskeletal components
As evidenced in the preceding sections, the subcellular dynamics of some components of the Toxoplasma cytoskeleton have been described, but we are just starting to tease out the mechanisms of development. To this end, targeted gene KOs, pharmacological manipulation of protein function or stability, dominant negative constructs, and disruptive overexpression are being applied to decipher the functions of certain cytoskeletal elements.
Initial research into the mechanisms of daughter budding focused on the differential contributions of the IMC and the subpellicular MT. When parasites are treated with the dinitroaniline herbicide oryzalin at low concentrations (0.5 µM), polymerization of the subpellicular MT is inhibited while spindle MT function and centrosome duplication remain unaffected, allowing for specific ablation of the subpellicular MT (Morrissette, Sibley, 2002b; Stokkermans et al., 1996). Under these conditions, centrosome duplication, DNA replication and karyokinesis continue but cytoskeletal elongation and cytokinesis are blocked (Morrissette, Sibley, 2002b; Shaw et al., 2000). In oryzalin treated cells, the alveoli-associated proteins ISP1 and ISP3 label numerous small rings reminiscent of early daughter buds in normally dividing parasites. Elongation beyond this early bud-ring stage does not occur, suggesting that IMC assemble begins but encounters an early block in the absence of subpellicular MT assembly.
Presumably, one or more IMC meshwork proteins provide the scaffold to generate these alveolar rings. In oryzalin treated parasites, IMC1 forms amorphous sheets, showing that some assembly of the cytoskeletal IMC protein meshwork still occurs in the absence of the subpellicular MT. However, these sheets do not co-localize or associate with the ISP1-positive early bud rings (Beck et al., 2010). While any of the remaining 13 IF-like IMC proteins might facilitate alveolar ring formation, IMC15 is a particularly attractive candidate since it is the earliest known bud marker and it enters the buds earlier than the ISPs.
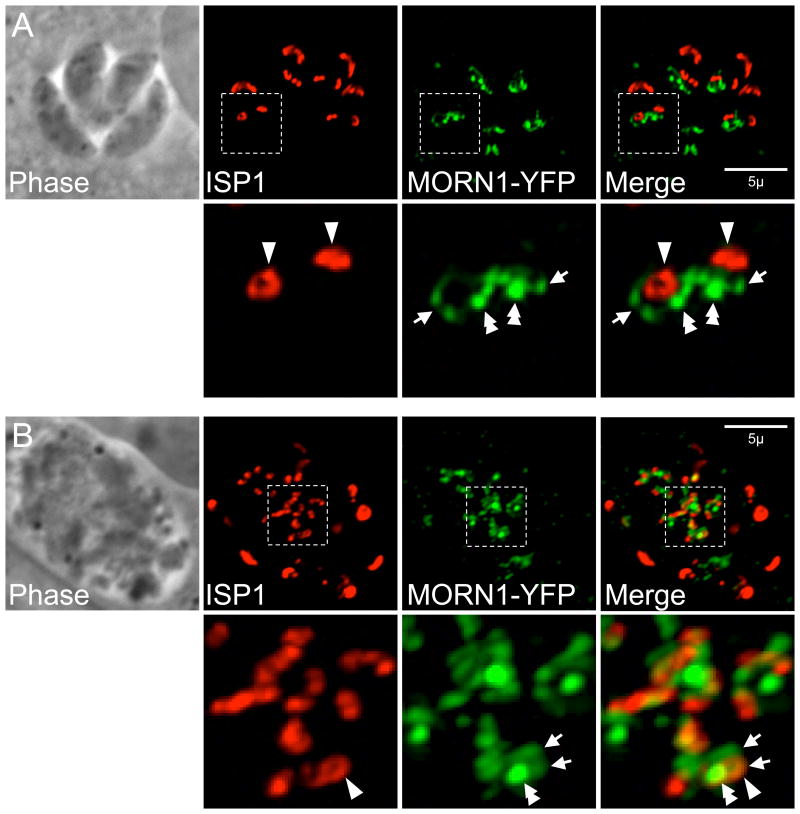
Live imaging of parasites during the first round of division following addition of oryzalin shows that the pair of MORN1 rings which assemble around the spindle pole in early budding are still formed without subpellicular MT polymerization (Hu, 2008). Following longer drug treatment, MORN1 is present in a variety of structures, some of which associate with IMC1 (Gubbels et al., 2006; Hu et al., 2006). To determine if MORN1 is present in the early bud rings labeled by ISP1, we directly compared MORN1 and ISP1 in oryzalin treated cells. ISP1-positive early bud rings appear to cluster around bright MORN1 punctata, which likely correspond to spindle poles (Fig. 7, double arrowheads). In addition, less signal-intense MORN1 structures also cluster around these puncta, some of which co-localize with ISP1 rings (Fig. 7, arrowheads) while others do not (Fig. 7, arrows). While MORN1 is able to form higher order structures, daughter initiation is largely unaffected in its absence indicating MORN1 is not critical for organizing the buds at the onset of endodyogeny (Heaslip et al., 2010; Lorestani et al., 2010). What then provides the ring template for these early buds? One attractive possibility is the apical polar ring. In support of this idea, RNG1 also labels rings in parasites treated with oryzalin showing that the apical polar ring is properly formed without the subpellicular MT polymerization. These RNG1-rings would be expected to associate with the alveolar rings marked by ISP1/3 if the apical polar ring serves as an initial bud template; however, a direct comparison has not been made (Beck et al., 2010; Gubbels et al., 2006; Hu, 2008; Tran et al., 2010). It is somewhat unusual that the number of centrosomes and RNG1 rings do not agree in oryzalin-treated parasites and this may indicate a breakdown of normal bud initiation control mechanisms (Tran et al., 2010). In addition, the timing is surprisingly different from the normal appearance of RNG1 at late stage daughter development, suggesting RNG1 association and possibly apical polar ring assembly, is controlled by cell cycle progression controls rather than by physical assembly cues (Tran et al., 2010). Taken together, these data suggest MT are not involved in the initiation of budding or in early bud formation but are required for proper IMC elongation in order to complete cytokinesis.
