Abstract
Human Ectrodactyly, Ectodermal dysplasia, Clefting (EEC) syndrome is an autosomal dominant developmental disorder defined by limb deformities, skin defects, and craniofacial clefting. Although associated with heterozygous missense mutations in TP63, the genetic basis underlying the variable expressivity and incomplete penetrance of EEC is unknown. Here we show that mice heterozygous for an allele encoding the Trp63 p.Arg318His mutation, which corresponds to the human TP63 p.Arg279His mutation found in patients with EEC, have features of human EEC. Using an allelic series, we discovered that whereas clefting and skin defects are caused by loss of Trp63 function, limb anomalies are due to gain- and/or dominant-negative effects of Trp63. Furthermore, we identify TAp63 as a strong modifier of EEC-associated phenotypes with regard to both penetrance and expressivity.
Keywords: TP63, TP53 homologue, TP63 p.Arg279His, Trp63, EEC syndrome, Cleft palate, Limb defects, Mouse model, Genetic modifier, TAp63
INTRODUCTION
Identifying genomic modifiers affecting disease severity in monogenic syndromes is challenging [Genin et al., 2008], and for rare diseases it is sometimes not possible to gain the statistical power required for accurate predictions. Ectrodactyly, Ectodermal dysplasia, Clefting (EEC) syndrome (OMIM 604292) is one such rare syndrome. This is an autosomal dominant syndrome characterized by anomalies of the hands and feet, ectodermal defects including those of the hair, teeth, and nails, and cleft lip and/or palate [Roelfsema and Cobben, 1996; Rinne et al., 2006]. The phenotype of Trp63-deficient mice [Mills et al., 1999; Yang et al., 1999] provided clues that led to the discovery that mutations in the DNA binding domain (DBD) of the TP53-related transcription factor TP63 cause EEC [Celli et al., 1999]. A persistent enigma is that EEC has highly variable expressivity [Fryns et al., 1990; Buss et al., 1995; Roelfsema and Cobben, 1996; Barrow et al., 2002]. However, due to the rarity of EEC syndrome, identifying genes that modify this feature has not been possible from human studies.
What has further confounded the correlation between EEC phenotypes and specific mutations is that TP63/Trp63 give rise to two classes of transcripts, TAp63 and ΔNp63, each of which is alternatively spliced at the C-terminus to further subdivide these classes into α, β, and γ isoforms, accounting for six distinct TP63/Trp63 proteins [Yang et al., 1998]. Mutations associated with EEC are located in the DNA-binding domain (DBD) of TP63, a region shared by all isoforms. The extent to which specific TP63 mutations cause EEC-related pathology, and the identity of genes that modulate the clinical features of EEC have not been explored.
Here we modeled human EEC by changing the codon for arginine (R) 318 of the Trp63 locus to histidine (H), thereby mimicking the human mutation TP63.Arg279His (herein referred to as the Trp63R279H mutation). We generated two mouse models with alleles encoding Trp63R279H (Trp63Aam1-R279HN and Trp63Aam2-R279H), and compared their phenotypes to mice deficient for either TAp63 isoforms, or all Trp63 isoforms. Using this allelic series, we established that mice heterozygous for the Trp63Aam1-R279HN allele have features of human EEC, and further we define TAp63 as a modifier of both penetrance and expressivity, providing functional evidence for these genetic characteristics of this syndrome.
MATERIALS AND METHODS
GENERATION OF AN ALLELIC SERIES OF Trp63 MUTATIONS
We used a gene targeted knock-in approach to model the TP63 p.Arg279His mutation found in human EEC by changing the arginine codon for amino acid 318 in exon 7 of Trp63 to histidine. Utilizing the pTVp63floxN construct we previously generated [Mills et al., 2002], the nucleotide sequence AGA was changed to CAT via site-directed mutagenesis to generate the pTVp63R279HN targeting construct (Fig. 1). The construct was sequence verified. The pTVp63R279HN targeting construct was electroporated into AB2.2 embryonic stem cells and positive selection with G418 was used to identify cells that had stably integrated the construct. Blastocyst injection and generation of chimeric animals was performed at the Cold Spring Harbor Laboratory Gene Targeting and Transgenic Shared Resource. Germline transmission generated the Trp63Aam1-R279HN model and the modified allele was detected via Southern blotting and PCR. Mating with B6.C-Tg (CMV-Cre) 1Cgn/J mice (The Jackson Laboratory) was used to excise the neomycin cassette, thereby establishing the Trp63Aam2-R279H and the Trp63 deficient lines (Trp63Aam3).
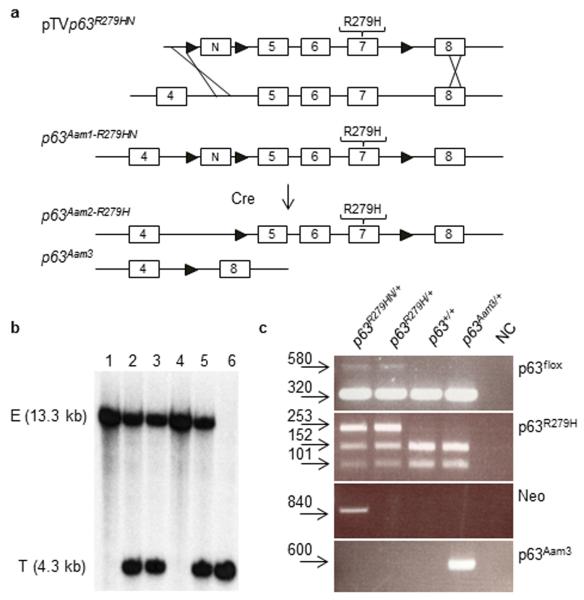
Figure 1. Generation of Trp63Aam1-R279HN, Trp63Aam2-R279N, and Trp63Aam3 mice.
