Pérez et al. address the mechanisms whereby duplicated transcription regulators differentiate from one another over evolutionary timescales. Combined large-scale biochemical measurements and genome-wide molecular biology methods show that four transcription regulators in the human commensal and pathogenic yeast Candida albicans that arose by successive duplications have differentiated from one another by acquiring different DNA-binding specificities, preferences for half-site spacing, and cofactor associations. The results illustrate how successive duplications and diversification of an ancestral transcription regulator can underlie major changes in an organism’s regulatory circuitry.
Keywords: transcription regulator, molecular evolution, Candida albicans, gene regulation, gene duplication, regulatory networks
Abstract
The duplication of transcription regulators can elicit major regulatory network rearrangements over evolutionary timescales. However, few examples of duplications resulting in gene network expansions are understood in molecular detail. Here we show that four Candida albicans transcription regulators that arose by successive duplications have differentiated from one another by acquiring different intrinsic DNA-binding specificities, different preferences for half-site spacing, and different associations with cofactors. The combination of these three mechanisms resulted in each of the four regulators controlling a distinct set of target genes, which likely contributed to the adaption of this fungus to its human host. Our results illustrate how successive duplications and diversification of an ancestral transcription regulator can underlie major changes in an organism’s regulatory circuitry.
Gene duplication is a major contributor to the emergence of new genetic functions in all three domains of life (Ohno 1970; Conant and Wolfe 2008). For this process to generate new gene functions, the two resulting duplicates, which start as identical copies right after duplication, must differentiate from each other. For instance, immediately after duplication of a transcription regulator (i.e., a sequence-specific DNA-binding protein), the two resulting copies will have identical DNA-binding profiles; hence, the two will control a common set of target genes. Population genetics predicts that this redundant state will be evolutionarily short-lived (Lynch 2007). Thus, if both copies are to be retained over evolutionary timescales, changes in one or both genes need to occur. Duplicated transcription regulator genes could undergo changes that modify the activities of the encoded proteins and/or their regulation.
To deduce the mechanisms whereby duplicated transcription regulators acquire new target gene repertoires and DNA-binding specificities, we examined a group of four closely related transcription regulators that arose by successive duplications in the lineage leading to the human commensal and pathogenic yeast Candida albicans. Using a combination of in vivo genome-wide molecular biology approaches and large-scale in vitro biochemical measurements, we show that these regulators have differentiated from one another by a combination of three mechanisms: (1) small changes in the intrinsic, monomer DNA-binding specificities; (2) different preferences for half-site arrangements; and (3) association with cofactors. It is the summation of these three mechanisms that confer on the proteins the specificity to regulate independent sets of target genes and control different aspects in the biology of the fungus. These changes in the duplicated transcription regulators led to large expansions in transcription circuitry and evolutionary novelty as they contributed to the ability of C. albicans to survive as part of the human microbiota.
Results and Discussion
Successive duplications of a LYS regulator homolog in the Candida lineage
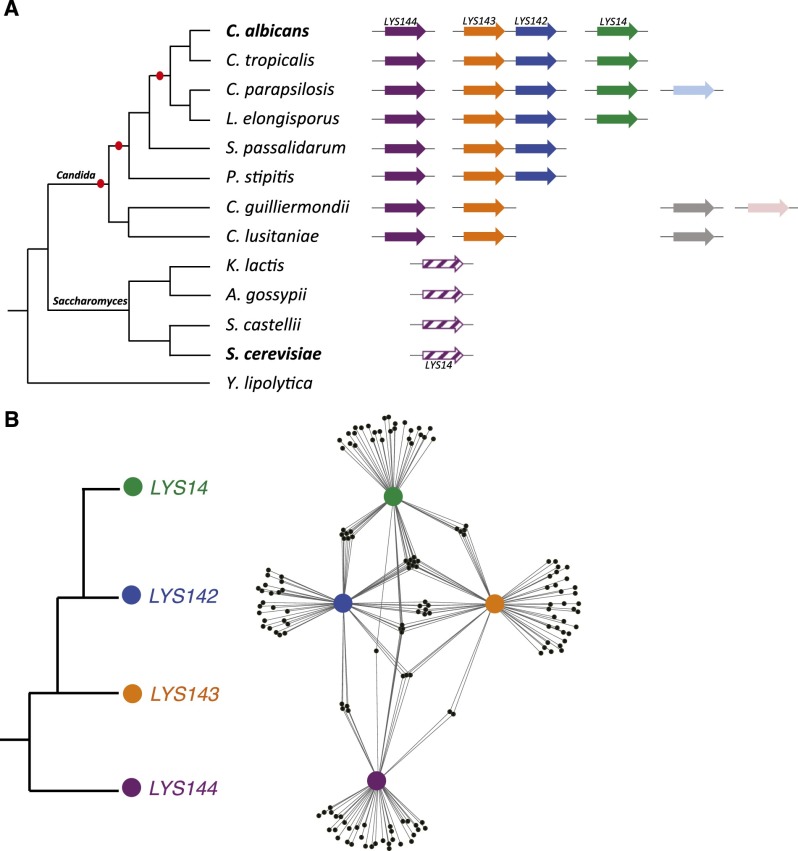
The C. albicans genome contains four homologs of LYS14, the key transcriptional regulator of lysine biosynthesis genes in the free-living, model yeast Saccharomyces cerevisiae (Ramos et al. 1988; Feller et al. 1994). When LYS14 is deleted, S. cerevisiae can no longer up-regulate the genes coding for the lysine biosynthetic enzymes, and, as a result, the mutant strain grows poorly on media lacking lysine (Ramos et al. 1988). Phylogenetic reconstructions (Supplemental Fig. 1) indicate that the ancestor of the LYS14 regulator underwent several successive duplications in the Candida clade lineage, which includes numerous human commensal and pathogenic species (Fig. 1A). Indeed, extant species in the Candida clade contain two to five homologs of LYS14. In contrast, there appears to be a single copy, that of LYS14 and its orthologs, in all of the extant species that belong to the Saccharomyces clade (Fig. 1A). The Candida and Saccharomyces clades last shared a common ancestor ∼300 million years ago (Taylor and Berbee 2006).
