dsRNA-dependent protein kinase R (PKR) plays a key role in innate immunity. PKR binds viral dsRNA and undergoes autophosphorylation, which leads to translational repression and signaling pathway modulation in infected cells. Kim et al. now show that PKR is activated during mitosis in uninfected cells. PKR interacts with dsRNAs formed by inverted Alu repeats, which become accessible to PKR during mitosis. Phosphorylated PKR then suppresses translation and coordinates mitosis. This study unveils a novel function of PKR and endogenous dsRNA mitosis in uninfected cells.
Keywords: dsRNA, PKR, translation, cell cycle
Abstract
dsRNA-dependent protein kinase R (PKR) is a ubiquitously expressed enzyme well known for its roles in immune response. Upon binding to viral dsRNA, PKR undergoes autophosphorylation, and the phosphorylated PKR (pPKR) regulates translation and multiple signaling pathways in infected cells. Here, we found that PKR is activated in uninfected cells, specifically during mitosis, by binding to dsRNAs formed by inverted Alu repeats (IRAlus). While PKR and IRAlu-containing RNAs are segregated in the cytosol and nucleus of interphase cells, respectively, they interact during mitosis when nuclear structure is disrupted. Once phosphorylated, PKR suppresses global translation by phosphorylating the α subunit of eukaryotic initiation factor 2 (eIF2α). In addition, pPKR acts as an upstream kinase for c-Jun N-terminal kinase and regulates the levels of multiple mitotic factors such as CYCLINS A and B and POLO-LIKE KINASE 1 and phosphorylation of HISTONE H3. Disruption of PKR activation via RNAi or expression of a transdominant-negative mutant leads to misregulation of the mitotic factors, delay in mitotic progression, and defects in cytokinesis. Our study unveils a novel function of PKR and endogenous dsRNAs as signaling molecules during the mitosis of uninfected cells.
Protein kinase R (PKR) was originally identified as a kinase that is activated by poliovirus dsRNA and inhibits translation (Ehrenfeld and Hunt 1971; Levin et al. 1980). PKR has since been recognized as an innate immune response factor and studied extensively for its role as a translational regulator during viral infection (Nallagatla et al. 2011; Dabo and Meurs 2012). Two dsRNA-binding domains located at the N terminus of the protein recognize a stretch of dsRNAs longer than ∼33 base pairs (bp), which leads to dimerization and subsequent autophosphorylation of the enzyme (Patel et al. 1995). One of the immediate consequences of phosphorylation/activation of PKR is the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) at Ser51 (Meurs et al. 1992). Phosphorylated eIF2α (peIF2α) blocks translational initiation by preventing the GDP-to-GTP exchange of eIF2 by eIF2B. This process depletes the pool of free eIF2 that is necessary to initiate a new round of translation (Sudhakar et al. 2000). peIF2α also prevents the dissociation of eIF2-GDP from the complete initiation complex, preventing the elongation of the 80S complex (Gross et al. 1987).
In addition to eIF2α, activated PKR is known to induce phosphorylation of a number of other substrates, including p53, inhibitor κB-β (IκB-β), and insulin receptor substrate 1 (IRS-1) (Zamanian-Daryoush et al. 2000; Yang et al. 2010; Bennett et al. 2012). Furthermore, different MAPK signaling pathways have been shown to be either positively or negatively regulated by PKR (Takada et al. 2007). Hence, in addition to its role as a regulator of translation, PKR can also act as a cue for multiple signal transduction pathways that respond to infection. Indeed, using PKR-null mice or PKR knockout mouse embryonic fibroblasts, it has been demonstrated that dsRNA-mediated induction of IFN-γ, which is expressed downstream from NF-κB signaling, was diminished in the absence of PKR (Yang et al. 1995; Kumar et al. 1997).
Recent evidence, however, suggested that the physiological function of PKR may extend beyond antiviral response. PKR might be involved in the control of cognition, although the mechanism remains unknown (Zhu et al. 2011). PKR activity is modulated in proliferation and cell cycle progression, with its peak activity at the G1/S transition in T98G glioblastoma cells (Zamanian-Daryoush et al. 1999). Long-term overexpression of PKR led to G2/M-phase arrest in CHO cells (Dagon et al. 2001). Furthermore, PKR has been shown to act as a tumor suppressor, as overexpression of transdominant-negative PKR (TN PKR) induced malignant transformation in NIH 3T3 cells (Koromilas et al. 1992). Consistent with this observation, ectopic expression of TAR RNA-binding protein (TRBP), a cellular inhibitor of PKR, also resulted in malignant transformation (Benkirane et al. 1997).
Although these studies implicated that PKR may regulate cellular proliferation and that its down-regulation may affect cell cycle progression, the underlying mechanism of PKR activation and the identity of its downstream targets in the cell cycle remain unknown. In this study, we found that PKR is specifically activated during early phases of mitosis by binding to a double-stranded structure formed by inverted Alu repeats (IRAlus) located in the 3′ untranslated region (UTR) of numerous mRNAs. Activated PKR then suppresses global translation by phosphorylating eIF2α and acts as an upstream kinase of c-Jun N-terminal kinase (JNK). Our findings demonstrate that PKR activation is tightly regulated during mitosis, and its activity is required for proper cell division.
Results
PKR and its downstream targets are phosphorylated during mitosis
While studying various dsRNA-binding proteins, including PACT, TRBP, and DICER, we unexpectedly found that PKR is phosphorylated specifically in mitotic HeLa cells (Fig. 1A, green). Note that mitotic cells could be distinguished from those in interphase by the round cell morphology and the condensed chromosomes visualized by DAPI staining (Fig. 1A, blue). In these mitotic cells, we could also detect strong phosphorylation of eIF2α (Fig. 1B, magenta) and JNK (pJNK) (Fig. 1C, yellow), two downstream targets of phosphorylated PKR (pPKR), indicating that PKR is indeed activated. This notion was further supported by Western blotting on cell cycle-arrested samples using another pPKR antibody (Fig. 1D). To arrest cells at either S or M phase, we used a thymidine–thymidine or thymidine–nocodazole double-block method, respectively. In cells treated with nocodazole, which disrupts polymerization of microtubule and arrests cells at prometaphase, phosphorylation of PKR and its downstream targets was enhanced (Fig. 1D). We further confirmed our results on cell cycle-arrested cells via immunocytochemistry. Nocodazole treatment significantly enriched cells that are positive for pPKR, peIF2α, and pJNK (Supplemental Fig. S1).
Figure 1.
M-phase-specific phosphorylation of PKR and its downstream targets. (A) Immunocytochemistry revealed a strong signal of pPKR in mitotic cells (green), while total PKR (red) is predominantly cytosolic. (B,C) In addition to pPKR, peIF2α (B) and pJNK (C) signals were observed in cells undergoing mitosis. Bars, 20 µm. (D) Western blotting on AS or S- or M-phase-arrested cells confirmed activation of PKR and phosphorylation of eIF2α and JNK during mitosis. (E) A subcellular fractionation experiment further suggested that pPKR is closely associated with nuclear/chromatin (N). (F) Pharmacological inhibition of PKR using 1 µM imidazole/oxindole (Imidazole) or 50 mM 2-aminopurine (2-AP) resulted in a decrease in peIF2α and pJNK. The numbers denote treatment time in minutes. (G) Close examination of patterns of pPKR (green), pJNK (yellow), and peIF2α (magenta) at different phases of mitosis. Bars, 20 µm.