(a) The codon for arginine (R) 318 within exons encoding the DBD of p63 was changed to the codon for histidine (H) to generate the pTVp63R279HN gene targeting vector that was used to establish the Trp63Aam1-R279HN mouse model. By crossing Trp63R279HN/+ to a Cre-expressing model, two independent alleles were generated: Trp63Aam2- R279H (lacking the Neomycin cassette) and Trp63Aam3 (lacking exons 5-7 that encode the DBD). Neomycin cassette, N; LoxP sites, triangle. (b) Characterization via Southern identified Trp63+/+ (lanes 1, 4), Trp63R279HN/+ (lanes 2, 3, 5) and Trp63R279HN/R279HN (lane 6) progeny. Endogenous allele, E; Targeted allele, T. (c) Trp63R279HN/ +, Trp63R279H/+, Trp63+/+, and Trp63Aam3/+ mice can be distinguished by PCR. The mutation in exon 7 of Trp63 disrupts a BsaHI site present in wild type Trp63. The p63R279H PCR assay used this alteration to distinguish mutant from wild type p63 alleles, as the PCR product digested with BsaHI generates either two wild type fragments (152 bp and 101 bp) or one mutant fragment (253 bp). The p63flox PCR assay detects the LoxP site present in intron 7 (580 bp) and the wild type allele (320 bp). The Neo PCR assay detects the Neomycin cassette (840 bp). The p63Aam3 PCR assay detects excised exons 5-7 (600 bp).
POLYMERASE CHAIN REACTION (PCR)
The AGA to CAT mutation disrupts a BsaHI restriction site present in exon 7 of Trp63. Primers REV exon 7 AGAATTCTGCTTGGTCCTTGG and FWD exon 7 GAAGTGGAGTTCTGGAGGGCAG, spanning exon 7 were used to amplify a 253 bp fragment. BsaHI digestion yielded two fragments of 152 and 101 bp for the wild type Trp63 allele, and a single 253 bp fragment for the Trp63R279HN and Trp63R279H alleles.
SOUTHERN BLOTTING
Southern hybridization was used to identify a diagnostic BamHI fragment resulting from accurate homologous recombination between the pTVp63R279HN targeting vector (which contained an exogenous BamHI restriction site) and the endogenous Trp63 locus. Southern hybridization identified the predicted 13.3 kb endogenous Trp63 allele and the 4.3 kb targeted Trp63R279HN allele.
KERATINOCYTE CULTURE
Primary keratinocyte cultures were prepared as described [Lin and Lowe, 2001] with several modifications. Mouse skin was floated on a layer of dispase (5 mg/ml)/Ca+2, Mg2+-free PBS at 4 °C (Quality Biological Inc. #130-057-161) overnight. Dermis and epidermis were separated and keratinocytes were collected by mincing the epidermis, placing it in DMEM (Cellgro #10-013-CV) and stirring for 15 minutes with a magnetic stirrer, thereafter filtering and seeding the cells with Keratinocyte EMEM (Lonza #06-174G) supplemented with 8% chelated fetal bovine serum/1% penicillin and streptomycin sulfate/0.05 mM Ca2+Cl2/10 ng/ml epidermal growth factor. For subsequent cell culture, keratinocytes were split once and seeded at 0.2×106/well (24 well plate).
QUANTITATIVE PCR (Q-PCR)
Total RNA was prepared with TRIzol reagent (Invitrogen #15596-018) and RT-PCR was performed according to SuperscriptTM III first-strand synthesis system (Invitrogen #18080-051) followed by Q-PCR using the LightCycler® 480 system (Roche #04 707 516 001). The following primers were used: TAp63-FWR CCAGAGGTCTTCCAGCATA and TAp63-REV TTTCGGAAGGTTCATCCAC, ΔNp63-FWR CTGGAAAACAATGCCCAGAC and ΔNp63-REV GAGGAGCCGTTCTGAATCTG, p63α-FWR CACAGACTGCAGCATTGTCA and p63α-REV CCCTGGGTCGTGAAATAGTC, p63β-FWR CATTGTCAGGATTTGGCA and p63β-REV GAGGTGAGGAGAAGTCGT, p63γ-FWR TACCTCCCTCAGCACACGA and p63γ-REV CTGAAGCAGGCTGAAAGGA, p16-FWR GTCACACGACTGGGCGATT and p16-REV CATGCTGCTCCAGATGGCT, p19-FWR TGAGGCTAGAGAGGATCTTGAGA and p19-REV GCAGAAGAGCTGCTACGTGAA, p53-FWR CGCTGCTCCGATGGTGAT and p53-REV TCGGGATACAAATTTCCTTCCA, Actin-FWR GATCTGGCACCACACCTTCT and Actin-REV GGGGTGTTGAAGGTCTCAAA.
ANTIBODIES
Primary antibodies used for immunofluorescence and western blot were: 4A4 anti-p63 (1:400, Santa Cruz #sc-8431), Keratin 14 (1:1000, Covance #PRB-155P), Fillagrin (1:100, Santa Cruz #sc-30230), Ki67 (1:200, Abcam #ab15580), Keratin 10 (1:500, Covance #PRB-159P), Keratin 6 (1:500). Alexa-conjugated secondary antibodies were: goat-α-rabbit A488 (1:2000, Invitrogen #A11008), donkey-α-mouse A568 (1:2000, invitrogen #A10037). Horseradish peroxidase-linked secondary antibodies were from GE Healthcare.
BROMODEOXYURIDINE (BrdU) INCORPORATION ASSAY
Primary keratinocytes were seeded at 0.2×106 cells/well (24 well plate) at day 1 and thereafter treated with 10 μM BrdU (BD Pharmingen #550891) for 2 hours at 37 °C at days 2 and 6 of culture. Bromodeoxyuridine-incorporation was assayed using an Invitrogen BrdU Staining Kit (#93-3943). Several representative areas were randomly selected for quantification of percentage of BrdU-positive cells.
SENESCENCE-ASSOCIATED-β-GALACTOSIDASE (SA-β-Gal) ASSAY
Primary keratinocytes were seeded at 0.2×106 cells/well (24 well plate) at day 1 and thereafter fixed with 0.5% glutaraldehyde (Sigma #G6403) in PBS, 15 minutes at RT and washed with 1 mM MgCl2 in PBS pH 5.5 before being assayed for SA-β-Gal activity, as described [Dimri et al., 1995] at days 2 and 6 of culture. Several representative areas were randomly selected for quantification of percentage SA-β-Gal-positive cells.
