Abstract
Objective
To evaluate the use of urine KIM-1 as a biomarker for supporting a diagnosis of kidney cancers before operation.
Methods
A total of 19 patients were enrolled in the study based on preoperative imaging studies.
Pre-operative and follow-up (1 month) uKIM-1 levels were measured and normalized with uCr levels and renal tumors were stained for KIM-1 using immunohistochemical techniques.
Results
The percentage of KIM-1 positive staining RCC cells ranged from 10 to 100% and the staining intensity ranged from 1+ to 3+. Based on the KIM-1 staining, 19 cases were divided into the KIM-1-negative staining group (n =7) and the KIM-1-positive group (n = 12).
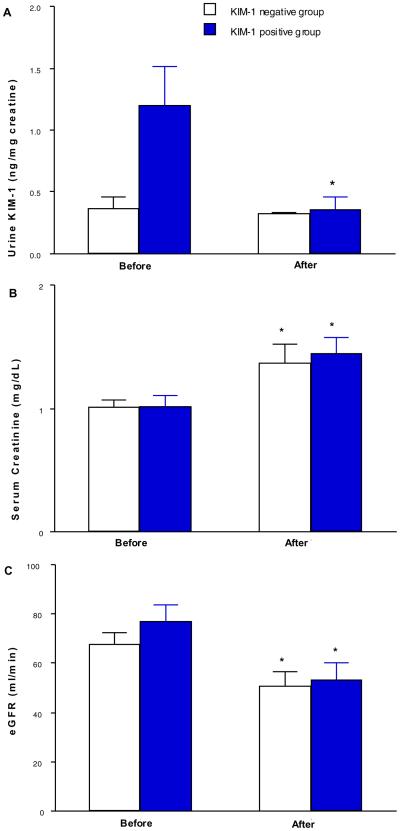
Serum creatinine (sCR) levels were significantly elevated after nephrectomy in both groups. In the KIM-1 negative group, uKIM-1/uCr remained at a similar level before (0.37 ± 0.1 ng/mg Cr) and after nephrectomy (0.32 ± 0.01 ng/mg Cr). However, in the KIM-1 positive group, elevated uKIM-1/uCr at 1.20 ± 0.31 ng/mg Cr was significantly reduced to 0.36± 0.1 ng/mg Cr, which was similar to the pre-operative uKIM-1/uCr (0.37 ± 0.1 ng/mg Cr) in the KIM-1 negative group.
Conclusion
Our study showed significant reduction in uKIM-1/uCr after nephrectomy, suggesting that urine KIM-1 may serve as a surrogate biomarker for kidney cancer and a non-invasive pre-operative measure to evaluate the malignant potential of renal masses.
Keywords: Kidney injury molecule-1, renal cell carcinoma, renal cystic lesions
INTRODUCTION
In the United States there were 58,000 cases and about 13,000 deaths from renal cell carcinoma in 2009 [1]. Renal cell carcinoma represents 3.9% of all US cancers and 2% of all cancer deaths. The majority of kidney tumors are discovered incidentally during investigation of unrelated complaints. However, nearly 30% of patients present with metastatic disease at the time of diagnosis and 30–40% of patients with clinically localized kidney cancer will have a recurrence. The diagnosis and monitoring of kidney cancer requires expensive and frequent imaging examinations. A reliable urinary assay for kidney cancer would have major implications for tumor screening in high-risk patients, risk stratification of patients, and potentially as a surrogate marker for response to therapy or post-treatment surveillance.
Since KIM-1 was discovered in 1998 [2], many studies from the Bonventre group and others indicate that KIM-1 is a sensitive and specific biomarker in detecting injury of proximal tubules in humans and other animals [3–6]. KIM-1 is a type I transmembranous glycoprotein containing an extracellular immunoglobulin-like domain topping a long mucin-like domain [2], and is found to be a phagocytic factor in repairing proximal tubules [7] Moreover, its ectodomain is released into the urine and serves as a sensitive urine marker for proximal renal tubule injury [8–11] Clear cell and papillary RCCs, derived from the proximal tubular cells, stain positively for KIM-1 [12–13]. Metastatic clear cell and papillary RCC are also positive for KIM-1 staining [13]. The renal tumors from distal nephron tubules, such as chromophobe RCC and oncocytoma stain negatively for KIM-1 [13]. Our previous study demonstrated that pre-operative urine KIM-1 (uKIM-1) are increased in the patients with major types of kidney tumors, but there are only five post-operative follow-up cases available in that study [12]. A recent study from another group reveals that pre-operative uKIM-1 in patients with RCC are significantly reduced after surgery but another urinary kidney injury marker, urinary neutrophil gelatinase-associated lipocalin (NGAL), does not change when comparing pre- and post-operative values [14]. The purpose of this prospective study was to compare pre-operative and post-operative uKIM-1 values to determine if renal tumors were the source for elevated uKIM-1 before nephrectomy.