Unlike the immunocytochemistry data, Western blot analysis showed a weak signal of pPKR and peIF2α even in the asynchronous (AS) and S-phase-arrested cells. To test the specificity of this signal, we incubated cell lysates with phosphatase for 30 min before loading them onto SDS-PAGE gel. While the enhanced signal of pPKR and peIF2α of the M-phase samples was diminished, the signal for AS and S-phase-arrested samples was unaffected by the treatment (Supplemental Fig. S2). Although we cannot rule out the possibility of background activation of pPKR in interphase, our data suggest that the pPKR and peIF2α bands detected in AS and S-phase-arrested cells most likely reflect the nonspecific binding of the antibodies to the unphosphorylated forms of the proteins.
Notably, we observed a strong signal of pPKR on the chromosomes of cells undergoing mitosis (Fig. 1A, green). pPKR was also detected in the cytosol, but stronger pPKR intensity was observed near the chromosomes. On the contrary, total PKR is more enriched in the cytosol and rather excluded from the chromosome area (Fig. 1A, red). A similar localization pattern was observed for pJNK, albeit to a lesser extent (Fig. 1C). Since the nuclear envelope is disintegrated during mitosis, pPKR must be closely associated with chromosomes in order to show such tight localization. To test this, we performed subcellular fractionation and examined the subcellular localization of pPKR and its downstream targets (Fig. 1E). Briefly, we lysed the S- or M-phase-arrested cells using a low percentage of detergent and separated cytosolic (C) and nuclear/chromatin (N) compartments (see the Materials and Methods for details). Subsequent Western blotting revealed that pPKR is present almost exclusively in the chromatin-associated fraction in the M-phase cells, in agreement with the immunocytochemistry data. The two downstream targets, peIF2α and pJNK, also showed a chromatin-associated pattern but were also detected in the cytosolic fraction. Considering the tight localization of pPKR, it is likely that these substrates are phosphorylated mainly at the chromatin and diffuse out to the cytosol. DROSHA and GAPDH were used as controls for soluble nuclear and cytosolic proteins, respectively. Chromatin-associated protein HISTONE H3 (H3) was detected exclusively in the nuclear/chromatin fraction.
Next, we treated M-phase-arrested cells with two different chemical inhibitors of PKR: imidazole/oxindole, denoted as Imidazole (Jammi et al. 2003), and 2-aminopurine, denoted as 2-AP (Hu and Conway 1993). Pharmacological inhibition of PKR for just 1 h resulted in a significant decrease in the level of peIF2α as well as pJNK (Fig. 1F), indicating that PKR may indeed be responsible for the phosphorylation of these proteins during mitosis.
We next examined the dynamic change of pPKR and its target phosphorylation status through mitosis (Fig. 1G). Since different mitotic phases show distinct morphological features, we searched for cells at different phases of mitosis in an AS population. The strong pPKR signal was first detected in the chromatin area at prometaphase, when condensed chromosomes were readily observed. A strong chromatin-associated signal persisted through metaphase and began to dissipate as chromosomes were separated at anaphase, but the cytosolic signal was still higher than the background. By the end of telophase, the pPKR signal was no longer detected. The pattern of pJNK was similar to that of pPKR, with a slight delay (Fig. 1G, yellow); the chromosome-associated pJNK signal was detected until metaphase and disappears gradually from the chromosomal region. peIF2α was also first detected at the prometaphase, but its dephosphorylation exhibited a clear time delay in which its phosphorylation lasted through telophase, and a weak signal still remained even at the end of cytokinesis (Fig. 1G, magenta).
To test whether the observed activation of PKR is specific to HeLa cells or occurs in general, we performed immunocytochemistry on K562 leukemia cells and MDA-MB-231 breast cancer cells and asked whether PKR and its downstream targets are phosphorylated specifically during mitosis. We indeed detected strong signals for pPKR, peIF2α, and pJNK in cells with condensed chromosomes that indicate cells in mitosis (Fig. 2A). Thus, the activation of PKR and the phosphorylation of its downstream targets also occur in different cancer cell lines. Going beyond cancer cells, we confirmed the expression of pPKR, peIF2α, and pJNK in human primary fibroblast cells (CCD-986sk) and mouse embryonic stem cells (R1) undergoing mitosis (Fig. 2A). Taken together, the observed phenomenon occurs in noncancer and nonhuman cells as well. The above results were also confirmed via Western blotting and immunostaining on cells arrested at S or M phase (Fig. 2B–D; Supplemental Fig. S3). We could not arrest CCD-986sk cells because fibroblast cells undergo mitotic slippage and adaptation (Lanni and Jacks 1998).
Figure 2.

Mitotic activation of PKR is a conserved phenomenon. (A) Mitotic activation of PKR and phosphorylation of its downstream targets were observed in multiple cell lines. Arrowheads indicate mitotic cells. Bars, 20 µm. (B–D) Western blotting on cell cycle-arrested K562 (B), MDA-MB-231 (C), or R1 (D) cells also confirmed the wide occurrence of the PKR regulatory module during mitosis.
Mitotic activation of PKR requires dsRNA
As a first step to understanding the function of PKR activation during mitosis, we sought to identify the upstream signal that leads to phosphorylation of PKR. PKR can be activated by binding to heparin, PACT, and a stretch of dsRNA greater than ∼33 bp (for review, see Cole 2007). Since the mechanism of PKR activation by heparin is not well characterized, we focused on the latter two candidates. First, we examined the subcellular localization and expression level of PACT and concluded that they remained constant through the cell cycle (Supplemental Fig S4A,B). Moreover, the interaction between PKR and PACT did not change between S- and M-phase-arrested cells (Supplemental Fig. S4C). Last, we knocked down PACT using siRNA and examined its effect on pPKR. Immunocytochemistry analysis showed that while PACT knockdown was successful, there was no difference in the pPKR signal between siLuc and siPACT transfected cells undergoing mitosis (Supplemental Fig. S4D).
On the contrary, treating cells with 5 µg/mL Actinomycin D (Act D) for 3 h resulted in a significant decrease in pPKR in mitotic cells (Fig. 3A), suggesting that cellular transcripts may be required for PKR activation. Note that the mitotic cells were distinguished by looking for round-shaped cells with a disrupted LAMIN A/C signal. As a control, we stained cells for NUCLEOPHOSMIN (NPM1) to visualize nucleoli whose structural integrity depends on the synthesis of RNA (Gontcharoff and Rao 1972). Disruption of the nucleolar structure in cells incubated with Act D confirmed that the drug treatment was successful (Fig. 3B).
Figure 3.
