Interactions between mRNA capping enzyme, RNA Pol2 CTD, and elongation factor Spt5 are thought to orchestrate cotranscriptional capping of nascent mRNAs. Structural and biophysical approaches now reveal that capping enzyme binds Spt5 and phosphorylated Pol2 CTD through interactions with distinct domains. The data suggest that Spt5 and Pol2 CTDs cooperate to recruit capping enzyme. Furthermore, a model is proposed in which capping enzyme reads a binary CTD code, simultaneously engaging Spt5 and Po2 during the earliest stages of transcription initiation and elongation.
Keywords: transcription, mRNA capping, regulation, Spt5, RNA polymerase II
Abstract
Interactions between RNA guanylyltransferase (GTase) and the C-terminal domain (CTD) repeats of RNA polymerase II (Pol2) and elongation factor Spt5 are thought to orchestrate cotranscriptional capping of nascent mRNAs. The crystal structure of a fission yeast GTase•Pol2 CTD complex reveals a unique docking site on the nucleotidyl transferase domain for an 8-amino-acid Pol2 CTD segment, S5PPSYSPTS5P, bracketed by two Ser5-PO4 marks. Analysis of GTase mutations that disrupt the Pol2 CTD interface shows that at least one of the two Ser5-PO4-binding sites is required for cell viability and that each site is important for cell growth at 37°C. Fission yeast GTase binds the Spt5 CTD at a separate docking site in the OB-fold domain that captures the Trp4 residue of the Spt5 nonapeptide repeat T1PAW4NSGSK. A disruptive mutation in the Spt5 CTD-binding site of GTase is synthetically lethal with mutations in the Pol2 CTD-binding site, signifying that the Spt5 and Pol2 CTDs cooperate to recruit capping enzyme in vivo. CTD phosphorylation has opposite effects on the interaction of GTase with Pol2 (Ser5-PO4 is required for binding) versus Spt5 (Thr1-PO4 inhibits binding). We propose that the state of Thr1 phosphorylation comprises a binary “Spt5 CTD code” that is read by capping enzyme independent of and parallel to its response to the state of the Pol2 CTD.
Essential eukaryal mRNA processing events are targeted to nascent transcripts made by RNA polymerase II (Pol2) via physical interactions of the processing machineries with the C-terminal domain (CTD) of Rpb1, the largest subunit of Pol2. The Pol2 CTD consists of tandemly repeated heptapeptides of consensus sequence Y1S2P3T4S5P6S7. The inherently plastic Pol2 CTD structure is sculpted by dynamic phosphorylation and dephoshorylation of the Tyr1, Ser2, Thr4, Ser5, and Ser7 residues and by cis–trans isomerization of the prolines. With up to 128n potential Pol2 CTD primary structures (where n is the number of heptads), the Pol2 CTD provides information about the state of the transcription complex (a CTD code) that is read by diverse Pol2 CTD receptor proteins that control transcription, modify chromatin structure, and catalyze or regulate mRNA capping, splicing, and polyadenylation (Buratowski 2009; Corden 2013; Eick and Geyer 2013; Geronimo et al. 2013).
The individual amino acids of the Pol2 CTD heptad and their phosphorylation marks are coding “letters” that have distinct outputs with respect to receptor recognition and impact on cellular gene expression (Schwer et al. 2014). The number of heptad repeats comprising the Pol2 CTD, the minimal number of repeats required for viability, and the prevalence of nonconsensus coding letters tend to increase with progression from unicellular to multicellular eukarya, presumably as a means to orchestrate the actions of increasing numbers of Pol2 CTD receptors. We envision a core Pol2 CTD code that governs events common to most eukarya; for example, the essential function of the Ser5-PO4–Pro6 coding “word” in recruiting mRNA capping enzymes to the Pol2 elongation complex (Schwer and Shuman 2011; Schwer et al. 2012), which is conserved in budding yeast, fission yeast, and mammals, notwithstanding major differences in the genetic and physical organization of the capping apparatus in these taxa (Lima et al. 1999; Changela et al. 2001; Pei et al. 2001; Fabrega et al. 2003; Ghosh et al. 2011).
The RNA capping enzymes are the first processing factors to act on the growing Pol2 transcript; indeed, capping can commence after synthesis of only a 19- to 22-mer nascent RNA, the point at which the 5′ triphosphate RNA end is extruded from the RNA-binding pocket of elongating Pol2 and becomes accessible to the capping enzymes (Chiu et al. 2002). The m7GpppN cap is formed by three successive enzymatic reactions: (1) RNA triphosphatase (TPase) hydrolyzes the RNA 5′ triphosphate end (pppRNA) to a diphosphate (ppRNA) plus inorganic phosphate, (2) GTP:RNA guanylyltransferase (GTase) converts ppRNA to GpppRNA via a covalent GTase-(lysyl-Nζ)–GMP intermediate, and (3) AdoMet:RNA(guanine-N7)-methyltransferase (MTase) converts GpppRNA to m7GpppRNA (Gu and Lima 2005). Whereas it had long been assumed that capping is a constitutive and efficient process for most mRNAs, this view may be too simplistic. The existence of factors that can stimulate capping of specific transcripts and the discovery of enzymes that mediate the decay of partially processed RNA intermediates in the cap synthetic pathway suggest that capping efficiency is a dynamic process (Chiu et al. 2002; Jiao et al. 2010, 2013; Chang et al. 2012).
One potential way to regulate capping is by influencing the timely recruitment of the capping enzymes to the Pol2 elongation complex. A conserved theme among eukarya is the direct binding of the GTase component to the Ser5-PO4 form of the Pol2 CTD (Ho and Shuman 1999; Fabrega et al. 2003; Ghosh et al. 2011). In mammals and budding yeast, the TPase components are recruited passively to the Pol2 CTD by virtue of their physical association with the GTase: in cis as a covalently fused TPase–GTase enzyme, Mce1, in mammals (Ho and Shuman 1999; Changela et al. 2001) or in trans as separately encoded subunits of a TPase•GTase complex in budding yeast (Ho et al. 1999; Gu et al. 2010). The fission yeast Schizosaccharomyces pombe has a distinctive strategy for targeting cap formation to Pol2 transcripts whereby the TPase (Pct1) and GTase (Pce1) enzymes are not associated physically but instead bind independently to the Ser5-phosphorylated Pol2 CTD (Pei et al. 2001).
Mammalian and fungal capping enzymes can also gain access to nascent Pol2 transcripts via physical interactions with the transcription elongation factor Spt5 (Wen and Shatkin 1999; Pei and Shuman 2002; Pei et al. 2006; Lidschreiber et al. 2013). Spt5 is a large polypeptide (∼1000–1200 amino acids) composed of multiple domain modules that associates with the Pol2 transcription complex shortly after initiation and can exert negative and positive effects on transcription elongation (Hartzog and Fu 2013). Fission yeast Spt5 has a distinctive C-terminal repeat domain (the “Spt5 CTD”) composed of 18 repeats of a nonapeptide motif (consensus: T1P2A3W4N5S6G7S8K9) that (1) binds the fission yeast RNA capping enzymes Pct1 and Pce1 (Pei and Shuman 2002) and (2) is targeted for threonine phosphorylation by the fission yeast Cdk9 kinase (Pei and Shuman 2002, 2003; Viladevall et al. 2009). Alignment of Spt5 CTD elements in metazoan and fungal species reveals substantive differences, although each contains a series of Thr–Pro or Ser–Pro dipeptide motifs followed by a hydrophobic side chain two residues downstream (Pei and Shuman 2002). The hydrophobic side chain is typically tryptophan in S. pombe; this position is substituted to a tyrosine or histidine in humans. Available genetic evidence points to overlapping roles of the CTDs of fission yeast Pol2 and Spt5 in recruiting the capping enzymes in vivo (Schneider et al. 2010). Unlike the capping enzyme•Pol2 CTD interactions, which stringently depend on the Ser5-PO4 mark, the binding of fission yeast Pct1 and Pce1 to the Spt5 CTD is independent of Thr1 phosphorylation (Pei et al. 2001; Pei and Shuman 2002).
The structural principles underpinning capping enzyme•Pol2 CTD interactions were initially elucidated via cocrystallization of Candida albicans GTase (Cgt1) and mammalian GTase (Mce1) bound to Ser5-phosphorylated Pol2 CTD peptide ligands (Fabrega et al. 2003; Ghosh et al. 2011). These two cellular GTases are structurally homologous enzymes composed of two domains: (1) an N-terminal nucleotidyltransferase (NTase) module that contains the guanylate-binding pocket and (2) a C-terminal OB-fold module. A comparison of the Candida and mammalian GTase•Pol2 CTD structures was most revealing because, although the docking sites for Pol2 CTD lie on the surface of the NTase domain in both cases, the docking sites are physically distinct and have little overlap, the atomic contacts to Pol2 CTD are different, and virtually none of the Pol2 CTD-interacting side chains of mammalian GTase are conserved in the Candida enzyme (Ghosh et al. 2011). Thus, capping enzymes from different taxa have evolved different strategies to read the Pol2 CTD code.