Figure 1.
Recently duplicated C. albicans LYS transcription regulators bind to largely nonoverlapping sets of target genes. (A) Cladogram depicting the phylogenetic relationships among extant species of the Candida and Saccharomyces clades. The arrows to the right of the tree represent the homologs of S. cerevisiae LYS14 found in each species’ genome. Gene orthology assignments (represented by arrows of the same color) are based on synteny and the reconstructed phylogeny of the gene family (Supplemental Fig. 1). Red ovals in the branches of the cladogram represent the inferred single-gene duplication events that gave rise to the four C. albicans homologs. No “strict orthology” can be inferred between a particular Candida LYS gene and Saccharomyces LYS14 based on phylogenetic reconstructions (Supplemental Fig. 1) or synteny (therefore, the annotation of one of the Candida genes as LYS14 is misleading in this respect). The similarity in color between LYS144 and Saccharomyces LYS14 depicts simply the fact that the DNA sequences recognized by these homologous proteins most closely resemble one another. (B) Inferred relationships among the four LYS regulators (to the left) in C. albicans and the gene network (to the right) formed by the four regulators (purple, orange, blue, and green circles) and their target genes (small black circles) as mapped by ChIP. The distances separating the four Lys proteins are inversely proportional to the number of shared target genes (fewer shared targets, greater separation). Although some target genes are bound by more than one regulator, most of the targets are specific to only one of the four LYS regulators.
C. albicans LYS regulators control largely nonoverlapping sets of target genes
A priori, there are two plausible explanations as to why C. albicans has four homologs of LYS14 while S. cerevisiae has a single gene: Either (1) the duplicated genes underwent “subfunctionalization” whereby each C. albicans gene now regulates a subset of lysine metabolic genes and the combined action of all four is equivalent to the function of the single ancestral gene or (2) at least one of the duplicated genes in C. albicans acquired new functions and no longer regulates the lysine biosynthetic enzymes. To distinguish between these two possibilities, we determined the set of genes regulated by each of the four LYS14 homologs in C. albicans by full-genome chromatin immunoprecipitation (ChIP) followed by array hybridization (ChIP–chip). Because some of the C. albicans LYS14 homologs are expressed only in a mammalian host (Perez et al. 2013), we ectopically expressed each regulator by placing its coding region under the control of the strong promoter TDH3 (Nobile et al. 2008; Perez et al. 2013). Using this strategy, we determined that each of the four transcription regulators binds to ∼50 target regions (Supplemental Table 1), with only partial overlap among them (Fig. 1B). None of the ChIP peaks was located in the intergenic region upstream of any lysine biosynthesis gene, consistent with our previous finding that none of the four C. albicans homologs of LYS14 is required for growth in the absence of lysine (Homann et al. 2009). Although none of them regulates lysine biosynthesis, we nevertheless retain the names originally assigned to them in the C. albicans genome annotation (Braun et al. 2005): LYS144, LYS143, LYS142, and LYS14. This nomenclature thus reflects the shared ancestry with the S. cerevisiae LYS14 gene rather than the functions in C. albicans.
Candida LYS regulators show different intrinsic DNA-binding specificities
Despite their recent expansion, the ChIP results show that the four C. albicans LYS regulators bind to largely independent sets of target genes, raising the question of how this differentiation in target gene recognition occured. We considered three nonexclusive models: (1) The proteins have different intrinsic (i.e., monomer) DNA-binding specificities. (2) The proteins function as dimers and have different preferences for the arrangement of their half-sites. (3) The proteins have acquired different cofactor interactions.
