Many questions remain about the relationship between chromatin loop formation and transcription. In erythroid cells, LDB1 is required for looping of the β-globin locus control region (LCR) to the active β-globin promoter. Dean and colleagues show that the LDB1 dimerization domain (DD) is necessary to restore LCR-promoter looping and transcription in LDB1-depleted cells. Deletion analysis reveals a conserved region of the LDB1 DD dispensable for dimerization and chromatin looping but necessary for transcription activation. The results thus uncouple enhancer–promoter looping from transcription at the β-globin locus.
Keywords: β-globin genes, chromatin, looping, enhancers, transcription
Abstract
Many questions remain about how close association of genes and distant enhancers occurs and how this is linked to transcription activation. In erythroid cells, lim domain binding 1 (LDB1) protein is recruited to the β-globin locus via LMO2 and is required for looping of the β-globin locus control region (LCR) to the active β-globin promoter. We show that the LDB1 dimerization domain (DD) is necessary and, when fused to LMO2, sufficient to completely restore LCR–promoter looping and transcription in LDB1-depleted cells. The looping function of the DD is unique and irreplaceable by heterologous DDs. Dissection of the DD revealed distinct functional properties of conserved subdomains. Notably, a conserved helical region (DD4/5) is dispensable for LDB1 dimerization and chromatin looping but essential for transcriptional activation. DD4/5 is required for the recruitment of the coregulators FOG1 and the nucleosome remodeling and deacetylating (NuRD) complex. Lack of DD4/5 alters histone acetylation and RNA polymerase II recruitment and results in failure of the locus to migrate to the nuclear interior, as normally occurs during erythroid maturation. These results uncouple enhancer–promoter looping from nuclear migration and transcription activation and reveal new roles for LDB1 in these processes.
Transcription of numerous genes regulated by distant enhancers depends on these elements establishing proximity through chromatin looping (Dekker 2008; Splinter and de Laat 2011). Indeed, whole-genome studies now suggest that this is a central mechanism to establish a cell type-specific transcriptome during development and differentiation. The mechanisms underlying the precise but dynamic apposition of enhancers and target genes are unclear. In one model, transcription factors bound to specific enhancers and required for their function contribute to chromatin looping through protein–protein interactions, and when they are reduced using RNAi, the looping and transcription diminish (Kadauke and Blobel 2009; Krivega and Dean 2012). In other instances, architectural factors such as CTCF and cohesin can be co-opted for cell type-specific enhancer looping by collaboration with cell-specific factors (Hadjur et al. 2009; Sekimata et al. 2009; Stadhouders et al. 2012). A third model involves transcription directly. For example, cohesin and the Mediator complex interact directly to loop enhancers to promoters (Kagey et al. 2010). It has also been suggested that RNA polymerase II (Pol II) density or transcription per se (Bulger and Groudine 2011) might lead to the observed enhancer/gene loops.
In the β-globin locus, reduction of the erythroid factors GATA1, FOG1, and EKLF (KLF1) or the more widely expressed lim domain binding 1 (LDB1) protein showed that they are required for β-globin activation and for looping between the gene and the β-globin locus control region (LCR) enhancer (Drissen et al. 2004; Vakoc et al. 2005; Song et al. 2007). In addition, reduction of LDB1 abrogates locus migration to the nuclear interior, where high levels of transcription are achieved in RNA Pol II transcription factories (Osborne et al. 2004). LDB1 is a non-DNA-binding protein with a conserved, 200-amino-acid N-terminal domain through which it can dimerize or multimerize in vitro (Breen et al. 1998; Jurata et al. 1998; Xu et al. 2003; Cross et al. 2010). The dimerization domain (DD) is required for rescue of LDB1 functions in the developing nervous system of flies and vertebrates in vivo (van Meyel et al. 1999; Thaler et al. 2002). The LDB1 dimerization domain alone, if tethered to the adult β-globin promoter by a Zn finger peptide, is able to force an LCR loop and partially rescue transcription (Deng et al. 2012).
In erythroid cells, the C-terminal LIM-interacting domain (LID) of LDB1 interacts with LMO2, which in turn provides association of LDB1 with chromatin through DNA-binding partners GATA1 and TAL1 (Wadman et al. 1997; Xu et al. 2003; Song et al. 2007). The LDB1 complex binds to a bipartite E-box/GATA motif that is common in regulatory regions of erythroid genes, including in the β-globin locus (Wadman et al. 1997; Soler et al. 2010). Genome-wide localization supports the idea that most shared TAL1 and GATA1 regulatory functions in mouse erythroid cells are carried out in concert with LDB1 (Fujiwara et al. 2009; Tripic et al. 2009; Yu et al. 2009; Soler et al. 2010; Li et al. 2013). Although GATA1 regulatory functions often require the cofactor and binding partner FOG1 (ZFPM1) and TAL1 and FOG1 have similar distribution patterns at select active erythroid genes, genome-wide studies have yet to establish whether and how frequently FOG1 co-occupies LDB1 complex sites (Tsang et al. 1997; Pal et al. 2004; Tripic et al. 2009).
How LDB1 dimerization participates in chromatin looping and transcription activation has not been explored. In this study, mutated or fused versions of LDB1 were expressed in the background of LDB1-depleted erythroid cells (Song et al. 2007), and their ability to rescue β-globin/LCR proximity and β-globin expression was investigated. Deletion of the LDB1 DD abrogated β-globin/LCR looping. Fusion of the DD to LMO2, but not to GATA1, was sufficient to completely rescue β-globin transcription and LCR looping. The LDB1 DD per se was required, as a heterologous DD fused to LMO2 failed to rescue. Deletion analysis revealed a small conserved region of the LDB1 DD that is dispensable for dimerization and chromatin looping but necessary for transcription activation, separating these processes. LDB1 interacts with FOG1 through this discrete region. Thus, LDB1 functions as a looping protein and has novel functions as a transcription coactivator.
Results
The LDB1 DD is necessary and sufficient, when fused to LMO2, to rescue β-globin gene activation and long-range enhancer looping in LDB1-depleted murine erythroleukemia (MEL) cells
LDB1 is required for erythroid differentiation, and therefore erythroid progenitor and embryonic stem cell-based systems cannot be used to address mechanistic questions about its function specifically in β-globin transcription activation and chromatin looping, which are late events. To circumvent this problem, we used MEL cells, which represent an early committed erythroid cell type frequently used as a model for terminal erythroid differentiation. Stable reduction of LDB1 in MEL cells using shRNA abrogates looping between the LCR and β-globin gene after DMSO induction and results in failure to activate β-globin transcription without affecting key genes involved in MEL cell differentiation (Song et al. 2007; Li et al. 2010). Notably, transcription of erythroid regulators GATA1, TAL1, and LMO2 are unaffected by LDB1 knockdown (Li et al. 2010; Song et al. 2010).
We tested the ability of LDB1 to rescue β-globin gene expression in the induced MEL cells with stable knockdown of LDB1 by expression of shRNA-immune HA-tagged full-length LDB1 (LDB1 FL) cDNA (Supplemental Fig. S1A). Stable cell lines were established that expressed LDB1 FL and total levels of LDB1 proteins similar to wild-type cells, as detected by Western blot analysis (Supplemental Fig. S1B). We wished to avoid massive overexpression of LDB1 FL, which might be expected to interfere with LCR-β-globin long-range interactions. RT-qPCR analysis after DMSO induction indicated that all lines expressed β-globin at levels similar to wild-type MEL cells and significantly higher than uninduced MEL cells or control LDB1 knockdown cells with an empty expression vector (Supplemental Fig. S1C). RT-qPCR with primers to the 3′ untranslated region (UTR) of LDB1 that do not amplify LDB1 FL indicated that expression of LDB1 FL had no effect on expression of residual endogenous LDB1 still detected in LDB1 knockdown cells due to incomplete reduction by shRNA (Supplemental Fig. S1C; Song et al. 2007, 2010). We conclude that β-globin expression in induced LDB1 knockdown cells can be fully rescued by stable expression of full-length shRNA-immune LDB1, confirming that the β-globin transcription defect in these cells is due to LDB1 reduction.