Figure 7.
ISP1 and MORN1-YFP localization (A) during early budding in normally dividing parasites or (B) after 48 hours growth in 0.5 µM oryzalin. In normal division, a pair of MORN1 rings marks the growing ends of the two daughter buds (arrows) and ISP1 labels each apical cap (arrowheads). MORN1 is also present in the spindle pole (double arrowhead) as well as in the maternal basal complex. (B) In the presence of oryzalin, subpellicular MT polymerization is blocked, preventing cytokinesis and resulting in a large amorphous cell. Numerous early bud rings are labeled by ISP1 (arrowhead). A number of bright MORN1 puncta (double arrowhead) are centrally located in the cell and may correspond with spindle poles (>2 spindle poles are expected as DNA replication and mitosis are not inhibited under these conditions). These puncta are surrounded by less signal intense MORN1 structures (arrows), a few of which co-localize with an ISP1-positive early bud ring, but most of which do not.
Rab11B plays a critical role in vesicular trafficking to the forming IMC. Experiments inducing a dominant negative phenotype of Rab11B halt alveoli biogenesis and bud formation, but not subpellicular MT polymerization (Agop-Nersesian et al., 2009). In parasites where IMC formation is disrupted in this way, IMC1, MORN1, and GAP50 all fail to assemble on the subpellicular MT, which still elongate but are misshapen (Agop-Nersesian et al., 2010). MORN1 overexpression leads to a similar uncoupling of MT formation from IMC formation (Gubbels et al., 2006). Therefore, proper assembly of the IMC is not required for MT polymerization but is required for proper bud morphology. Interestingly, knockdown or dominant negative expression of clathrin (CHC1) disrupts the whole secretory pathway including the IMC (Breinich, Meissner et al, in preparation), whereas a dominant negative of the dynamin-like protein DrpB specifically ablates the rhoptry, microneme, and dense granule secretory organelles, but not the Golgi apparatus or IMC (Breinich et al., 2009). This illustrates that IMC formation traffics through a dedicated branch of the secretory pathway. Furthermore, overexpression of the MyoA-tail results in defects of IMC biogenesis, indicating a role of a Rab11A-MLC1-Myosin motor in daughter cell assembly. Alternatively overexpression of MyoA could lead to depletion of MLC1 affecting the activity of another myosin (Agop-Nersesian et al., 2009). Expression of dominant negative constructs of MyoB or C, which are located in the basal complex, leads to a larger residual body, suggesting a role in cytokinesis, but how exactly these myosins act in this process is unclear (Delbac et al., 2001). It should be noted that this step does not necessarily require polymerized actin (Shaw et al., 2000), and the role of myosin in the process is therefore still mysterious. The myosin family has several additional members whose roles in cytoskeletal development and cell division have yet to be evaluated (Foth et al., 2006; Heintzelman, Schwartzman, 2001; Polonais et al., 2011a; Santos et al., 2009). In addition, a role for IMC assembly was suggested for ALP1 (Gordon et al., 2008), which is also a member of an extensive family (Gordon, Sibley, 2005). Although the function of these myosins and actin-like proteins has not been determined, it is likely that some of these function in cell division. In support of this hypothesis is the recent description of actin-related protein 4a (ARP4a) with a role in chromosome segregation (Suvorova et al., 2012).
Multiple lines of evidence indicate that nuclear division does not require functional cytoskeletal development. For instance, disruption of ISP2, which affects the earliest stages of daughter budding, still presents properly divided nuclei in the majority of affected cells (Beck et al., 2010). The dominant negative Rab11B phenotype that completely inhibits IMC formation does not affect nuclear division (Agop-Nersesian et al., 2010). Even when contraction of the daughter buds malfunctions in the MORN1 KD parasites, nuclear division is unaffected (Lorestani et al., 2010). When parasites are treated with a high dose of oryzalin (2.5 μM) the nucleus fails to divide properly; however, this is most likely due to interference with the spindle MT (Morrissette, Sibley, 2002b; Stokkermans et al., 1996). When the concentration of oryzalin is kept low (0.5 μM), the spindle forms and the nucleus divides, but cytokinesis is blocked due to the specific ablation of the subpellicular MT (Morrissette, Sibley, 2002b). In summary, nuclear division is not linked to IMC formation and it is unlikely to be linked to subpellicular MT formation. Ultrastructural studies do not highlight a candidate division mechanism as no contractile collar of sorts can be distinguished (Gubbels et al., 2008b). Therefore, it is currently unknown which structure or mechanism facilitates karyokinesis.
The hierarchical organization of the ISP proteins within the alveoli was dissected by genetic ablation of ISP1 (Beck et al., 2010). In Δisp1 parasites, ISP2/3 relocalize into the apical cap, demonstrating that ISP1 serves a gate-keeping role, preventing access of other family members into the apical cap. This relocalization of paralogous family members may explain the lack of any obvious phenotype in Δisp1 parasites. While the mechanism of relocalization is not known, it specifically requires a C-terminal domain of ISP1 as an ISP1 truncation loses the ability to prevent entry of other ISPs into the apical cap and the homologous domain from ISP2 fails to restore this activity.