METHODS
Patient Selection
Following approval from the Human Investigation Committee of William Beaumont Hospitals patients were prospectively enrolled based on specific diagnostic criteria.
To qualify for enrollment patients had to have a solitary renal mass suspicious for renal cell carcinoma seen on computed tomography (CT) scan or magnetic resonance imaging (MRI) imaging. Only masses that appeared to be less than stage T3 according to the current Staging System of the American Joint Committee on Cancer (AJCC) criteria, and demonstrated no evidence of lymph node or metastatic involvement on cross-sectional imaging, were included in the study. In addition patients were required to have a normal serum creatinine and a normal-appearing contralateral kidney. Patients with any history of prior therapy for RCC (including surgery, immunotherapy, chemotherapy, radiation, hormonal, or investigational therapy) were excluded.
Study Protocol
On the day of surgery patients were admitted to the operating room according to standard hospital protocols. Urine samples for KIM-1 and creatinine levels were obtained in the preoperative area. A radical or partial nephrectomy by laparoscopic, robotic or open approach was performed by one of the study investigators. Pre-operative blood samples were also collected to measure serum creatinine [15]. Urine KIM-1 levels were measured in the Renal Division, Brigham and Women's Hospital, Boston, MA using a bead-based ELISA technique. Both serum and urine creatinine were measured in clinical laboratory of William Beaumont Hospital as clinical samples. Thirty days after the operation, follow-up urine and blood samples were obtained for post-operative measurements for urine KIM-1, urine creatinine and serum creatinine measurements, respectively. The estimated glomerular filtration rate (eGFR) rates pre and post surgery were calculated with the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) equation [15].
Urine ELISA for KIM-1
The KIM-1 levels in urine were measured using microbead based sandwich ELISA assay as described previously [16]. The same human renal ELISA kit for analyzing urine KIM-1 is also available commercially (Bioassay Works, LLC, Ijamsville, MD). Briefly, 30 μl of urine sample, standards (KIM-1 recombinant protein), and positive and negative controls were incubated with ~5000 polystyrene beads that were conjugated with KIM-1 capture antibody (AF 1750, for 1 hour on an orbital shaker. Beads were washed three times with PBST (0.5%-Tween-PBS) solution and incubated with detection antibody (BAF 1750, RnD Systems) for 45 minutes on an orbital shaker. The amount of KIM-1 antigen bound was measured by incubating the bead complex with streptavidin conjugated with Phycoerythrin for 15 minutes and the amount of the fluorescence was measured by Bio-Plex 100 machine. KIM-1 levels in the urine samples were interpolated based on 13-point standard curve using logistic regression model. All the measurements were made in duplicate and the % coefficient variation is less than 15%. We normalized uKIM-1 to the spot urinary creatinine concentration (uCr) to compensate for the differences in relative amounts of water removed along the nephron.
Pathology Examination and Immunohistochemical Stains of Renal Lesions
Pathological evaluation of the mass was performed in the standard fashion. Tumors near renal collecting system (<1cm) or involving renal sinus fat were denoted as near the collecting system. Renal tissue was fixed in formalin and paraffin embedded and five steps used for single antibody staining were as follows. Tissues were sectioned at 3-μm and dried for 60 min at 60°C. Slides were then dewaxed in 3 xylene baths for 3 min each, 3 100% alcohol baths, 3 min each, followed by 30 seconds of running water. Third, antigen retrieval was carried out in a Tris EDTA Buffer at pH-8.0, 20 min at 99°C, 20 min cool down at room temperature, followed by a quick water rinse. Fourth, slides were placed in H2O2 for 15 min followed by a quick water rinse and then placed in Tris buffer pH-7.6. Finally, slides were placed into a programmed Dako Autostainer (DakoCytomation, Carpinteria, CA) using a Thermo Scientific UltraVision LP Detection System (Kalamazoo, MI). The program consisted of 5 min Ultra V block, 30 min incubation with a primary KIM-1 monoclonal antibody (AKG7 clone at 1:10 dilution), 8 min Primary Antibody Enhancer, 10 min HRP polymer (equivalent to secondary antibody) and 5 min of the chromagen DAB to achieve a brown color staining. The renal mass was examined microscopically for tumor type, grade and association to surrounding renal parenchyma, renal capsule, renal sinus, renal vessels and resection margins. Immunohistochemical stains for KIM-1 were performed on histologic sections of the mass. Staining was also carried out for CD68, a phagocytic factor [17, 18], using a CD68 antibody (KP1 clone) from Dako Cytomation and the same protocol as for KIM-1 staining.
The staining results were recorded as follows: negative = absence of staining; 1+ = any tumor cell staining to 10% of the tumor stained; 2+ = 11% to 50% of the tumor stained; and 3+ = over 50% of the tumor stained. The staining intensity was also assessed and recorded as: 1+ = light-brown, fine granular membranous/cytoplasmic staining, 2+ = moderate granular membranous/cytoplasmic staining and 3+ = dark-brown, coarse granular membranous/cytoplasmic staining.