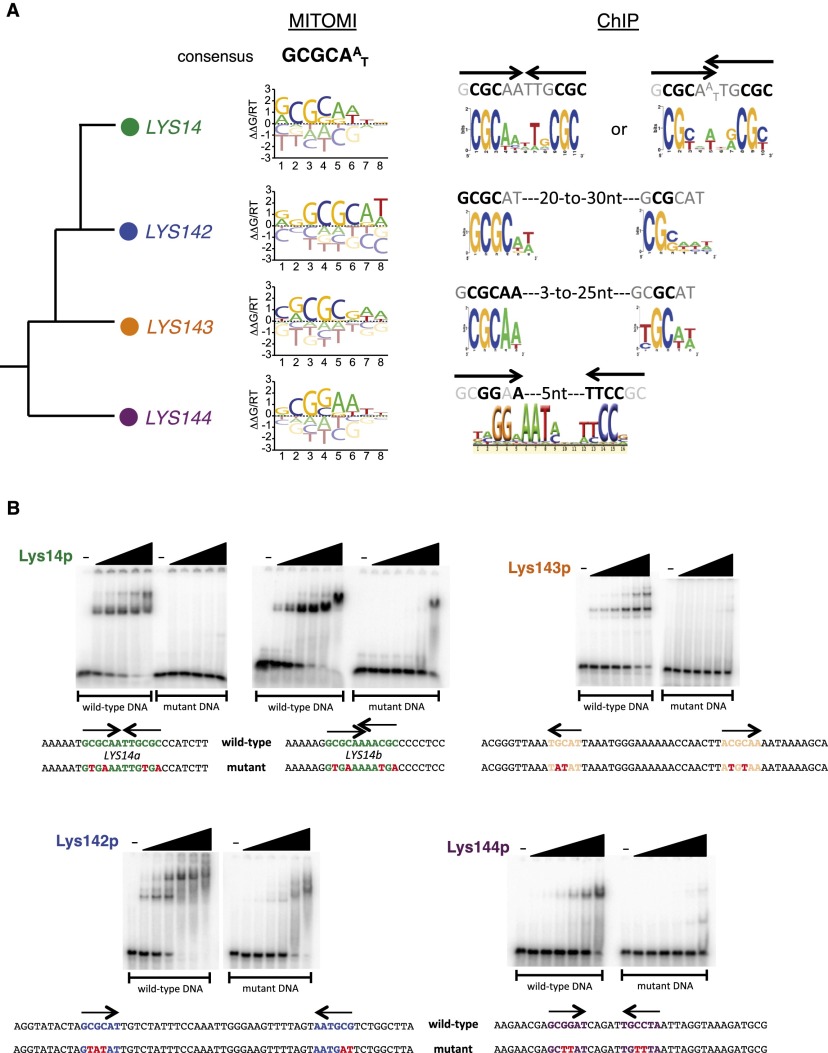
To address the first possibility, we used a microfluidics-based approach (MITOMI 2.0) (Fordyce et al. 2010, 2012) that measured the DNA sequence preferences of each of the four LYS regulators in vitro. The proteins belong to the zinc cluster class of regulators (for review, see MacPherson et al. 2006), which typically consist of three domains—DNA-binding, dimerization, and a large “activation” domain—all connected by linker regions of variable size (Supplemental Fig. 2A). The core 35-amino-acid DNA-binding domain (based on crystal structures of other members of the family) (e.g., see Fitzgerald et al. 2006) shows 60% amino acid identity across the four Lys proteins (Supplemental Fig. 2B) and includes six cysteine residues, all of them conserved, which are predicted to interact with two zinc atoms (MacPherson et al. 2006). For MITOMI experiments, we used truncated versions of the Lys proteins that included either the DNA-binding domain alone (LYS143) or the DNA-binding domain in conjunction with the putative dimerization domains (LYS14, LYS142, and LYS144); full-length proteins were not efficiently transcribed/translated in the assay. By measuring binding affinities for a library containing all possible DNA 8-mers with each of the four proteins, the MITOMI experiments revealed that each Lys protein binds to a set of DNA sequences that were similar, but not identical, to one another (Supplemental Fig. 3; Supplementary Data Set 1). For each protein, we then analyzed this pattern of oligonucleotide binding to determine a consensus binding specificity, represented here as a position-specific affinity matrix (PSAM) (Fig. 2A). The consensus sequence that we identified for each protein, GCGCAAT, represents the monomer-binding sequence and resembles the monomer-binding sequences described for other regulators of the zinc cluster class (MacPherson et al. 2006). While broadly similar, these PSAMs also revealed small but significant differences in binding preferences. The MITOMI-derived motifs were significantly overrepresented in the regions bound by the Lys proteins in vivo (ChIP peaks compared with a set of random intergenic sequences) (Supplemental Fig. 4); however, differences between these MITOMI motifs per se were insufficient to explain why each of the regulators bound in vivo to a unique set of target genes (Supplemental Fig. 4). We note that Lys144p shows the most distinct MITOMI-derived motif compared with the other three (Fig. 2A; Supplemental Figs. 3, 4); this motif most closely resembles the DNA sequence bound by S. cerevisiae Lys14p (Becker et al. 1998). These results indicate that there must be determinants in addition to the monomer-binding specificity that contribute to the distinct in vivo DNA-binding profiles of each Lys protein.