RESULTS
GENERATION OF AN ALLELIC SERIES OF Trp63 MUTATIONS
To model human EEC syndrome, we introduced the Trp63R279H missense mutation corresponding to the TP63 p.Arg279His mutation associated with human EEC into the endogenous Trp63 locus using gene targeted knock-in (Fig. 1). This initial allele, designated Trp63Aam1-R279HN (herein referred to as Trp63R279HN), carried a floxed neomycin cassette, which was subsequently removed in vivo by crossing to Cre-expressing mice, thereby establishing the Trp63Aam2-R279H allele (herein referred to as Trp63R279H). This approach also generated the Trp63Aam3 null allele, in which exons encoding the DBD of Trp63 were removed. Thus, we generated three distinct mouse models: two models with alleles encoding the Trp63R279H point mutation, one containing and the other lacking the neomycin cassette (Trp63R279HN and Trp63R279H, respectively), and the corresponding Trp63Aam3 null model, which served as an iso-allelic control mimicking the established Trp63Brdm3 null model [Mills et al., 2002]. Along with the Trp63Aam4-TA null allele we established in which mice homozygous for this allele (herein referred to as Trp63TA−) are deficient specifically for TAp63 isoforms [Guo et al., 2009], this allelic series provided novel models for defining the underlying genetic basis of EEC.
THE Trp63R279H ALLELE RETAINS PARTIAL FUNCTION IN THE ABSENCE OF WILD TYPE Trp63
To compare these newly established Trp63 alleles, we analyzed the phenotype of homozygous mice. Homozygosity for Trp63R279HN, Trp63R279H, or Trp63Aam3 caused limb truncations and defects in epithelial morphogenesis that were similar to the reported Trp63-deficient models [Mills et al., 1999; Yang et al., 1999] at the gross level (Supplemental Fig. 1a). However, we found that Trp63R279H/R279H embryos were distinct from both Trp63R279HN/R279HN and Trp63Aam3/Aam3 embryos, as the epithelial markers keratin 6 and 14 were readily detected in the epidermis of the skin at embryonic day 19.5 (E19.5) (Supplemental Fig. 1b). This indicated that skin morphogenesis, although incomplete, proceeded to a later stage in Trp63R279H/R279H embryos. These findings indicate that the Trp63R279H allele is unique in that it is sufficient to initiate and to maintain the program of epidermal morphogenesis until late stages of embryogenesis—a characteristic not observed in embryos homozygous for either the Trp63R279HN or the Trp63Aam3 allele.
HETEROZYGOSITY FOR Trp63R279HN CAUSES PHENOTYPES SIMILAR TO CLINICAL FEATURES OF HUMAN EEC
Since EEC is caused by heterozygous mutations in TP63, we asked whether mice heterozygous for mutant Trp63 alleles had the features of human EEC, including cleft palate, ectrodactyly and/or ectodermal dysplasia. We first noticed that transmission of the mutant Trp63R279HN allele caused substantial lethality (18%), as Trp63R279HN/+ weanlings were underrepresented (Table I). This lethality did not occur in Trp63R279H/+ weanlings, indicating that the two alleles were distinct with regards to survival. We also noted some lethality (~8%) in Trp63Aam3/+ weanlings. However, in contrast to the lethality of Trp63R279HN/+ mice, this lethality only affected males (Tables I and Supplemental Table I). For the Trp63R279HN/+ cohort, lethality was apparent at birth and was associated with severe cleft palate; indeed, Trp63R279HN/+ newborns with cleft palate were easily identified by the absence of a visible milk pouch in the stomach (Fig. 2a), as well as by visual examination of the upper palate. Scanning electron microscopy of E18.5 embryos indicated that the palatal shelves of Trp63R279HN/+ embryos had not fused (Fig. 2b). We therefore analyzed the palates at E15.0, a stage in which palate fusion has been initiated [Bush and Jiang, 2012]. At this stage, adhesion and fusion of the secondary palate had stalled in a subset of Trp63R279HN/+ embryos, appearing similar to the palates of E13.5 wild type mice (Fig. 2c). Additionally, Trp63 expression was detected in intact epithelia covering the palatal shelves of both wild type and Trp63R279HN/+ embryos. In contrast to the high percentage of clefting in Trp63R279HN/+ neonates (15.4%), cleft palate was not detected in Trp63Aam3/+ neonates, and occurred so rarely in Trp63R279H/+ neonates (2.3%) that it did not significantly impact survival (Table II). These findings provide functional evidence that cleft palate has high penetrance in mice heterozygous for the Trp63R279HN allele, has low penetrance in mice heterozygous for the Trp63R279H allele, and does not occur in mice heterozygous for the Trp63Aam3 allele, underscoring the distinction between these Trp63 alleles with regards to this feature of EEC syndrome.
Table 1.
Transmission of the p63 Aam1-R279HN, p63 Aam2-R279H, and p63 Aam3 alleles at weaning
| Mating | Total | Expected % | Observed % (total number) | P-value | ||
|---|---|---|---|---|---|---|
| +/+ | +/− | +/+ | +/− | |||
| p63Aam1-R279HN/+ × p63+/+ a | 218 | 50 | 50 | 68.8 (150) | 31.2 (68) | <0.0001* |
| p63Aam1-R279HN/+ × p63+/+ b | 26 | 50 | 50 | 76.9 (20) | 23.1 (6) | 0.006* |
| p63Aam1-R279HN+ × p63+/+ c | 59 | 50 | 50 | 64.4 (38) | 35.6 (21) | 0.0269* |
| p63Aam2-R279H/+ × p63+/+ a | 285 | 50 | 50 | 51.9 (148) | 48.1 (137) | 0.5147 |
| p63Aam2-R279H/+ × p63+/+ b | 152 | 50 | 50 | 56.6 (86) | 43.4 (66) | 0.1048 |
| p63Aam2-R279H/+ × p63+/+ c | 208 | 50 | 50 | 56.3 (117) | 43.7 (91) | 0.0714 |
| p63Aam3/+ × p63+/+ b | 205 | 50 | 50 | 58.0 (119) | 42.0 (86) | 0.0212* |
mixed background (50% C57BL/6N, 50% 129Sv)
C57BL/6N background
129Sv background
significant, two tailed P-values calculated using Fisher’s exact test
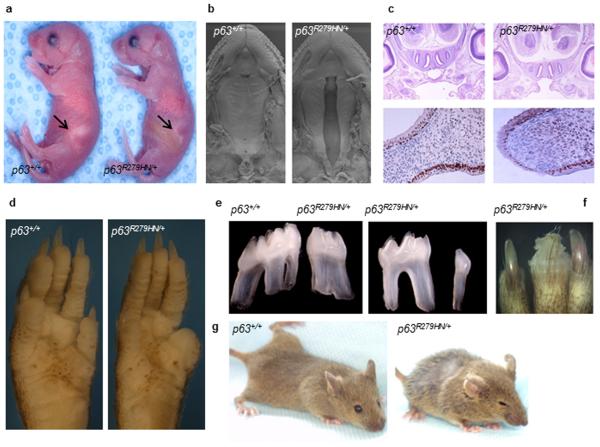
Figure 2. Trp63Aam1-R279HN/+ mice display features of human EEC syndrome.
