Abstract
The non-classical zinc finger protein, Neural Zinc Finger Factor-1, contains six Cys2His2Cys domains. All three cysteines and the second histidine directly bind Zn(II). Using a combination of mutagenesis, metal coordination and DNA binding studies, we report that the first histidine is involved in a functionally important hydrogen bonding interaction.
Zinc finger (ZF) proteins are zinc co-factored metalloproteins that are critical for a myriad of biological processes, most notably transcriptional regulation.1, 2 There are at least fourteen distinct classes of ZFs.3 All ZFs contain one or more discrete ZF domains that include cysteines and/or histidines, which serve as ligands for zinc coordination.1, 4, 5 One ZF family, collectively known as the ‘CCHHC’ type ZF family, is unique in that it contains five potential zinc ligands: Cys2His2Cys within each ZF domain.1 Three members of this family have been identified to date: Neural Zinc Finger Factor-1 (NZF-1), Myelin Transcription Factor 1 (MyT1) and Suppression of Tumorgenicity 18 (ST18).6-8 NZF-1 and MyT1 are essential for the development of the central nervous system where they function to control the development of neurons and oligodendrocytes via regulation of the β-retinoic acid receptor (βRAR) and the proteolipid protein, respectively.6, 7 ST18 is associated with controlling tumour development, although its specific role in this process has not yet been established.9
All CCHHC family ZFs contain multiple, highly homologous, CCHHC domains arranged in clusters (Fig. 1a).1 A bona fide DNA target has only been identified for the cluster of NZF-1 made up of ZF domains #2 and #3 (named NZF-1-F2F3).6, 10 We have shown that the few non-conserved amino acid residues present in NZF-1-F2F3 are critical for sequence specific DNA binding.11 This model of DNA recognition contrasts the paradigm for classical ZFs: classical ZFs utilize a Cys2His2 ligand set for Zn(II) coordination and there are only a few additional conserved amino acids that are critical for the fold and function of the protein.1, 11-13
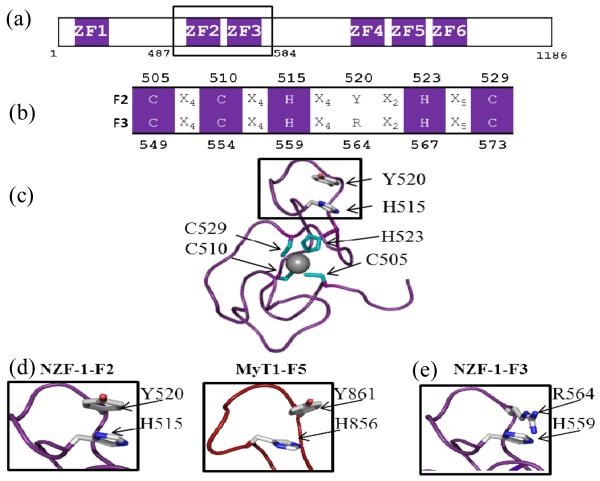
Fig. 1.
(a) Cartoon depicting the arrangement of the ZF domains of NZF-1 with NZF-1-F2F3 boxed. (b) Alignment of ZF domains of NZF-1-F2 and NZF-1-F3 with conserved CCHHC residues shown in purple and non-conserved residues denoted in black. (c) NMR structure of NZF-1-F2, with metal coordinating ligands in cyan (PDBID: 1PXE). (d) A zoomed in view of the local structure surrounding the non-coordinating histidines found in NZF-1-F2 (PDBID: 1PXE) and MyT1-F5 (PDBID: 2JYD). (e) Zoomed in view of the predicted structure of NZF-1-F3 centred at the non-coordinating histidine. All structures were generated using PyMOL.