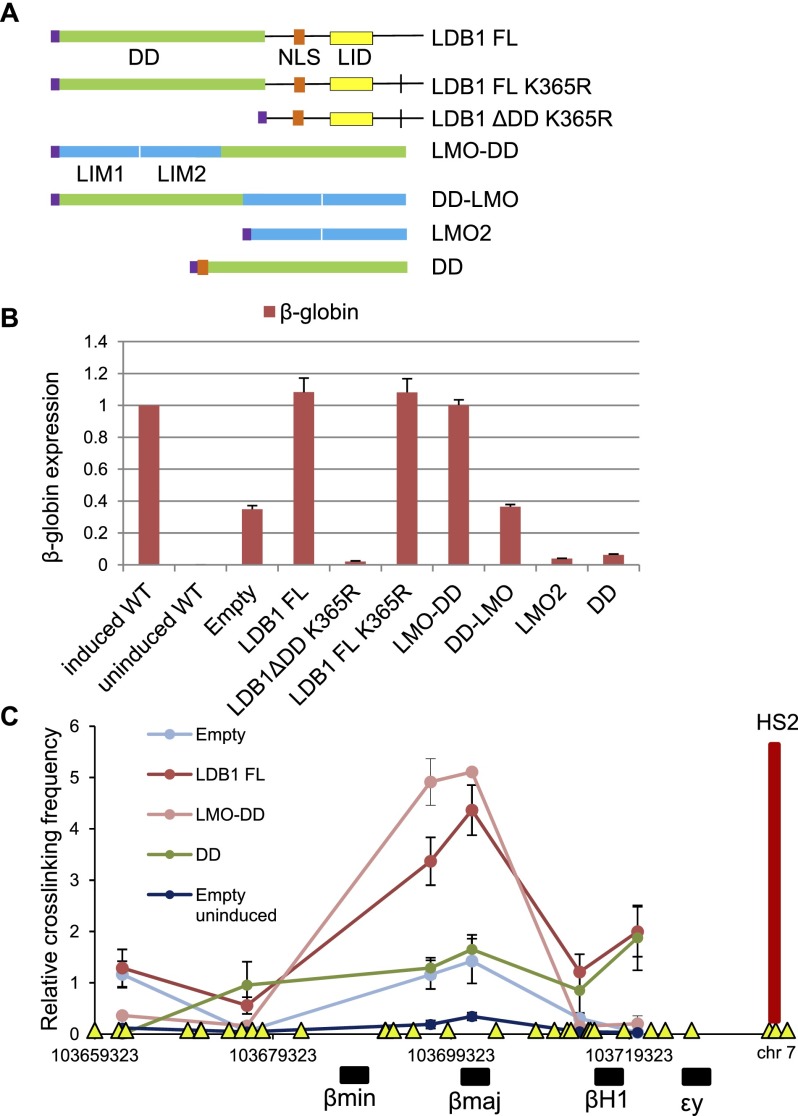
This system allowed functional testing of different domains of LDB1 (Fig. 1A). We first sought to test the effect of deleting the LDB1 DD (1–200, DD). LDB1ΔDD was highly unstable in MEL cells; however, we were able to produce a stable version of the protein by mutation of a potential ubiquitinylation site (K365R) (Supplemental Fig. S2). In the context of LDB1 FL, the K365R mutation had no effect on the ability of the protein to fully rescue β-globin transcription to the same extent as LDB1 FL (Fig. 1B). In contrast, LDB1ΔDD K365R could not rescue β-globin transcription and acted as a dominant-negative inhibitor of β-globin transcription that persisted after incomplete LDB1 knockdown (Song et al. 2007). This is expected for a protein that may interact with LMO2 but cannot dimerize (Thaler et al. 2002). We conclude that the DD is necessary for LDB1 dimerization and β-globin transcription rescue.
Figure 1.
β-Globin gene transcription activation requires the LDB1 DD. (A) Diagram of cDNAs expressed in induced LDB1 knockdown MEL cells. (Purple) HA tag; (green) DD; (orange) nuclear localization signal (NLS); (yellow) LID; (blue) LMO2 LIM1 and LIM2 domains. The small vertical bar indicates K365R mutation. (B) β-Globin gene expression in representative induced LDB1 knockdown MEL cells expressing the indicated LDB1-related proteins at levels similar to LDB1 FL. (C) 3C-qPCR relative cross-linking frequencies observed for induced LDB1 knockdown MEL cell lines expressing LDB1 FL, LMO-DD, or DD or with an empty vector (both induced and uninduced) using LCR HS2 as the viewpoint (red vertical bar). The X-axis shows genomic coordinates and the location of globin genes. (Yellow triangles) BglII restriction sites. Error bars in B and C indicate SEM; n = 3 biological replicates.
We hypothesized that fusion of the LDB1 DD missing the 3′ LID-containing domain directly to LMO2 may produce a protein capable of participating in complex formation with DNA-binding partners GATA1 and TAL1 and of β-globin transcription rescue in LDB1 knockdown cells. Two HA-tagged proteins were produced with the DD fused to LMO2 FL at either the C-terminal end of LMO2 (LMO-DD) or the N-terminal end (DD-LMO) (Fig. 1A). When stably expressed in LDB1 knockdown MEL cells, LMO-DD can activate β-globin expression after induction with DMSO to the same extent as LDB1 FL and to levels seen in wild-type induced MEL cells, while, interestingly, DD-LMO could not (Fig. 1B). Expression of LMO2 or the DD alone caused repression of residual β-globin expression in LDB1 knockdown MEL cells, as expected, presumably because they sequester residual endogenous LDB1 away from productive complex formation and dimerization (Thaler et al. 2002; Terano et al. 2005).
Next, we carried out chromosome conformation capture (3C) to assess chromatin looping upon β-globin transcription rescue of induced LDB1 knockdown MEL cells expressing LDB1 FL or LMO-DD. We observed a robust signal for proximity between the anchor LCR fragment and the β-globin genes in both cases that was very similar to the pattern seen in fetal liver cells (Tolhuis et al. 2002), indicating complete restoration of long-range LCR/β-globin interaction (Fig. 1C). Looping was significantly lower in cells with an empty vector, although it was slightly elevated over uninduced cells, consistent with an incomplete knockdown of LDB1 protein or independent function of other looping factors such as EKLF (Drissen et al. 2004). Thus, the LDB1 DD alone, when fused to the LIM2 domain of LMO2, is sufficient to fully restore β-globin locus conformation and function in LDB1 knockdown MEL cells.
Interestingly, heterologous dimerizing proteins such as LexA could not rescue β-globin expression when fused to LMO2, nor could an LMO2–LMO2 fusion protein (Supplemental Fig. S3A), suggesting that the DD has additional functions beyond dimerization. Likewise, a fusion of the DD to GATA1 FL was unable to rescue β-globin transcription, indicating that LMO2 is not dispensable for this function. None of these fusion proteins was capable of supporting long-range LCR looping (Supplemental Fig. S3B).
Functional dissection of the LDB1 DD
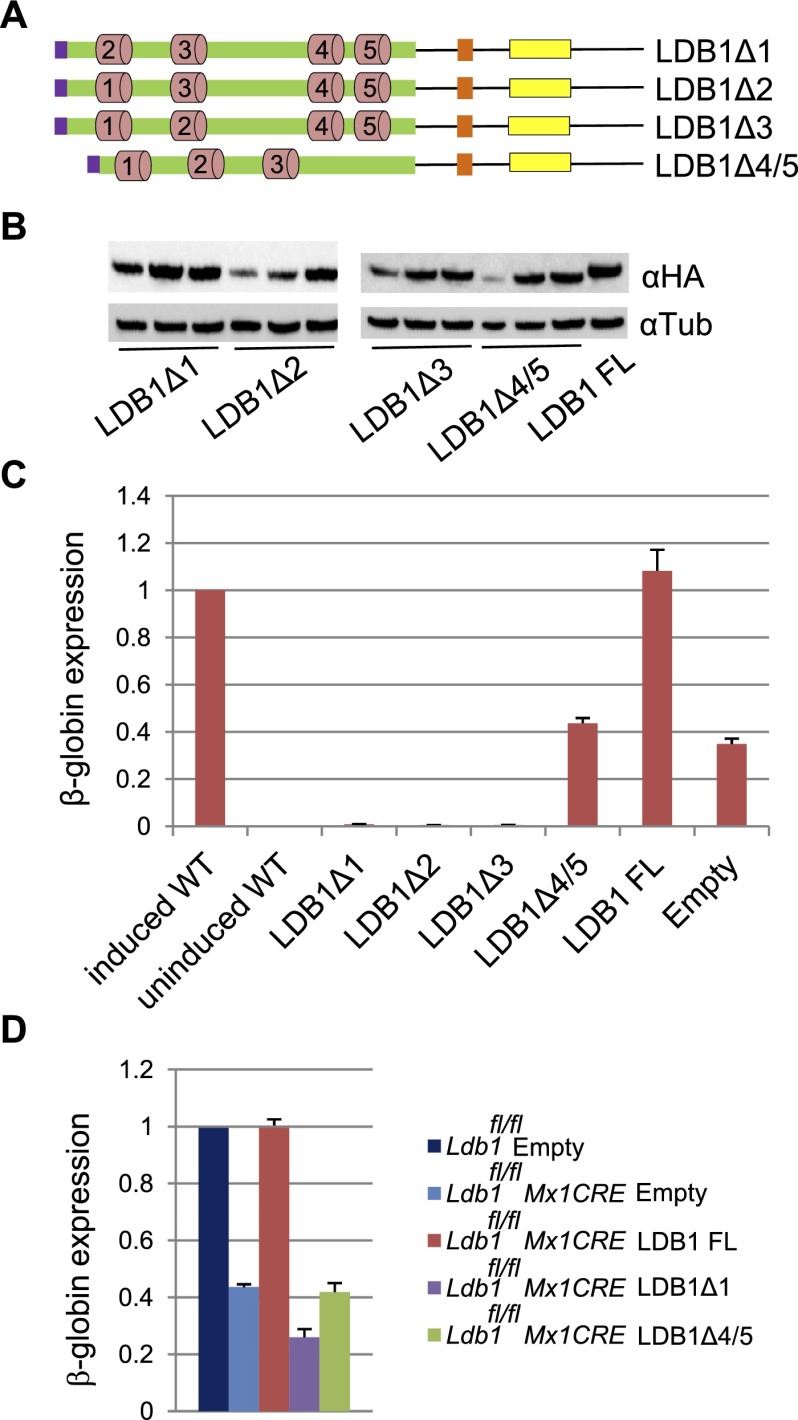
Several small, conserved regions of the DD (Supplemental Fig. S4) predicted to form α-helical structures were discerned using PSIpred (McGuffin et al. 2000; Cross et al. 2010). To explore the role of these regions in LDB1 dimerization, we stably expressed HA-tagged versions of LDB1 without sequences encoding potential helix 1 (LDB1Δ1), helix 2 (LDB1Δ2), helix 3 (LDB1Δ3), or helices 4 and 5 (LDB1Δ4/5) (Fig. 2A) in the background of LDB1 knockdown MEL cells.