Here we extend the structural analysis to the GTase enzyme of fission yeast and its interactions with the CTDs of Pol2 and Spt5. We report crystal structures of Pce1 bound to Pol2 CTD and Spt5 CTD ligands. Key findings are that (1) the CTDs of Pol2 and Spt5 interact with completely distinct sites on the NTase and OB domains of the fission yeast GTase, respectively, and (2) whereas the interface of GTase with the Pol2 CTD is dependent on Ser5 phosphorylation, GTase binding to Spt5 CTD is antagonized by Thr1 phosphorylation. We probed by structure-guided mutagenesis the contributions of the GTase•CTD interfaces to CTD binding in vitro and capping enzyme function in vivo. Our findings cohere into a model in which the capping enzyme recognizes and responds to distinct coding cues and phosphorylation marks in the Pol2 and Spt5 CTDs.
Results
Crystallization of the Pce1–GMP and Pce1•CTD complexes
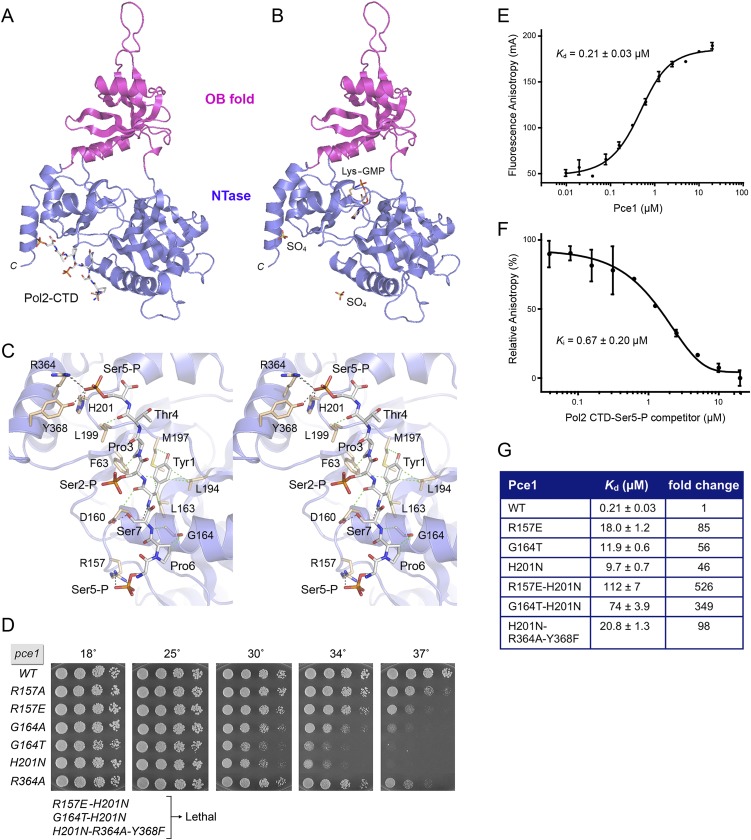
Purified fission yeast GTase Pce1 was reacted with GTP and Mg2+ to form the covalent enzyme–GMP intermediate. After removing free GTP and Mg2+, we grew crystals of Pce1–GMP that had been premixed with either a 21-amino-acid Pol2 CTD phosphopeptide comprising three heptad repeats in which Ser2 and Ser5 were both phosphorylated or an N-terminal fluorescein-labeled 22-amino-acid Spt5 CTD peptide composed of 2.5 nonamer repeats. We determined the structure of the Pce1•Pol2 CTD complex in space group P21 by molecular replacement using the NTase domain of C. albicans Cgt1 as the search model followed by docking of the OB domain into electron density. The asymmetric unit contained two Pce1 protomers. An 8-amino-acid peptide segment, S5pP6S7Y1S2pP3T4S5p, of the input 21-amino-acid Pol2 CTD ligand was placed into electron density at the base of each NTase domain (Fig. 1A). The Pce1•Pol2 CTD model was refined at 2.4 Å resolution with R/Rfree values of 0.180/0.235 (Supplemental Table S1). Crystals grown from the mixture of Pce1 and 22-amino-acid fluorescein-Spt5 CTD peptide were also in space group P21, with two protomers in the asymmetric unit. Molecular replacement yielded a 2.1 Å model of the Pce1–GMP intermediate with R/Rfree values of 0.186/0.232 (Fig. 1B; Supplemental Table S1), albeit with no evidence of the fluorescein-Spt5 CTD ligand in electron density.
Figure 1.
Structural and functional analysis of the Pce1•Pol2 CTD-PO4 interface. The structures of the Pce1•Pol2 CTD complex (A) and the covalent Pce1–GMP intermediate (B) are depicted as cartoon models with the NTase domain colored blue and the OB-fold domain colored magenta. (A) The Pol2 CTD is rendered as a stick model with gray carbons. (B) The lysyl-N-GMP adduct is shown as a stick model with beige carbons. Two sulfate anions shown in B superimpose on the two Pol2 CTD Ser5 phosphates in A. (C) A detailed stereo view of the interface of the 8-amino-acid Pol2 CTD peptide (gray carbons) with the NTase domain (beige carbons). Electrostatic and hydrogen-bonding contacts are denoted by black dashed lines, and van der Waals contacts are indicated by green dashed lines. (D) Mutational analysis of the Pce1•Pol2 CTD interface. Growth of S. pombe strains with the indicated chromosomal pce1 alleles in which amino acids at the Pol2 CTD interface were mutated individually or in combination was assessed as follows. Cells were inoculated in YES broth and grown at 30°C. Exponentially growing cultures were adjusted to A600 of 0.1, and aliquots (3 μL) of serial fivefold dilutions were spotted to YES agar and incubated at the indicated temperatures. The three alleles denoted as “Lethal” were those for which no haploid progeny were recovered after sporulation of heterozygous diploids. (E) Pce1 binding to a 28-mer fluorescent-tagged Pol2 CTD–Ser5-PO4 ligand was assayed by a change in anisotropy as described in the Materials and Methods. The reaction mixtures contained 0.4 μM BODIPY-FL-Pol2 CTD–Ser5-PO4 and increasing concentrations of Pce1. Anisotropy is plotted as a function of input Pce1. Each datum is the average of three independent experiments ± SEM. The Kd was calculated by data fitting in Prism. (F) Competition between labeled and unlabeled Pol2 CTD–Ser5-PO4 for binding to Pce1. Pce1 (0.25 μM) was preincubated with increasing concentrations of untagged 28-amino-acid Pol2 CTD–Ser5-PO4 prior to addition of 0.4 μM BODIPY-FL-Pol2 CTD–Ser5-PO4. The relative anisotropy is plotted as a function of competitor concentration. Each datum is the average of three independent experiments ± SEM. The Ki was calculated by data fitting in Prism. (G) Wild-type Pce1 and the indicated mutants were assayed for binding to 28-mer fluorescent-tagged Pol2 CTD–Ser5-PO4. The Kd values are shown next to the fold decrements in affinity of mutant versus wild-type Pce1.
Overview of the Pce1–GMP structure
As expected, Pce1 comprises an N-terminal NTase domain and a C-terminal OB-fold domain (Fig. 1B). The N-terminal 5-amino-acid peptide extends across the interdomain cleft to make contact with the OB fold. Distal to the OB fold, the C-terminal α helix (Gly350 to Lys372) extends across the cleft, where it is integrated as a lateral component of the NTase domain. A DALI search of the protein database with the Pce1–GMP covalent intermediate recovered budding yeast, human, and Chlorella virus GTases as the most closely related homologs (Z-scores of 39.4–29.1), followed by exemplary archaeal, bacterial, and eukaryal ATP-dependent DNA ligases (Z-scores of 16.0–13.9) and bacterial NAD+-dependent DNA ligases (Z-scores of 10.2–9.8) (Supplemental Table S2). GTases and DNA ligases belong to the covalent NTase superfamily (Shuman and Lima 2004); they share a mechanism of catalysis via an enzyme-(lysyl-Nζ)-NMP intermediate and a catalytic core composed of NTase and OB domains. Conserved peptide motifs I, Ia, III, IV, and V, containing essential active site constituents, form the NMP-binding pocket of the Pce1 NTase domain, which is organized as a six-strand anti-parallel β sheet and a three-strand anti-parallel β sheet flanking the GMP nucleotide.
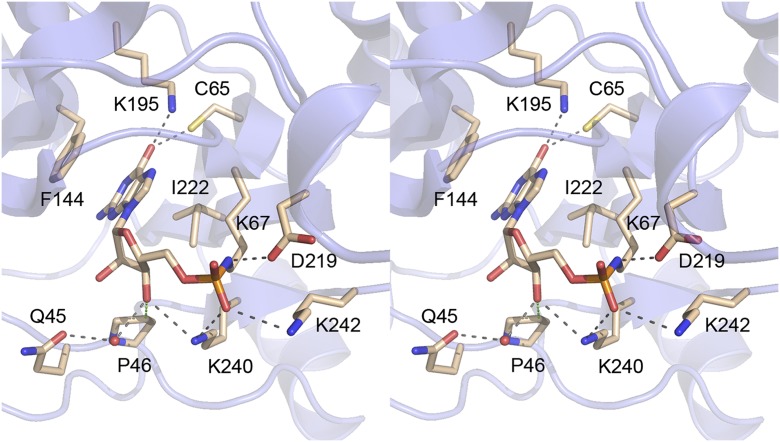
The Pce1 electron density revealed a covalent linkage between Lys67-Nζ (motif I, 65CEKSDGIR72) and the guanylate 5′ phosphorus in the active site. The guanosine nucleoside adopts an anti-conformation, with the guanine nucleobase sandwiched by Phe144 (motif IIIa) and Ile222 (motif IV, 219DGLIF223) (Fig. 2). Guanine specificity is enforced by hydrogen bonds to the O6 atom from Cys65-Sγ (motif I) and Lys195-Nζ. The phosphate is contacted by motif V residues Lys240 and Lys242. The ribose O3′ makes a hydrogen bond with Lys240, a water-mediated contact with Gln45, and a van der Waals contact with Pro46 (Fig. 2).