Although the CCHHC ZFs have five potential Zn(II) coordinating ligands per domain (3 Cys, 2 His; Fig. 1b), studies have shown that only four ligands directly coordinate Zn(II) (Fig 1c). These ligands are the three cysteines and the second conserved histidine resulting in a CCHC ligand set.14-16 The role of the first conserved (non-Zn(II)-coordinating) histidine is not understood. The NMR structure of just finger 2 of NZF-1 (NZF-1-F2) has been reported and the non-coordinating histidine appears to be involved in a pi-stacking interaction with a tyrosine residue to stabilize the fold around the zinc ion such that the protein can bind to DNA (Fig. 1d).15 However, we found that when this histidine is mutated to a phenyalanine in a functional construct of NZF-1 (NZF-1-F2F3), DNA binding was completely abolished, indicating that the role of the histidine is not a simple pi-stacking interaction.11 Moreover, in the structure of F5 of MyT1, which has 80% sequence homology to F2 of NZF-1, the histidine and tyrosine are not oriented in a position that allows for pi-stacking (Fig. 1d).14, 17 Both structures, NZF-1-F2 and MyT1-F5, lack significant secondary structure and principally fold into a series of flexible loops. Circular dichroism studies on different ZFs of this family confirm these results.11, 18 Thus, the differences in orientations between the histidine and tyrosine residues observed in the two NMR structures indicate that the overall folds of these domains are highly flexible.
Careful inspection of the sequences of F2 and F3 of NZF-1 revealed that the tyrosine residue that sometimes appears to pi-stack with the non-coordinating histidine is absent in F3. An arginine is found in place of this residue. To better understand the interactions taking place in F3, a model of NZF-1-F3 was generated in PyMOL with the NZF-1-F2 structure as a scaffold (Fig 1e; ESI†).19 Arginine residues often participate in hydrogen bonding interactions, but are not involved in pi stacking interactions.20 Thus, the non-coordinating histidine may participate in hydrogen bonding, rather than pi-stacking, as histidine can form hydrogen bonds with either tyrosine or arginine but can only form pi stacking interactions with tyrosine.
Mutants of NZF-1-F2F3 were prepared to assess the role of hydrogen bonding in function (ESI, Fig. S1†). The non-coordinating histidines from F2 (H515) and F3 (H559) of NZF-1 were mutated to glutamine producing three mutants: NZF-1-F2F3-H515Q, NZF-1-F2F3-H559Q, and NZF-1-F2F3-H515/559Q (called H515Q, H559Q and H515/559Q). The mutations are designed to retain hydrogen-bonding interactions, but disrupt pi-stacking.
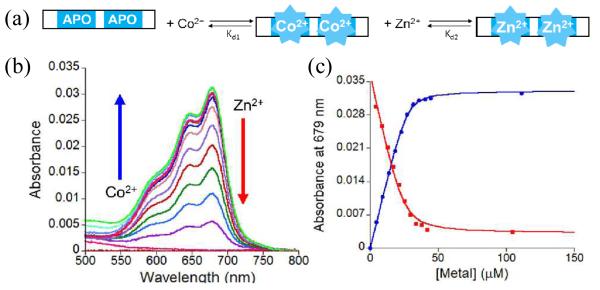
To determine whether the mutants bound Zn(II), optical titrations using Co(II) as a spectroscopic probe for Zn(II) were performed (ESI†). This is a common strategy used to investigate metal ion coordination to ZF sites.21-23 Co(II), like Zn(II), can coordinate cysteine and histidine ligands in a tetrahedral geometry but has the advantage of having a partially filled d-shell (d7) which allows for optically observable d-d transitions. The optical spectrum for a Cys2HisCys site is expected to exhibit transitions between 550 – 750 nm.1, 24 All three mutant peptides were titrated with Co(II) and the expected d-d bands were observed (Fig. 2; Fig. S2†). The intensity of the d-d transitions became saturated after 2 equivalents of Co(II) were added, indicating the expected 2:1 Co(II):peptide binding stoichiometry. No intermediate spectra were observed during the course of the titrations, which suggests that both ZFs within each domain bind to Co(II) with equal affinity. The data were fit to a 1:1 binding equilibrium, treating the two sites equally.23 Upper limit dissociation constants (Kds) of 6.9 ± 0.3 × 10−7, 5.2 ± 1.2 × 10−7, and 3.9 ± 1.0 × 10−7 M were determined for Co(II) binding to H515Q, H559Q, and H515/559Q, respectively. To determine the affinities of the peptides for Zn(II), solutions of the peptides with 20 excess molar equivalents of Co(II) were titrated with Zn(II) and the decrease in the d-d absorbance bands monitored. These data were fit a competitive binding model23, 24 and upper limit Kds for Zn of 3.0 ± 0.9 × 10−9, 5.1 ± 2.1 × 10−10, and 1.2 ± 0.6 × 10−11 M were determined for H515Q, H559Q, and H515/559Q, respectively. These Kds are in the same range as those reported for other wild type (WT) and mutant NZF-1 constructs indicating that the mutations have not affected the peptides’ ability to bind metal ions (Table S1†).25
Fig. 2.