Figure 2.
The LDB1 DD can be functionally subdivided. (A) Diagram of cDNAs that were expressed in LDB1 knockdown MEL cells. Predicted DD helical regions 1–5 are indicated. Other designations are the same as in Figure 1A. (B) Western blots of protein extracts from three induced LDB1 knockdown MEL cell lines expressing the indicated proteins. α-Tubulin served as a loading control. (C) β-Globin gene expression in representative induced cell lines expressing LDB1Δ1–LDB1Δ4/5. Expression in induced wild-type (WT) MEL cells was set to 1. (D) Expression level of the β-globin gene in IFN-β-treated E14.5 Ldb1fl/fl fetal liver cells without MX1Cre and with an empty vector or with MX1Cre and expressing LDB1 FL, LDB1Δ1, or LDB1Δ4/5 or with an empty vector. Expression level in E14.5 Ldb1fl/fl was set to 1. Error bars in C and D indicate SEM; n = 3 biological replicates.
Small deletions in the DD did not compromise the stability of LDB1 protein (Fig. 2B). None of the DD mutant proteins rescued β-globin expression above the level seen in cells with an empty vector (Fig. 2C). LDB1Δ1–Δ3 reduced background β-globin expression similar to LDB1 lacking the DD (LDB1ΔDDK356R), suggesting that they had lost the ability to self-interact. However, in LDB1Δ4/5 clones, transcription of β-globin remained at levels seen with an empty vector. These results suggest that while the complete DD is required for β-globin transcription rescue, the domain may be functionally subdivided. Deletion of DD regions 1, 2, or 3 compromises LDB1 dimerization; however, LDB1Δ4/5 may retain some ability to self-interact, albeit without β-globin transcription rescue.
To test this possibility and overcome the limitation that LDB1 is required for erythroid differentiation, we repeated this experiment in a mouse model of conditional Ldb1 deletion (Li et al. 2010). Embryonic day 14.5 (E14.5) fetal livers were collected from animals homozygous for a floxed Ldb1 allele and carrying Cre recombinase driven by the Mx1 promoter. E14.5 fetal livers with floxed Ldb1 but without Cre served as a control. Cells were cultured over a period of 72 h after induction of Cre expression by INF-β treatment (Supplemental Fig. S5A). Cre expression resulted in >50% deletion of Ldb1 and decreased Ldb1 and β-globin mRNA (Supplemental Fig. S5A,B). After 24 h, cells were transduced with retroviral vectors expressing LDB1 FL or LDB1Δ1 or LDB1Δ4/5 versions. LDB1 FL fully rescued β-globin expression in the background of reduced endogenous Ldb1 (Fig. 2D). LDB1Δ1 and LDB1Δ4/5 failed to rescue β-globin expression, but LDB1Δ1 exhibited dominant-negative behavior, while LDB1Δ4/5 did not. These results recapitulate those seen in LDB1 knockdown MEL cells and further support the contention that LDB1Δ4/5 can dimerize but fails to rescue β-globin transcription.
If LDB1Δ4/5 can dimerize, we predicted that DDΔ4/5 absent the C-terminal LIM-containing domain should be able to pull down endogenous LDB1 in wild-type MEL cells. To test this prediction, we stably expressed the DD or deleted versions in the background of wild-type LDB1-replete MEL cells (Supplemental Fig. S6). Coimmunoprecipitation (co-IP) experiments using an antibody against the HA tag indeed showed that both DD and DDΔ4/5 successfully interacted with endogenous LDB1, consistent with the ability to dimerize, while DDΔ1 did not (Supplemental Fig. S6C). Furthermore, DD and DDΔ4/5 inhibited β-globin transcription in induced wild-type cells, which is the expected result because, absent the LID, species that can dimerize with endogenous LDB1 will sequester it away from productive long-range interactions (Supplemental Fig. S6D). DDΔ1 did not repress β-globin expression, consistent with the inability to dimerize. Together, the results lead us to conclude that LDB1Δ4/5 is capable of dimerization even though such interaction does not rescue β-globin transcription in the LDB1 knockdown MEL cell background.
Dimerization of LDB1 rescues proximity between the β-globin LCR and gene even though transcription is not activated
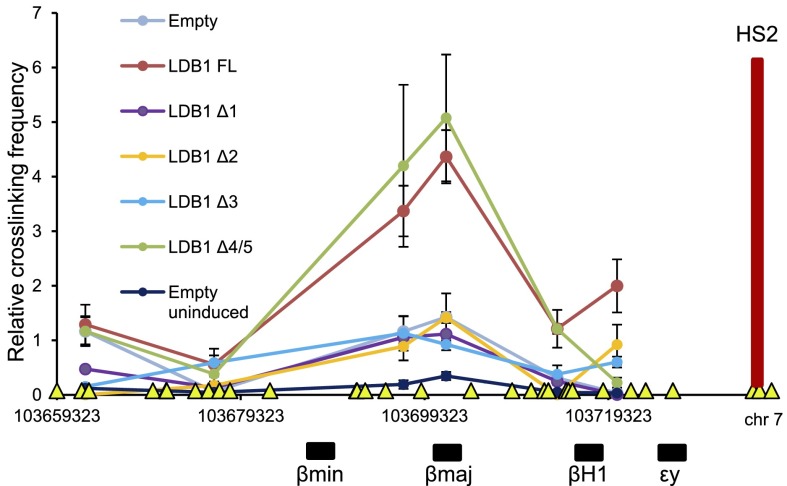
Dimerization of LDB1 is expected to underlie the long-range interaction between the LCR and β-globin gene (Song et al. 2007; Deng et al. 2012). To further probe the dimerization potential of LDB1 proteins with small DD deletions, we carried out 3C using HS2 of the LCR as the anchor. Induced LDB1 knockdown MEL cells expressing LDB1 FL exhibited a robust signal for proximity between the LCR and β-globin genes (Fig. 3), as expected. The β-globin gene and LCR were not in proximity in cells expressing LDB1Δ1, LDB1Δ2, or LDB1Δ3 deletion mutants, similar to cells containing an empty vector. Strikingly, LDB1Δ4/5 fully restored looping between the LCR and the β-globin gene, similar to LDB1 FL. We interpret this result to indicate that LDB1 dimerization is sufficient for β-globin looping but not for transcription activation, separating these processes and implicating a novel function of the DD4/5 region in β-globin transcription rescue.
Figure 3.
Dimerization of LDB1 is required for chromatin looping. Relative cross-linking frequencies were determined by 3C-qPCR using the LCR HS2 as the viewpoint (red vertical bar) for induced LDB1 knockdown MEL cell lines expressing the indicated proteins or with an empty vector (both induced and uninduced cells). The X-axis shows genomic coordinates and globin gene locations. (Yellow triangles) BglII restriction sites. Error bars indicate SEM; n = 3 biological replicates.