Mitotic activation of PKR depends on dsRNA. (A) Treating cells with 5 µg/mL Act D for 3 h resulted in a significant decrease in pPKR signal. (B) As a positive control, disruption of nucleolus localization of NPM1 was observed under the same treatment conditions. (C) The effect of different enzyme treatments on the pattern of pPKR and its downstream targets. We used enzymes that can digest dsDNA (DNase I); ssRNA (RNase T1); ssRNA and dsRNA (RNase A); and dsDNA, ssRNA, and dsRNA (MNase). H3 and NPM1 were used as controls. Bars, 20 µm.
To further assess the role of RNA in PKR activation during mitosis, we used different enzymes that can digest dsDNA, ssRNA, and/or dsRNA and examined their effects on the pattern of pPKR and its downstream targets (Fig. 3C). We treated permeabilized cells with DNase I (for dsDNA), RNase T1 (for ssRNA), RNase A (for ssRNA and dsRNA), or MNase (for dsDNA, ssRNA, and dsRNA) for 10 min prior to fixation. Interestingly, cytosolic pPKR signal was increased at the expense of the chromatin-associated pPKR signal when dsDNA was digested (Fig. 3C). RNase T1 only slightly decreased the pPKR signal intensity, while a stronger effect was observed when RNase A, which can digest both ssRNA and dsRNA, was used (Fig. 3C). pPKR signal was completely abolished when cells were treated with MNase (Fig. 3C). The pattern of pJNK showed a change of patterns very similar to those of pPKR in response to different enzyme treatments (Fig. 3C, yellow). The cytosolic pattern of peIF2α was unaffected, but its intensity showed changes correlated to those of pPKR (Fig. 3C, magenta). H3 and NPM1 were used as controls whose localization depends on DNA or RNA, respectively (Fig. 3C). Overall, these results suggest dsRNA as a likely class of inducers that is responsible for PKR activation. Phosphorylation of PKR may be a dynamic process in which constant influx is needed to maintain strong expression of pPKR.
PKR binds to IRAlu elements during mitosis
One possible class of RNA activators of PKR is IRAlus residing in the 3′ UTR of mRNAs. Alu elements, which comprise the most abundant type of short interspersed elements (SINEs), constitute >10% of the human genome (Lander et al. 2001). Some Alu elements in the 3′ UTR are transcribed as part of an mRNA and can induce post-transcriptional regulation (Brosius 1999). More importantly, when an Alu element is followed by another Alu element in reverse complement orientation within a single transcript, the two, denoted as IRAlus, can form an intramolecular dsRNA that can be recognized by dsRNA-binding protein ADAR to undergo A-to-I editing (Athanasiadis et al. 2004; Blow et al. 2004). Considering the close homology between dsRNA-binding domains of ADAR and those of PKR (Chang and Ramos 2005), it is likely that these intramolecular dsRNAs are also recognized by PKR. Furthermore, Alu elements are typically 300 nucleotides (nt) long and show high sequence similarity to each other. Thus, IRAlus will form dsRNA that is long enough to bind to and activate PKR (Elbarbary et al. 2013; Ricci et al. 2014). Consistent with this, STAUFEN-1 (STAU) and PKR compete for binding to IRAlus, and IRAlus can induce PKR phosphorylation and subsequent translational suppression when the level of STAU is decreased (Elbarbary et al. 2013).
Another line of evidence favoring IRAlus as likely mitosis-specific activators of PKR is their subcellular localization. Previously, it had been shown that intramolecular dsRNAs formed by IRAlu elements are retained in the nucleus by binding to p54nrb, which sequesters the RNA in hNEAT1-containing paraspeckles (Chen et al. 2008). We hypothesized that, during mitosis, these nuclear RNAs will diffuse out and interact with PKR as the nuclear envelope breaks down. Together, this evidence suggest that in interphase, IRAlu elements are sequestered in the nucleus and segregated from cytosolic PKR, but, during mitosis, mixing of nuclear and cytosolic components allows the two to interact, which can lead to mitosis-specific phosphorylation of PKR.
To test the above hypothesis, we first examined the subcellular localization of reporter mRNAs via fluorescent in situ hybridization (FISH). These reporters contain either a single Alu (for Alu) or a pair of Alus in reverse complement orientation (for IRAlus) from NICOLIN 1 (NICN1) 3′ UTR (Fig. 4A; Chen et al. 2008). We designed an antisense RNA probe that targets the EGFP region that is commonly present in both reporters. We observed that in interphase cells, the IRAlu reporter shows nuclear accumulation, while a single Alu reporter is predominantly cytosolic (Fig. 4B). The low EGFP protein signal from the IRAlu reporter is also consistent with the previous data that IRAlus can suppress translation via nuclear retention of the hosting mRNA (Fig. 4B; Chen et al. 2008), but, during mitosis, the nuclear IRAlu mRNAs become dispersed throughout the cell, which largely overlaps with the pattern of total PKR at this time, as illustrated in Figure 1A.
Figure 4.
PKR binds to IRAlus during mitosis. (A) Schematics of Alu and IRAlu EGFP reporters used in this study, modified from Chen et al. (2008). (B) Using FISH, subcellular localization of Alu or IRAlu reporters was visualized. In interphase cells, the presence of IRAlus results in nuclear retention of the mRNA (red) and subsequent translational suppression, indicated by a low EGFP protein signal (green). However, during mitosis, both Alu and IRAlu reporter mRNAs are distributed throughout the cytosol. (C) Normalized log2 fold enrichment of the indicated RNAs relative to the GAPDH mRNA in a PKR RNA immunoprecipitation experiment. The RNAs were quantified by qRT–PCR. The left seven genes are predicted to contain IRAlu elements in their 3′ UTRs based on Chen et al. (2008). The averages of at least four replicates for each gene are plotted, with error bars indicating standard error of the mean. (D) FISH on MRTO4, MCM4, PSMB2, RPL15, and RPL13 mRNAs that are predicted to contain IRAlu elements. GAPDH and β-actin (ACTB) mRNAs were used as control mRNAs without IRAlus.
We next tested whether PKR physically associates with the IRAlu-containing mRNAs in a cell cycle-specific manner. We transfected HeLa cells with IRAlu reporter and arrested them at either S or M phase. The interaction between PKR and RNA is reduced upon phosphorylation (Jammi and Beal 2001), indicating that the binding between PKR and its activating RNAs may be transient. Thus, we cross-linked cells by using formaldehyde before cell lysis to preserve any protein–RNA interactions and then performed immunoprecipitation using anti-PKR antibody. Cross-linking also allows harsh washing that reduces artificial interactions formed after cell lysis. Coimmunoprecipitated RNAs were extracted, and the enrichment of the reporter mRNA was examined via quantitative RT–PCR (qRT–PCR). We found that the IRAlu reporter mRNA was significantly enriched in the M-phase-arrested cells (Fig. 4C, EGFP-NICN1). Furthermore, IRAlu-containing EGFP mRNAs did not show any enrichment in S-phase-arrested cells, indicating that IRAlus may be M-phase-specific activators of PKR.