(a) Cartoon of ZF titrations. (b) Plot of the change in the absorption spectrum between 500-800 nm as Co(II) and Zn(II) are titrated with 25 μM H515Q. (c) Plot of the absorbance at 679 nm as a function of added Co(II) (blue) or Zn(II) (red). The solid line represents non-linear least squares fits to 1:1 and competitive binding equilibria, respectively.
The Co(II)-peptides were further characterized by X-band Electron paramagnetic resonance (EPR) spectroscopy at 12 K (ESI†). Co(II)-WT exhibited a broad spectrum indicative of high spin Co(II). Rhombic symmetry was observed with g values of 5.36, 2.96, and 2.31 (Fig. 3a). No observable hyperfine splitting was noted. The spectrum is consistent with spectra reported for other tetrahedral Co(II) coordination complexes.26-29 The EPR spectrum of H515/559Q was identical to that of the WT peptide (Fig. S3a†), suggesting that the mutations did not affect the geometry at the metal centres. The EPR spectrum of a previously reported mutant in which the metal coordinating histidine had been mutated such that the non-coordinating histidine now bound Co(II) (named NZF-1-F2F3-H523/567F)11 was also recorded to determine how switching the coordinating ligands affects the metal centre. The spectrum showed only slight differences, indicating that NZF-1 forms high-spin Co(II) species when four coordinating ligands are present (Fig. S3b†).
Fig. 3.
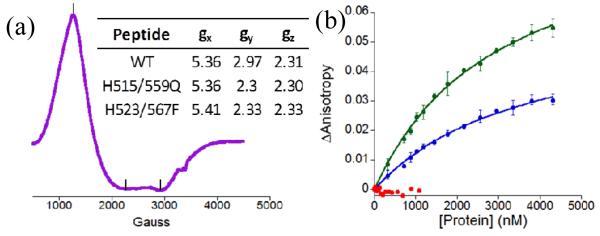
(a) EPR spectrum of Co(II)NZF-1-F2F3, table of g values for WT and variants (inset). (b) Comparison of anisotropy as H515Q (blue), H559Q (green), and H515/559Q (red) are titrated with βRAR.
Fluorescence anisotropy (FA) of the mutants with βRAR DNA, which is NZF-1’s physiological target, and a random segment of DNA was performed to measure the effects of the mutations on DNA binding (ESI†)11, 24 H515Q and H559Q bound the βRAR DNA specifically with Kds of 3.0 ± 0.2 × 10−6 M and 3.7 ± 0.3 × 10−6 M, respectively; whereas, the double mutant, H515/559Q, did not bind to DNA with measurable affinity (Table S1, Fig. 3b). In comparison, WT binds βRAR DNA with an affinity of 1.4 ± 0.2 × 10−8 M.11
There are several possible interpretations of these results. One explanation is that for the single ZF domain mutants, only the non-mutated domain is binding DNA and by inference when both domains are mutated no binding would be observed. There are some examples of single ZF domains that can bind to DNA or RNA targets.30-32 These single ZFs typically exhibit Kds in the micromolar regime.32 To investigate this, a single ZF domain of NZF-1, NZF-1-F2, was prepared using published protocols15 and DNA binding was measured. NZF-1-F2 did not bind to either the βRAR or the random DNA (data not shown). Thus, the explanation that mutation of a single ZF domain merely abrogates binding from that mutated domain was ruled out.