LCR long-range looping interactions are independent of β-globin locus nuclear relocalization
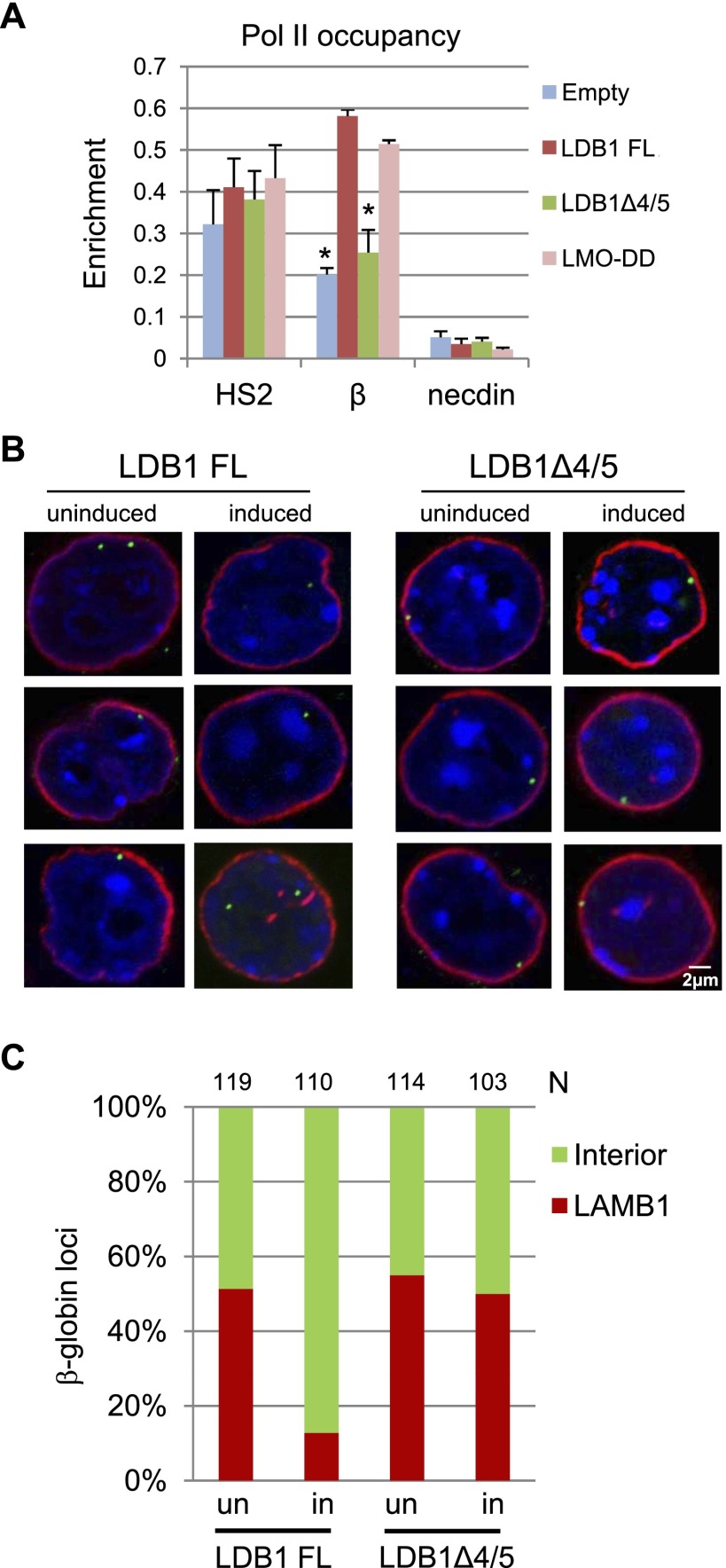
Our observations so far allow us to conclude that enhancer/gene looping in the β-globin locus does not require transcription. Earlier data suggested that the LCR and possibly LCR/β-globin looping exert a primary effect on Pol II elongation rather than promoter occupancy (Sawado et al. 2003; Deng et al. 2012). Therefore, we next asked whether RNA Pol II resided at the β-globin gene and LCR in the looped but transcriptionally inactive locus in induced LDB1 knockdown MEL cells stably expressing LDB1Δ4/5. Pol II chromatin immunoprecipitation (ChIP) revealed similar levels at LCR/HS2 in cells expressing LDB1Δ4/5, LDB1 FL, or LMO-DD or with an empty vector, consistent with previous work showing no effect of LDB1 reduction on HS2 Pol II occupancy (Fig. 4A; Song et al. 2010). However, Pol II occupancy at the β-globin promoter in cells expressing LDB1Δ4/5 was low, similar to cells with an empty vector, compared with cells expressing LDB1 FL or LMO-DD that actively transcribe the β-globin gene: Some Pol II signal at the β-globin promoter in cells with the empty vector is expected due to incomplete reduction of LDB1 protein by shRNA. These results show that robust recruitment of Pol II to the β-globin promoter requires 4/5 and also suggest that Pol II recruitment is a post-chromatin looping step in transcription activation.
Figure 4.
Chromatin looping is not sufficient for β-globin gene activation and β-globin locus intranuclear migration. (A) ChIP was performed using an RNA Pol II antibody and chromatin from induced LDB1 knockdown MEL cells expressing LDB1FL, LDB1Δ4/5, or LMO-DD or with an empty expression vector. Necdin served as a negative control. Error bars indicate SEM; n = 3 biological replicates. Values are compared with the value for LDB1 FL. (*) P < 0.05 by Student’s t-test. (B) Three-dimensional immuno-FISH analysis of uninduced and induced LDB1 knockdown MEL cells expressing LDB1 FL or LDB1Δ4/5. (Red) LAMB1 immunofluorescence; (green) β-globin locus probe; (blue) DAPI stain. (C) The graph shows association of β-globin loci with the nuclear lamina. n = nuclei scored.
β-Globin loci relocate to the nuclear interior, where they associate with transcription factories to achieve high levels of transcription (Osborne et al. 2004; Ragoczy et al. 2006). The separation of looping and transcription mediated by LDB1Δ4/5 allowed us to address whether looping occurs before or after nuclear migration. Looping might be a prerequisite for locus migration or, alternatively, might occur in transcription factories as a consequence of Pol II density or of other factors contributing to genome higher-order organization for transcription.
We carried out confocal microscopy for LDB1 knockdown MEL cells expressing LDB1 FL or LDB1Δ4/5 after performing DNA immuno-FISH for the β-globin locus and immunostaining for LAMB1 at the nuclear periphery. β-Globin loci in uninduced cells are visualized as contacting the nuclear lamina (Fig. 4B,C). Upon induction, β-globin loci in cells expressing LDB1 FL migrate away from the nuclear periphery, with only ∼12% of loci retaining a minimal overlap of two pixels with LAMB1 (P < 0.001 by Fisher’s exact test, compared with uninduced cells) (Fig. 4B,C). However, induced cells expressing LDB1Δ4/5 show a pattern of lamina association indistinguishable from uninduced cells (P = 0.496). A ChIP for LAMB1 at the β-globin promoter reflected this pattern (Supplemental Fig. S7; Handoko et al. 2011). Robust LAMB1 detection at the β-globin promoter in uninduced cells is greatly reduced after induction for cells expressing LDB1 FL but is retained for cells expressing LDB1Δ4/5. Interestingly, LAMB1 is not detected at the LCR, suggesting a primary role for the β-globin promoter in nuclear lamina association. These results suggest a scenario in which looping occurs before locus migration and before transcription activation in nuclear Pol II factories. The results emphasize the important role of the protein–protein interactions that are involved in looping.