In addition to the reporter, we examined endogenous mRNAs with at least one pair of IRAlu elements. Previously, bioinformatics analysis predicted that at least 333 human genes contain IRAlus in their 3′ UTRs (Chen et al. 2008). The majority of these transcripts undergo partial editing by ADAR, suggesting that these transcripts will likely form dsRNA structures in cells (Chen et al. 2008). We found that, similarly to IRAlu reporter mRNA, most of the transcripts with predicted IRAlu elements also interact with PKR (Fig. 4C). Two of the transcripts (MRTO4 and CYCS) showed enrichment comparable with that of IRAlu reporter EGFP mRNA. Importantly, enrichment of these mRNAs was observed only in the M-phase-arrested cells. Furthermore, among the seven control RNAs that were not predicted to contain IRAlus (no-IRAlus), none of them shows significant binding to PKR. Although the enrichment levels were variable, the mRNAs with predicted IRAlus tend to bind more frequently to PKR compared with those without IRAlus.
We then examined the subcellular localization of endogenous mRNAs with IRAlus using FISH. We found that three of the predicted IRAlu mRNAs, MRTO4, MCM4, and PSMB2, show strong nuclear signals during interphase that diffuse out to cytosol during mitosis (Fig. 4D). The nuclear retention pattern of IRAlu-containing transcripts is clearly different from that of other transcripts such as GAPDH and β-ACTIN mRNA, which are predominantly cytosolic (Fig. 4D). Notably, not all IRAlu-containing mRNAs were localized in the nucleus. Two ribosomal mRNAs, RPL15 and RPL13, showed highly cytosolic localization despite the predicted IRAlu elements. One possible explanation is that multiple isoforms with different 3′ UTRs are generated by alternative processing. For example, one of the isoforms of NICN1 mRNA localizes in the cytosol because it contains only one of the two Alu repeats required to form the double-stranded structure (Chen et al. 2008). We also examined the localization of CYCS mRNA, but its signal was too weak for reliable detection (data not shown). Thus, the intramolecular dsRNAs formed by IRAlu elements may bind to and activate PKR in mitosis, a period during which the nuclear envelope is absent.
PKR suppresses bulk translation during mitosis
It is well established that mitotic cells synthesize proteins at a much slower rate compared with cells in interphase (Prescott and Bender 1962; Tarnowka and Baglioni 1979). This decrease in translation has been attributed to the inhibition of 5′ cap-dependent translation by 14-3-3σ (Wilker et al. 2007) and hypophosphorylation of eIF4E-binding proteins (4E-BPs) (Pyronnet et al. 2001), which result in suppression of translation in most mRNAs, while certain mRNAs with internal ribosomal entry sites (IRESs) show increased translational activity. This switch to IRES-dependent translation is thought to be necessary for mitotic progression and to initiate interphase of the daughter cells. However, another line of evidence suggests that 4E-BPs are hyperphosphorylated instead of hypophosphorylated during mitosis and that hyperphosphorylated 4E-BPs cannot inhibit cap-dependent translation because they no longer interact with eIF4E (Heesom et al. 2001; Ramirez-Valle et al. 2010). Our result is also consistent with the latter observation that mTOR, an upstream kinase of 4E-BPs, is still phosphorylated and that 4E-BP1 is hyperphosphorylated during mitosis in HeLa and other cell lines examined (Figs. 1D, 2B–D; Supplemental Fig. S2). At the same time, it has been shown that eIF2α also contributes to translational regulation during mitosis, as it is phosphorylated in G2/M phase of U2-OS osteosarcoma cells (Datta et al. 1999), and its phosphorylation can up-regulate the efficiency of IRES-mediated translation (Gerlitz et al. 2002).
Considering that eIF2α is the most well-characterized substrate of PKR, we asked whether PKR contributes to the suppression of general translation during mitosis. We started by characterizing mitotic translation of HeLa cells. We performed metabolic labeling and assessed protein synthesis rates by quantifying 35S-methionine incorporation rates. We found that during mitosis, the rate of protein synthesis was ∼35% of that of the S phase, which is consistent with earlier studies (Fig. 5A; Prescott and Bender 1962; Tarnowka and Baglioni 1979). Ribosome fractionation using a sucrose gradient also showed a significant increase in the monosome peak at the expense of polyribosome peaks in M-phase-arrested cells (Supplemental Fig. S5A,B). This increase in the 80S monosome peak is consistent with the characteristics of the translation block at the initiation step (Ricci et al. 2014). A similar pattern of ribosomes was observed when eIF2α phosphorylation was induced by nutritional stress (Tonelli et al. 2011). We then treated M-phase-arrested cells with two different PKR inhibitors at multiple time points and examined the changes in the peIF2α signal. The peIF2α signal was significantly down-regulated within 30 min after the drug treatment and nearly completely abolished by 2 h (Fig. 5B,C). A decrease in the peIF2α signal was accompanied by an increase in general translation; pharmacological inhibition of PKR for 1 h in M-phase-arrested cells increased the methionine incorporation rate to ∼60% of that of the S phase (Fig. 5D; Supplemental Fig. S5C).
Figure 5.
PKR controls mitotic translation. (A) Protein synthesis rate in S- or M-phase-arrested cells can be assessed using 35S-methionine incorporation rates. An average of three biological replicates is shown. (*) P < 0.01. (B,C) Pharmacological inhibition of PKR using 1 µM imidazole/oxindole (Imidazole) (B) or 50 mM 2-aminopurine (2-AP) (C) resulted in a marked decrease in the peIF2α (magenta) signal within 30 min. Bars, 20 µm. (D) Metabolic labeling on M-phase-arrested cells treated with the PKR inhibitors for 1 h reveals that the decrease in peIF2α leads to an increase in the general translation rate (n = 6). (*) P < 0.01. (E) A similar increase in protein synthesis rate was observed when PKR was knocked down in M-phase-arrested cells (n = 6). (*) P < 0.01. (F) Knockdown of PKR in synchronized cells resulted in a large increase in the protein synthesis rate at mitosis and early G1 phase (n = 3). The numbers at the bottom indicate hours after release from the double-thymidine block. (*) P < 0.01.
In order to rule out possible side effects of PKR inhibitors, we used RNAi to further confirm our results. When PKR was depleted in M-phase-arrested cells, the general translation was increased by ∼50% (Fig. 5E; Supplemental Fig. S5D). In addition, we knocked down PKR in cells synchronized at S phase via the thymidine double-block method and performed metabolic labeling upon releasing them from the block. Both the control and siPKR transfected cells progressed through the cell cycle in a similar manner, although the PKR-depleted cells showed moderate accumulation of the G2/M fraction (Supplemental Fig. S6). A metabolic labeling experiment revealed that down-regulation of PKR led to an increase in the protein synthesis rate, with the most prominent effect occurring when the cells undergo mitosis (Fig. 5F; Supplemental Fig. S5E). Upon release from the double-thymidine block (see Supplemental Fig. S6 for FACS analysis), siPKR transfected cells showed increased translation by <10% during S and G2 phases. However, 10 and 12 h after the release, which corresponds to M and early G1 phase, PKR knockdown resulted in an at least 25% increase in the protein synthesis rate in three biological replicates. The magnitude of the effect of PKR knockdown was variable in each biological replicate, but we reproducibly observed the strongest increase in the protein synthesis rate during this period. Combined, these data indicate that PKR activation significantly contributes to the suppression of translation during mitosis.