(a) A subset of Trp63R279HN/+ neonates lack a visible milk pouch (arrow) have cleft palate, and die shortly after birth. (b) Scanning electron microscopy of the upper palate at E18.5 visualizes the clefting in Trp63R279HN/+ mice. (c) Histological analyses of E15.0 indicates that the palatal shelves are not fused in clefted Trp63R279HN/+ mice. Further, Trp63 is expressed in the epithelial layer of both wild type fused palate and Trp63R279HN/+ animals with cleft palate. Coronal sections of the upper palate are shown. (d) Unilateral defects of the distal limbs were noted in 1/40 Trp63R279HN/+ mice at weaning. Digit truncation (right) and unaffected limb (left) from a littermate are shown. (e) Tooth morphogenesis is defective in a subset of Trp63R279HN/+ mice. Abnormal root structure of upper molars (left panel) and hyperdontia of lower molar (right panel) have been noted. (f) Dystrophic nails are seen in Trp63R279HN/+ mice. (g) With age, Trp63R279HN/+ are distinguishable from their wild type littermates by their sparse coat and eye squinting.
Table 2.
p63Aam1-R279HN and p63Aam2-R279H neonates are born with cleft palate
| Mating | Total | +/+ | +/− | % cleft palate | |
|---|---|---|---|---|---|
| +/+ | +/− | ||||
| p63Aam1-R279HN/+ × p63+/+ a | 48 | 22 | 26 | 0% | 15.4% (4/26) |
| p63Aam2-R279H/+ × p63+/+ a | 81 | 37 | 44 | 0% | 2.3% (1/44) |
| p63Aam3/+ × p63+/+ b | 41 | 24 | 17 | 0% | 0% |
mixed background (50% C57BL/6N, 50% 129Sv)
C57BL/6N background
In addition to cleft palate, other features of EEC were noted in Trp63R279HN/+ mice, including anomalies of the distal limbs (Fig. 2d), defective tooth morphogenesis (abnormal root structures and hyperdontia) (Fig. 2e), dystrophic nails (Fig. 2f), and with age, alopecia, ruffled coat, and squinted eyes (Fig. 2g), features that were not detected in the Trp63R279H/+ or Trp63Aam3/+ cohorts. These data demonstrate that the Trp63R279HN allele is distinct from both the Trp63R279H and the Trp63Aam3 alleles, and further indicate that as for cleft palate, these EEC-associated pathologies of Trp63R279HN/+ mice are not simply due to haploid levels of Trp63.
Features of ectodermal dysplasia in patients with EEC are sparse hair, alopecia, and a thin, dry skin [Rinne et al., 2006]. Since Trp63R279HN/+ mice developed ruffled coats and alopecia (Fig. 2g), we analyzed the Trp63 allelic series for skin defects. Surprisingly, no major skin abnormalities were detected in Trp63R279HN/+ or Trp63R279H/+ mice at E16.5 or postnatal day 0 (Supplemental Fig. 2). To more closely analyze epidermal phenotypes, we harvested primary keratinocytes from Trp63R279HN/+ and Trp63R279H/+ mice. While our analyses of intact skin had not identified proliferative defects (as visualized using markers K6 and Ki67 or in histopathology of the dermis or epidermis) (Supplemental Fig. 2 and data not shown), keratinocytes from Trp63R279HN/+ and Trp63R279H/+ mice had reduced proliferation and a corresponding increase in cellular senescence, as seen by decreased BrdU incorporation, augmented SA-β-gal expression, and activation of senescence-inducing pathways (Fig. 3). These analyses indicate that keratinocytes from Trp63R279HN/+ and Trp63R279H/+ mice have augmented senescence. Altogether, our findings from this Trp63 allelic series indicate that the Trp63R279HN/+ model is unique in its ability to accurately recapitulate phenotypes reminiscent of the clinical spectrum of human EEC syndrome.
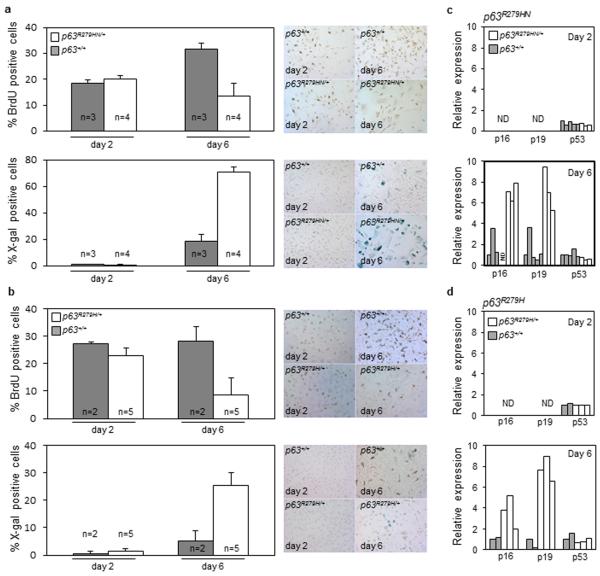
Figure 3. Primary keratinocytes from Trp63Aam1-R279HN/+ and Trp63Aam2-R279H/+ cease proliferate and enter senescence.
As seen via BrdU incorporation and SA-β-Gal detection, Trp63R279HN/+ (a) and Trp63R279H/+ (b) primary keratinocytes have decreased proliferation and increased senescence after 6 days in culture. Data are presented as mean and bars represent standard deviation. Transcripts for p16 and p19 are upregulated in Trp63R279HN/+ (c) and Trp63R279H/+ (d) keratinocytes after 6 days of culture, indicating an augmented senescent response. All assays were conducted with littermates; one representative experiment out of 2-4 independent assays is shown.