Differing SWI/SNF complex occupancy in looped but inactive β-globin loci compared with active loci
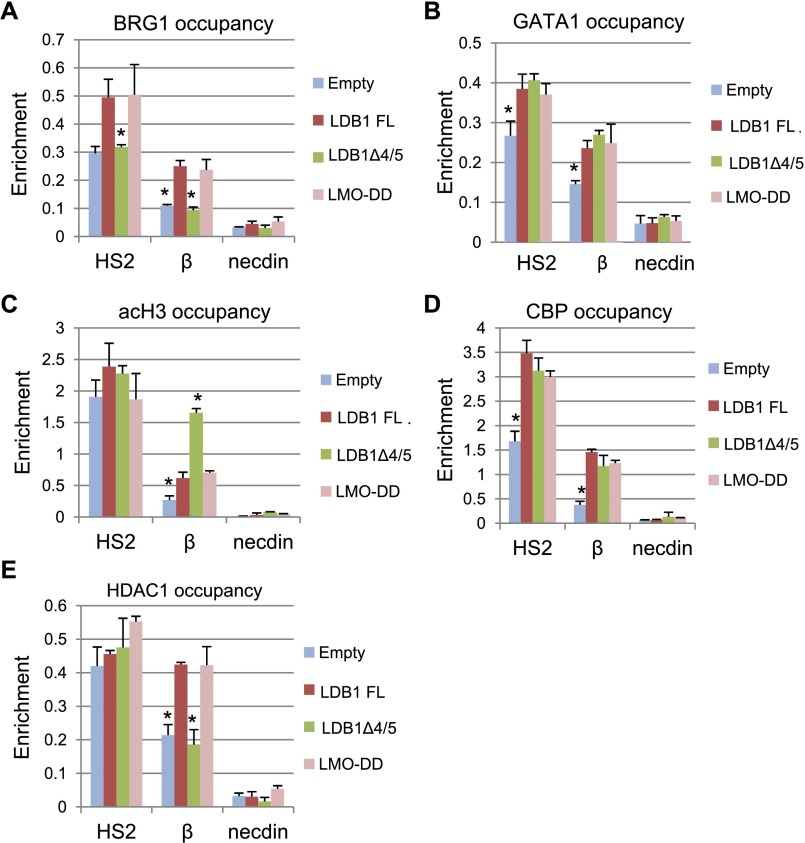
To explore the difference between looped and transcriptionally active versus inactive β-globin loci, we performed ChIP using antibodies to the SWI/SNF ATPase BRG1. BRG1 is recruited to the β-globin promoter at an early time point after GATA1 restoration in GATA1-deficient G1E cells, preceding maximal looping and promoter occupancy by coactivators and Pol II (Kim et al. 2009). Surprisingly, we found that BRG1 occupancy at HS2 and at the β-globin promoter was similar in cells expressing LDB1 FL or LMO-DD, while occupancy in the looped but inactive locus in LDB1Δ4/5-expressing cells was no greater than in cells with an empty vector (Fig. 5A). Failure to recruit BRG1 is reflected in lower sensitivity of the locus to DNase I (Supplemental Fig. S8). Reduced GATA1 might result in failure to recruit BRG1 (Kim et al. 2009); however, ChIP for GATA1 revealed similar occupancy at both HS2 and β-globin in cells expressing LDB1Δ4/5, LDB1 FL, or LMO-DD (Fig. 5B). This result is expected, since these versions of LDB1 all occupied β-globin locus chromatin (data not shown). These data indicate that BRG1 promoter occupancy is associated with transcription activation but not with unproductive loop formation in the β-globin locus.
Figure 5.
The LDB1 4/5 region is required for SWI/SNF and NuRD complex occupancy at the β-globin promoter. ChIP was performed using chromatin from induced LDB1 knockdown MEL cells expressing LDB1FL, LDB1Δ4/5, or LMO-DD or containing an empty expression vector. Antibodies used were to BRG1 (A), GATA1 (B), acH3 (C), CBP (D), and HDAC1 (E). The data for acH3 were normalized to H3 and actin signals. Necdin served as a negative control. Error bars indicate SEM; n = 3 biological replicates. Values are compared with the value for LDB1 FL. (*) P < 0.05 by Student’s t-test.
Acetylated histone tails also recruit BRG1 through the bromodomain (Hassan et al. 2001). This mark is enriched at both the LCR and β-globin gene in mature erythroid cells (Forsberg et al. 2000). ChIP assays showed that H3 acetylation (acH3) at the LCR was similar for induced LDB1 knockdown cells expressing LDB1 FL, LDB1Δ4/5, or LMO-DD or with an empty vector (Fig. 5C). Surprisingly, acH3 was abnormally high at the β-globin promoter in cells expressing LDB1Δ4/5 compared with cells expressing LDB1 FL or LMO-DD that rescue β-globin transcription. Since histone acetylation levels reflect a balance between acetylase and deacetylase activities that is important for regulated transcription (Wang et al. 2009; Perissi et al. 2010), we next determined CBP and HDAC1 occupancy. CBP occupancy in the β-globin locus was similarly restored by all tested versions of LDB1, consistent with recruitment of CBP by GATA1 (Fig. 5D; Blobel et al. 1998). In contrast, HDAC1 occupancy at the β-globin promoter was low in cells expressing LDB1Δ4/5, similar to cells with an empty vector (Fig. 5E). We interpret these results to indicate a direct or indirect role of the 4/5 region of the LDB1 DD in recruiting or stabilizing HDAC1.
The DD 4/5 region of LDB1 is important for FOG1 and nucleosome remodeling and deacetylating (NuRD) complex recruitment to the β-globin locus
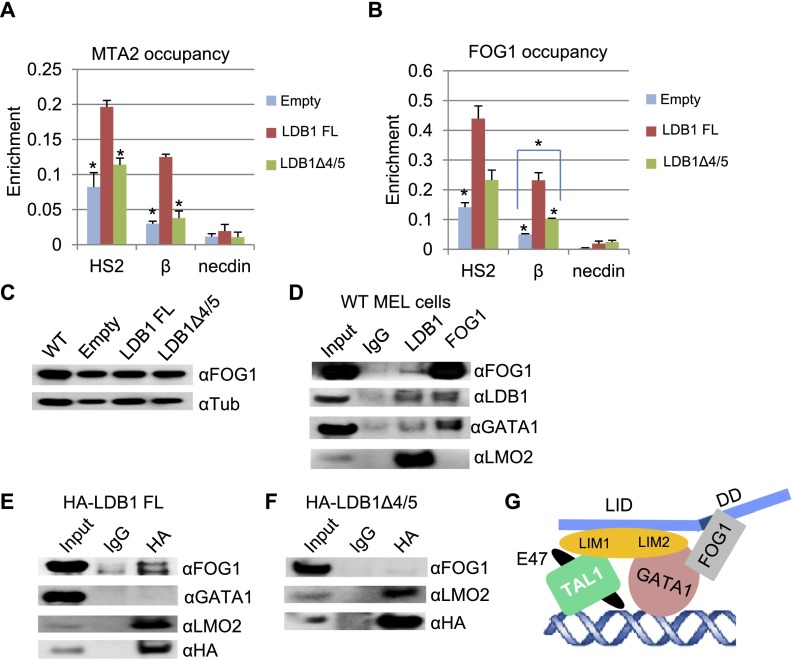
HDAC1 is part of the NuRD complex that is recruited to the β-globin gene by the GATA1 cofactor FOG1 (Hong et al. 2005). NuRD is required for repression as well as activation of certain FOG1-dependent GATA1 targets, including the β-globin gene (Miccio et al. 2010). Other components of the NuRD complex in erythroid cells include CHD4 (also known as Mi-2β) and MTA2 (Hong et al. 2005; Rodriguez et al. 2005). These observations led us to determine MTA2 and FOG1 occupancy at looped but transcriptionally inactive β-globin loci in cells expressing LDB1Δ4/5. ChIP experiments using antibodies against MTA2 or FOG1 revealed reduced occupancy of these proteins in the β-globin locus in cells expressing LDB1Δ4/5 compared with LDB1 FL (Fig. 6A,B). Differences in FOG1 occupancy were not due to differences in FOG1 protein levels, as indicated by Western blotting (Fig. 6C).
Figure 6.
Interaction between the LDB1 DD and FOG1. ChIP was performed with chromatin from induced LDB1 knockdown MEL cells expressing LDB1 FL or LDB1Δ4/5 or with an empty vector. Antibodies were against MTA2 (A) or FOG1 (B). Necdin served as a negative control. Error bars indicate SEM; n = 3. Values are compared with the value for LDB1 FL. (*) P < 0.05 by Student’s t-test. (C) Western blots of protein extracts from induced wild-type (WT) MEL cells and LDB1 knockdown MEL cell lines with an empty vector or expressing LDB1 FL or LDB1Δ4/5 with FOG1 antibodies. α-Tubulin served as a loading control. (D) Immunoprecipitation was performed with antibodies to LDB1 or FOG1 using nuclear extracts from induced wild-type MEL cells. Immunoprecipitation material was analyzed by Western blot with LDB1, FOG1, GATA1, and LMO2 antibodies. (E,F) Immunoprecipitation was performed with an HA antibody and nuclear extracts from induced LDB1 knockdown MEL cells expressing LDB1 FL (E) or LDB1 DDΔ4/5 (F). Immunoprecipitation material was analyzed by Western blot with FOG1, GATA1, and LMO2 antibodies. HA antibodies served as a positive control. (G) A model depicting protein–protein interactions within the LDB1 complex, including FOG1. Colored shapes representing factors are depicted as touching when such interaction is supported by biochemical and/or structural data (see the text). LMO2 N-terminal LIM1 and C-terminal LIM2 regions are indicated. The DD4/5 location in LDB1 is shaded.
FOG1 is a multi-Zn-finger protein that interacts with the N-terminal Zn finger of GATA1 (GATA1NF) (Tsang et al. 1997). Thus, FOG1 and LDB1 have at least an indirect association through the LDB1 complex at FOG-dependent GATA1 target genes. Weak interaction between LDB1 and GATA1 has been observed in a biotin-LDB1 pull-down or α-GATA1 immunoprecipitation carried out under mild wash conditions (Rodriguez et al. 2005; Meier et al. 2006). Using nuclear extracts of induced MEL cells, we observed that LDB1 protein could be pulled down reciprocally and efficiently with antibodies to FOG1 and LDB1 (Fig. 6D). However, LDB1 immunoprecipitation could be secondary to GATA1 pull-down, which was also observed.