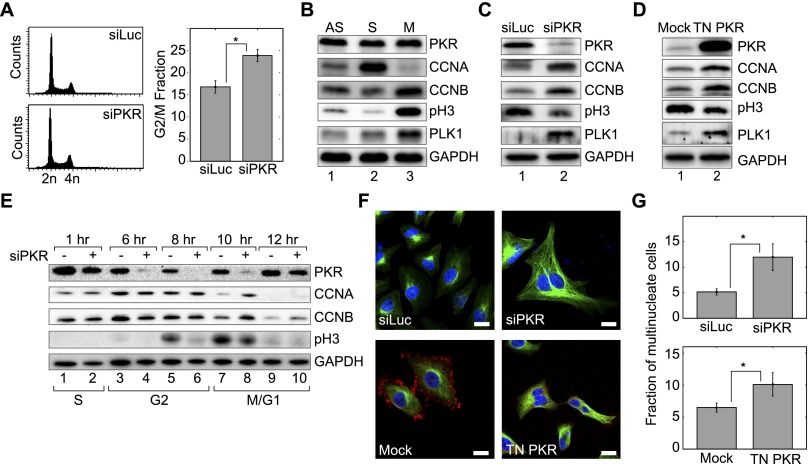
PKR regulates mitotic factors
To further characterize PKR activation and its effect on cell cycle progression, we performed FACS analysis after transfecting AS cells with siPKR for 48 h. We consistently observed a small but statistically significant increase in the number of cells in the G2/M fraction (Fig. 6A). We then examined whether PKR can control the expression patterns of mitotic factors that are specifically regulated in G2/M phase (Fig. 6B). The levels of CYCLIN A (CCNA), CYCLIN B (CCNB), and POLO-LIKE KINASE 1 (PLK1) and the phosphorylation status of HISTONE H3 (pH3) were monitored by Western blotting. In control cells, CCNA expression reaches the peak level in G2 phase and is quickly degraded upon entering mitosis (Fig. 6B,E). The rest of the proteins are known to be highly expressed during G2 and M phases. In PKR-depleted cells, we observed a decrease in the pH3 level, while the levels of the other proteins were elevated (Fig. 6C).
Figure 6.
PKR regulates expression of multiple mitotic factors. (A) FACS analysis on cells transfected with siPKR revealed an increase in the G2/M population. An average of seven biological replicates is shown. (*) P < 0.01. (B) Western blotting of the expression patterns of proteins that are highly regulated during G2/M phase. (C) Knockdown of PKR in AS cells led to an increased level of CCNA, CCNB, and PLK1, while the level of pH3 was decreased. (D) Overexpression of TN PKR resulted in increased expression of CCNA, CCNB, and PLK1, while the level of pH3 was decreased. (E) Western blotting on synchronized cells also showed that the two cyclins are increased, while pH3 was decreased in PKR-depleted cells. The numbers at the top indicate hours after release from the double-thymidine block. (F) Disruption of PKR activation via RNAi (top) or ectopic expression of TN PKR (bottom) resulted in improper cytokinesis and accumulation of multinucleate cells. Green denotes α-tubulin, and blue shows DAPI. For TN PKR, the red signal denotes successful expression of TN PKR, which is tagged with Flag. Bars, 20 µm. (G) An average percentage of multinucleate cells from six biological replicates when PKR was knocked down using RNAi (top) or ectopic expression of TN PKR (bottom). For each biological replicate, the fraction of multinucleate cells was determined by counting ∼1000 cells in total. (*) P < 0.01.
To rule out possible off-target effects of siRNAs, we confirmed our results by transfecting cells with TN PKR. This PKR variant has two amino acid substitutions in its catalytic domain, making the enzyme inactive. Since the mutant PKR still contains intact dsRNA-binding domains, it is expected to compete with endogenous PKR for binding to dsRNAs and thereby acts as a transdominant negative. Consistent with the knockdown experiments, ectopic expression of TN PKR also showed similar effects: a small but reproducible increase of the G2/M fraction (data not shown); up-regulation of CCNA, CCNB, and PLK1; and down-regulation of pH3, albeit to a lesser extent compared with PKR knockdown (Fig. 6D).
To understand the kinetics of molecular changes during the G2/M transition, we synchronized cells at S phase using the thymidine double-block method and collected the cells at different time points after releasing them from the block. Both CCNA expression and CCNB expression persisted longer in PKR-depleted cells, as they are still detected strongly 10 h after the release, unlike the control cells (Fig. 6E). In addition, pH3 appeared later, with a decreased expression level in siPKR-treated cells (Fig. 6E). In normal cells, CCNA and CCNB are expressed during G2 phase and degraded during M phase, while pH3 appears specifically in M phase. Expression of pH3 at the 8-h control sample likely reflects a small population of cells that have already entered mitosis. Therefore, our data suggest that PKR knockdown affects mitotic progression and that PKR is required to properly turn off G2-phase genes and induce M-phase factors.
Our current data suggest that PKR may exert some of its effects through JNK phosphorylation during mitosis. The mitotic activation of JNK has been documented previously (Ribas et al. 2012), but the identity of the upstream cue for JNK during mitosis remains unknown. We showed that JNK is phosphorylated in mitotic HeLa cells in a PKR-dependent manner (Figs. 1F, 3C). We also noticed that many of the effects of PKR knockdown are very similar to those reported for JNK inhibition: increased expression of CCNB but a decreased pH3 level (Oktay et al. 2008). Furthermore, the activation of JNK has been implicated to promote mitotic entry (Oktay et al. 2008). This is also consistent with our observation that PKR knockdown resulted in the abolition of pJNK expression and misregulation of G2/M factors (Figs. 1F, 6E). Hence, PKR may act as a signaling cue that is responsible for JNK activation during mitosis.
To determine the physiological role of PKR, we asked whether the abnormal expression of mitotic factors could lead to defects in cell division. Previous reports have suggested that modulation of the PLK1 level by either overexpression or knockdown resulted in the significant increase in the population of cells with more than one nucleus (Meraldi et al. 2002), indicative of defects in cytokinesis following separation of chromosomes. Hence, we tested whether PKR can affect cytokinesis by transfecting cells with siPKR two consecutive times over a 90-h period and then counting the number of multinucleate cells. In comparison with control transfected samples (siLuc), we observed a significant increase in the number of cells with more than one nucleus (Fig. 6F,G). We also observed a similar effect when we introduced TN PKR (Fig. 6F,G). Thus, PKR is necessary for the regulation of multiple mitotic factors, signaling, and, ultimately, proper cell division.