Figure 2.
DNA motifs preferred by C. albicans LYS transcription regulators. (A) DNA motifs derived from MITOMI and ChIP data sets. (B) Gel shift assays showing binding of the Lys proteins to their predicted binding sites. 32P-labeled DNA fragments (∼0.4 nM) containing the predicted wild-type or mutant LYS-binding sites were incubated with increasing concentrations of purified Lys protein (0, 0.039, 0.156, 0.625, 2.5, 10, and 40 nM) for 30 min at room temperature in standard EMSA buffer and resolved in 6% polyacrylamide gels run with 0.5× TGE. Point mutations introduced in the binding sites are indicated in red.
Candida LYS regulators bind to different arrangements of DNA-binding sites
To address the second model (i.e., the potential contribution of different arrangements of DNA monomer-binding sites to the overall affinity of the proteins), we more closely analyzed the DNA sequences enriched in each ChIP data set. The DNA sequences occupied in vivo by Lys14p and Lys144p were clearly composed of two repeats arranged in specific configurations that for Lys14p consisted of inverted repeats located directly adjacent to each other or overlapping by 1 nucleotide (nt) (Fig. 2A). In contrast, the sequences occupied by Lys144p were comprised of inverted repeats separated by 5 nt (Fig. 2A). We refer to each repeat as a half-site, which is the sequence bound by a monomer of the protein. The half-site motifs independently derived from the ChIP data closely resembled the corresponding MITOMI-generated motifs. Additional DNA-binding experiments using gel mobility shift assays confirmed that the regulators preferentially bound to the predicted arrangement of DNA sequences; the introduction of point mutations in key positions of the DNA motifs abolished this binding (Fig. 2B). In contrast to the specific configurations of the Lys14p and Lys144p half-sites, the sequences occupied in vivo by Lys142p and Lys143p indicated that these proteins bind to flexible arrangements between a “strong” and a “weak” half-site separated by 20–30 nt for Lys142p and 3–25 nt for Lys143p (Fig. 2A). The motifs representing the “strong” sites are in close agreement with the corresponding MITOMI-derived motif.