Trp63 EXPRESSION IS ENHANCED SPECIFICALLY IN THE Trp63R279HN/+ MODEL
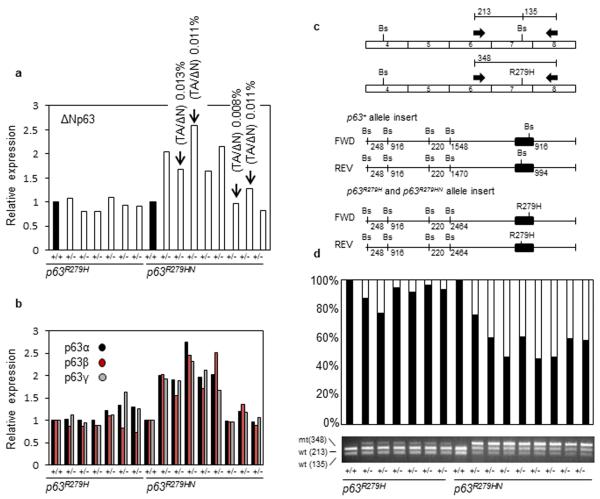
Phenotypes associated with EEC were much more frequent in the Trp63R279HN/+ model, although Trp63R279H/+ mice had the same EEC-causing mutation. To assess whether we could detect alterations in the skin, we analyzed neonatal skin of the two Trp63R279H encoding models for Trp63 expression using immunofluorescence. This showed that Trp63 protein was noticeably more abundant in the epidermis of Trp63R279HN/+ mice relative to that of Trp63R279H/+ mice (Fig. 4a-b). Western analyses confirmed this Trp63 accumulation specifically in the epidermis of Trp63R279HN/+ neonates, and also implicated ΔNp63α, β and γ (the three major isoforms transcribed from the ΔNp63 promoter) as being co-upregulated (Fig. 4c). Patients with EEC that are heterozygous for TP63R279H alleles also accumulate TP63 protein in the epidermis [Browne et al., 2011]. This finding defines a clear molecular distinction between the Trp63R279HN/+ and Trp63R279H/+ models. To determine whether Trp63 was being regulated at the transcriptional level, we assessed Trp63 expression using real-time PCR. For Trp63R279HN/+ mice, we noted a marked variation in Trp63 transcript level amongst individual pups from the same litter, with ΔNp63 transcript being upregulated (Fig. 5a). This transcriptional variation was not seen in Trp63R279H/+ mice, as ΔNp63 expression levels were similar overall and similar to littermate controls. Trp63R279HN/+ neonates with enhanced expression of ΔNp63 had increased levels of α, β, and γ transcripts, indicating a general upregulation of all ΔNp63 isoform subtypes (Fig. 5b). To assess the relative abundance of mutant Trp63 transcripts, we took advantage of the fact that the nucleotides altered to generate the arginine to histidine mutation disrupted a BsaHI restriction enzyme site, which provided a straightforward PCR assay that could differentiate wild type from mutant Trp63 transcripts (Fig. 5c). This allowed us to calculate the ratio of mutant to wild type Trp63 transcripts in the epidermis of Trp63R279HN/+ and Trp63R279H/+ mice. We discovered that Trp63R279HN/+ mice expressed a higher proportion of mutant Trp63 transcript compared to Trp63R279H/+ mice (p=0.00082) (Fig. 5d). Whereas mutant transcript accounted for ~50% of the total Trp63 transcript pool in Trp63R279HN/+ epidermis, mutant transcript accounted for only ~8% of the total Trp63 transcript pool in Trp63R279H/+ epidermis. These findings indicate that the EEC-like model Trp63R279HN/+ expresses more of the Trp63R279H-encoding transcript and has enhanced Trp63 protein accumulation.
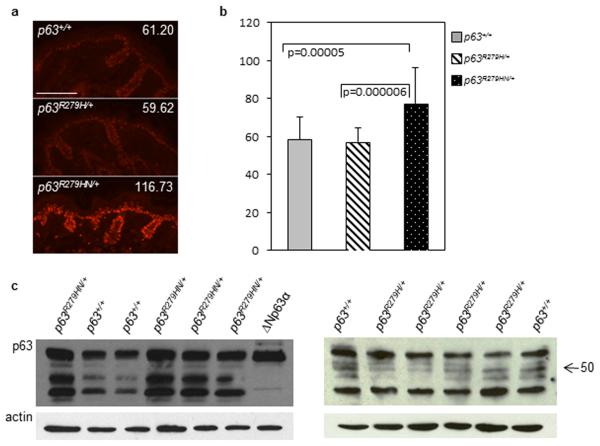
Figure 4. Trp63Aam1-R279HN/+ accumulate Trp63 protein in the epidermis.
Trp63R279HN/+ animals have an increased Trp63 intensity within the epidermis shown via (a) immunofluorescence for Trp63 (red) in Trp63R279HN/+ and Trp63R279H/+ neonatal skin and (b) calculation of the intensity score via Velocity 6.2 (PerkinElmer). The intensity score is shown as an arbitrary unit. Data are presented as mean and bars represent standard deviation. White bar, 100 μm. (c) Western shows accumulation of several Trp63 isoforms within the epidermis of Trp63R279HN/+ mice, which is not detected in Trp63R279H/+ mice.
Figure 5. Trp63Aam1-R279HN/+ mice express a higher proportion of mutant Trp63 transcript in comparison to Trp63Aam2-R279H/+mice.
(a) Detection of ΔNp63 transcript in epidermis of newborn mice. ΔNp63 transcript is expressed more robustly in the epidermis of Trp63R279HN/+ mice. Four of eight Trp63R279HN/+ samples had detectable TAp63 transcript. (b) Using the same samples as in panel (a), similar transcriptional patterns were detected for α, β, and γ transcripts, indicating a general up regulation of each C-terminally spliced class of transcripts. Results are shown as a ratio of wild type littermate control to the respective mutant sample. (c) Schematic diagram of the assay used to differentiate wild type from mutant Trp63 transcripts. Wild type and mutant transcripts can be differentiated by the presence or absence, respectively, of a BsaHI site in exon 7. Primers specific for exons 6 and 8 (which flank the codon for R279) were used for PCR amplification of wild type and mutant Trp63 transcripts in Trp63R279HN and Trp63R279H heterozygotes from an RT-PCR reaction. The PCR reaction was digested with BsaHI to generate either two wild type fragments (213 bp and 135 bp) or one mutant fragment (348 bp). (d) The undigested PCR-reaction was also used for TOPO-cloning and subsequent transformation. For each individual sample, 100 clones were picked and digested with BsaHI to determine the ratio of mutant to wild type transcript. Trp63R279H/+ mice express a lower proportion of mutant transcript in relation to Trp63R279HN/+ mice (p=0.00082). Black, wild type transcript; White, mutant transcript.