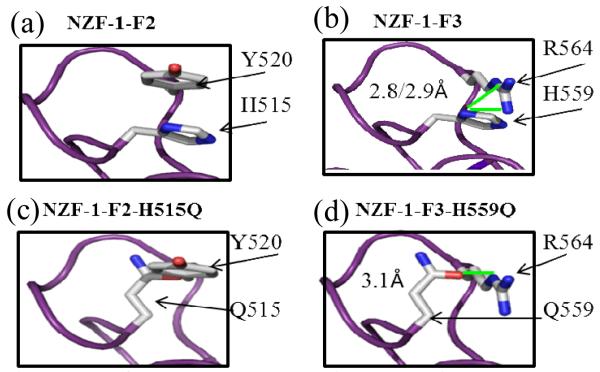
A second explanation is that hydrogen bonding is but one of several properties required for proper fold and function of NZF-1-F2F3. To further understand the structural consequences of the H to Q mutants, the structures of the mutant ZFs were generated in PyMOL using the NZF-1-F2 structure as a scaffold (ESI†).19 These structures were compared to the structures of WT NZF-1-F2 and the modelled structure of NZF-1-F3. The likelihood of H-bonding interactions between H and Y (for WT F2), Q and Y (for the F2 mutant, H515Q), H and R (for WT F3), or between Q and R (for the F3 mutant, H559Q) were determined by estimating the bond distances between the potential hydrogen bond donor and acceptor atoms (Fig. 4). For F2 and H515Q, the H/Q likely serves as the H-bond acceptor while Y serves as the H-bond donor. The closest calculated bond distances for these constructs are 4.5 and 4.6 Å20, respectively. These distances are too long for a H-bond interaction to occur.20 For WT F3 and H559Q, where H/Q serves as the H-bond donor and R as the acceptor, these distances are 2.8 and 3.1 Å, which are typical distances for a hydrogen bonding interactions.20 This analysis suggests that hydrogen bonding is important for the fold and function of F3, but not for F2. The DNA binding results can be interpreted to support this conclusion. For F2, where hydrogen bonding does not appear to be important for function, the H to Q mutation disrupted another key feature that is important for fold and function, thus explaining the decrease in affinity of the mutant for DNA. For F3, where H-bonding appears to be important, the mutation of H to Q (H559Q) retains the hydrogen bond, but DNA binding is still negatively impacted. This can be explained by the observation that Q does not completely mimic the hydrogen bonding of the native H. The acceptor atoms of H and Q (N and O, respectively) are not only in different orientations, but could be accepting the H-bond from different amides on the donor arginine (Fig. 4). Therefore, we propose that these differences in WT and mutant F3 affect folding and DNA binding. Alternatively, Y520 and R564 could directly contact DNA, thus any mutation would result in loss of binding.
Fig. 4.
Close up of (a) F2: H515 and Y520; (b) F3: H559 and R564; (c) H515Q and Y520; (d) H559Q and R564. Distances of potential H bond shown adjacent to bond (shown in green).
Taken together, these results underscore the complexity of the DNA binding interactions mediated by NZF-1. The published structures of single ZF domains contain minimal secondary structure and how these structures change upon DNA binding is not known. Attempts to obtain the structure of NZF-1-F2F3 bound to its DNA partner are currently in progress in our lab to further unravel the complex zinc mediated NZF-1/DNA recognition event.
Supplementary Material
Acknowledgments
This work was supported by NSF (SLJM: CHE-0747863, CHE-1306208) and NIH, NINDS (ANB:F31NS074768). Thanks to David Goldberg and Alison McQuilken (JHU) for EPR assistance and Holly Cymet of Stevenson Univ. for the NZF-1-F2 construct.
Footnotes
Electronic Supplementary Information (ESI) available: [Experimental details and characterization data, UV-visible and EPR spectra, table of dissociation constants]. See DOI: 10.1039/c000000x/
References
- 1.Michalek JL, Besold AN, Michel SL. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- 2.Klug A. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 3.Matthews JM, Sunde M. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 4.Laity JH, Lee BM, Wright PE. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 5.Andreini C, Bertini I, Cavallaro G. PLoS One. 2011;6:e26325. doi: 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Yu VC, Buchholz F, O’Connell S, Rhodes SJ, Candeloro C, Xia YR, Lusis AJ, Rosenfeld MG. J. Biol. Chem. 1996;271:10723–10730. doi: 10.1074/jbc.271.18.10723. [DOI] [PubMed] [Google Scholar]