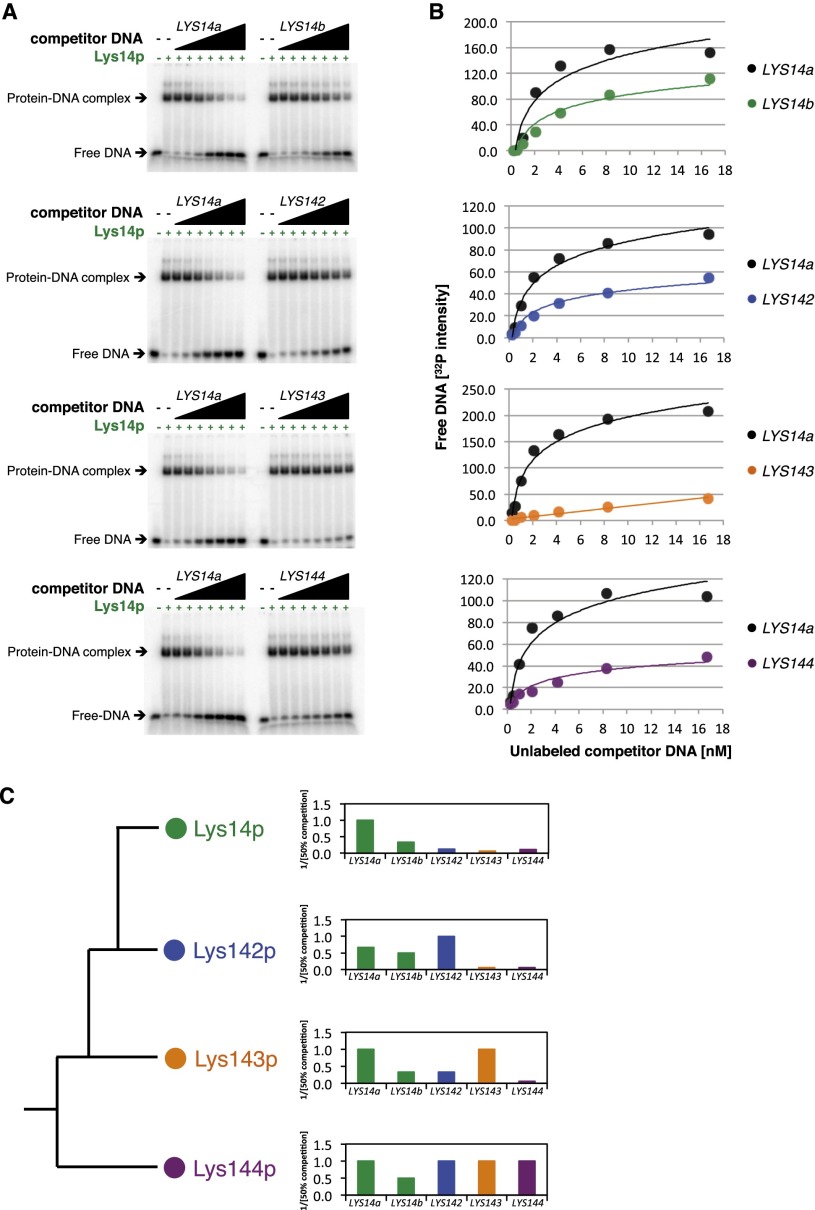
To experimentally determine whether the Lys proteins are able to discriminate among their different half-site arrangements by themselves, we carried out competition binding assays in which we incubated each one of the four proteins with its “preferred” arrangement of DNA-binding sequences. Upon binding, we competed each reaction with unlabeled DNA fragments containing the other regulator’s DNA-binding sites. In general, Lys14p, Lys142p, and Lys143p showed strong preferences for their specific arrangements, as described above (twofold to 20-fold higher affinity) (Fig. 3; Supplemental Figs. 5, 6). In contrast, Lys144p showed no strong preference for its “own” site arrangement compared with the others (Fig. 3C; Supplemental Fig. 7), suggesting that additional factors must contribute to the binding profile of Lys144p in vivo.
Figure 3.
DNA-binding preferences among the four C. albicans Lys proteins. (A) Determination of the DNA-binding preferences of Lys14p through competition experiments. The purified C. albicans Lys14 protein was incubated with a radiolabeled DNA fragment containing its preferred binding site (LYS14a; i.e., the sequence consisting of repeats adjacent to each other but not overlapping). Increasing amounts of unlabeled competitor DNA fragments containing LYS14a-, LYS14b- (sequence with the repeats overlapping by 1 nt), LYS142-, LYS143-, or LYS144-binding sites were added to the reactions, and the mixtures were resolved by polyacrylamide gel electrophoresis. (B) Quantification of the assays shown in A and best-fit curves. (C) Summary of the binding preferences of the four proteins. (Top panel) The Lys14p-binding preferences are a summary of the data displayed in A and B. The bars show the inverse of the concentration of competitor DNA needed to achieve a 50% reduction in binding to the radiolabeled sequence, with the concentration of the cognate fragment set to a value of 1. Therefore, values <1 correspond to weaker binding. The middle and bottom panels summarize the corresponding experiments for Lys142p, Lys143p, and Lys144p; the images, quantification, and best-fit curves for these three proteins are shown in Supplemental Figures 5–7.