TAp63 IS A POTENT MODIFIER OF PENETRANCE AND EXPRESSIVITY OF EEC PHENOTYPES
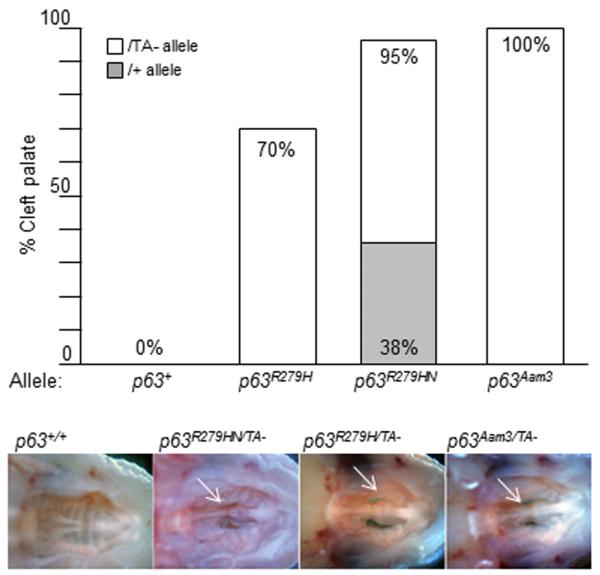
We hypothesized that TAp63 might function as a modifier of EEC. To test this idea genetically, we established our Trp63 allelic series in a TAp63-deficient background and assessed the penetrance of phenotypes already investigated in Trp63R279HN/+ mice (Supplemental Table II). Trp63R279HN/TA− and Trp63R279H/TA− mice were generated by crossing Trp63R279HN/+ and Trp63R279H/+ to Trp63+/TA− mice, a TAp63 deficient model we previously established using chromosome engineering [Guo et al., 2009]. The Trp63TA− allele expresses ΔNp63, but not TAp63 isoforms. As a control, we established Trp63Aam3/TA− mice. Intriguingly, the penetrance of clefting in Trp63R279HN/+ and Trp63R279H/+ neonates was enhanced when wild type TAp63 was absent, culminating in 95% and 70% of Trp63R279HN/TA− and Trp63R279H/TA− pups, respectively (Fig. 6). Importantly, 100% of Trp63Aam3/TA− neonates had cleft palate, which indicated that the clefting phenotype was due to loss of wild type Trp63 rather than being unique to the presence of the Trp63R279H mutation. Of note is that neither Trp63TA+/− nor Trp63TA−/− mice had cleft palate, which indicated that loss of wild type TAp63 alone was not sufficient for this phenotype. Thus, our findings indicate that Trp63-related cleft palate is due to absence of wild type TAp63 in the context of haploid levels of wild type ΔNp63.
Figure 6. The penetrance of cleft palate is increased in a TAp63 deficient background.
The penetrance of cleft palate seen in Trp63R279H/+ and Trp63R279HN/+ is increased when the wild type TAp63 allele is absent. The lower jaw was removed and the upper palate visualized in Trp63R279H, Trp63R279HN and Trp63Aam3 TAp63-deficient neonates. Cleft is shown with an arrow.
In addition to TAp63 being a potent modifier of cleft palate, we also discovered that TAp63 modifies the penetrance of skin defects. The epidermis was abnormal in both Trp63R279HN/TA− and Trp63R279H/TA− mice, with the stratum basale and stratum spinosum being similarly affected in both models (Supplemental Fig. 3). This phenotype was seen as a thickening of Trp63-, K14-, and K10-expressing layers in patches of the epidermis covering the skin surface. In addition, the dermis was thicker in large areas of the skin of both models. As we had observed for cleft palate, this increase in epidermal thickness was also seen in Trp63Aam3/TA− neonates (Supplemental Fig. 3). These findings indicate that TAp63 deficiency increases the penetrance of both cleft palate and skin defects, showing that these phenotypes are caused by loss of Trp63 function, and furthermore, that wild type TAp63 protects from these phenotypes.
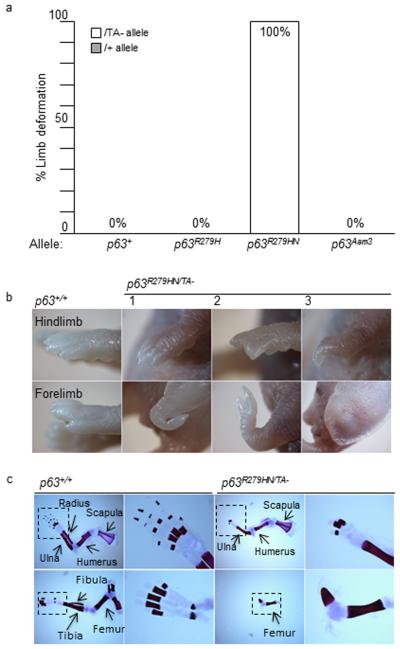
Absence of wild type TAp63 not only augmented the occurrence of palate and skin phenotypes, it also increased the penetrance of limb anomalies (Fig. 7). Further, the phenotype ranged from lack of hind limbs to single ectrodactyly, thereby displaying variable expressivity. However, in contrast to cleft palate and skin defects, the ability of TAp63 loss to enhance the penetrance of limb anomalies was specific to the Trp63R279HN allele; that is, limb defects occurred in 100% of Trp63R279HN/TA− neonates, but did not occur in either Trp63R279H/TA− or in Trp63Aam3/TA− neonates (Fig. 7). This again establishes Trp63R279HN as the model in our allelic series that most reflects the features of human EEC, and while TAp63 modifies the penetrance of each of the EEC phenotypes studied, limb anomalies are unique in that they are not simply due to loss of Trp63 activity, as is the case for both cleft palate and skin defects.