Discussion
Previous studies on PKR have been focused on its role in innate immunity against exogenous genetic materials (Nallagatla et al. 2011; Dabo and Meurs 2012). Here, we uncover the cellular function of PKR as a mitotic regulator that responds to cellular dsRNAs. According to our model illustrated in Figure 7, PKR is predominantly localized in the cytosol during interphase, where it is segregated from nuclear RNAs with double-stranded secondary structure such as those containing IRAlu elements. At the onset of mitosis, the nuclear envelope disintegrates, which allows mixing of nuclear and cytosolic components. PKR gains access to these dsRNAs and undergoes dimerization and autophosphorylation. Phosphorylated PKR then suppresses general translation and acts as an upstream regulator of JNK, coordinating the global network of the mitotic program.
Figure 7.
A model for the regulation of PKR in the cell cycle. (A) In interphase, cytosolic PKR remains inactive, as PKR-activating dsRNAs are sequestered in the nucleus. (B) However, during mitosis, disintegration of the nuclear envelope allows interaction between PKR and dsRNAs. Activated PKR then catalyzes phosphorylation of eIF2α and JNK to regulate mitotic translation and coordinate mitotic processes.
In addition to PKR, cells express other dsRNA sensors, such as Toll-like receptor 3 (TLR3), retinoic acid-inducible gene-1 (RIG-I), and melanoma differentiation-associated gene 5 (MDA5), as a defense mechanism against viral infection (for review, see Peisley and Hur 2013). However, the ligand recognition domain of TLR3 faces the endosomal lumen and will not be able to interact with cytosolic IRAlus. In addition, RIG-I recognizes the 5′-triphosphate end of dsRNA (Hornung et al. 2006), which is not present in IRAlus that form intramolecular dsRNA. Last, MDA5 signaling activity is correlated with the length of the dsRNA and typically requires an ∼1- to 7-kb stretch for sufficient activity (Kato et al. 2008), while the Alu element is only ∼300 nt long. Therefore, PKR provides a unique opportunity for cells to use these cellular dsRNAs as a regulatory element to control important cellular processes such as mitosis.
When PKR activation is impaired by transfection of siRNA or ectopic expression of TN PKR, one of the most prominent effects is defects in cytokinesis, resulting in increased multinucleate cell population. One possible explanation is the increased PLK1 expression. Not mutually exclusively, increased general translation from dephosphorylation of eIF2α might contribute to the defect. In support of this, cells lacking 14-3-3σ show an increased protein synthesis rate during mitosis, which results in cytokinesis defects (Wilker et al. 2007). Considering the chromatin-associated localization of pPKR, it is also possible that PKR may play a more direct role in chromosome segregation. In the future, detailed high-throughput analysis will be necessary to identify molecular players, map possible PKR-binding sites on chromosomes, and elucidate the mechanism behind the effects of PKR depletion.
The mitotic activation of PKR and phosphorylation of its downstream targets are observed in several cell lines of different origins, suggesting that the proposed regulation of mitosis by PKR might be a general conserved phenomenon. It is particularly interesting that PKR and its downstream targets are activated in R1 embryonic stem cells. In both mouse and human embryonic stem cells, interferons are not induced in response to viral infection or the introduction of long dsRNAs (Chen et al. 2010; Wang et al. 2013). However, these cells still express functional PKR capable of catalyzing phosphorylation of eIF2α. Our current results suggest that PKR may play a role in stem cells not for its ability as an antiviral agent but for proper cell division and cell cycle progression.
It is noteworthy that a decreased expression of PKR is one of the hallmarks in many types of cancer, although the mechanism behind PKR misregulation remains unknown (Jagus et al. 1999). Our current study shows that PKR depletion resulted in a significant number of cells failing to undergo proper cytokinesis and accumulation of a multinucleated population. Multinucleated cells can lead to aneuploidy and mark early events in tumorigenesis (Fujiwara et al. 2005). In addition, down-regulation of PKR by ectopic expression of TN PKR or TRBP resulted in malignant transformation (Koromilas et al. 1992; Benkirane et al. 1997). PKR knockout mice were viable and did not show spontaneous tumor formation when monitored for a year after birth (Abraham et al. 1999). It is plausible that a different eIF2α kinase might have compensated for the loss of PKR in these mice. The compensation among different eIF2α kinases has been demonstrated in yeast, where expression of mammalian eIF2α kinases PKR and heme-regulated inhibitor can partially rescue the phenotypes of GCN2 knockout, a conserved eIF2α kinase (Dever et al. 1993). In addition, these PKR knockout mice could have survived by modulating other pathways to compensate for the loss of PKR. This difference between knockdown and knockout has been observed in multiple systems (for review, see Sherr and Roberts 2004). Further detailed characterization of PKR will help us to understand the contribution and mechanism of PKR in tumorigenesis. Our findings now provide an opportunity to probe the role of the dsRNA–PKR pathway in normal and aberrant cell cycles.
In this study, we identified IRAlus as a possible class of PKR activators during mitosis. A recent report suggested that IRAlus can also activate PKR in interphase. Elbarbary et al. (2013) showed that STAU binds to and allows the export of IRAlu-containing mRNAs to cytosol. This nuclear export of IRAlu mRNAs mediated by STAU (in addition to mRNA isoforms with alternative polyadenylation sites) can be one of the mechanisms by which these mRNAs are translated. More importantly, STAU still binds to IRAlus in the cytosol and limits the access of PKR to these dsRNAs (Elbarbary et al. 2013). This competition between STAU and PKR might account for the lack of pPKR in the nucleus of interphase cells despite the presence of unphosphorylated PKR, as shown in Figure 1E. Consequently, STAU knockdown resulted in increased phosphorylation of PKR and global suppression of translation (Elbarbary et al. 2013). Hence, a low level of pPKR might be present in the cytosol of interphase cells, which can account for the small increase in general translation when PKR is knocked down in interphase (Fig. 5F).
In addition to intramolecular dsRNAs, two Alu elements in two different transcripts may form intermolecular dsRNAs that can be recognized by PKR. Such regulation has been shown to induce STAU-mediated mRNA decay (Gong and Maquat 2011). In this case, an Alu element in the 3′ UTR of SERPINE1 mRNA forms an imperfect dsRNA with another Alu element in 1/2-sbsRNA1 long noncoding RNA (lncRNA) (Gong and Maquat 2011). Thermodynamic analysis predicted that SERPINE1 mRNA can potentially form intermolecular dsRNA with >300 lncRNAs (Gong and Maquat 2011). Considering the wide occurrence of Alu elements in the human genome, virtually any mRNA with Alu in its 3′ UTR can potentially activate PKR. In addition, functional IRAlu elements may reside in introns (Chen et al. 2008) that will also be exposed to cytosolic PKR during mitosis. Furthermore, there might exist other types of dsRNAs that can be recognized by PKR. Recent studies have shown that mRNAs contain multiple internal stem–loop structures that can be recognized by dsRNA-binding proteins such as STAU (Cho et al. 2012; Ricci et al. 2014). Thus, it will be interesting to perform a genome-wide search for RNA activators of PKR and study how they escape PKR activation in interphase.