While disruption of ISP3 also results in no apparent phenotype, parasites lacking ISP2 regularly assemble more than two daughters per round of endodyogeny, sometimes generating as many as eight buds in a single mother cell (Beck et al., 2010). Less frequently, severe and fatal defects in replication are also observed including missegregation of organelles. Together, these data indicated that the carefully orchestrated arrangement of the ISP proteins within the alveoli sub-domains plays a key role in the proper assembly of daughter parasites.
6. Toxoplasma cell division in other life cycle stages
In the intermediate host, Toxoplasma tachyzoites and bradyzoites divide by endodyogeny. However, when Toxoplasma enters a definite host, i.e. any cat of the felid family, the parasite switches to a polyendodygenic division mode in the epithelial cells of the small intestine and produces merozoites, which then differentiate into the sexual stages. In this alternative division mode the parasite goes through several rounds of DNA synthesis and nuclear divisions without budding daughter cytoskeletons. In a final round of coordinated mitosis, all the nuclei trigger the formation of two daughter buds per nucleus, which form within the cytoplasm (see (Gubbels et al., 2008b) for a recent review). How Toxoplasma is able to uncouple nuclear division from budding in this process is unknown, but there is likely a soluble factor in the cytoplasm that triggers all nuclei to go through the final mitotic round which is coupled to daughter budding. In this respect it is interesting to note that many of the pharmacologically or genetically induced mutant phenotypes result in large polyploid cells with multiple nuclei. From this we can conclude that DNA synthesis and karyokinesis can easily be uncoupled from daughter budding, but at the moment we do not understand how the switch back to daughter budding is made (Ferguson et al., 2008; Gubbels et al., 2008b). Unfortunately, these pre-sexual life stages are poorly experimentally accessible as they cannot be reproduced in vitro and require experimental cat infections, which are cumbersome and inefficient. It is interesting that in the intermediate host approximately one percent of the parasite division rounds results in 3-4 daughter parasites, indicating that two rounds of DNA synthesis and karyokinesis can occur before onset of cell division (Choi-Rhee et al., 2004). Taking into account that polyploid intermediates are the most frequently found division mode in apicomplexans, a model wherein the number of daughter nuclei is actively controlled by a signaling pathway is the most attractive model. Consistent with this model, mutations in Rab6 (involved in retrogade transport from the Golgi (Stedman et al., 2003)), Niemann-Pick type C1-related protein 1 (TgNRC1, residing the IMC with a role in lipid metabolism (Lige et al., 2011)), and ISP2 (Beck et al., 2010) have been shown to result in dramatic increases in the incidences of viable, multi-daughter division rounds. This again supports the presence of a signaling pathway, which nature is at presence unknonw.
Finally, a forward genetic approach identified several genes with roles in the cell division cycle of Toxoplasma (Gubbels et al., 2008a). Among these were several transcription factors and somewhat surprisingly two RCC1 domain proteins, which are regulators of chromosome condensation that control nuclear transport and mitotic progression through nucleotide exchange of Ran-GTPases (Frankel, Knoll, 2009). RCC1 domain proteins have also been associated with virulence in Toxoplasma (Frankel et al., 2007). Moreover, several kinases were also identified in the forward genetic mutant screen (Gubbels et al., 2008a). Inventories of apicomplexan kinomes have identified multiple unique and conserved kinases (Peixoto et al., 2010; Solyakov et al., 2011) suggesting that these could have a function in assembly of the unique cytoskeleton. Therefore it appears that progression through the cell division cycle combines several post-translational modification mechanisms some of which may differ from the well-studied control mechanisms of cell division playing the most prominent roles in higher eukaryotes.
7. Conclusions and open questions
Advances in the tools and technology for the Toxoplasma systems have in recent years revealed numerous molecules involved in the internal budding process (Figs. 2, 8). The sequenced genome of Toxoplasma eases the identification of protein families and the robust genetic and cell biological tools make their characterization possible. It has become clear that budding is driven by the assembly of numerous cytoskeleton components, and that many of these components are unique to the Apicomplexa and/or Alveolata. We are still in the early stages of studying cell division, which consist mostly of gene discovery and descriptions of their subcellular localization and temporal behavior throughout cytokinesis. The rate of discovery is currently very high and therefore there are still many unknowns to be filled in. For instance, only a handful of the proteins identified in proteomical analysis of cytoskeletal fractions have entered this discovery pipeline (Gould et al., 2011; Hu et al., 2006; Xia et al., 2008). In order to better define cytoskeletal development it is necessary to identify and characterize all the proteins of the cytoskeleton. As shown in this review, defining the sequence of events is critical, especially to analyze phenotypes upon manipulation of the cytoskeleton components to dissect their function. Furthermore, determination of the sequence of events will permit testing of the dependence of one event on another. The summary of our current understanding of the time line of Toxoplasma endodyogeny is provided in Figure 8.