Figure 2.
The GTase active site. A stereo view is shown of the covalent lysyl-GMP intermediate and surrounding components of the active site. Atomic contacts to lysyl-GMP are depicted as dashed lines, with hydrogen bonds colored black and van der Waals contacts colored green. The guanine nucleobase is sandwiched between Phe144 and Ile222.
Prior studies had suggested that the position of the OB module with respect to the NTase domain changes in sync with the GTase reaction cycle, whereby the GTase adopts an open conformation with the NTase and OB domains splayed apart to permit ingress of the GTP substrate followed by closure of the OB domain over the triphosphate moiety to properly orient the pyrophosphate leaving group during attack of the lysine nucleophile at the α-phosphorus (Håkansson et al. 1997). In the Pce1 structure, the OB domain adopts a partially open conformation intermediate to that of the wide-open state seen for Candida Cgt1 (Fabrega et al. 2003) and the catalytically poised closed conformation of Chlorella virus GTase (Håkansson et al. 1997).
Pce1•Pol2 CTD-PO4 complex
The Pce1–Pol2 CTD complex crystallized in the same space group as Pce1–GMP. Whereas both protomers in the asymmetric unit superimposed closely on their counterparts in the Pce1–GMP crystal, there was no apparent density for nucleotide in the active site of the A protomer; the active site in the B protomer had density for guanosine base, but the phosphate could not be modeled. We surmise that the preformed covalent Pce1–GMP adduct was hydrolyzed during crystallization.
The Pol2 CTD octapeptide ligand S5pP6S7Y1S2pP3T4S5p is engaged on the basal surface of the NTase domain. The Pol2 CTD-docking site of Pce1 is composed mainly of three separate segments of the primary structure that are brought together in the tertiary structure: a loop–helix segment, 156SRLLDKRLG164; a loop segment, 194LKKMELGH201; and the C-terminal crossover helix segment 364REAYY368 (Fig. 1A). The direct participation of the C-terminal helix in Pol2 CTD binding explains a prior finding that deletion of the C-terminal 42 amino acids of the 402-amino-acid Pce1 protein abolished Pce1 interaction with the Pol2 CTD in vivo, as gauged by yeast two-hybrid assay (Pei and Shuman 2002).
The Pol2 CTD is anchored to Pce1 at both ends by ionic and polar contacts to the terminal Ser5 phosphate groups (Fig. 1C). The N-terminal Ser5-PO4 is held by a bidentate salt bridge from the Arg157 guanidinium nitrogens to the O1P and O3P oxygens. The C-terminal Ser5-PO4 O3P is coordinated by His201, Arg364, and Tyr368. It is noteworthy that the two surface anion-binding sites for the Ser5 phosphates seen in the Pce1•Pol2 CTD complex are occupied by sulfate anions in Pce1–GMP crystal (Fig. 1, cf. A and B). Thus, the spacing between the anion sites is geared to accommodate the Ser5-PO4 marks on consecutive heptad repeats. Between the Ser5-PO4 “bookends,” the Pol2 CTD adopts an extended β-like conformation. The Pro3 and Pro6 residues are both in the trans-isomer configuration.
The atomic contacts of the Pol2 CTD amino acids with Pce1, proceeding from N to C termini of the bound peptide, are as follows: The Pro6 side chain and Ser7 main chain amide make van der Waals contacts with Gly164; the Ser7-Oγ donates a hydrogen bond to the Asp160 carboxylate; the Tyr1 side chain makes van der Waals contacts to Leu163, Leu194, Met197, and Phe63; the Tyr1 main chain amide donates a hydrogen bond to the Asp160 main chain carbonyl, while the Tyr1 main chain carbonyl makes van der Waals contact with Asp160-Cβ; the Pro3 side chain, which stacks on the Pol2 CTD Tyr1 side chain, makes a van der Waals contact with Phe63; and the Thr4 main chain carbonyl makes van der Waals contact with Leu199 (Fig. 1C). The salient theme is that the majority of the interactions at the Pce1•Pol2 CTD interface involve Ser5-PO4 and Tyr1. This is in accord with previous findings that Pce1 (1) fails to bind the Pol2 CTD in the absence of Ser5 phosphorylation and (2) fails to bind to the Ser5-phosphorylated Pol2 CTD when Tyr1 is mutated to alanine (Pei et al. 2001). Pce1 contacts with Pro6 are minimal in comparison, consistent with the observation that changing Pro6 to alanine does not compromise Pce1 binding to Ser5-phosphorylated Pol2 CTD in vitro (Pei et al. 2001). We infer that the essentiality of Pro6 for capping enzyme function in fission yeast in vivo reflects the critical role of Pro6 in directing the phosphorylation of Ser5 (Schwer et al. 2012). Pro3 also makes minimal direct contact with Pce1 in the crystal structure, yet mutating Pro3 to alanine eliminated Pce1 binding to Ser5-phosphorylated Pol2 CTD in vitro; we speculate that the stacking of the Pro3 side chain on Tyr1 is the pertinent contribution of Pro3 to the Pce1–Pol2 CTD interaction. It is noteworthy that the Ser2-PO4 and Thr4 side chains project away from the protein surface and make no contacts with Pce1 (Fig. 1C). This is consistent with the findings that Ser2 phosphorylation and mutation of Thr4 to alanine have no effect on Pce1 binding in vitro to the Ser5-phosphorylated Pol2 CTD (Pei et al. 2001) and that neither Ser2 phosphorylation nor Thr4 is essential for fission yeast vegetative growth (Schwer and Shuman 2011).
Functional probing of the Pce1•Pol2 CTD interface
Mutations of individual Pce1 amino acids comprising the Pol2 CTD-docking site were introduced by marked allelic replacement into one chromosomal pce1 locus of a diploid S. pombe strain. The diploids were sporulated, and viable haploids were genotyped by sequencing to verify the allelic replacement. Serial dilutions of the wild-type and mutant strains containing equal numbers of cells were spot tested for growth on agar medium at 18°C–37°C (Fig. 1D). Arg157 is the sole ligand for the Pol2 CTD N-terminal Ser5-PO4 group. The R157A and R157E mutants were viable and grew as well as wild-type cells at 18°C–30°C (as gauged by colony size) but displayed temperature-sensitive (ts) growth defects at 37°C; the glutamate change was more deleterious than alanine in this regard. The C-terminal Ser5-PO4 group is coordinated by three side chains: His201, Arg364, Tyr368. The singly mutated R364A strain thrived at 18°C–34°C but grew slowly at 37°C (Fig. 1D). The H201N strain grew well at 18°C–25°C, slowly at 30°C–34°C, and not at all at 37°C. These results signify that weakening either of the two Ser5-PO4-binding sites of Pce1 elicits a hypomorphic growth phenotype.
An instructive finding was that subtraction of all ionic and polar contacts with the C-terminal Ser5-PO4 group by a H201N–R364A–Y368F triple mutation was lethal in vivo, as judged by the inability to recover viable mutant haploids after sporulation of a pce1+ pce1-(H201N–R364A–Y368F) diploid. These results indicate that (1) the C-terminal Ser5-PO4-docking site is essential for capping enzyme function, and (2) the N-terminal Ser5-PO4 site defined by Arg157 cannot sustain capping enzyme function by itself. Nevertheless, the two Ser5-PO4 sites do appear to functionally cooperate, insofar as combining the R157E and H201N changes resulted in lethality (Fig. 1D). The genetics of the two sites of Pce1 for Ser5-PO4 sites in adjacent heptads resonate with the genetics of the Pol2 CTD itself, whereby fission yeasts are viable when the Pol2 CTD is composed of alternating Ser5 and Ser5Ala heptads (Schwer and Shuman 2011).
To perturb the interface of Pce1 with Pol2 CTD side chain Pro6 that packs against Gly164, we installed G164A and G164T mutations, both of which elicited ts growth phenotypes, with threonine being more severe than alanine (Fig. 1D). Combining the G164T and H201N mutations in the C-terminal Ser5-PO4 site was lethal, underscoring the additive contributions of the Pol2 CTD interfacial residues to capping function in vivo.
Although the Pol2 CTD interfacial residues are located on the surface of the NTase domain away from the active site, we were concerned in interpreting the in vivo mutational effects to test whether interfacial mutations affected GTase activity. We produced and purified recombinant versions of each of the mutated Pce1 proteins (Supplemental Fig. S1) and assayed for their ability to react with [α32P]GTP in vitro to form the Pce1–[32P]GMP intermediate. None of the mutations at the Pol2 CTD interface affected adduct formation (Supplemental Fig. S1).