To ask whether a direct interaction between FOG1 and LDB1 exists, specifically through the DD 4/5 region, co-IP experiments were performed using an antibody against the HA tag (Fig. 6E,F). As expected, both HA-LDB1 FL and HA-LDB1Δ4/5 efficiently pulled down LMO2 from nuclear extracts of induced LDB1 knockdown MEL cells expressing these proteins. Under the stringent conditions we used for the anti-HA pull-down, HA-LDB1 FL did not pull down GATA1 (Song et al. 2007). Importantly, the panels in which Western blotting was performed with antibodies to FOG1 clearly indicate that a fraction of FOG1 can be immunoprecipitated by LDB1 FL but not by LDB1Δ4/5, supporting direct interaction between FOG1 and LDB1 through the DD 4/5 region that is independent of GATA1. Such a proposed model is depicted in Figure 6G.
LDB1-regulated genes that are sensitive to the deletion of LDB1 4/5 are FOG-dependent
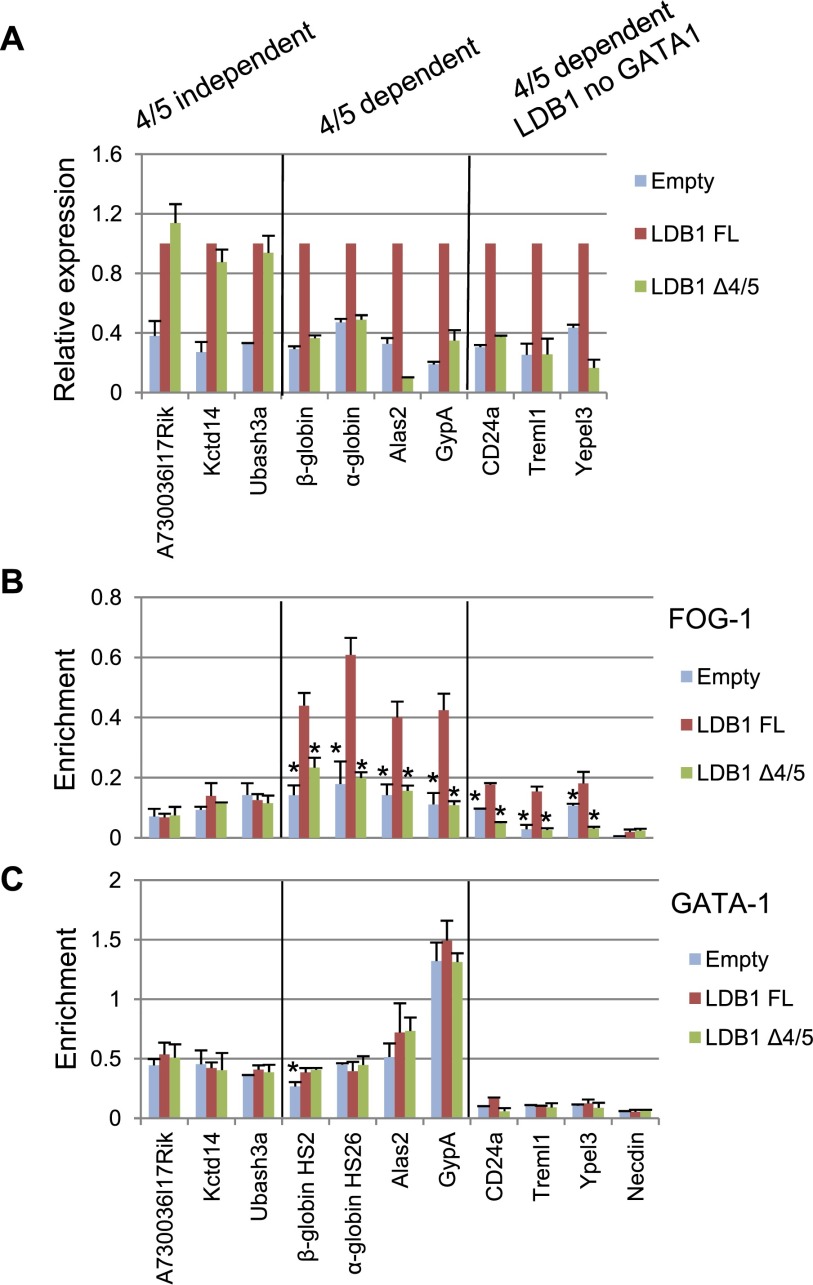
To test whether FOG1–LDB1 interaction has functional consequences, we carried out RNA sequencing (RNA-seq) for induced wild-type and LDB1 knockdown MEL cells and for knockdown cells expressing either LDB1 FL or LDB1Δ4/5. Differential expression analysis of single-end 51-base-pair (bp) polyA+ RNA-seq reads identified 496 genes that were significantly repressed (Padj < 0.05) more than twofold by LDB1 knockdown and rescued by LDB1 FL expression in knockdown cells. Of these genes, transcription of 349 was rescued by both LDB1 FL and LDB1Δ4/5 (hereafter referred to as 4/5-independent genes) (Supplemental Fig. S9; see the Supplemental Material). We validated the RNA-seq results for three 4/5-independent genes by qRT–PCR (Fig. 7A, left; Supplemental Fig. S10A). ChIP assays showed that the promoters of Kctd14 and Ubash3a and an intron of A730036I17Rik are occupied by GATA1 with very low FOG1 occupancy (Fig. 7B,C, left). Rescue of FOG1-independent genes by LDB1Δ4/5 attests to the functionality of the mutant protein.
Figure 7.
The 4/5 region of LDB1 is required for regulation of FOG1-dependent genes. Depicted are representative LDB1 4/5-independent (left panels) and 4/5-dependent (middle panels) LDB1 complex-occupied genes or regulatory elements and 4/5-dependent genes occupied by LDB1 but not GATA1 (right panels) in induced LDB1 knockdown MEL cells expressing LDB1 FL or LDB1 Δ4/5 or with an empty vector. (A) qRT–PCR validation of gene expression. FOG1 (B) and GATA1 (C) occupancy determined by ChIP. X-axis designations for B and C are the same. For representative genome browser screen shots, see Supplemental Figure S9. Error bars indicate SEM; n = 3 biological replicates. Values are compared with the value for LDB1 FL. (*) P < 0.05 by Student’s t-test.
In contrast, there were 147 genes that were rescued by LDB1 FL but not by LDB1Δ4/5 (4/5-dependent genes). Results for a subset of these genes were verified by RT–PCR (Fig. 7A, middle; Supplemental Fig. S10B), including α-globin, β-globin, Alas2, and Gypa, whose transcription is known to be FOG1-dependent (Crispino et al. 1999; Anguita et al. 2004; Letting et al. 2004; Pal et al. 2004; Campbell et al. 2013). FOG1 occupied these genes or their regulatory regions in cells expressing LDB1 FL but was significantly decreased in cells expressing LDB1Δ4/5 (Fig. 7B, middle). GATA1 occupancy was similar in cells expressing LDB1 FL or LDB1Δ4/5, suggesting that LDB1 may contribute to FOG1 stabilization through the 4/5 region independent of GATA1 (Fig. 7C, middle). We further identified 35 4/5-dependent genes with LDB1 peaks that did not overlap with GATA1 peaks (Fig. 7A–C, right; Supplemental Fig. S10C) using ChIP-seq data for induced MEL cells (Soler et al. 2010). FOG1 occupancy at these sites was 4/5-dependent, lending further support to LDB1–FOG1 interaction independent of GATA1. These results identify LDB1 as a novel regulator of FOG1 function.
Mutations that prevent FOG1 interaction with GATA1 are deleterious in humans (Crispino et al. 1999). For example, the GATA1 V205A mutation is known to cause dyserythropoietic anemia (Nichols et al. 2000). To investigate whether mutation of genes whose regulation is affected by LDB1 are similarly deleterious, we queried the Online Mendelian Inheritance in Man (OMIM) database for human homologs of LDB1-activated and occupied mouse genes (Supplemental Fig. S11A,B). Human disease-associated homologs were significantly enriched in the LDB1 4/5-dependent group over all other human homologs (P = 0.001, Fisher’s exact test), while 4/5-independent genes were not (P = 0.93). Interestingly, almost half of the LDB1 4/5-dependent genes were blood disease-related, in contrast to <15% of the LDB1 4/5-independent genes (Supplemental Fig. S11C). We conclude that regulation of a significant cohort of blood disease-associated genes depends on interaction with and stabilization of FOG1 chromatin association by LDB1 through the 4/5 DD region.