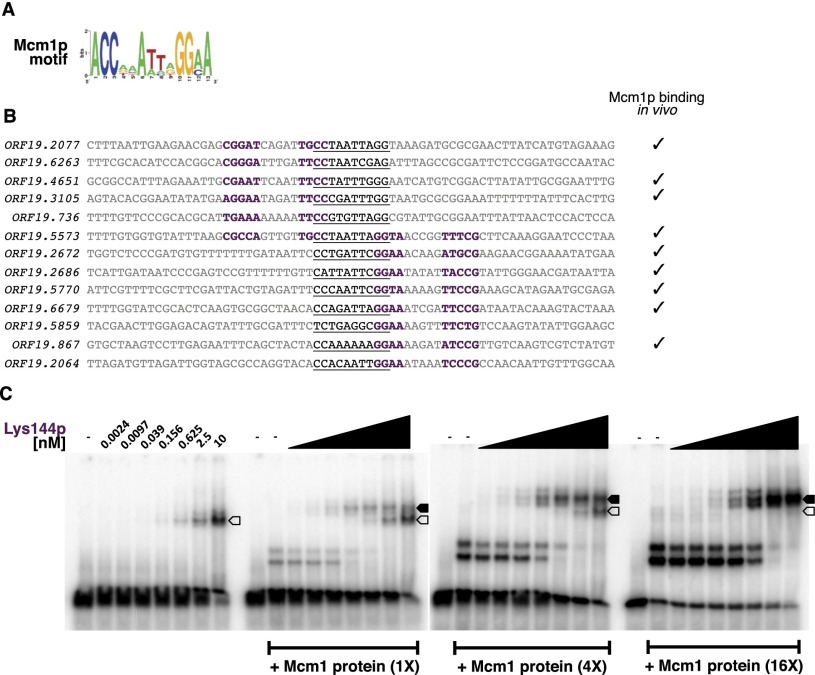
Lys144p binds to DNA cooperatively with Mcm1p
As described above, a third model that could account for the discrimination of Lys144p in binding in vivo is the interaction with one or more cofactors. To test this possibility, we searched the Lys144p ChIP data set for additional overrepresented sequences. This analysis revealed a DNA sequence (Fig. 4A) that was reminiscent of the DNA motif recognized by Mcm1p in C. albicans (Tuch et al. 2008; Askew et al. 2011). A significant proportion of these sites is occupied by Mcm1p in vivo (P = 3.4 × 10−6) (Fig. 4B; Tuch et al. 2008). Moreover, purified Mcm1 protein binds to one of these sites in vitro (Supplemental Fig. 8). The Lys144p and Mcm1p sites are arranged in a strict configuration (Fig. 4B), suggesting that binding of one protein may facilitate or, conversely, interfere with the binding of the other. We tested this possibility and found that Lys144 protein preferentially binds to its DNA sites when the Mcm1 protein is present (Fig. 4C); thus, the two proteins bind cooperatively. The binding of Mcm1p to promoters in combination with a second regulator is a common mechanism to regulate transcription of specific sets of genes in S. cerevisiae (for example, see Carr et al. 2004) and C. albicans (Tuch et al. 2008; Askew et al. 2011). Although Lys14p in S. cerevisiae does not appear to cooperate with Mcm1p, Lys144p in C. albicans clearly has this ability.
Figure 4.
Mcm1p promotes Lys144p binding to DNA. (A) Putative Mcm1p motif identified in the set of DNA sequences occupied in vivo by Lys144p. (B) Distribution of Lys144p and Mcm1p DNA-binding sites in a subset of the sequences occupied by Lys144p. Putative Lys144p sites are shown in purple (half-sites), whereas the predicted Mcm1p sites are underlined. Check marks to the right indicate whether Mcm1p has been found to bind in vivo to the respective target gene (Tuch et al. 2008). (C) Gel shift assays carried out with one of the sequences shown in B (ORF19.2077) and purified Lys144 and Mcm1 proteins. The open arrow corresponds to the DNA+Lys144p-bound complex, whereas the solid arrow indicates the location of the DNA+Lys144+Mcm1 tripartite complex.
LYS transcription regulators contribute to Candida’s colonization of mammalian hosts
We described here how the four LYS transcription regulators have diversified with respect to their DNA-binding specificities and cofactor interactions and therefore target gene selection. What, however, is the overall biological function of each protein? Our ChIP data indicated that Lys143p binds upstream of WOR1 (Zordan et al. 2006) and AHR1 (Askew et al. 2011; Wang et al. 2011) (Supplemental Fig. 9), two key regulators of white–opaque switching in C. albicans, a phenomenon whereby two distinct, heritable cell types (white and opaque) arise from the same genome (for a review, see Soll 2004; Lohse and Johnson 2009). White and opaque cells interact differently with the host immune system, prefer different host niches, and may differ in their suitability for commensal or invasive growth (Pande et al. 2013). The ectopic expression of LYS143 also resulted in increased transcript levels of WOR3 (Supplemental Fig. 10), another DNA-binding protein that can affect this phenotypic switch (Lohse et al. 2013). These observations raised the possibility that LYS143 may affect white–opaque switching as well. Consistent with this hypothesis, a C. albicans lys143 mutant strain showed an approximately fivefold increase in the frequency of white-to-opaque switching (Supplemental Fig. 9). LYS143 clearly regulates genes in addition to WOR1, but it remains to be seen whether and how these other genes are related to white–opaque switching.
We recently reported that two of the LYS regulators, LYS14 and LYS144, are required for C. albicans to proliferate in its mammalian host, with each regulator being required for a separate niche (Perez et al. 2013): While the lys144 mutant displays impaired ability to colonize the mammalian gut, the lys14 mutant colonizes the intestine at wild-type levels. The converse is true when we measured their fitness after bloodstream infections; that is, only the lys14 mutant displays a phenotype in this niche. Further evidence of nonredundancy among the four C. albicans regulators is the finding that the lys143 mutant is the only one of the four regulator mutants affecting white–opaque switching (Supplemental Fig. 9). Likewise, the ectopic expression of different LYS regulators in C. albicans results in distinct genome-wide transcriptional profiles (Supplemental Fig. 10). Taken together, these findings demonstrate that each of the duplicated LYS regulators has taken a largely distinct role in C. albicans: The phenotypes resulting from their deletion are different, and their targets of regulation are, for the most part, nonoverlapping.