Currently, it is unclear how pPKR is localized and associated to the chromosome region. Nuclear localization of pPKR has been observed in limited cases, including in CD34+ cells from high-risk myelodysplastic syndrome patients (Follo et al. 2008; Blalock et al. 2011). One possibility is that pPKR binds to an unknown protein factor that tethers the enzyme to the chromatin. For example, substrates of pPKR might be enriched near the chromosome region and recruit PKR. A recent mass spectrometry analysis on PKR interactome revealed that pPKR interacts with proteins involved in chromatin modification and cell division, such as BUB3 and NPM1 (Blalock et al. 2014). Alternatively, it is possible that there may be different classes of dsRNAs that are localized to the chromosome region during mitosis and tether PKR. In this case, the chromosome might serve as a center for activating PKR as well as catalyzing phosphorylation of PKR targets such as eIF2α and JNK. In the future, it will be interesting to identify PKR-interacting proteins and dsRNAs and examine their localized activities at various stages of the cell cycle.
Materials and methods
Cell cycle arrest and PKR inhibitor treatment
To arrest cells at S or M phase, the double-thymidine block method was used. Briefly, cells were treated with 2 mM thymidine for 18 h and then released into fresh medium. After 9 h, 2 mM thymidine (for S-phase arrest) or 100 ng/mL nocodazole (for M-phase arrest) was added to the medium, and cells were incubated for an additional 17 h. When indicated, M-phase-arrested cells were treated with 1 μM imidazole/oxindole (EMD Millipore) or 50 mM 2-aminopurine (Sigma-Aldrich) for 1 h.
Immunocytochemistry
Cells cultured on a coverslip were fixed with 4% paraformaldehyde for 10 min at room temperature. For enzyme treatment, cells were permeabilized with 0.02% Triton and incubated with different enzymes for 10 min at 37°C before fixation. The following enzymes were used in this study: 200 U/mL DNase I (Takara), 1 mg/mL RNase A (Invitrogen), 40 U/mL RNase T1 (Life Technologies), and 40,000 U/mL MNase (New England Biolabs). Fixed cells were then permeabilized in 0.3% Triton and blocked in 1% BSA for 1 h. Cells were incubated with primary antibodies diluted in 1% BSA for 2 h. Alexa fluor-conjugated secondary antibodies were used to label the primary antibodies. The primary antibodies used in this study were PKR (Santa Cruz Biotechnology), pPKR (Santa Cruz Biotechnology), pJNK (Promega), peIF2α (Cell signaling), NPM1 (Abcam), α-Tubulin (Abcam), LAMIN A/C (Santa Cruz Biotechnology), and Flag (Sigma-Aldrich). Stained cells were imaged with a Zeiss LSM 700 confocal microscope using a C-Apochromat 40× lens with an NA of 1.20.
Subcellular fractionation
For the subcellular fractionation experiment, a protocol modified from Wysocka et al. (2001) was used. Cell pellets were lysed by 10 min of incubation on ice in buffer A (10 mM Tris-HCl at pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, Protease inhibitor cocktail [Calbiochem]) supplemented with 0.1% NP-40. Cells were centrifuged at 1300g for 5 min. The supernatant (cytosolic fraction) was cleaned by high-speed centrifugation at 20,000g for 15 min. The pellet fraction was washed once with buffer A and resuspended in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, Protease inhibitor cocktail) for 30 min on ice. The lysate was then sonicated using a Bioruptor and analyzed via Western blotting.
Western blotting
Total cell lysates were prepared using Hepes buffer (20 mM Hepes at pH 7.4, 100 mM NaCl, 20 mM KAc, 10 mM MgCl2, 10 mM ZnCl2, 1% NP-40, 1 mM Na3VO4, 50 mM NaF). For phosphatase treatment, λ phosphatase (Santa Cruz Biotechnology) was used following the manufacturer’s instruction. Thirty micrograms to 50 μg of each protein sample was separated by 10% SDS-PAGE gel and transferred to PVDF membrane using Amersham semidry transfer system. The following primary antibodies were used in this study: anti-PKR, anti-GAPDH, anti-CCNA, anti-CCNB, and anti-PLK1 were purchased from Santa Cruz Biotechnology; anti-pPKR was purchased from Epitomics; anti-pJNK was purchased from Promega; anti-DROSHA was purchased from Abcam; and anti-H3, anti-pH3, anti-peIF2α, anti-pmTOR, anti-mTOR, anti-p4E-BP1, and anti-4E-BP1 were purchased from Cell Signaling.
Formaldehyde cross-linking and immunoprecipitation
Cells were cross-linked using 0.75% formaldehyde in 1× PBS for 10 min at room temperature and quenched with 1 M glycine for 5 min. Cross-linked cells were lysed in 10 mM Tris-HCl (pH 7.4), 500 mM NaCl, 2 mM EDTA, and 0.1% NP-40 and immunoprecipitated for 3 h at 4°C. Normal rabbit IgG from Santa Cruz Biotechnology was used as a negative control, and PKR antibody from Millipore EMD was used.
RNA extraction and qRT–PCR
Total RNA was extracted using Trizol (Life Technologies) following the manufacturer’s protocol. Purified RNA was treated with DNase I (Takara) and reverse-transcribed using RevertAid reverse transcriptase (Fermentas). cDNA was amplified by SYBR Green PCR master mix (Applied Biosystems) and analyzed by the StepOnePlus real-time PCR system. Primers used in this study are provided in Supplemental Table T2.
Acknowledgments
We thank Dr. Ling-Ling Chen and Dr. Gordon G. Carmichael for kindly providing Alu and IRAlu reporters and appreciate Dr. Lynne Maquat’s thoughtful discussion. We are also grateful to the members of our laboratory for their input and helpful discussions. This research was supported by Research Center Program EM1302 of the Institute for Basic Science, the T.J. Park Post-doctoral Fellowship (to Y.K.), and the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2012-Fostering Core Leaders of the Future Basic Science Program) (to J.-E.P.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242644.114.