Analysis in vitro of Pce1 binding to Pol2 CTD–Ser5-PO4
We implemented a fluorescence polarization assay to study the binding of purified Pce1 to a 28-amino-acid synthetic Pol2 CTD peptide composed of four heptad repeats, each with Ser5-PO4, that was labeled at the N terminus with BODIPY-FL. Fluorescence anisotropy increased with Pce1 concentration; the data fit well to a single-site binding model with an apparent Kd of 0.21 ± 0.03 μM (Fig. 1E). Inclusion of unlabeled competitor Pol2 CTD–Ser5-PO4 peptide displaced the bound fluorescent Pol2 CTD from Pce1 in a concentration-dependent manner with an apparent Ki of 0.67 ± 0.20 μM (Fig. 1F). We proceeded to determine the effects of mutations of amino acids at the Pce1•Pol2 CTD interface on Pce1 binding to N-BODIPY-FL-Pol2 CTD–Ser5-PO4; the results are shown in Figure 1G. Mutations that were consequential in vivo (ts or lethal) elicited significant decrements in Pol2 CTD affinity, ranging from ∼50-fold to 500-fold. The greatest decrements were seen for the R157E–H201N and G164T–H201N mutations, which were lethal in vivo; these two double mutations had additive effects on Pol2 CTD binding in vitro compared with the component R157E, H201N, and G164T single changes.
Fission yeast capping enzyme reads the Pol2 CTD code in a distinctive manner
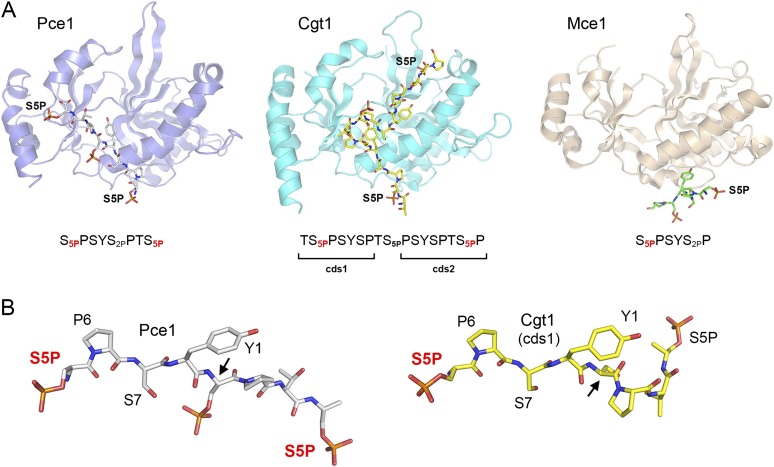
Previous structures showed that mammalian GTase Mce1 and budding yeast GTase Cgt1 exploit distinct NTase domain surfaces to interact with a Pol2 CTD–Ser5-PO4 ligand (Fabrega et al. 2003; Ghosh et al. 2011). The present structure of the Pce1•Pol2 CTD–Ser5-PO4 complex illuminates how fission yeast Pce1 has evolved a unique structural basis for its reading of the same Pol2 CTD Ser5-PO4 coding letter. The foremost distinction of the Pce1•Pol2 CTD interaction is that it comprises a single docking site for an 8-amino-acid Pol2 CTD peptide, with Ser5-PO4 residues from adjacent heptads at the N and C termini. In contrast, mammalian Mce1 and Candida Cgt1 recognize 6-amino-acid and 7-amino-acid Pol2 CTD segments, respectively, each with only one Ser5-PO4 mark (Fig. 3A). Cgt1 has two distinct Pol2 CTD-docking sites (cds1 and cds2) on the surface of its NTase domain that recognize Ser5-PO4-containing heptads in differently phased registers: cds1 binds register TS5PPSYSP, and cds2 binds PSYSPTS5PP (Fig. 3A). Both cds1 and cds2 feature ionic and polar contacts to the Ser5-PO4 groups.
Figure 3.
Fission yeast capping enzyme reads the Pol2 CTD code in a distinctive manner. (A) The structures of the NTase domains of capping enzymes S. pombe Pce1 (blue), C. albicans Cgt1 (cyan; Protein Data Bank [PDB] 1P16), and mammalian Mce1 (beige; PDB 3RTX) in complexes with Pol2 CTD phosphopeptide ligands were superimposed and then offset horizontally. The Pol2 CTDs are depicted as sticks models, and the phospho-Ser5 side chains docked on the NTase surface are labeled (S5P). The amino acid sequences of the engaged Pol2 CTD peptides are indicated below the structures, and the S5P residues contacted by the GTases are denoted by red subscripts. (B) The Pol2 CTD–Ser5-PO4 ligands from the Pce1 and Cgt1 structures were aligned and then offset horizontally. The sites at which the trajectory of the Pol2 CTD diverges in the two structures are highlighted by arrows. The S5P residues contacted by the GTases are labeled in red.
The Pce1 Pol2 CTD-binding site overlaps cds1 of Candida Cgt1 with respect to its location on the NTase domain surface (Fig. 3A) and its atomic interactions with the S5PPSY segment (Fig. 3A). Indeed, the conformations of this tetrapeptide segment of the Pol2 CTD ligand are nearly identical in Pce1 and Cgt1(cds1) (Fig. 3B). In addition, the Pce1 Arg157 side chain that makes contact with the N-terminal Ser5 phosphate group is conserved as Arg157 in Cgt1, where it too coordinates the Ser5 phosphate oxygens. The trajectory of the Pol2 CTD peptide diverges in the Pce1 and Cgt1 structures by virtue of an acute rotation of the distal peptide around the Ser2 N–Cα bond (Fig. 3B). In Pce1, the distal Pol2 CTD segment remains in an extended conformation that places the C-terminal Ser5-PO4 in an anion-binding pocket formed by His201, Arg364, and Tyr368. In Cgt1, the equivalent of Pce1 Arg364 is a serine, thereby eliminating this potential ionic contact to a Ser5-PO4. Instead, the Cgt1-bound Pol2 CTD deviates and undergoes a sharp kink that lifts the TS5P dipeptide off the protein surface before placing the distal PSYSPTS5PP heptad in cds2, anchored by a Ser5-PO4-binding pocket formed by Cgt1 side chains Arg140, Lys178, and Lys193. Because Cgt1 Lys178 and Lys193 correspond to Thr178 and Leu191 in Pce1, the “two-cds” binding mode does not appear to be an option for the fission yeast enzyme. Rather, our findings here highlight that each capping enzyme surveyed to date has evolved a unique structural solution to read the Ser5-PO4 letter of the Pol2 CTD code.
Structure of Pce1 bound to the Spt5 CTD
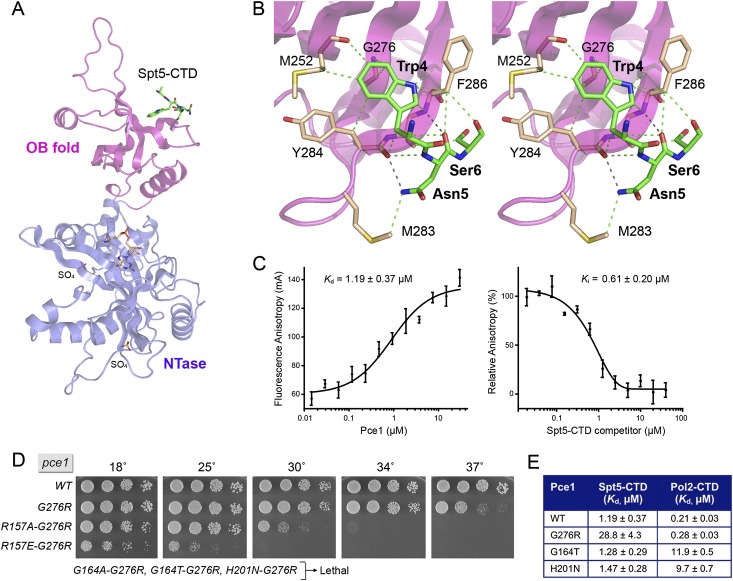
We cocrystallized Pce1–GMP with an 18-amino-acid Spt5 CTD peptide comprising two nonamer repeats. Molecular replacement yielded a 3.1 Å structure of the Pce1•Spt5 CTD complex in space group I222, with one Pce1–GMP protomer in the asymmetric unit engaged with a 4-amino-acid segment, A3W4N5S6, of the input Spt5 CTD peptide (R/Rfree 0.201/0.253) (Supplemental Table S1). The AWNS peptide was docked on the surface of the Pce1 OB fold (Fig. 4A). This location of the Spt5 CTD in the crystal is consistent with yeast two-hybrid studies demonstrating that the OB-containing segment of Pce1 from amino acids 235 to 350 sufficed for interaction with the Spt5 CTD in vivo (Pei and Shuman 2002). Two sulfate anions demarcating the Pol2 CTD–Ser5-PO4 site were engaged on the surface of the NTase domain of the Pce1•Spt5 CTD complex.
Figure 4.