The next step in characterizing the role of cytoskeletal proteins in cell division is to define their mode of action, e.g. the coordination and mechanism underlying its assembly in the cytoskeleton. As discussed, gene KOs, KDs, or overexpression of dominant negative alleles are useful tools in this pursuit. This will tell how critical the role of a certain gene is. However, functionality usually comes down to only a handful of critical amino acids, which can either form the active site of enzymatic activity, or are the site of post-translational modification (e.g. phosporylation or acylation, proteolytic cleavage) (Beck et al., 2010; Frenal et al., 2010; Lorestani et al., 2010). For example, GAP, Hsp20, ISP, and IMC proteins contain predicted palmitoylation sites, some of which have been validated. Taking other cell division systems as a guide, phosphorylation very likely drives the progression of steps, which is in line with the presence of cyclin dependent protein kinases and mitotic kinases (Gubbels et al., 2008b; Peixoto et al., 2010). Moreover, post-translational modifications are likely also a key factor in subcellular localization and assembly and disassembly of protein complexes. The nature and extent of these modifications is currently barely understood and even less is known about the identity of the enzymes responsible. For example, no palmitoyl acyltransferases have yet been firmly identified. Although it has been shown that IMC1 is proteolytically cleaved in the maturation process (Mann et al., 2002), the protease responsible is unknown. A role was suspected for aspartic protease 1 (TgAsp1) in IMC biogenesis, but a KO of this gene has no affect on IMC development (Polonais et al., 2011b; Shea et al., 2007). IMC2 is a predicted phosphatase, with one paralog encoded in the genome, but it has not been studied in detail (Mann, Beckers, 2001). Finally, we recently identified a kinase localizing to the IMC (Chen and Gubbels, unpublished data) as well as two related phosphatases, one of which localizes to the apical complex and one localizing to the basal complex of the daughter buds (Lorestani, Ivey, and Gubbels, unpublished data). Taken together, these aspects warrant more work as the enzymes controlling the progression of budding as well as the transitions throughout the budding process will make the best new drug targets.
Besides considering the unique cytoskeleton as composed of promising drug targets, from a biological perspective several questions are emerging. One of the most intriguing aspects is how the centrosome provides the spatio-temporal localization cue for the formation of new daughter buds. At first sight, Toxoplasma daughter bud assembly appears to resemble ascospore formation in yeast, which is coupled to meiosis II (Neiman, 2005). Here the functional ortholog of the centrosome, the spindle pole body, provides the platform for an internally budding spore composed of membrane supported by septin filaments and a leading complex at the growing end, which superficially resemble the Toxoplasma alveoli, IMC filaments and MORN1-containing basal complex, respectively. About a half dozen proteins have been identified with specific roles in spore formation. First, proteins assemble into a platform on top of the spindle pole body (outer plaque) and comprise proteins recruiting vesicles that form the start of the daughter bud. This is reminiscent of the very early appearance of IMC15 and Rab11B. However, no proteins have been identified yet in Toxoplasma that could be functional orthologs of the yeast proteins that recruit the vesicles. Currently there are no direct functional or structural orthologs of the yeast machinery encoded in the Toxoplasma genome, indicating that this is a case of convergent evolution. However, since yeast has been studied in significantly more detail, at least we have an idea for what the likely nature is of proteins we can expect to play a role at these early events.
Another puzzling aspect is the exact function of TgCentrin2. Besides its suggested role in basal complex constriction (Hu, 2008), it is also present in the annuli on the apical cap. This immediately also brings up the question of why the apical cap is different from the rest of the cytoskeleton? Many proteins are unique to the apical cap, such as GAP70, PhIL1, ISP1, and IMC11, but there is no clear understanding of why this is the case. It is possible that this structure is especially reinforced to withstand the forces during the host cell invasion process, or possibly it serves another specialized role in the invasion process.
Currently, new questions are emerging about the role of the cytoskeleton in cell division at an exponential rate with each novel cytoskeletal discovery. Since the pathogenesis of Toxoplasma is closely linked to its rapid rate of replication and replication hinges on proper formation of its cytoskeleton, this is an aspect of parasite biology that demands increased attention. Furthermore, this structure provides an optimal target for improved therapeutic treatments as it is largely constructed of proteins not found in the mammalian host cell.
Acknowledgments
Cell division work in the Gubbels lab is sponsored by National Institutes of Health grants R01AI081924 and U54AI057159 (New England Regional Center of Excellence in Biodefense and Emerging Infectious Disease, development grant), a March of Dimes Basil O'Connor Starter Award 5-FY09-98, and a Smith Family Foundation New Investigator Grant. Work in the Meissner lab is sponsored by Wellcome Trust Senior Fellowship (Grant number: 087582/Z/08/Z) Work in the Bradley is sponsored by National Institutes of Health grant RO1AI064616. J.R.B was funded by the Microbial Pathogenesis Training Grant T32-AI07323.