Figure 7. The penetrance of limb defects is increased in a TAp63 deficient background.
(a) The limb defects (1/40) seen in Trp63R279HN/+ mice is 100% penetrant in a TAp63-deficient background (Trp63R279HN/TA− mice). (b) Analysis of three Trp63R279HN/TA− neonates (#1-3) exemplifies the range of limb deformities from a complete lack of hind limbs (#3) to single ectrodactyly of the fore limb (#1). (c) Skeletal preparations show hindlimb and forelimb abnormalities in a TA-deficient background for Trp63R279NH mice. The boxed area is magnified to visualize the phalanges.
DISCUSSION
In this study we established a unique allelic series of Trp63 mutations, including two distinct alleles encoding mutant Trp63 proteins corresponding to TP63 p.Arg279His associated with human EEC syndrome. Although these two EEC models are heterozygous for Trp63 alleles encoding the same single amino acid mutation, only one of them, Trp63R279HN/+, displayed the symptoms characteristic of EEC syndrome. By analyzing these two models, we exposed a fundamental difference between them: the Trp63R279HN/+ model that most closely mimics human EEC is unique in that it accumulates Trp63 protein in the epidermis. This feature also occurs in the epidermis of patients with EEC and the TP63 p.Arg279His alteration [Browne et al., 2011]. Further, Trp63R279HN/+ mice have a generalized increase in Trp63 transcript abundance, and express mutant and wild type transcripts at essentially equivalent levels. On the other hand, the Trp63R279H/+ model—which does not mimic human EEC nearly as frequently as does the Trp63R279HN/+ model—expresses only low levels of mutant Trp63 transcript. The underlying cause for the distinction between the two EEC-encoding models is not clear, but could be due to differences in nonsense-mediated decay, splicing efficiency, or in recruitment of transcription factors. The only difference that we engineered when designing the two EEC-encoding alleles was the presence or absence of the neomycin cassette within intron 4. Although the Trp63R279H model is the genetically correct model since it best reflects the TP63 allele encoding TP63 p.Arg279His in patients with EEC, it is the Trp63R279HN model that displayed the wide range of characteristic EEC features. Since the two EEC-encoding alleles caused distinct degrees of disease severity, a feature that models the clinical symptoms in human patients with EEC [Roelfsema and Cobben, 1996; Rinne et al., 2006], they have provided a unique opportunity for investigating EEC-associated features with regards to development, penetrance, and severity.
The TP63 p.Arg279His protein has a longer half-life than wild type TP63, partially accounting for the protein accumulation seen in patients with EEC [Browne et al., 2011]. Furthermore, degradation of DBD mutant TP63 protein can be induced by co-expression of wild type ΔNp63α [Browne et al., 2011] or TAp63γ [Ying et al., 2005]. These lines of evidence are in agreement with our findings. First, the increased half-life of mutant TP63, coupled with our finding that the mutant Trp63 transcript is expressed more robustly in the Trp63R279HN/+ model, likely accounts for the observed protein accumulation. In contrast, Trp63 protein does not accumulate in Trp63R279H/+ mice; this model expresses a much lower proportion of mutant p63 transcript compared to the Trp63R279HN/+ model, accounting for an absence of Trp63 protein accumulation. Thus, a threshold of mutant Trp63 seems to be required to evoke the phenotypes characteristic of EEC, a condition that is met more easily in the high expressing EEC model, Trp63R279HN/+. Since Trp63 functions as a tetramer, our working model is that the ratio of mutant to wild type protein affects the formation of functional Trp63 tetramer, abrogation of which is the determining factor behind the development of EEC phenotypes in our animals. Second, given that wild type TP63 enforces degradation of mutant TP63, coupled with our finding that TAp63 is a potent modifier of the features of EEC (discussed below), we hypothesize that wild type TAp63 normally keeps levels of mutant TP63 in check, thereby protecting against features of EEC syndrome in some patients, thus accounting for the variable expressivity and incomplete penetrance.
A second way of modulating Trp63 levels is by Irf6. Under normal circumstances, ΔNp63 induces Irf6 expression [Moretti et al., 2010; Thomason et al., 2010], which functions in a negative feedback loop to facilitate degradation of ΔNp63 [Moretti et al., 2010]. Importantly, the Trp63R279H mutation abrogates this feedback loop, as Irf6 cannot efficiently facilitate degradation of mutant ΔNp63 [Moretti et al., 2010]. Furthermore, human keratinocytes heterozygous for TP63R279H express compromised levels of IRF6 [Thomason et al., 2010]. This blockade in IRF6-mediated TP63 degradation could culminate in accumulation of TP63 p.Arg279His protein, thereby reaching the threshold needed to evoke EEC pathology.
The notion that there are genetic modifiers for EEC syndrome that affect the variability of the clinical features in patients has long been acknowledged in the field. However, the lack of robust genetic studies due to the small number of patient samples available has hampered identification of modifying loci. One study, which was focused on the allele encoding TP63 p.Arg280Cys, found linkage to human chromosomes 4 and 14 [Ray et al., 2004], however no candidate genes at these loci have been suggested. Since mutant TP63 proteins have been found to interact either directly or indirectly with wild type TP63 [Ying et al., 2005; Khokhar et al., 2008; Browne et al., 2011], we took the straightforward genetic approach of assaying for the ability of TAp63 to modify the penetrance of EEC-associated phenotypes by eliminating wild type TAp63 in vivo. By also removing wild type TAp63 in the corresponding null model Trp63Aam3, we were able to directly determine whether specific phenotypes were due to loss-of-function effects on Trp63, or conversely by a gain-of-function effect of the Trp63R279H protein.
The shift in penetrance for each of the three cardinal features of EEC was increased by the absence of TAp63. For the Trp63R279HN/+ model, the penetrance of cleft palate was only 38% in a wild type background, but increased to 95% in a TAp63-deficient background. For the Trp63R279H/+ model, cleft palate was almost non-existent in a wild type background, but its penetrance increased to 70% in a TAp63-deficient background. While cleft palate occurs in mice deficient for all known Trp63 isoforms [Thomason et al., 2008] clefting has not been reported in TAp63 deficient mice [Suh et al., 2006; Guo et al., 2009; Su et al., 2009]. In contrast, cleft palate was 100% penetrant in Trp63Aam3/TA− mice. This indicates that cleft palate is due to absence of wild type TAp63 in the context of haploid levels of ΔNp63, and that the cleft palate seen in our EEC models, although strongly affected by wild type TAp63 levels, is not due to a gain-of-function of Trp63R279H, but rather is due to the generation of an inactive form of Trp63. Thus, these findings indicate that the cleft palate observed in our EEC models is due to loss of Trp63 function.