Structural and functional analysis of the Pce1•Spt5 CTD interface. (A) The structure of the Pce1•Spt5 CTD complex is depicted as a cartoon model with the NTase domain colored blue and the OB-fold domain colored magenta. The Spt5 CTD peptide bound to the OB domain is rendered as a stick model with green carbons. The lysyl-N-GMP adduct is shown as a stick model with beige carbons. Two sulfates on the surface of the NTase domain that mimic the two Pol2 CTD phosphate groups are shown as stick models. (B) A detailed stereo view of the interface of the Spt5 CTD peptide (green carbons) with the OB domain (beige carbons). Hydrogen bonds are denoted by black dashed lines, and van der Waals contacts are indicated by green dashed lines. (C, left panel) Pce1 binding to a fluorescent-tagged Spt5 CTD-(801–898) ligand was assayed by a change in anisotropy as described in the Materials and Methods. The reaction mixtures contained 0.3 μM BODIPY-FL-Spt5 CTD-(801–898) and increasing concentrations of Pce1. Anisotropy is plotted as a function of input Pce1. Each datum is the average of three independent experiments ± SEM. The Kd was calculated by data fitting in Prism. (Right panel) Competition between labeled and unlabeled Spt5 CTD for binding to Pce1. Pce1 (1 μM) was preincubated with increasing concentrations of untagged Spt5 CTD-(801–898) prior to addition of 0.3 μM BODIPY-FL-Spt5 CTD-(801–898). The relative anisotropy is plotted as a function of competitor concentration. Each datum is the average of three independent experiments ± SEM. The Ki was calculated by data fitting in Prism. (D) The separate Pce1 Pol2 and Spt5 CTD sites collaborate in vivo. Growth of S. pombe strains with the indicated chromosomal pce1 alleles in which amino acids at the Spt5 CTD and Pol2 CTD interfaces were mutated individually or in combination was assessed as described in the legend for Figure 1. The three alleles denoted as “Lethal” were those for which no haploid progeny were recovered after sporulation of heterozygous diploids. (E) Mutations that disrupt binding to the Spt5 CTD do not affect binding to the Pol2 CTD and vice versa. Wild-type Pce1 and the indicated mutants were assayed for binding to BODIPY-FL-Spt5 CTD-(801–898) and BODIPY-FL-Pol2 CTD–Ser5-PO4 ligands. The Kd values are shown.
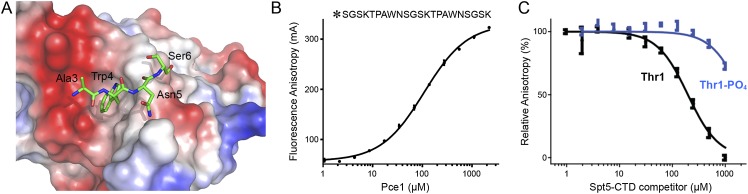
The Trp4 residue of the Spt5 CTD is lodged in a hydrophobic pocket formed by Pce1 residues Met252, Asn274, Gly276, Tyr284, and Phe286 (Fig. 4B). Trp4 makes myriad van der Waals contacts: The indole ring stacks over the Gly264 Cα and N atoms and the Phe286 Cα and N atoms; the indole edges contact Met252 Cγ and O, Tyr284 Cβ, and Phe286 Cβ and Cδ1; and the tryptophan Cβ contacts the Tyr284 carbonyl. The centrality of Trp4 to the crystallographic Pce1•Spt5 CTD interface rationalizes previous findings that Pce1 failed to bind in vitro to an Spt5 CTD nonamer repeat array in which every Trp4 was replaced by alanine (Schneider et al. 2010). The Asn5 residue donates a hydrogen bond from Nδ2 to the Pce1 Tyr284 main chain carbonyl and receives a hydrogen bond to the Asn5 main chain carbonyl from the Phe286 main chain amide. Asn5 also makes a van der Waals contact with Met283 (Fig. 4B). The Ser6 residue of the Spt5 CTD makes van der Waals interactions with Ser285 and Phe286. The Spt5 CTD Ala3 residue makes no contacts with Pce1.
The Pce1 Pol2 and Spt5 CTD sites function independently in vitro
A fluorescence polarization assay was applied to analyze the binding of Pce1 to purified recombinant Spt5-(801–898)—composed of 10 nonamer repeats (Pei and Shuman 2002)—that was labeled at the N terminus with BODIPY-FL. Fluorescence anisotropy increased with Pce1 concentration, with an apparent Kd of 1.19 ± 0.37 μM (Fig. 4C). Unlabeled competitor Spt5-(801–898) displaced the bound fluorescent Spt5-(801–898) from Pce1 in a concentration-dependent manner, with an apparent Ki of 0.61 ± 0.20 μM (Fig. 4C). We perturbed the Spt5 CTD-docking site by replacing Gly276, against which Trp4 packs, with a bulky arginine side chain. The G276R lesion did not affect GTase activity in vitro (Supplemental Fig. S1), but the Pce1-G276R protein displayed a 24-fold decrement in binding affinity for N-BODIPY-FL-Spt5-(801–898), with a Kd of 28.8 ± 4.3 μM (Fig. 4E). This effect was specific for the Spt5 CTD, insofar as the binding affinity of the G276R protein for the N-BODIPY-FL-Pol2 CTD–Ser5-PO4 ligand (0.28 μM) was virtually the same as wild-type Pce1 (0.21 μM) (Fig. 4E). We found that the G164T and H201N changes that elicited ∼50-fold decrements in Pol2 CTD–Ser5-PO4 affinity had no effect on Pce1 binding to the Spt5 CTD (Fig. 4E). These results affirm that the physically separate Pol2 and Spt5 CTD sites on Pce1 are functionally independent.
A key question is whether Pce1 can bind simultaneously to the Pol2 and Spt5 CTDs. To address this point, we performed the experiment shown in Supplemental Figure S2. Pce1 (0.25 μM) was preincubated with N-BODIPY-FL-Pol2 CTD–Ser5-PO4 (0.4 μM) to form a binary complex. This mixture was supplemented with increasing concentrations of unlabeled Spt5-(801–898) and assayed for a change in fluorescence anisotropy of the labeled Pol2 CTD peptide. We observed an Spt5 concentration-dependent increase in anisotropy (Supplemental Fig. S2A), which we take to signify that the Spt5 CTD binds to the Pce1•Pol2 CTD binary complex to form a Pce1•Pol2 CTD•Spt5 CTD ternary complex (Supplemental Fig. S2B). The apparent Kd for binding of Spt5-(801–898) to the Pce1•Pol2 CTD binary complex was 0.18 ± 0.06 μM.
The Pce1 Pol2 and Spt5 CTD sites collaborate in vivo
We replaced the chromosomal wild-type pce1+ gene with a G276R mutant with an enfeebled Spt5-docking site in the OB domain. The G276R strain grew as well as the wild-type strain on agar medium at 18°C–34°C but grew slowly at 37°C (Fig. 4D), signifying that the interaction of Pce1 with Spt5 is not essential per se. This conclusion is consistent with the prior finding that fission yeast cells deleted for the entire Spt5 CTD segment are also viable, albeit sick (Schneider et al. 2010). The salient finding here was that combining G276R with the otherwise viable G164A, G164T, or H201N lesions in the Pol2 CTD-binding site of Pce1 was synthetically lethal (Fig. 4D). Moreover, the combination of G276R with the ts R157A and R157E mutations in the N-terminal Pol2 CTD–Ser5-PO4 site of Pce1 resulted in a significant downward shift in their restrictive growth temperatures (cf. Figs. 4D and 1D). These genetic results fortify the hypothesis, based initially on the effects of truncating the S. pombe Rpb1 CTD heptad array and deleting the Spt5 CTD nonamer array (Schneider et al. 2010), that the two CTDs play overlapping roles in vivo in promoting the capping of nascent Pol2 transcripts by Pce1.
Effect of threonine phosphorylation of the Spt5 CTD on interaction with Pce1
Thr1 of the S. pombe Spt5 nonamer repeat TPAWNSGSK is targeted for phosphorylation by Cdk9. Here and in previous studies, we found that Pce1 can bind to the unphosphorylated form of the Spt5 CTD. However, it is unclear whether the Pce1•Spt5 CTD interaction is affected by threonine phosphorylation. The Thr1 residue was not visualized in our cocrystal structure of Pce1 bound to the tetrapeptide AWNS. The view in Figure 5A of the Spt5 CTD peptide (as a green stick model) docked on an electrostatic surface model of the Pce1 OB domain highlights a plausible trajectory of the Thr1 and Pro2 residues flanking Ala3 that could place Thr1 over a negatively charged surface, in which case threonine phosphorylation might elicit a charge repulsion with this negative surface patch.
Figure 5.
Threonine phosphorylation of the Spt5 CTD weakens interaction with Pce1. (A) The image shows the Spt5 CTD peptide (green carbons) docked on a surface qualitative electrostatic model of the Pce1 OB domain (generated in PyMOL using vacuum eletrostatic protein contact potential; red [−86.5] to blue [86.5]). Positive and negative surface potential are colored blue and red, respectively. The Trp4 side chain inserts into a hydrophobic pocket. (B) Pce1 binding to a fluorescent-tagged 22-amino-acid Spt5 CTD peptide (primary structure as shown, with the N-terminal tag denoted by an asterisk) was assayed by a change in anisotropy as described in the Materials and Methods. The reaction mixtures contained 25 nM fluorescein-Spt5 CTD and increasing concentrations of Pce1. Anisotropy is plotted as a function of input Pce1. Each datum is the average of three independent experiments ± SEM. (C) The effect of Thr1 phosphorylation on competition for the Spt5-binding site. Pce1 (100 μM) was preincubated with increasing concentrations of untagged Spt5 CTD peptide or untagged Spt5–Thr1-PO4 peptide prior to addition of 25 nM fluorescein-Spt5 CTD peptide. The relative anisotropy is plotted as a function of competitor concentration. Each datum is the average of three independent experiments ± SEM.