Conclusions
Our results indicate that the four copies that resulted from successive duplications of the LYS ancestor in the lineage giving rise to C. albicans have differentiated from one another by a combination of three mechanisms: (1) small changes in the intrinsic, monomer DNA-binding specificities (i.e., variations in the core GCGCAAT motif); (2) different preferences for half-site arrangements (direct vs. inverted repeats; preferred distances between repeats); and (3) association with cofactors (e.g., Lys144p with Mcm1p). These three differences together give each C. albicans Lys protein the specificity to regulate a separate set of target genes and thereby control different aspects in the biology of the fungus. Although we cannot rule out other changes (e.g., differences in the potency of the activation domains or other cofactor interactions), we believe that the changes that we documented are sufficient to explain how each duplicated regulator has acquired a distinct group of target genes.
Our findings illustrate the prominent role that changes in the binding specificity of transcription regulators can play in the rewiring of gene regulatory networks. While the mechanisms uncovered here have been invoked to account for observed differences in DNA-binding preferences among members of monophyletic groups of transcription regulators (Ptashne 2004; Jolma et al. 2013; Nakagawa et al. 2013), our results demonstrate (largely through direct biochemical experiments) that it is the interplay of all three mechanisms—not any single mechanism on its own—that is responsible for the diversification of the LYS regulators. This diversification took place during the evolution of the C. albicans lineage and is closely linked to the ability of this clade to proliferate in mammalian hosts. Since a significant proportion of transcription regulators found in eukaryotic genomes arose through gene duplications (Weirauch and Hughes 2011), we propose that similar combinations of biochemical changes underlie the diversification of duplicated transcription regulators, the consequent expansion of transcription networks, and the generation of novel phenotypes.
Materials and methods
C. albicans strains
All strains used in this study are derivatives of the clinical isolate SC5314 (Noble and Johnson 2005) and are listed in Supplemental Table 2. Gene tagging and construction of TDH3 promoter-driven overexpression strains followed standard fusion PCR strategies (Nobile et al. 2008; Hernday et al. 2010). Oligos used in the study are listed in Supplemental Table 3.
Phylogenetic reconstructions
Three maximum likelihood phylogenies were generated by aligning (1) full-length amino acid sequences, (2) activation domain sequences, and (3) DNA-binding domain sequences using the software MUSCLE (Edgar 2004). Multiple phylogenetic models were then tested using RAxML (Stamatakis 2006) in order to find the best-fitting phylogenetic model according to the Akaike Information Criteria (Akaike 1973). The best-fitting models were (1) PROTGAMMAJTT, (2) PROTGAMMALG, and (3) PROTCATLG, respectively. Approximate likelihood ratio (aLR) branch supports were computed using PhyML (Anisimova et al. 2011) by first computing the aLR test statistics and then manually converting the statistic to aLRs using the following relationship: aLR = exp(aLRT/2.0). The Candida Gene Order Browser (Maguire et al. 2013) was used to assess synteny.
Full-genome ChIP
Each transcription regulator was tagged with a GFP tag at the N-terminal end of the protein in a wild-type reference strain background. The tagged strains along with untagged controls were grown to mid-logarithmic phase in YPD at 30°C, and ChIP was carried out as described (Hernday et al. 2010) with the following modifications: GFP-tagged regulators were immunoprecipitated with an anti-rGFP polyclonal antibody (Clontech); the DNA recovered after cross-link reversal was purified with QIAquick PCR purification columns (Qiagen) and amplified using the GenomePlex complete whole-genome amplification kit (Sigma). Input and immunoprecipitated DNA were fluorescently labeled and competitively hybridized to custom full-genome oligonucleotide tiling microarrays (Agilent) as previously described (Perez et al. 2013). MochiView (Homann and Johnson 2010) was used for data visualization, identification of binding events, and DNA motif analysis.
ChIP–chip data analysis
The microarray data were normalized using the global lowess method. A full description of the data analysis can be found in the Supplemental Material.
ChIP DNA motif analysis
Sequences of 500 nt centered on the midpoint of ∼15–20 of the top-scoring peaks for each regulator were used to derive motifs in Scope (Chakravarty et al. 2007) or MochiView. The software’s default parameters were used.
MITOMI 2.0
Experiments for transcription regulator target site discovery were performed largely as published previously (Fordyce et al. 2010, 2012) and are described in the Supplemental Material.