References
- Abraham N, Stojdl DF, Duncan PI, Methot N, Ishii T, Dube M, Vanderhyden BC, Atkins HL, Gray DA, McBurney MW, et al. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem 274: 5953–5962 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT 1997. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J 16: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RL, Pan Y, Christian J, Hui T, May WS Jr 2012. The RAX/PACT–PKR stress response pathway promotes p53 sumoylation and activation, leading to G(1) arrest. Cell Cycle 11: 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock WL, Bavelloni A, Piazzi M, Tagliavini F, Faenza I, Martelli AM, Follo MY, Cocco L 2011. Multiple forms of PKR present in the nuclei of acute leukemia cells represent an active kinase that is responsive to stress. Leukemia 25: 236–245 [DOI] [PubMed] [Google Scholar]
- Blalock WL, Piazzi M, Bavelloni A, Raffini M, Faenza I, D’Angelo A, Cocco L 2014. Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. J Cell Physiol 229: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR 2004. A survey of RNA editing in human brain. Genome Res 14: 2379–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J 1999. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115–134 [DOI] [PubMed] [Google Scholar]
- Chang KY, Ramos A 2005. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J 272: 2109–2117 [DOI] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG 2008. Alu element-mediated gene silencing. EMBO J 27: 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Yang L, Carmichael GG 2010. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 9: 3552–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN 2012. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151: 765–777 [DOI] [PubMed] [Google Scholar]
- Cole JL 2007. Activation of PKR: an open and shut case?. Trends Biochem Sci 32: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabo S, Meurs EF 2012. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses 4: 2598–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagon Y, Dovrat S, Vilchik S, Hacohen D, Shlomo G, Sredni B, Salzberg S, Nir U 2001. Double-stranded RNA-dependent protein kinase, PKR, down-regulates CDC2/cyclin B1 and induces apoptosis in non-transformed but not in v-mos transformed cells. Oncogene 20: 8045–8056 [DOI] [PubMed] [Google Scholar]
- Datta B, Datta R, Mukherjee S, Zhang Z 1999. Increased phosphorylation of eukaryotic initiation factor 2α at the G2/M boundary in human osteosarcoma cells correlates with deglycosylation of p67 and a decreased rate of protein synthesis. Exp Cell Res 250: 223–230 [DOI] [PubMed] [Google Scholar]
- Dever TE, Chen JJ, Barber GN, Cigan AM, Feng L, Donahue TF, London IM, Katze MG, Hinnebusch AG 1993. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci 90: 4616–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld E, Hunt T 1971. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc Natl Acad Sci 68: 1075–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Li W, Tian B, Maquat LE 2013. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev 27: 1495–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follo MY, Finelli C, Mongiorgi S, Clissa C, Bosi C, Martinelli G, Blalock WL, Cocco L, Martelli AM 2008. PKR is activated in MDS patients and its subcellular localization depends on disease severity. Leukemia 22: 2267–2269 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Gerlitz G, Jagus R, Elroy-Stein O 2002. Phosphorylation of initiation factor-2α is required for activation of internal translation initiation during cell differentiation. Eur J Biochem 269: 2810–2819 [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontcharoff M, Rao B 1972. Dependence of the nucleolar structure on DNA and RNA synthesis. Chromosoma 38: 441–457 [DOI] [PubMed] [Google Scholar]
- Gross M, Wing M, Rundquist C, Rubino MS 1987. Evidence that phosphorylation of eif-2(α) prevents the eif-2B-mediated dissociation of Eif-2·GDP from the 60 S subunit of complete initiation complexes. J Biol Chem 262: 6899–6907 [PubMed] [Google Scholar]
- Heesom KJ, Gampel A, Mellor H, Denton RM 2001. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr Biol 11: 1374–1379 [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Hu YH, Conway TW 1993. 2-Aminopurine inhibits the double-stranded RNA-dependent protein-kinase both in vitro and in vivo. J Interferon Res 13: 323–328 [DOI] [PubMed] [Google Scholar]
- Jagus R, Joshi B, Barber GN 1999. PKR, apoptosis and cancer. Int J Biochem Cell Biol 31: 123–138 [DOI] [PubMed] [Google Scholar]
- Jammi NV, Beal PA 2001. Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity. Nucleic Acids Res 29: 3020–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammi NV, Whitby LR, Beal PA 2003. Small molecule inhibitors of the RNA-dependent protein kinase. Biochem Biophys Res Commun 308: 50–57 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257: 1685–1689 [DOI] [PubMed] [Google Scholar]
- Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams BR 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J 16: 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol 18: 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DH, Petryshyn R, London IM 1980. Characterization of double-stranded-RNA-activated kinase that phosphorylates α subunit of eukaryotic initiation factor 2 (eIF-2α) in reticulocyte lysates. Proc Natl Acad Sci 77: 832–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J 21: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs EF, Watanabe Y, Kadereit S, Barber GN, Katze MG, Chong K, Williams BR, Hovanessian AG 1992. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol 66: 5805–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, Bevilacqua PC 2011. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol 21: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktay K, Buyuk E, Oktem O, Oktay MH, Giancotti FG 2008. The c-Jun N-terminal kinase (JNK) functions upstream of Aurora B to promote entry into mitosis. Cell Cycle 7: 533–541 [DOI] [PubMed] [Google Scholar]
- Patel RC, Stanton P, Mcmillan NMJ, Williams BRG, Sen GC 1995. The interferon-inducible double-stranded RNA-activated protein-kinase self-associates in vitro and in vivo. Proc Natl Acad Sci 92: 8283–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Hur S 2013. Multi-level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci 70: 1949–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 26: 260–268 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N 2001. Suppression of cap-dependent translation in mitosis. Genes Dev 15: 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Valle F, Badura ML, Braunstein S, Narasimhan M, Schneider RJ 2010. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol Cell Biol 30: 3151–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas VT, Goncalves BS, Linden R, Chiarini LB 2012. Activation of c-Jun N-terminal kinase (JNK) during mitosis in retinal progenitor cells. PLoS ONE 7: e34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev 18: 2699–2711 [DOI] [PubMed] [Google Scholar]
- Sudhakar A, Ramachandran A, Ghosh S, Hasnain SE, Kaufman RJ, Ramaiah KV 2000. Phosphorylation of serine 51 in initiation factor 2α (eIF2α) promotes complex formation between eIF2α(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry 39: 12929–12938 [DOI] [PubMed] [Google Scholar]
- Takada Y, Ichikawa H, Pataer A, Swisher S, Aggarwal BB 2007. Genetic deletion of PKR abrogates TNF-induced activation of IκBα kinase, JNK, Akt and cell proliferation but potentiates p44/p42 MAPK and p38 MAPK activation. Oncogene 26: 1201–1212 [DOI] [PubMed] [Google Scholar]
- Tarnowka MA, Baglioni C 1979. Regulation of protein synthesis in mitotic HeLa cells. J Cell Physiol 99: 359–367 [DOI] [PubMed] [Google Scholar]
- Tonelli RR, Augusto Lda S, Castilho BA, Schenkman S 2011. Protein synthesis attenuation by phosphorylation of eIF2α is required for the differentiation of Trypanosoma cruzi into infective forms. PLoS ONE 6: e27904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RX, Wang JD, Paul AM, Acharya D, Bai FW, Huang FQ, Guo YL 2013. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem 288: 15926–15936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MATM, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. 2007. 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W 2001. Loss of HCF-1–chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol 21: 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J 14: 6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Nath A, Opperman MJ, Chan C 2010. The double-stranded RNA-dependent protein kinase differentially regulates insulin receptor substrates 1 and 2 in HepG2 cells. Mol Biol Cell 21: 3449–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, Der SD, Williams BR 1999. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene 18: 315–326 [DOI] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BR 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol Cell Biol 20: 1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PJ, Huang W, Kalikulov D, Yoo JW, Placzek AN, Stoica L, Zhou H, Bell JC, Friedlander MJ, Krnjevic K, et al. 2011. Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell 147: 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]