To evaluate the impact of Spt5 CTD phosphorylation, we tested the ability of the unphosphorylated and Thr1-phosphorylated 22-amino-acid synthetic peptides SGSKTPAWNSGSKTPAWNSGSK and SGSK(Tp)PAWNSGSK(Tp)PAWNSGSK to compete with a fluorescein-tagged 22-mer Spt5 CTD peptide for binding to Pce1 in a fluorescence polarization assay. An initial Pce1 titration showed that the Kd for the 22-mer fluorescent Spt5 CTD containing two complete nonamer repeats was 109 ± 5 μM (Fig. 5B); this value was 90-fold higher than the Kd of Pce1 for the much longer Spt5-(801–898) ligand containing 10 nonamer repeats (Fig. 4C). This acute dependence on the number of nonamer repeats for high-affinity binding of Pce1 to Spt5 CTD in the fluorescence assay is in accord with results obtained using a His tag affinity chromatography assay for protein–protein interaction, which showed that Pce1 bound well to an Spt5 CTD composed of 10 nonamer repeats, less well to a six-nonamer array, and not detectably to a Spt5 CTD with only four nonamer elements (Pei and Shuman 2002). Notwithstanding the relatively low affinity of Pce1 for the 22-mer fluorescent Spt5 CTD, this ligand proved suitable for competition studies, which revealed that whereas unlabeled 22-mer Spt5 CTD displaced the fluorescent Spt5 CTD with a Ki of 198 ± 22 μM, the Thr1-phosphorylated Spt5 CTD did so with a Ki of 3680 ± 780 μM (Fig. 5C). Thus, Thr1 phosphorylation elicited an 18-fold decrement in affinity of the fission yeast GTase for the Spt5 CTD.
Discussion
Structure-inspired model for trafficking of capping enzyme with the transcription elongation complex
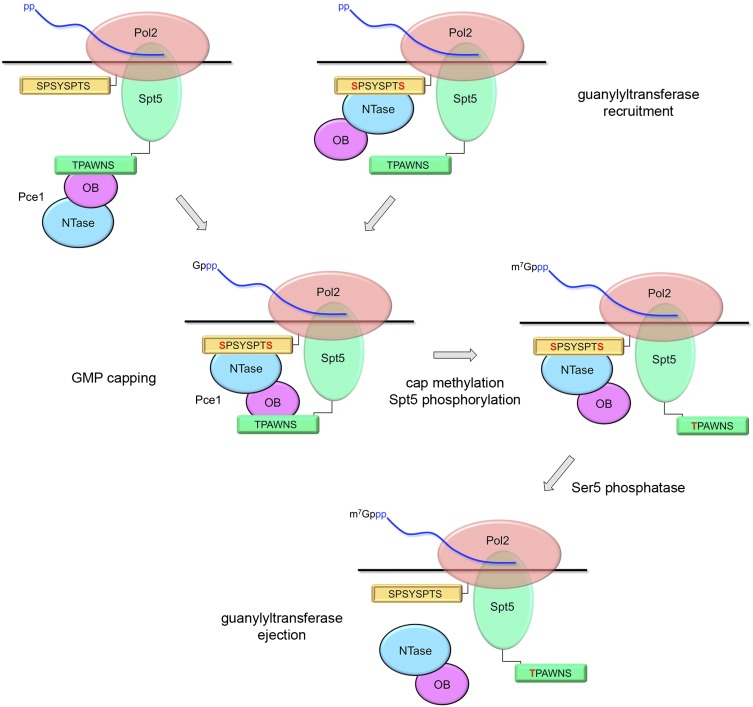
Proof that capping enzyme recruitment is a chief function of the Pol2 CTD Ser5-PO4 mark in fission yeast stemmed from the finding that the lethality incurred by changing all Ser5 positions to alanine could be reversed by covalently fusing an mRNA capping apparatus to the Rpb1 CTD-S5A mutant subunit (Schwer and Shuman 2011). The present study establishes the unique structural basis for the interaction of fission yeast GTase Pce1, via its NTase domain, with an 8-amino-acid Pol2 CTD segment bracketed by two Ser5-PO4 marks. We show that at least one of the two Ser5-PO4-binding sites is required for cell viability and that both sites are important for cell growth at 37°C. We also provide the first structural insights concerning the interaction of a capping enzyme with the unmodified Spt5 CTD at a novel binding site in the OB-fold domain of Pce1 that captures the Trp4 residue of the Spt5 nonamer repeat. Thus, there are two parallel routes for ingress of Pce1 to the early transcription elongation complex: via phosphorylation-dependent binding to the Pol2 CTD or phosphorylation-independent binding to the Spt5 CTD (Fig. 6, top row).
Figure 6.
Model for trafficking of capping enzyme with the transcription elongation complex. Two routes of GTase recruitment are posited, differing in whether Pce1 engages initially with the Ser5-phosphorylated Pol2 CTD or the unmodified Spt5 CTD. See the Results for a detailed description. Phosphorylated CTD residues are colored red.
Our crystal structures and biochemical analysis of CTD binding by mutated enzymes indicate that occupancies of the Pol2 and Spt5 CTD sites on the NTase and OB domains are independent events. Moreover, our biochemical evidence avers that Pce1 can form a ternary complex in which the Pol2 and Spt5 CTDs are engaged simultaneously. This suggests a scenario in which the Ser5-phosphorylated Pol2 CTD and unphosphorylated Spt5 CTD cooperate in vivo to stabilize Pce1 on the elongation complex and thereby target the 5′ end of the nascent transcript for GMP capping (Fig. 6, middle row, left). Several lines of evidence highlight the importance of the capping enzyme–Spt5 interactions in vivo; for example, (1) the slow growth and abnormal morphology of fission yeast cells lacking the Spt5 CTD was alleviated by overexpression of Pct1 and Pce1 or mammalian Mce1 (Schneider et al. 2010); (2) mutating Trp4 (the critical residue sensed by Pce1) to alanine in all nonamer repeats of the Spt5 CTD mimicked the growth defects of a complete Spt5 CTD deletion, but growth was restored by overexpression of capping enzyme Mce1 (Schneider et al. 2010); and (3) as shown presently, a viable hypomorphic mutation in the Pce1 Spt5 CTD-binding site was synthetically lethal with any of several mutations in the Pol2 CTD-binding site (Fig. 4).
Early during transcription elongation and presumably subsequent to capping, the fission yeast Spt5 CTD is phosphorylated on Thr1 by the cyclin-dependent kinase Cdk9. Because fission yeast Cdk9 exists in vivo as a stable ternary complex with its cyclin partner, Pch1, and the cap (guanine-N7)-MTase Pcm1 (Pei et al. 2006; St Amour et al. 2012), the action of Cdk9 on the transcription complex has multiple important consequences. First, it prompts cap methylation by corecruited Pcm1 to yield a mature m7GpppN 5′ end of the nascent RNA. Second, it installs on the Spt5 CTD a Thr-PO4 mark that is apparently inimical to Pce1 binding, thereby weakening the interaction of Pce1 with the elongation complex (Fig. 6, middle row, right). In addition, the Thr1-PO4 mark is important for the positive transcription elongation activity of fission yeast Spt5, insofar as changing all Spt5 CTD Thr1 positions to alanine sensitized S. pombe cells to growth inhibition by 5-azauracil, whereas changing Thr1 to a phosphomimetic glutamate did not (Schneider et al. 2010).
We infer that the state of Thr1 phosphorylation comprises a binary “Spt5 CTD code” that is read by Pce1 independent of and parallel to its response to the state of the Pol2 CTD. CTD phosphorylation has opposite effects on Pce1’s interactions with Pol2 (required for binding) versus Spt5 (which inhibits binding). Finally, we envision that ejection of Pce1 from the transcription elongation complex is triggered by removal of the Ser5-PO4 mark from the Pol2 CTD via the action of one or more of the several fission yeast CTD phosphatases (Fig. 6, bottom row).
Materials and methods
Purification of S. pombe capping enzyme Pce1
The pce1 ORF was inserted into pET-based bacterial expression plasmid pSMT3 (Mossessova and Lima 2000) so as to encode the Pce1 protein fused to an N-terminal His6Smt3 domain. Mutations were introduced into pSMT3-Pce1 with a QuickChange kit (Stratagene). The mutated Pce1 ORFs were sequenced to verify the absence of unwanted coding changes. The expression plasmids were transformed into Escherichia coli BL21(DE3) codon plus RIL (Stratagene). Cultures of E. coli containing the expression plasmids were grown in super broth in baffled flasks at 37°C to an A600 of 1.5, after which the cells were cold-shocked by placing the flasks for 30 min on ice. The cultures were adjusted to 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) and then incubated overnight at 18°C with constant shaking. Cells were harvested by centrifugation at 16,000g and resuspended in buffer containing 50 mM Tris-HCl (pH 8.0) and 20% (w/v) sucrose. All subsequent procedures were performed at 4°C.