- 7.Kim JG, Hudson LD. Mol. Cell. Biol. 1992;12:5632–5639. doi: 10.1128/mcb.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yee KS, Yu VC. J. Biol. Chem. 1998;273:5366–5374. doi: 10.1074/jbc.273.9.5366. [DOI] [PubMed] [Google Scholar]
- 9.Jandrig B, Seitz S, Hinzmann B, Arnold W, Micheel B, Koelble K, Siebert R, Schwartz A, Ruecker K, Schlag PM, Scherneck S, Rosenthal A. Oncogene. 2004;23:9295–9302. doi: 10.1038/sj.onc.1208131. [DOI] [PubMed] [Google Scholar]
- 10.Berkovits HJ, Berg JM. Biochemistry. 1999;38:16826–16830. doi: 10.1021/bi991433l. [DOI] [PubMed] [Google Scholar]
- 11.Besold AN, Oluyadi AA, Michel SL. Inorg. Chem. 2013;52:4721–4728. doi: 10.1021/ic4003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael SF, Kilfoil VJ, Schmidt MH, Amann BT, Berg JM. Proc. Natl. Acad. Sci. U. S. A. 1992;89:4796–4800. doi: 10.1073/pnas.89.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhanasekaran M, Negi S, Sugiura Y. Acc. Chem. Res. 2006;39:45–52. doi: 10.1021/ar050158u. [DOI] [PubMed] [Google Scholar]
- 14.Gamsjaeger R, Swanton MK, Kobus FJ, Lehtomaki E, Lowry JA, Kwan AH, Matthews JM, Mackay JP. J. Biol. Chem. 2008;283:5158–5167. doi: 10.1074/jbc.M703772200. [DOI] [PubMed] [Google Scholar]
- 15.Berkovits-Cymet HJ, Amann BT, Berg JM. Biochemistry. 2004;43:898–903. doi: 10.1021/bi035159d. [DOI] [PubMed] [Google Scholar]
- 16.Blasie CA, Berg JM. Inorg. Chem. 2000;39:348–351. doi: 10.1021/ic990913y. [DOI] [PubMed] [Google Scholar]
- 17.Gamsjaeger R, O’Connell MR, Cubeddu L, Shepherd NE, Lowry JA, Kwan AH, Vandevenne M, Swanton MK, Matthews JM, Mackay JP. J. Biol. Chem. 2013;288:35180–35191. doi: 10.1074/jbc.M113.482075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rich AM, Bombarda E, Schenk AD, Lee PE, Cox EH, Spuches AM, Hudson LD, Kieffer B, Wilcox DE. J. Am. Chem. Soc. 2012;134:10405–10418. doi: 10.1021/ja211417g. [DOI] [PubMed] [Google Scholar]
- 19.The PyMOL Molecular Graphics System, Version 1.1. Schrödinger, LLC; [Google Scholar]
- 20.Berg J, Tymoczko JL, Stryer L. Biochemistry. 6 edn. W. H. Freeman and Company; New York, N.Y.: 2007. pp. 1–59. ch. 1 - 2. [Google Scholar]
- 21.Bertini I, Luchinat C. Adv. Inorg. Biochem. 1984;6:71–111. [PubMed] [Google Scholar]
- 22.Berg JM, Godwin HA. Annu. Rev. Biophys. Biomol. Struct. 1997;26:357–371. doi: 10.1146/annurev.biophys.26.1.357. [DOI] [PubMed] [Google Scholar]
- 23.Berg JM, Merkle DL. J. Am. Chem. Soc. 1989;111:3759–3761. [Google Scholar]
- 24.Besold AN, Lee SJ, Michel SL, Sue NL, Cymet HJ. JBIC, J. Biol. Inorg. Chem. 2010;15:583–590. doi: 10.1007/s00775-010-0626-1. [DOI] [PubMed] [Google Scholar]
- 25.Magyar JS, Godwin HA. Anal. Biochem. 2003;320:39–54. doi: 10.1016/s0003-2697(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 26.Kang PC, Eaton GR, Eaton SS. Inorg. Chem. 1994;33:3660–3665. [Google Scholar]
- 27.Makinen MW, Kuo LC, Yim MB, Wells GB, Fukuyama JM, Kim JE. J. Am. Chem. Soc. 1985;107:5245–5255. [Google Scholar]
- 28.Kuo LC, Makinen MW. J. Am. Chem. Soc. 1985;107:5255–5261. [Google Scholar]
- 29.Walsby CJ, Krepkiy D, Petering DH, Hoffman BM. J. Am. Chem. Soc. 2003;125:7502–7503. doi: 10.1021/ja034589o. [DOI] [PubMed] [Google Scholar]
- 30.Dathan N, Zaccaro L, Esposito S, Isernia C, Omichinski JG, Riccio A, Pedone C, Di Blasio B, Fattorusso R, Pedone PV. Nucleic Acids Res. 2002;30:4945–4951. doi: 10.1093/nar/gkf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parraga G, Horvath SJ, Eisen A, Taylor WE, Hood L, Young ET, Klevit RE. Science. 1988;241:1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- 32.Michel SL, Guerrerio AL, Berg JM. Biochemistry. 2003;42:4626–4630. doi: 10.1021/bi034073h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.