References
- Agop-Nersesian C, Egarter S, Langsley G, Foth BJ, Ferguson DJ, Meissner M. Biogenesis of the inner membrane complex is dependent on vesicular transport by the alveolate specific gtpase rab11b. PLoS Pathog. 2010;6:e1001029. doi: 10.1371/journal.ppat.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agop-Nersesian C, Naissant B, Ben Rached F, Rauch M, Kretzschmar A, Thiberge S, Menard R, Ferguson DJ, Meissner M, Langsley G. Rab11a-controlled assembly of the inner membrane complex is required for completion of apicomplexan cytokinesis. PLoS Pathog. 2009;5:e1000270. doi: 10.1371/journal.ppat.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, Ferguson DJ, Sahoo N, Gubbels MJ. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of toxoplasma gondii. Cell Microbiol. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhuff WD, Gilk SD, Whitmarsh R, Tilley LD, Hunter C, Ward GE. Targeted disruption of tgphil1 in toxoplasma gondii results in altered parasite morphology and fitness. PLoS ONE. 2011;6:e23977. doi: 10.1371/journal.pone.0023977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, Cruz de Leon J, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of toxoplasma imc proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R, Musoke A, Morzaria S, Gardner M, Nene V. Theileria: Intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parasitology. 2004;129 Suppl:S271–283. doi: 10.1017/s0031182003004748. [DOI] [PubMed] [Google Scholar]
- Bock R, Jackson L, de Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. 2004;129 Suppl:S247–269. doi: 10.1017/s0031182004005190. [DOI] [PubMed] [Google Scholar]
- Bommer W, Hofling KH, Heunert HH. [in vivo observations on the penetration of toxoplasma into the host cell] Dtsch Med Wochenschr. 1968;93:2365–2367. doi: 10.1055/s-0028-1110941. passim. [DOI] [PubMed] [Google Scholar]
- Breinich MS, Ferguson DJ, Foth BJ, van Dooren GG, Lebrun M, Quon DV, Striepen B, Bradley PJ, Frischknecht F, Carruthers VB, Meissner M. A dynamin is required for the biogenesis of secretory organelles in toxoplasma gondii. Curr Biol. 2009;19:277–286. doi: 10.1016/j.cub.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, Cowman AF, Crabb BS, Gilson PR. A novel family of apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J Biol Chem. 2009;284:25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M, Kudryashev M, Leis A, Leonard K, Baumeister W, Menard R, Meissner M, Frischknecht F. Cryoelectron tomography reveals periodic material at the inner side of subpellicular microtubules in apicomplexan parasites. J Exp Med. 2007;204:1281–1287. doi: 10.1084/jem.20062405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miguel N, Lebrun M, Heaslip A, Hu K, Beckers CJ, Matrajt M, Dubremetz JF, Angel SO. Toxoplasma gondii hsp20 is a stripe-arranged chaperone-like protein associated with the outer leaflet of the inner membrane complex. Biology of the cell / under the auspices of the European Cell Biology Organization. 2008;100:479–489. doi: 10.1042/BC20080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbac F, Sanger A, Neuhaus EM, Stratmann R, Ajioka JW, Toursel C, Herm-Gotz A, Tomavo S, Soldati T, Soldati D. Toxoplasma gondii myosins b/c: One gene, two tails, two localizations, and a role in parasite division. J Cell Biol. 2001;155:613–623. doi: 10.1083/jcb.200012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski JM, Niesman IR, Sibley LD. Actin in the parasite toxoplasma gondii is encoded by a single copy gene, act1 and exists primarily in a globular form. Cell Motil Cytoskeleton. 1997;37:253–262. doi: 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- Dubremetz JF, Elsner YY. Ultrastructural study of schizogony of eimeria bovis in cell cultures. J Protozool. 1979;26:367–376. doi: 10.1111/j.1550-7408.1979.tb04639.x. [DOI] [PubMed] [Google Scholar]
- Dubremetz JF, Torpier G. Freeze fracture study of the pellicle of an eimerian sporozoite (protozoa, coccidia) J Ultrastruct Res. 1978;62:94–109. doi: 10.1016/s0022-5320(78)90012-6. [DOI] [PubMed] [Google Scholar]
- Ferguson DJ, Sahoo N, Pinches RA, Bumstead JM, Tomley FM, Gubbels MJ. Morn1 has a conserved role in asexual and sexual development across the apicomplexa. Eukaryot Cell. 2008 doi: 10.1128/EC.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci U S A. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryotic cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Knoll LJ. The ins and outs of nuclear trafficking: Unusual aspects in apicomplexan parasites. DNA Cell Biol. 2009;28:277–284. doi: 10.1089/dna.2009.0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Mordue DG, Knoll LJ. Discovery of parasite virulence genes reveals a unique regulator of chromosome condensation 1 ortholog critical for efficient nuclear trafficking. Proc Natl Acad Sci U S A. 2007;104:10181–10186. doi: 10.1073/pnas.0701893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. Identification of the membrane receptor of a class xiv myosin in toxoplasma gondii. J Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilk SD, Raviv Y, Hu K, Murray JM, Beckers CJ, Ward GE. Identification of phil1, a novel cytoskeletal protein of the toxoplasma gondii pellicle, through photosensitized labeling with 5-[125i]iodonaphthalene-1-azide. Eukaryot Cell. 2006;5:1622–1634. doi: 10.1128/EC.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M, Carver RK, Sulzer AJ. Reproduction of toxoplasma gondii by internal budding. J Parasitol. 1958;44:161–171. [PubMed] [Google Scholar]