For purification of the His6Smt3-tagged proteins, cells were resuspended in lysis buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol (BME), 0.1% IGEPAL (Fluka), and 1 mM phenylmethanesulfonylfluoride (PMSF) and then disrupted by sonication. Lysates were clarified by centrifugation at 45,000g, and the resulting supernatants were applied to a Ni-NTA superflow resin (Qiagen) that had been equilibrated in buffer A (20 mM Tris-HCl at pH 8.0, 350 mM NaCl, 1 mM BME) containing 20 mM imidazole. Bound proteins were eluted with 250 mM imidazole in buffer A. The His6Smt3 tags were removed by incubating the 250 mM eluate fractions with Smt3 protease Ulp1 (Mossessova and Lima 2000) at a protein:Ulp1 ratio of 1000:1 (w/w) for 6–18 h at 4°C. The tag-free S. pombe proteins were separated from His6Smt3 by gel filtration through a Superdex 75 or Superdex 200 column (GE Healthcare) equilibrated in buffer A. The peak Pce1 fractions were pooled, dialyzed against buffer B (20 mM Tris-HCl at pH 8.0, 50 mM NaCl, 1 mM BME), and concentrated by centrifugal ultrafiltration to 3–10 mg/mL.
Pce1 was further purified by anion exchange chromatography. The Superdex fraction was adsorbed to a MonoQ 10/10 column (GE Healthcare) and eluted with a 20-column-volume linear gradient of 0.05–1 M NaCl in 20 mM Tris-HCl (pH 8.0) and 1 mM BME. The MonoQ preparation was guanylylated by incubation with 1 mM GTP and 5 mM MgCl2 for 30 min on ice. Free GTP and MgCl2 were removed by several rounds of dilution and concentration by centrifugal ultrafiltration in buffer containing 20 mM Tris-HCl (pH 8.0) and 1 mM BME. Pce1–GMP was finally concentrated to 10 mg/mL in 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 1 mM TCEP.
Pce1 crystallizations and structure determinations
The Pce1•Pol2 CTD complex
A mixture of 220 μM Pce1–GMP and 260 μM 21-amino-acid Pol2 CTD phosphopeptide, YS2PPTS5PPSYS2PPTS5PPSYS2pPTS5PPS comprising three heptad repeats with Ser2-PO4 and Ser5-PO4 in each repeat, was incubated for 1 h at 4°C and then mixed on a coverslip with an equal volume of precipitant solution containing 0.1 M sodium acetate (pH 7.0) and 23% (v/v) PEG-400. Crystals were grown at 18°C by hanging drop vapor diffusion against the precipitant solution and flash-frozen directly in liquid nitrogen. Diffraction data from a single crystal to 2.4 Å resolution were collected at the Advanced Photon Source (APS) Northeastern Collaborative Access Team (NE-CAT) beamline 24-IDE equipped with an ADSC Quantum 315 detector. The data were indexed, integrated, and scaled using HKL2000 (Otwinowski and Minor 1997). The crystal was in space group P21 with two Pce1 protomers in the asymmetric unit. Initial phases were obtained by molecular replacement in PHASER (McCoy et al. 2007) using the coordinates of the NTase domain of CaCgt1 (Protein Data Bank [PDB] 1P16) as the search model. The CaCgt1 OB domain was then docked manually into density. After a single round of refinement in CNS (Brunger et al. 1998), electron density corresponding to the Pol2 CTD peptide was evident. Iterative rounds of refinement and model building in CNS, PHENIX (Adams et al. 2010), and COOT (Emsley et al. 2010) yielded a model with R/Rfree of 0.202/0.248 at 2.40 Å resolution with good geometry, as assessed by MolProbity (Chen et al. 2010), with 97.9%, 100%, and 0% in favored, allowed, and disallowed regions of Ramachandran space, respectively. The A protomer was a continuous polypeptide from the residual N-terminal serine at the Ulp1 cleavage site to Lys372. The B protomer was a continuous polypeptide from the residual N-terminal serine to Lys371. A guanosine nucleoside was modeled in the active site of the B protomer near the Lys67 nucleophile. Both protomers were associated with an 8-amino-acid Pol2 CTD phosphopeptide fragment, S5pP6S7Y1S2pP3T4S5p.
The Pce1•Spt5 CTD complex
A mixture of 220 μM Pce1–GMP and 260 μM 18-amino-acid Spt5 CTD peptide, TPAWNSGSRTPAWNSGSK, comprising two nonamer repeats, was incubated for 1 h at 4°C and then mixed on a coverslip with an equal volume of precipitant solution containing 0.1 M sodium citrate (pH 6.0), 0.5 M ammonium sulfate, and 1.35 M lithium sulfate. Crystals were grown at 18°C by hanging drop vapor diffusion against the precipitant solution. The crystals were cryoprotected with precipitant solution supplemented with 10% (w/v) xylitol. Diffraction data from a single crystal to 3.1 Å were collected at APS NE-CAT beamline 24-IDE. All data were indexed, integrated, and scaled using HKL2000. The crystal was in space group I222 with one Pce1 protomer in the asymmetric unit. The structure was solved by molecular replacement in PHASER (McCoy et al. 2007) using the coordinates of the Pce1 protein from the Pce1•Pol2 CTD-PO4 structure as the search model. After a single round of refinement in CNS, electron density corresponding to a Spt5 CTD peptide fragment was evident. Iterative rounds of refinement and model building in PHENIX and COOT yielded a model with R/Rfree of 0.20/0.25 at 3.1 Å resolution with good geometry, as assessed by MolProbity, with 95.6%, 100%, and 0% in favored, allowed, and disallowed regions of Ramachandran space, respectively. The Pce1 polypeptide extended from Ala2 to Pro373 with a 4-amino-acid gap between Glu5 and Glu10. The active site nucleophile Lys67 was covalently guanylylated. Pce1 was associated with a 4-amino-acid Spt5 CTD peptide fragment, A3W4N5S6.
The Pce1–GMP intermediate
A mixture of 220 μM Pce1–GMP and 1 mM N-terminal fluorescein-labeled 22-amino-acid Spt5 CTD peptide, SGSKTPAWNSGSKTPAWNSGSK, comprising 2.5 nonamer repeats, was incubated for 1 h at 4°C and then mixed on a coverslip with an equal volume of precipitant solution containing 21% (w/v) PEG3350 and 0.2 M ammonium sulfate. Crystals were grown at 18°C by hanging drop vapor diffusion against the precipitant solution. The crystals were cryoprotected in buffer containing 21% (w/v) PEG3350, 0.2 M ammonium sulfate, and 20% (v/v) glycerol. Diffraction data from a single crystal to 2.1 Å resolution were collected at APS NE-CAT beamline 24-IDC equipped with Pilatus detector. The data were indexed, integrated, and scaled using HKL2000. The crystal was in space group P21 with two Pce1 protomers in the asymmetric unit. The structure was solved by molecular replacement in PHASER using the coordinates of the Pce1 protein from the Pce1•Spt5 CTD structure as the search model. Iterative refinement and adjustment in PHENIX and COOT yielded a model with R/Rfree of 0.19/0.23 at 2.1 Å resolution and good geometry, as assessed by MolProbity, with 97.7%, 100%, and 0% in favored, allowed, or disallowed regions of Ramachandran space, respectively. Both protomers were continuous polypeptides from the N-terminal serine to Lys372. In both protomers, Lys67 was covalently guanylylated. No electron density corresponding to the Spt5 CTD peptide was observed.
Purification of S. pombe Spt5 CTD proteins
An expression plasmid encoding His6Smt3-Spt5-(801–898) under the control of a bacteriophage T7 promoter was introduced into E. coli BL21(DE3) codon plus RIL. A 1-L culture was grown in super broth in a baffled flask at 37°C to an A600 of 1.0, at which time the flask was placed for 30 min on ice. The culture was adjusted to 0.5 mM IPTG and then incubated overnight at 18°C with constant shaking. Cells were harvested by centrifugation at 16,000g and resuspended in 50 mM Tris-HCl (pH 8.0) and 20% (w/v) sucrose. All subsequent procedures were performed at 4°C. His6Smt3-Spt5-(801–898) was isolated from a soluble bacterial extract by Ni-NTA chromatography and then incubated with Ulp1 to cleave the tag. The tag-free Spt5-(801–898) protein, with a nonnative N-terminal serine and leucine derived from the Smt3 fusion junction, was separated from His10Smt3 and recovered in the flow-through fraction during a second round of Ni-NTA chromatography. Spt5 CTD was then gel-filtered through a Superdex 75 column; the peak fractions were pooled and concentrated by centrifugal ultrafiltration to 17 mg/mL in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM BME.
A variant tag-free Spt5 CTD with an N-terminal cysteine (for use in fluorescence labeling) was obtained by mutating the tag junction in the His6Smt3-Spt5-(801–898) expression plasmid. The N(Cys)-Spt5-(801–898) protein was purified by sequential Ni affinity, Ulp1 cleavage, Ni flow-through, and gel filtration steps and then dialyzed against buffer containing 50 mM Tris-HCl (pH 7.0), 100 mM NaCl, and 1 mM TCEP.