Mice deficient for all Trp63 isoforms and those deficient specifically for ΔNp63 isoforms display severely altered epidermal morphology and are born lacking a mature skin barrier [Mills et al., 1999; Yang et al., 1999; Romano et al., 2012]. In contrast, Trp63R279HN/+ and Trp63R279H/+ mice are born with functional skin, and even after combined TAp63 loss, the skin barrier is intact, although an increase in epidermal and dermal thickness occurs. Skin defects are present in Trp63R279HN/TA−, Trp63R279H/TA−, and Trp63Aam3/TA− mice, again indicating that the Trp63R279H mutation generates an inactive form of Trp63, which culminates in skin defects that are due to TAp63 deficiency in combination with haploid levels of ΔNp63. Thus, as we found for cleft palate, the skin defects observed in EEC models are due to loss of Trp63 activity.
Whereas the cleft palate and skin defects described above are due to loss of wild type Trp63 function, limb defects are not detected in mice co-heterozygous for inactivating alleles of Trp63 and TAp63. The striking limb defects in Trp63R279HN/TA− mice are synthetic phenotypes not detected until the wild type TAp63 allele is lost in the context of the Trp63R279HN allele. This indicates that the limb defects observed in the EEC models are due to either a gain-of-function or a dominant-negative effect of the Trp63R279H protein, and that this effect is normally masked by wild type TAp63. Limb anomalies are rarely present in Trp63R279HN/+ mice, and have never been described in TAp63 deficient mice [Suh et al., 2006; Guo et al., 2009; Su et al., 2009]. The Trp63R279HN/TA− mice have severe limb alterations, similar to what has been described for both p63 [Mills et al., 1999; Yang et al., 1999] and ΔNp63 deficient mice [Romano et al., 2012]. However, our control Trp63Aam3/TA− mice, which lacked wild type TAp63 and have haploid levels of ΔNp63, have normal limbs. Further, Trp63R279H/TA− mice do not have limb abnormalities, again highlighting the differences seen between the two different EEC-encoding alleles. This genetic evidence provides a clear connection between R279H mutant Trp63 protein and wild type TAp63, providing an explanation for the variable expressivity characteristic of human EEC syndrome. We suggest that efforts be made to determine whether TAp63 modifies EEC-associated missense mutations in the human population.
In conclusion, we have generated an allelic series of Trp63 mutations, including an animal with a mutation that models key pathological features of human EEC syndrome. In addition, this work uncovers a previously unknown role for wild type TAp63 in affecting the penetrance and expressivity of EEC-associated phenotypes, suggesting that TAp63 is a modifier of human EEC syndrome.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Mills laboratory for helpful suggestions and critical reading of the manuscript, and the Cold Spring Harbor Laboratory Gene Targeting & Transgenic and Animal Shared Resources for assistance. This project was supported by research grant #6-FY03-69 from March of Dimes Birth Defects Foundation (A.A.M and E.L.G.), grant # RSG-06-190-01-MGO from the American Cancer Society (A.A.M.), the Swedish Research Council (E.V.L.), and the Lauri Strauss Leukemia Foundation (E.V.L.).
REFERENCES
- Barrow LL, van Bokhoven H, Daack-Hirsch S, Andersen T, van Beersum SE, Gorlin R, Murray JC. Analysis of the p63 gene in classical EEC syndrome, related syndromes, and non-syndromic orofacial clefts. J Med Genet. 2002;39:559–566. doi: 10.1136/jmg.39.8.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne G, Cipollone R, Lena AM, Serra V, Zhou H, van Bokhoven H, Dotsch V, Merico D, Mantovani R, Terrinoni A, Knight RA, Candi E, Melino G. Differential altered stability and transcriptional activity of DeltaNp63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124:2200–7. doi: 10.1242/jcs.079327. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss PW, Hughes HE, Clarke A. Twenty-four cases of the EEC syndrome: clinical presentation and management. J Med Genet. 1995;32:716–723. doi: 10.1136/jmg.32.9.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Legius E, Dereymaeker AM, Van den Berghe H. EEC syndrome without ectrodactyly: report of two new families. J Med Genet. 1990;27:165–168. doi: 10.1136/jmg.27.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124:357–368. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhar SK, Kommagani R, Kadakia MP. Differential effects of p63 mutants on transactivation of p53 and/or p63 responsive genes. Cell Res. 2008;18:1061–1073. doi: 10.1038/cr.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Lowe SW. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Nat Acad Sci USA. 2001;98:5025–5030. doi: 10.1073/pnas.091100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32:138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, Ballaro C, Alema S, Chimenti S, Guerrini L, Costanzo A. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray AK, Marazita ML, Pathak R, Beever CL, Cooper ME, Goldstein T, Shaw DF, Field LL. TP63 mutation and clefting modifier genes in an EEC syndrome family. Clin Genet. 2004;66:217–222. doi: 10.1111/j.1399-0004.2004.00287.x. [DOI] [PubMed] [Google Scholar]
- Rinne T, Spadoni E, Kjaer KW, Danesino C, Larizza D, Kock M, Huoponen K, Savontaus ML, Aaltonen M, Duijf P, Brunner HG, Penttinen M, van Bokhoven H. Delineation of the ADULT syndrome phenotype due to arginine 298 mutations of the p63 gene. Eur J Hum Genet: EJHG. 2006;14:904–910. doi: 10.1038/sj.ejhg.5201640. [DOI] [PubMed] [Google Scholar]
- Roelfsema NM, Cobben JM. The EEC syndrome: a literature study. Clin Dysmorphol. 1996;5:115–127. doi: 10.1097/00019605-199604000-00003. [DOI] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, Sinha S. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Dixon MJ, Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 2010;120:1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, Mckeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Ying H, Chang DL, Zheng H, McKeon F, Xiao ZX. DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol Cell Biol. 2005;25:6154–6164. doi: 10.1128/MCB.25.14.6154-6164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.