Protein purification
The N-terminal portions of the C. albicans Lys144, Lys143, Lys142, and Lys14 proteins (amino acids 1–279, 1–142, 1–280, and 1–236, respectively) were N-terminally fused to 6His and to the maltose-binding protein, expressed in Escherichia coli as described (Fitzgerald et al. 2006), and purified with Ni-NTA columns (Qiagen). Protein concentrations were estimated in Coomassie blue-stained gels using known concentrations of bovine serum albumin as standards.
Gel mobility shift assays
EMSAs were carried out as described previously (Cain et al. 2012).
White-to-opaque switching assay
Plate-based, quantitative white–opaque switching assays were performed as described (Miller and Johnson 2002) with the following modifications. Either three (in case of the mutant deletion strains) or five (reference strain) biological replicates were performed for each strain. Each biological replicate used cells taken from three to five independent colonies.
Accession numbers
The ChIP–chip data reported in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE52203.
Acknowledgments
We thank Jiashun Zheng (Hao Li’s laboratory) for providing some of the DNA templates used in MITOMI. J.C.P. was supported by a Post-doctoral Fellowship from the American Heart Association and a Post-doctoral Research Award from the University of California at San Francisco’s Program for Breakthrough Biomedical Research (PBBR). P.M.F. was supported by a K99 Pathway to Independence award from the US National Institute of General Medical Sciences (5K99GM099848). This work was supported by US National Institute of Allergy and Infectious Diseases grant RO1 AI049187 (to A.D.J.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.242271.114.
References
- Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. In Proceedings of the second international symposium on information theory (ed. Petrov BN, Caski F), pp. 267–281. Akademiai Kiado, Budapest. [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60: 685–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A, Nantel A, Whiteway M 2011. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol 79: 940–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Feller A, el Alami M, Dubois E, Pierard A 1998. A nonameric core sequence is required upstream of the LYS genes of Saccharomyces cerevisiae for Lys14p-mediated activation and apparent repression by lysine. Mol Microbiol 29: 151–163 [DOI] [PubMed] [Google Scholar]
- Braun BR, van Het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, et al. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet 1: 36–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD 2012. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190: 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr EA, Mead J, Vershon AK 2004. α1-induced DNA bending is required for transcriptional activation by the Mcm1-α1 complex. Nucleic Acids Res 32: 2298–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty A, Carlson JM, Khetani RS, DeZiel CE, Gross RH 2007. SPACER: identification of cis-regulatory elements with non-contiguous critical residues. Bioinformatics 23: 1029–1031 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Dubois E, Ramos F, Pierard A 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol Cell Biol 14: 6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MX, Rojas JR, Kim JM, Kohlhaw GB, Marmorstein R 2006. Structure of a Leu3-DNA complex: recognition of everted CGG half-sites by a Zn2Cys6 binuclear cluster protein. Structure 14: 725–735 [DOI] [PubMed] [Google Scholar]
- Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR 2010. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol 28: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce PM, Pincus D, Kimmig P, Nelson CS, El-Samad H, Walter P, DeRisi JL 2012. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc Natl Acad Sci 109: E3084–E3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday AD, Noble SM, Mitrovich QM, Johnson AD 2010. Genetics and molecular biology in Candida albicans. Methods Enzymol 470: 737–758 [DOI] [PubMed] [Google Scholar]
- Homann OR, Johnson AD 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152: 327–339 [DOI] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD 2009. White-opaque switching in Candida albicans. Curr Opin Microbiol 12: 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Hernday AD, Fordyce PM, Noiman L, Sorrells TR, Hanson-Smith V, Nobile CJ, DeRisi JL, Johnson AD 2013. Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc Natl Acad Sci 110: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sinauer Associates, Sunderland, MA. [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70: 583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SL, OhEigeartaigh SS, Byrne KP, Schroder MS, O’Gaora P, Wolfe KH, Butler G 2013. Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol 30: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302 [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML 2013. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci 110: 12349–12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol 10: 2180–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, New York, NY. [Google Scholar]
- Pande K, Chen C, Noble SM 2013. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet 45: 1088–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, Kumamoto CA, Johnson AD 2013. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11: e1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. 2004. A genetic switch, third edition, phage λ revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ramos F, Dubois E, Pierard A 1988. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur J Biochem 171: 171–176 [DOI] [PubMed] [Google Scholar]
- Soll DR 2004. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. BioEssays 26: 10–20 [DOI] [PubMed] [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML 2006. Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98: 838–849 [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol 6: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song W, Huang G, Zhou Z, Ding Y, Chen J 2011. Candida albicans Zcf37, a zinc finger protein, is required for stabilization of the white state. FEBS Lett 585: 797–802 [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR 2011. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell Biochem 52: 25–73 [DOI] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci 103: 12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]