- Gonzalez Del Carmen M, Mondragon M, Gonzalez S, Mondragon R. Induction and regulation of conoid extrusion in toxoplasma gondii. Cell Microbiol. 2009;11:967–982. doi: 10.1111/j.1462-5822.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Beatty WL, Sibley LD. A novel actin-related protein is associated with daughter cell formation in toxoplasma gondii. Eukaryot Cell. 2008;7:1500–1512. doi: 10.1128/EC.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Sibley LD. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genomics. 2005;6:179. doi: 10.1186/1471-2164-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Kraft LG, van Dooren GG, Goodman CD, Ford KL, Cassin AM, Bacic A, McFadden GI, Waller RF. Ciliate pellicular proteome identifies novel protein families with characteristic repeat motifs that are common to alveolates. Mol Biol Evol. 2011 doi: 10.1093/molbev/msq321. [DOI] [PubMed] [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. Alveolins, a new family of cortical proteins that define the protist infrakingdom alveolata. Mol Biol Evol. 2008;25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Lehmann M, Muthalagi M, Jerome ME, Brooks CF, Szatanek T, Flynn J, Parrot B, Radke J, Striepen B, White MW. Forward genetic analysis of the apicomplexan cell division cycle in toxoplasma gondii. PLoS Pathog. 2008a;4:e36. doi: 10.1371/journal.ppat.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A morn-repeat protein is a dynamic component of the toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, White M, Szatanek T. The cell cycle and toxoplasma gondii cell division: Tightly knit or loosely stitched? Int J Parasitol. 2008b;38:1343–1358. doi: 10.1016/j.ijpara.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Hager KM, Striepen B, Tilney LG, Roos DS. The nuclear envelope serves as an intermediary between the er and golgi complex in the intracellular parasite toxoplasma gondii. Journal of cell science. 1999;112(Pt 16):2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- Haldar K, Mohandas N. Malaria, erythrocytic infection, and anemia. Hematology Am Soc Hematol Educ Program. 2009:87–93. doi: 10.1182/asheducation-2009.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann J, Hu K, He CY, Pelletier L, Roos DS, Warren G. Golgi and centrosome cycles in toxoplasma gondii. Mol Biochem Parasitol. 2006;145:125–127. doi: 10.1016/j.molbiopara.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Heaslip AT, Dzierszinski F, Stein B, Hu K. Tgmorn1 is a key organizer for the basal complex of toxoplasma gondii. PLoS Pathog. 2010;6:e1000754. doi: 10.1371/journal.ppat.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaslip AT, Ems-McClung SC, Hu K. Tgicmap1 is a novel microtubule binding protein in toxoplasma gondii. PLoS ONE. 2009;4:e7406. doi: 10.1371/journal.pone.0007406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzelman MB, Schwartzman JD. Myosin diversity in apicomplexa. J Parasitol. 2001;87:429–432. doi: 10.1645/0022-3395(2001)087[0429:MDIA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Herm-Gotz A, Agop-Nersesian C, Munter S, Grimley JS, Wandless TJ, Frischknecht F, Meissner M. Rapid control of protein level in the apicomplexan toxoplasma gondii. Nat Methods. 2007;4:1003–1005. doi: 10.1038/nmeth1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herm-Gotz A, Delbac F, Weiss S, Nyitrai M, Stratmann R, Tomavo S, Sibley LD, Geeves MA, Soldati D. Functional and biophysical analyses of the class xiv toxoplasma gondii myosin d. Journal of muscle research and cell motility. 2006;27:139–151. doi: 10.1007/s10974-005-9046-1. [DOI] [PubMed] [Google Scholar]
- Hu K. Organizational changes of the daughter basal complex during the parasite replication of toxoplasma gondii. PLoS Pathog. 2008;4:e10. doi: 10.1371/journal.ppat.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Johnson J, Florens L, Fraunholz M, Suravajjala S, DiLullo C, Yates J, Roos DS, Murray JM. Cytoskeletal components of an invasion machine - the apical complex of toxoplasma gondii. PLoS Pathog. 2006;2:121–138. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Mann T, Striepen B, Beckers CJ, Roos DS, Murray JM. Daughter cell assembly in the protozoan parasite toxoplasma gondii. Mol Biol Cell. 2002a;13:593–606. doi: 10.1091/mbc.01-06-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Roos DS, Murray JM. A novel polymer of tubulin forms the conoid of toxoplasma gondii. J Cell Biol. 2002b;156:1039–1050. doi: 10.1083/jcb.200112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a toxoplasma gondii strain lacking ku80. Eukaryotic cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TM, Rajfur Z, Jacobson K, Beckers CJ. Immobilization of the type xiv myosin complex in toxoplasma gondii. Mol Biol Cell. 2007;18:3039–3046. doi: 10.1091/mbc.E07-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley A, Soldati D. The glideosome: A molecular machine powering motility and host-cell invasion by apicomplexa. Trends Cell Biol. 2004;14:528–532. doi: 10.1016/j.tcb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Lau RK, Kwok AC, Chan WK, Zhang TY, Wong JT. Mechanical characterization of cellulosic thecal plates in dinoflagellates by nanoindentation. J Nanosci Nanotechnol. 2007;7:452–457. [PubMed] [Google Scholar]