Discussion
Looping and transcription activation can be separated
The temporal relationship between chromatin looping to distant enhancers and transcription activation of genes has long been enigmatic. Here we functionally dissected the DD of LDB1, which is required for LCR/β-globin looping, and discovered that amino acids 173–192 are dispensable for loop formation but are required for transcription activation. Earlier work in which β-globin transcription initiation and elongation were inhibited by α-amanitin or DRB had provided evidence that a loop already established between the β-globin LCR and gene can be maintained in the absence of ongoing transcription (Mitchell and Fraser 2008; Palstra et al. 2008). Our results establish that enhancer–promoter loops can be initially formed in the absence of transcription activation over background levels (Figs. 1C, 3) and argue against the idea that enhancer–gene proximity is a consequence of association of Pol II with both elements or of transcription (Bulger and Groudine 2011).
Many enhancers are associated with Pol II and, indeed, Pol II occupies the LCR in induced LDB1 knockdown MEL cells expressing LDB1Δ4/5, but despite proximity between the LCR and gene, Pol II is not increased at the β-globin promoter. This is consistent with data showing that LDB1 is required for efficient recruitment of Pol II to the β-globin promoter but not to HS2 in induced MEL cells (Song et al. 2010). Furthermore, β-globin promoter occupancy by the LDB1 complex and Pol II occurred normally after deletion of the LCR from the endogenous mouse globin locus, indicating that promoter Pol II recruitment is independent of LCR occupancy (Sawado et al. 2003; Song et al. 2010). A role for the LCR and possibly LCR/β-globin looping in Pol II elongation rather than promoter occupancy has been reported (Sawado et al. 2003; Deng et al. 2012). In addition, our results suggest an important role for LDB1, specifically the DD4/5 region, in Pol II promoter recruitment.
Both LCR/β-globin looping and locus migration away from the nuclear periphery fail to occur in induced LDB1 knockdown MEL cells (Song et al. 2007, 2010), but the relationship between these processes was unclear. Rescue of LDB1 knockdown cells by LDB1Δ4/5 now indicates that LCR looping likely precedes but is not sufficient for migration. Failure of the locus to migrate away from the nuclear periphery is most likely the major determinant of failure to activate transcription. The results raise the possibility that interactions mediated by the DD 4/5 region may be involved in nuclear migration either directly or indirectly. Sequences or factors that might be involved in migration away from the periphery of active loci are unknown, although actin is implicated (Chuang et al. 2006; Dundr et al. 2007). Rescue of LCR/β-globin looping but not intranuclear migration by LDB1Δ4/5 may provide an important tool to begin to unravel how intranuclear migration occurs.
LDB1 is a transcription coactivator
The inability to replace the LDB1 DD with heterologous DDs first suggested that the DD might have additional functions beyond looping (Supplemental Fig. S3). This conclusion is reinforced by the observation that LDB1Δ4/5 can dimerize and support LCR/β-globin looping but not transcription activation or locus migration. Our ChIP studies comparing active and inactive looped β-globin loci revealed differences in cofactor recruitment. In particular, BRG1 recruitment to the β-globin locus was deficient in cells expressing LDB1Δ4/5, although the locus was in a looped conformation and occupied by the LDB1 complex. In agreement with our results, kinetic studies in which the INF-β enhancer was artificially relocalized at a distance from its target promoter found that looping between them was dependent on transcription factors and preceded recruitment of coactivators and Pol II (Nolis et al. 2009). Moreover, it is known that GATA1 chromatin occupancy is not dependent on BRG1 on a genome-wide scale (Hu et al. 2011). Nevertheless, our data seem to contrast with the finding of early recruitment of BRG1 to globin loci before maximal GATA1 occupancy and looping in G1E cells (Kim et al. 2009). Thus, certain aspects of cofactor recruitment and chromatin looping remain unclear.
Our results point to the importance of LDB1, particularly the 4/5 region, for BRG1 recruitment. Possibly, this is a direct effect, as evidence has been presented in Drosophila that the LDB1 ortholog Chip interacts directly with Osa, a component of the Brahma SWI/SNF remodeling complex, although the region of Chip that is implicated is distinct from LDB1 DD4/5 (Heitzler et al. 2003). Alternatively, BRG1 recruitment might be compromised by reduced NuRD recruitment to the β-globin locus in LDB1Δ4/5-expressing cells. Evidence has been presented for interaction between BRG1 and NuRD, and, furthermore, SWI/SNF and NuRD can each influence the occupancy of the other in different contexts (Datta et al. 2005; Ramirez-Carrozzi et al. 2006; Yildirim et al. 2011).
In LDB1Δ4/5-expressing cells, while there was decreased occupancy in the β-globin locus of the NuRD complex component HDAC1, CBP occupancy was equivalent to that in cells expressing LDB1 FL. These activities maintain a dynamic cycle of acetylation/deacetylation (Wang et al. 2009), and their imbalance, leading to histone hyperacetylation, has been suggested to destabilize chromatin to the extent that transcription is inhibited (Perissi et al. 2010). NuRD is recruited by FOG1 to carry out downstream effects of GATA1 (Hong et al. 2005; Miccio et al. 2010). Consistent with low levels of NuRD, FOG1 was also reduced at the LCR and inactive β-globin promoter in cells expressing LDB1Δ4/5 compared with LDB1 FL. Co-IP data directly comparing HA-tagged LDB1 FL and LDB1Δ4/5 suggest that FOG1 can interact with the 4/5 region of LDB1. Interestingly, reduced FOG1 and NuRD occupancy at the β-globin gene were observed in other studies in which globin loci failed to migrate away from the nuclear periphery (Lee et al. 2011).
FOG1 is required for LCR/β-globin looping presumably through GATA1 stabilization or recruitment of other factors (Letting et al. 2004; Pal et al. 2004; Vakoc et al. 2005). Therefore, a challenging question that remains is how looping occurs in cells expressing LDB1Δ4/5 in which FOG1 recruitment is reduced compared with cells expressing LDB1 FL. We note that FOG1 recruitment in LDB1Δ4/5-expressing cells was, nevertheless, significantly elevated compared with cells with an empty vector (Fig. 6B). We speculate that partial recruitment of FOG1 suffices for stabilization of the LDB1 complex, including GATA1, allowing LDB1 dimerization and looping, but does not adequately support recruitment of NuRD, which compromises downstream processes, including Pol II recruitment and intranuclear migration. Alternatively, LDB1 may contribute to stabilization of GATA1 chromatin occupancy in a FOG1-independent fashion (Song et al. 2010). Together, these experiments show that LDB1 DD4/5 is involved in transcription activation beyond dimerization and looping and suggest that FOG1/LDB1 interaction through DD4/5 is central to this role.
Our results lead to a model in which the LDB1 LID interacts across LMO2 in such a way that the DD rests along the GATA1/FOG1 interface of the complex (Fig. 6G). Interestingly, structural data reported for interacting fragments of proteins within the LDB1 complex are consistent with this orientation. First, both nuclear magnetic resonance (NMR) and crystal structure studies place the LDB1 LID along the length of LMO2 LIM1 and LIM2 domains with N-terminal LDB1 sequences, including 4/5, oriented near the LIM2/GATA1NF/FOG1 interface (El Omari et al. 2011; Wilkinson-White et al. 2011). Second, the DNA-binding GATA1NF interacts simultaneously with a Zn finger of FOG1 and with LMO2 LIM2 using different surfaces (Liew et al. 2005; Wilkinson-White et al. 2011). This orientation of protein interfaces within the LDB1 complex suggests a reason why LDB1 DD fusion to LMO2 LIM2 (LMO-DD) successfully rescued β-globin locus conformation and function in LDB1 knockdown MEL cells, but further structural studies will be needed.
Homologs of mouse LDB1 4/5-dependent genes are significantly associated with human disease
Our biochemical evidence describing LDB1/FOG1 interaction is supported by RNA-seq experiments. First, we observed rescue of numerous LDB1-dependent genes by both LDB1 FL and LDB1Δ4/5, attesting to the function of the mutant protein. Second, a subset of LDB1 knockdown-repressed genes rescued by LDB1 FL was shown to be LDB1 DD4/5-dependent (Fig. 7). These genes or their regulatory regions are not rescued by LDB1Δ4/5 and are known to be FOG1-dependent, suggesting that in addition to GATA1, LDB1 normally stabilizes FOG1 at these sites. Moreover, mutations affecting stabilization of FOG1 in the complex cause disease-associated phenotypes (Ciovacco et al. 2008). By RNA-seq analysis, we identified that the 4/5 region of LDB1 is required for proper expression of a significant number of blood disease-associated genes that are FOG1-dependent. This observation further supports the importance of FOG1 protein in providing a healthy red blood cell phenotype and provides a new explanation of mechanisms underlying congenital hematologic diseases.
Materials and methods
Cell lines
Wild-type and LDB1 knockdown MEL cells were cultured and induced as described (Song et al. 2007).
Primary erythroid progenitor culture, genotyping, and deletion analysis
Mouse fetal livers were extracted from E14.5 embryos homozygous for the floxed LDB1 gene with or without CRE under control of the Mx1 gene promoter (Li et al. 2010). Cells were cultured as described (von Lindern et al. 2001). CRE expression was induced by 250 U/mL IFN-β (Millipore) in culture medium. Genotyping and Cre-mediated deletion were as described (Li et al. 2010).