Fluorescent-tagged Pol2 and Spt5 CTDs
A 28-amino-acid synthetic Pol2 CTD–Ser5-PO4 peptide (YSPTS5pPSYSPTS5pPSYSPTS5pPSYSPTS5pPS) was labeled with BODIPY-FL at the N terminus via N-hydroxy succinimidyl ester chemistry as follows. Ten microliters of 1 mM Pol2 CTD peptide was added to 90 μL of 0.35 M sodium bicarbonate (pH 8.1). BODIPY-FL linked to an N-hydroxy succinimidyl ester group by a five-carbon-atom spacer was dissolved in DMSO at 10 mg/mL. Ten microliters of the BODIPY-FL solution was added to the Pol2 CTD solution 1 μL at a time at room temperature, followed by gentle mixing after each addition. The reaction was allowed to proceed overnight at 4°C. The labeled peptide was separated from the excess BODIPY-FL by buffer exchange during concentration. The final BODIPY-FL-Pol2 CTD–Ser5-PO4 peptide was concentrated to 60 μM in water. The peptide concentration was determined by measuring the absorbance of the sample at 274 nm, taking into account the contribution of the BODIPY-FL tag.
N(Cys)-Spt5-(801–898) was labeled with BODIPY-FL at the N terminus via maleimide chemistry as follows. One-hundred-forty microliters of 10 mM BODIPY-FL, linked to a maleimide group by a five-carbon-atom spacer and dissolved in DMSO, was added 10 μL at a time to 1 mL of a 70 μM solution of N(Cys)-Spt5-(801–898) with mixing after each addition. After overnight incubation at 4°C, the BODIPY-FL-(Cys)-Spt5-(801–898) was separated from free BODIPY-FL by gel filtration through a Sephadex G-25 PD-10 column. Residual unconjugated BODIPY-FL was removed by buffer exchange during centrifugal ultrafiltration to attain a concentration of 178 μM Spt5-(801–898) in buffer B. The protein concentration was determined by measuring the absorbance of the sample at 274 nm.
Assays of CTD•capping enzyme interactions by fluorescence polarization
The Pce1–GMP proteins used for fluorescence polarization assays were guanylylated in vitro by incubating the Superdex enzyme preparations with 1 mM GTP and 5 mM MgCl2 for 30 min on ice followed by dialysis against buffer B and concentration to 10–60 mg/mL protein.
Binding reaction mixtures (17–20 μL) containing 20 mM Tris-HCl (pH 8.0); 50 mM NaCl; 4 mM dithiothreitol (DTT); either 300 nM BODIPY-FL-N(Cys)-Spt5-(801–898), 400 nM labeled BODIPY-FL-Pol2 CTD-Ser5-PO4, or 25 nM fluorescein-labeled 22-amino-acid synthetic Spt5 CTD peptide SGSKTPAWNSGSKTPAWNSGSK (purchased from New England Peptides); and increasing concentrations of Pce1–GMP as specified were incubated for 15 min on ice. Aliquots (15 μL) were transferred to wells of a 384-well microplate, and fluorescence polarization was measured at 23°C with a SpectraMax M5 microplate reader (Molecular Devices) using an excitation wavelength of 485 nm, an emission wavelength of 538 nm, and a cutoff of 530 nm for BODIPY-FL-labeled peptides. Fluorescence polarization using fluorescein-labeled peptide was measured using an excitation wavelength of 485 nm, an emission wavelength of 525 nm, and a cutoff of 515 nm. Experiments were performed in triplicate. In experiments where the Pce1 concentration that achieved 50% saturation was at least three times greater than the concentration of the labeled ligand, Kd values were derived by fitting the data in Prism to a single-site binding model (Lundblad et al. 1996). In experiments where the Pce1 concentration that achieved 50% saturation was less than or similar to the concentration of labeled ligand, the Kd values were derived by fitting the data in Prism to a single-site binding model that accounted for receptor depletion (Lundblad et al. 1996). For competition experiments, Pce1–GMP was incubated with serial dilutions of the unlabeled competitor for 15 min on ice. Fluorescent-labeled CTDs were then added, and the mixtures were incubated for another 15 min on ice. Aliquots (15 μL) were transferred to the wells of a 384-well microplate, and measurements were obtained as described above. In these assays, the fluorescent ligand and Pce1 concentrations were as follows: 0.3 μM BODIPY-FL-Spt5 CTD-(801–898) and 1 μM Pce1; 0.4 μM 28-mer BODIPY-FL-Pol2 CTD–Ser5-PO4 and 0.25 μM Pce1; or 25 nM 22-mer fluorescein–Spt5 CTD and 100 μM Pce1. Experiments were performed in triplicate, and data were fit in Prism to a one-site binding model for competitor.
Effects of CTD interfacial mutations on Pce1 function in vivo
Allelic replacement at chromosomal pce1+ locus was performed as follows. We constructed series of integration cassette plasmids containing, in the 5′-to-3′ direction, (1) a 1675-base-pair (bp) segment encompassing the pce1 ORF (wild type or mutated) and 559 bp of 5′ flanking chromosomal DNA, (2) a 268-bp DNA segment harboring the nmt1+ transcription termination signal, (3) a kanMX cassette conferring resistance to G418, and (4) a 316-bp segment of 3′ flanking pce1 chromosomal DNA. The 5′-pce1-kanMX-pce1-3′ integration cassettes were excised from the plasmids and transfected into S. pombe diploids. kanMX transformants selected for growth on YES agar containing 0.15 mg/mL G418 were genotyped by Southern blotting to confirm correct integration at one of the pce1 loci. The pce1-kanMX allele was then PCR-amplified and sequenced to confirm the presence of the desired mutations. The heterozygous pce1+ pce1-kanMX strains were sporulated. A random population of haploid progeny (∼3000, as gauged by plating on YES agar) was plated on YES-G418 agar. A pce1 mutant allele was classified as lethal when no G418-resistant haploid progeny were recovered after growth for 6 d at 30°C. Viable pce1-kanMX haploid strains formed colonies on selective G418 agar at frequencies consistent with random segregation. To gauge the effect of the pce1 mutations on vegetative growth, cultures of haploid S. pombe pce1-kanMX strains (all of mating type h−) were grown in liquid medium at 30°C until A600 reached 0.6–0.9. The cultures were adjusted to a final A600 of 0.1, and aliquots (3 μL) of serial fivefold dilutions were spotted on YES agar. The plates were incubated at 18°C, 25°C, 30°C, 34°C, and 37°C.
Acknowledgments
We thank NE-CAT beamlines (APS) supported by RR-15301 (National Institutes of Health National Center for Research Resources). APS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract number DE-AC02-06CH11357. Research reported in this study was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers GM061906 (to C.D.L.) and GM052470 (to B.S. and S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S.S. is an American Cancer Society Research Professor. C.D.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242768.114.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chang JH, Jiao X, Chiba K, Oh C, Martin CE, Kiledjian M, Tong L 2012. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat Struct Mol Biol 19: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A, Ho CK, Martins A, Shuman S, Mondragon A 2001. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J 20: 2575–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell 10: 585–597 [DOI] [PubMed] [Google Scholar]
- Corden JL 2013. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem Rev 113: 8423–8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Geyer M 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113: 8456–8490 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Geronimo C, Bataille AR, Robert F 2013. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev 113: 8491–8522 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD 2011. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell 43: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Lima CD 2005. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol 15: 99–106 [DOI] [PubMed] [Google Scholar]
- Gu M, Rajashankar KR, Lima CD 2010. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 18: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson K, Doherty AJ, Shuman S, Wigley DB 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89: 545–553 [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Fu J 2013. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta 1829: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser2 and Ser5 phosphorylation in the recruitment and allosteric activation of mammalian capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Ho CK, Lehman K, Shuman S 1999. An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res 27: 4671–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M 2013. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell 50: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CD, Wang LK, Shuman S 1999. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99: 533–543 [DOI] [PubMed] [Google Scholar]
- Lidschreiber M, Leike K, Cramer P 2013. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol 33: 3805–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad JR, Laurance M, Goodman RH 1996. Fluorescence polarization analysis of protein–DNA and protein–protein interactions. Mol Endocrinol 10: 607–612 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD 2000. Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem 277: 19639–19648 [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S 2003. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation and mutational analysis. J Biol Chem 278: 43346–43356 [DOI] [PubMed] [Google Scholar]
- Pei Y, Hausmann S, Ho CK, Schwer B, Shuman S 2001. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J Biol Chem 276: 28075–28082 [DOI] [PubMed] [Google Scholar]
- Pei Y, Du H, Singer J, St Amour C, Granitto S, Shuman S, Fisher RP 2006. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol Cell Biol 26: 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Pei Y, Shuman S, Schwer B 2010. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol 30: 2353–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Shuman S 2011. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 43: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Sanchez AM, Shuman S 2012. Puncuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci 109: 18024–18029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bitton DA, Sanchez AM, Bähler J, Shuman S 2014. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc Natl Acad Sci 111: 4185–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Lima CD 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol 14: 757–764 [DOI] [PubMed] [Google Scholar]
- St Amour CV, Sansó M, Bösken CA, Lee KM, Larochelle S, Zhang C, Shokat KM, Geyer M, Fisher RP 2012. Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) Rpb1 carboxyl-terminal domain substrate recognition. Mol Cell Biol 32: 2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP 2009. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell 33: 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes & Dev 13: 1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]