- Lige B, Romano JD, Bandaru VV, Ehrenman K, Levitskaya J, Sampels V, Haughey NJ, Coppens I. Deficiency of a niemann-pick, type c1-related protein in toxoplasma is associated with multiple lipidoses and increased pathogenicity. PLoS Pathog. 2011;7:e1002410. doi: 10.1371/journal.ppat.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorestani A, Sheiner L, Yang K, Robertson SD, Sahoo N, Brooks CF, Ferguson DJ, Striepen B, Gubbels MJ. A toxoplasma morn1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- Mann T, Gaskins E, Beckers C. Proteolytic processing of tgimc1 during maturation of the membrane skeleton of toxoplasma gondii. J Biol Chem. 2002;277:41240–41246. doi: 10.1074/jbc.M205056200. [DOI] [PubMed] [Google Scholar]
- Meissner M, Schluter D, Soldati D. Role of toxoplasma gondii myosin a in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Mondragon R, Frixione E. Ca(2+)-dependence of conoid extrusion in toxoplasma gondii tachyzoites. J Eukaryot Microbiol. 1996;43:120–127. doi: 10.1111/j.1550-7408.1996.tb04491.x. [DOI] [PubMed] [Google Scholar]
- Montagna GN, Buscaglia CA, Munter S, Goosmann C, Frischknecht F, Brinkmann V, Matuschewski K. Critical role for heat shock protein 20 (hsp20) in migration of malarial sporozoites. J Biol Chem. 2012;287:2410–2422. doi: 10.1074/jbc.M111.302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Morrissette NS, Murray JM, Roos DS. Subpellicular microtubules associate with an intramembranous particle lattice in the protozoan parasite toxoplasma gondii. J Cell Sci. 1997;110(Pt 1):35–42. doi: 10.1242/jcs.110.1.35. [DOI] [PubMed] [Google Scholar]
- Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002a;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette NS, Sibley LD. Disruption of microtubules uncouples budding and nuclear division in toxoplasma gondii. J Cell Sci. 2002b;115:1017–1025. doi: 10.1242/jcs.115.5.1017. [DOI] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BA, Chiappino ML. Cytoskeleton of toxoplasma gondii. J Protozool. 1987;34:217–226. doi: 10.1111/j.1550-7408.1987.tb03162.x. [DOI] [PubMed] [Google Scholar]
- Nishi M, Hu K, Murray JM, Roos DS. Organellar dynamics during the cell cycle of toxoplasma gondii. J Cell Sci. 2008;121:1559–1568. doi: 10.1242/jcs.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino N, Yoneda C. The fine structure and mode of division of toxoplasma gondii. Arch Ophthalmol. 1966;75:218–227. doi: 10.1001/archopht.1966.00970050220015. [DOI] [PubMed] [Google Scholar]
- Peixoto L, Chen F, Harb OS, Davis PH, Beiting DP, Brownback CS, Ouloguem D, Roos DS. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe. 2010;8:208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, He CY, Hu K, Toomre D, Coppens I, Roos DS, Joiner KA, Warren G. Golgi biogenesis in toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- Plattner H, Klauke N. Calcium in ciliated protozoa: Sources, regulation, and calcium-regulated cell functions. Int Rev Cytol. 2001;201:115–208. doi: 10.1016/s0074-7696(01)01003-8. [DOI] [PubMed] [Google Scholar]
- Polonais V, Javier Foth B, Chinthalapudi K, Marq JB, Manstein DJ, Soldati-Favre D, Frenal K. Unusual anchor of a motor complex (myod-mlc2) to the plasma membrane of toxoplasma gondii. Traffic. 2011a;12:287–300. doi: 10.1111/j.1600-0854.2010.01148.x. [DOI] [PubMed] [Google Scholar]
- Polonais V, Shea M, Soldati-Favre D. Toxoplasma gondii aspartic protease 1 is not essential in tachyzoites. Experimental parasitology. 2011b;128:454–459. doi: 10.1016/j.exppara.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Porchet E, Torpier G. Freeze fracture study of toxoplasma and sarcocystis infective stages. Z Parasitenkd. 1977;54:101–124. doi: 10.1007/BF00380795. [DOI] [PubMed] [Google Scholar]
- Poupel O, Tardieux I. Toxoplasma gondii motility and host cell invasiveness are drastically impaired by jasplakinolide, a cyclic peptide stabilizing f-actin. Microbes Infect. 1999;1:653–662. doi: 10.1016/s1286-4579(99)80066-5. [DOI] [PubMed] [Google Scholar]
- Radke JR, Striepen B, Guerini MN, Jerome ME, Roos DS, White MW. Defining the cell cycle for the tachyzoite stage of toxoplasma gondii. Mol Biochem Parasitol. 2001;115:165–175. doi: 10.1016/s0166-6851(01)00284-5. [DOI] [PubMed] [Google Scholar]
- Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int J Parasitol. 2007;37:1599–1607. doi: 10.1016/j.ijpara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG, Burns RG. The polar ring of coccidian sporozoites: A unique microtubule-organizing centre. J Cell Sci. 1984;65:193–207. doi: 10.1242/jcs.65.1.193. [DOI] [PubMed] [Google Scholar]
- Santos JM, Lebrun M, Daher W, Soldati D, Dubremetz JF. Apicomplexan cytoskeleton and motors: Key regulators in morphogenesis, cell division, transport and motility. Int J Parasitol. 2009;39:153–162. doi: 10.1016/j.ijpara.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Scholtyseck E. [fine structure of the zoites from mature cysts of the so-called m-body (frenkelia spec)] Z Parasitenkd. 1970;33:252–261. doi: 10.1007/BF00259495. [DOI] [PubMed] [Google Scholar]
- Scholtyseck E. Ultrastructure. In: Hammond DM, Long PL, editors. The coccidia: Eimeria, isospara, toxoplasma, and related genera. University Park Press; Baltimore, MD: 1973. pp. 81–144. [Google Scholar]