Plasmid construction
All HA-tagged proteins used in these studies were expressed from the pMY-IRES-Neo vector (Cell Biolabs). Plasmid construction details are provided in the Supplemental Material.
Retrovirus production and gene transduction
Platinum-A packaging cells (Cell Biolabs) were transfected with LDB1 and control expression vectors by Lipofectamine 2000 (Invitrogen). Virus supernatant was collected after 3 d. Wild-type or LDB1 knockdown MEL cells were incubated with viral supernatant in the presence of 10 μg/mL polybrene. After 1 d, the medium was changed to MEL medium. Neomycin was added 2 d after transduction, and cells were selected during 1 wk. Stable clones were isolated, and transgene expression was checked by Western blot hybridization. Fetal liver cells were infected by RetroNectin-bound virus infection method (TaKaRa) on the day following CRE expression. Cells were harvested 48 h after infection.
Western blotting
Proteins were isolated by resuspending cells in RIPA buffer (50 mM Tris-HCl at pH 7.6, 150 mM NaCl, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS), with the concentration determined by BCA kit after removal of debris (Thermo Scientific). Sample preparation, electrophoresis, transfer, and hybridization followed Nupage protocols (Invitrogen). See the Supplemental Material for antibodies. Blots were developed by ECL Plus (Thermo Scientific).
RNA extraction and RT–PCR
RNA was isolated with the RNeasy kit (Qiaqen). Two micrograms of RNA was treated with RNase-free DNase I (Life Technologies) for 15 min at 25°C. RNA was reverse-transcribed using the Superscript III first strand synthesis kit with random hexamers (Life Technologies). RT–PCR was performed as described (Song et al. 2007). Data were normalized to the actin or hprt signal. See the Supplemental Material for primers.
ChIP
ChIP was performed as described (Song et al. 2010) with some modifications. EGS (Pierce) followed by formaldehyde was used for cross-linking chromatin for HA, FOG1, MTA, HDAC1, and BRG1 ChIP (Zeng et al. 2006). See the Supplemental Material for antibodies and primers. Details of data normalization, if any, are included in the figure legends.
Co-IP
Nuclear extract was prepared as described (Yusufzai et al. 2004). Co-IP was performed as described (Brand et al. 2004), with some modifications. Experimental details and antibodies used are provided in the Supplemental Material.
3C
3C assays were performed as described (Hagege et al. 2007) using BglII (New England Biolabs) cleavage. Relative cross-linking between HS2 and fragments of interest was analyzed by real-time qPCR with published TaqMan probes and primers (Deng et al. 2012). Data were normalized to an interaction in the ERCC gene and, for Supplemental Figure S3, were further normalized to interaction of HS2 with a fragment outside the β-globin locus containing the Olfr64 gene.
DNase I sensitivity assay
DNase I sensitivity was assayed by real-time qPCR as described (Kiefer and Dean 2012).
Three-dimensional (3D) immuno-FISH
3D immuno-FISH was performed as described (Bolland et al. 2013). The probe was prepared by the FISH Tag DNA Green kit (Life Technology). Images were collected using a confocal microscope (LSM 510, Carl Zeiss). FISH spots were judged to contact the lamina if at least several pixels overlapped the LAMB1 stain. See the Supplemental Material for antibodies.
RNA-seq library construction, sequencing, and computational analysis
RNA from induced wild-type MEL cells, LDB1 knockdown cells, and knockdown cells expressing either LDB1 FL or LDB1Δ4/5 was isolated with the RNeasy kit (Qiaqen). RNA-seq libraries were constructed using TruSeq RNA sample prep kit version 2 (Illumina) according to the manufacturer’s protocol. Three biological replicates of each cell type were sequenced on a HiSeq 2000. Further details appear in the Supplemental Material. Raw and processed data are available from Gene Expression Omnibus at accession number GSE54549.
Acknowledgments
We thank Dr. Elissa Lei, Dr. Gerd Blobel, Dr. Paul Love, and Dr. Liqi Li for helpful comments on the manuscript. We thank Dr. Paul Love and Dr. Liqi Li for supplying floxed Ldb1 animals. We thank Dr. Tom Misteli and Dr. Karen Meaburn for help with the FISH studies, and Francine Katz for help in MEL cell cloning. Work in the laboratory of A.D. is supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (ZIA DK015508-25).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.239749.114.
References
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci 95: 2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland DJ, King MR, Reik W, Corcoran AE, Krueger C 2013. Robust 3D DNA FISH using directly labeled probes. J Vis Exp 78: 50587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M 2004. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 11: 73–80 [DOI] [PubMed] [Google Scholar]
- Breen JJ, Agulnick AD, Westphal H, Dawid IB 1998. Interactions between LIM domains and the LIM domain-binding protein Ldb1. J Biol Chem 273: 4712–4717 [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Wilkinson-White L, Mackay JP, Matthews JM, Blobel GA 2013. Analysis of disease-causing GATA1 mutations in murine gene complementation systems. Blood 121: 5218–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831 [DOI] [PubMed] [Google Scholar]
- Ciovacco WA, Raskind WH, Kacena MA 2008. Human phenotypes associated with GATA-1 mutations. Gene 427: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, Mackay JP, Orkin SH 1999. Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell 3: 219–228 [DOI] [PubMed] [Google Scholar]
- Cross AJ, Jeffries CM, Trewhella J, Matthews JM 2010. LIM domain binding proteins 1 and 2 have different oligomeric states. J Mol Biol 399: 133–144 [DOI] [PubMed] [Google Scholar]
- Datta J, Majumder S, Bai S, Ghoshal K, Kutay H, Smith DS, Crabb JW, Jacob ST 2005. Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer Res 65: 10891–10900 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dekker J 2008. Gene regulation in the third dimension. Science 319: 1793–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W 2004. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179: 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Vyas P, Patient R, Porcher C, Mancini EJ 2011. Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex. Blood 117: 2146–2156 [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci 97: 14494–14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Heitzler P, Vanolst L, Biryukova I, Ramain P 2003. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev 17: 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA 2005. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J 24: 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN 1998. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem 273: 3152–3157 [DOI] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA 2009. Chromatin loops in gene regulation. Biochim Biophys Acta 1789: 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer CM, Dean A 2012. Monitoring the effects of chromatin remodelers on long-range interactions in vivo. Methods Mol Biol 833: 29–45 [DOI] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH 2009. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci 106: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I, Dean A 2012. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev 22: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Johnson KD, Boyer ME, Bresnick EH 2011. Relocalizing genetic loci into specific subnuclear neighborhoods. J Biol Chem 286: 18834–18844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci 101: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lee JY, Gross J, Song SH, Dean A, Love PE 2010. A requirement for Lim domain binding protein 1 in erythropoiesis. J Exp Med 207: 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Freudenberg J, Cui K, Dale R, Song SH, Dean A, Zhao K, Jothi R, Love PE 2013. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood 121: 4575–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CK, Simpson RJ, Kwan AH, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP 2005. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci 102: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT 2000. The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development 133: 4913–4923 [DOI] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, et al. 2010. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J 29: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev 22: 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet 24: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D 2009. Transcription factors mediate long-range enhancer–promoter interactions. Proc Natl Acad Sci 106: 20222–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci 101: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W 2008. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS ONE 3: e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG 2010. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 11: 109–123 [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M 2003. The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB 2009. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity 31: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van IJcken W, et al. 2010. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S-H, Hou C, Dean A 2007. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell 28: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S-H, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A 2010. Multiple functions of Ldb1 required for β-globin activation during erythroid differentiation. Blood 116: 2356–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Laat W 2011. The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J 30: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Thongjuea S, Andrieu-Soler C, Palstra RJ, Bryne JC, van den Heuvel A, Stevens M, de Boer E, Kockx C, van der Sloot A, et al. 2012. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J 31: 986–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terano T, Zhong Y, Toyokuni S, Hiai H, Yamada Y 2005. Transcriptional control of fetal liver hematopoiesis: dominant negative effect of the overexpression of the LIM domain mutants of LMO2. Exp Hematol 33: 641–651 [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL 2002. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein–protein interactions. Cell 110: 237–249 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA 2009. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- van Meyel DJ, O’Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB 1999. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol Cell 4: 259–265 [DOI] [PubMed] [Google Scholar]
- von Lindern M, Deiner EM, Dolznig H, Parren-Van AM, Hayman MJ, Mullner EW, Beug H 2001. Leukemic transformation of normal murine erythroid progenitors: v- and c-ErbB act through signaling pathways activated by the EpoR and c-Kit in stress erythropoiesis. Oncogene 20: 3651–3664 [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 16: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM 2011. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci 108: 14443–14448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol 23: 7585–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41: 694–698 [DOI] [PubMed] [Google Scholar]