- Shaw MK, Compton HL, Roos DS, Tilney LG. Microtubules, but not actin filaments, drive daughter cell budding and cell division in toxoplasma gondii. J Cell Sci. 2000;113(Pt 7):1241–1254. doi: 10.1242/jcs.113.7.1241. [DOI] [PubMed] [Google Scholar]
- Shaw MK, Tilney LG. Induction of an acrosomal process in toxoplasma gondii: Visualization of actin filaments in a protozoan parasite. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9095–9099. doi: 10.1073/pnas.96.16.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea M, Jakle U, Liu Q, Berry C, Joiner KA, Soldati-Favre D. A family of aspartic proteases and a novel, dynamic and cell-cycle-dependent protease localization in the secretory pathway of toxoplasma gondii. Traffic. 2007;8:1018–1034. doi: 10.1111/j.1600-0854.2007.00589.x. [DOI] [PubMed] [Google Scholar]
- Sheffield HG, Melton ML. The fine structure and reproduction of toxoplasma gondii. J Parasitol. 1968;54:209–226. [PubMed] [Google Scholar]
- Shirley MW, Smith AL, Tomley FM. The biology of avian eimeria with an emphasis on their control by vaccination. Adv Parasitol. 2005;60:285–330. doi: 10.1016/S0065-308X(05)60005-X. [DOI] [PubMed] [Google Scholar]
- Solyakov L, Halbert J, Alam MM, Semblat JP, Dorin-Semblat D, Reininger L, Bottrill AR, Mistry S, Abdi A, Fennell C, Holland Z, Demarta C, Bouza Y, Sicard A, Nivez MP, Eschenlauer S, Lama T, Thomas DC, Sharma P, Agarwal S, Kern S, Pradel G, Graciotti M, Tobin AB, Doerig C. Global kinomic and phospho-proteomic analyses of the human malaria parasite plasmodium falciparum. Nat Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- Stedman TT, Sussmann AR, Joiner KA. Toxoplasma gondii rab6 mediates a retrograde pathway for sorting of constitutively secreted proteins to the golgi complex. J Biol Chem. 2003;278:5433–5443. doi: 10.1074/jbc.M209390200. [DOI] [PubMed] [Google Scholar]
- Stelly N, Mauger JP, Claret M, Adoutte A. Cortical alveoli of paramecium: A vast submembranous calcium storage compartment. J Cell Biol. 1991;113:103–112. doi: 10.1083/jcb.113.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkermans TJ, Schwartzman JD, Keenan K, Morrissette NS, Tilney LG, Roos DS. Inhibition of toxoplasma gondii replication by dinitroaniline herbicides. Exp Parasitol. 1996;84:355–370. doi: 10.1006/expr.1996.0124. [DOI] [PubMed] [Google Scholar]
- Striepen B, Crawford MJ, Shaw MK, Tilney LG, Seeber F, Roos DS. The plastid of toxoplasma gondii is divided by association with the centrosomes. The Journal of cell biology. 2000;151:1423–1434. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Jordan CN, Reiff S, van Dooren GG. Building the perfect parasite: Cell division in apicomplexa. PLoS Pathog. 2007;3:e78. doi: 10.1371/journal.ppat.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Lehmann MM, Kratzer S, White MW. Nuclear actin-related protein is required for chromosome segregation in toxoplasma gondii. Mol Biochem Parasitol. 2012;181:7–16. doi: 10.1016/j.molbiopara.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, Hu K, Andrews PD, Roos DS, Murray JM. Measuring tubulin content in toxoplasma gondii: A comparison of laser-scanning confocal and wide-field fluorescence microscopy. Proc Natl Acad Sci U S A. 2002;99:2014–2019. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Tilney MS. The cytoskeleton of protozoan parasites. Curr Opin Cell Biol. 1996;8:43–48. doi: 10.1016/s0955-0674(96)80047-0. [DOI] [PubMed] [Google Scholar]
- Tran JQ, de Leon JC, Li C, Huynh MH, Beatty W, Morrissette NS. Rng1 is a late marker of the apical polar ring in toxoplasma gondii. Cytoskeleton (Hoboken) 2010;67:586–598. doi: 10.1002/cm.20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran JQ, Li C, Chyan A, Chung L, Morrissette NS. Spm1 stabilizes subpellicular microtubules in toxoplasma gondii. Eukaryotic cell. 2011 doi: 10.1128/EC.05161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipori S, Ward H. Cryptosporidiosis: Biology, pathogenesis and disease. Microbes Infect. 2002;4:1047–1058. doi: 10.1016/s1286-4579(02)01629-5. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Morrison DP, Gaji RY, Murray JM, Entzeroth R, Howe DK, Striepen B. Plastid segregation and cell division in the apicomplexan parasite sarcocystis neurona. J Cell Sci. 2005;118:3397–3407. doi: 10.1242/jcs.02458. [DOI] [PubMed] [Google Scholar]
- Vivier E, Petitprez A. [the outer membrane complex and its development at the time of the formation of daughter cells in toxoplasma gondii] J Cell Biol. 1969;43:329–342. [PMC free article] [PubMed] [Google Scholar]
- White MW, Jerome ME, Vaishnava S, Guerini M, Behnke M, Striepen B. Genetic rescue of a toxoplasma gondii conditional cell cycle mutant. Mol Microbiol. 2005;55:1060–1071. doi: 10.1111/j.1365-2958.2004.04471.x. [DOI] [PubMed] [Google Scholar]
- White MW, Radke J, Conde de Felipe M, Lehmann M. Cell cycle control/parasite division. Horizon Scientific Press; Norwich, UK: 2007. [Google Scholar]
- Xia D, Sanderson SJ, Jones AR, Prieto JH, Yates JR, Bromley E, Tomley FM, Lal K, Sinden RE, Brunk BP, Roos DS, Wastling JM. The proteome of toxoplasma gondii: Integration with the genome provides novel insights into gene expression and annotation. Genome Biol. 2008;9:R116. doi: 10.1186/gb-2008-9-7-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]