Tissue Microarray and Immunohistochemical Stains
To compare KIM-1 staining with CD68 staining in various renal tumors, a tissue microarray block containing 22 clear RCC, 20 papillary RCC, 25 chromophobe RCC and 23 oncocytoma was sectioned and stained for both KIM-1 and CD68. The staining pattern was evaluated as described above.
Experimental Procedures of KIM-1 DNA Sequencing
To determine the integrity of DNA sequence of KIM-1 in renal tumors, genomic DNA was prepared from paraffin-embedded tumor tissue (10 to 15 sections of 10 μm thick cuts) containing four subtypes of archival renal tumors (3 clear cell RCC, 3 papillary RCC, 2 chromophobe RCC and 2 oncocytoma). Using the purified DNAs as the template, one μl of the initial PCR reaction was used as template for a second PCR using primer sets that were nested within the sequence amplified by the initial primers and PCR was performed to amplify each of the nine major coding exons of the KIM-1 gene. Each PCR product then went through a cloning and sequencing (GENEWIZ) process. DNA was prepared and analyzed for presence of the insert by an EcoR I restriction digest and agarose gel/ethidium bromide separation and detection. Two to five clones containing exon 4 inserts were sequenced and analyzed for each specimen. Vector NTI software was used to compare exon DNA sequence to the published sequence of HAVCR-1 (NM_012206), in order to identify altered DNA sequence in renal tumors.
Statistics
Results were expressed as the mean ± SEM. Pre-operative values of uKIM-1, serum creatinine and eGFR were compared with respective post-operative values using student's t test (Statview). A p value less than 0.05 was considered statistically significant.
RESULTS
Compare urine KIM-1 before and after nephrectomy
A total of 23 patients were initially enrolled in the study. Four cases were excluded from the study. Two patients initially enrolled withdrew after the operation (patient #1 and #11). The third patient (patient #23) never made a follow-up visit, thus no post-operative data could be obtained. The fourth patient (# 15) underwent a laproscopic cryoablation instead of partial or radical nephrectomy as initially intended and was excluded.
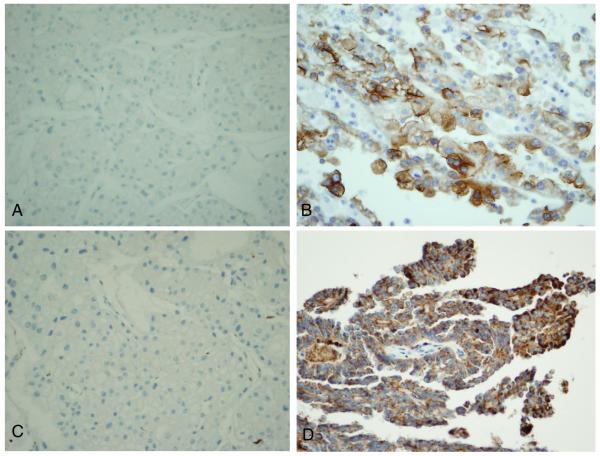
The remaining 19 patients were divided into two groups based on the lesions' staining for KIM-1. The two groups consisted of masses that demonstrated negative staining of KIM-1 (group 1) or positive staining of KIM-1 (group 2) (Table-1). Group 1 included two chromophobe RCC (Figure 1A) and 5 benign renal lesions. Group 2 included 12 RCC cases: four papillary type RCC and eight clear cell type RCC tumors. These renal tumors ranged from 2.4 to 10.5 cm in size without extra-renal extension or sinus invasion. All tumors were reported as Fuhrman Grade 2 to 3 out of 4, except one (patient #20) reported as Fuhrman grade 4. All the RCC tumors were limited within the kidneys except one with renal vein invasion (patient #20, T3b). Tumor pathological data are listed in Table 2. Clear cell type RCC stained mild to moderately positive for KIM-1 while papillary RCC stained strongly positive (Figure 1B) as seen in our previous studies.
Table 1.
Age (years), gender, urine KIM-1 Levels (ng/mg urine creatinine), serum Creatinine levels (mg/dL) and CKD-EPI (ml/min per 1.73m2) in negative and positive stained KIM-1 cases.
| KIM-1- Negative Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases # |
Age/ Gender |
Preoperative KIM-1 |
Postoperative KIM-1 |
Change in KIM-1 |
Preoperative sCr |
Postoperative sCr |
Change in sCr |
Preoperative CKD-EPI |
Postoperative CKD-EPI |
| 4 | 56F | 0.74 | 0.59 | −0.15 | 0.97 | 1.54 | 0.57 | 65 | 37 |
| 5 | 66M | 0.37 | 0.15 | −0.19 | 0.74 | 1.38 | 0.64 | 96 | 53 |
| 7 | 54F | 0.00 | 0.19 | 0.19 | 1.05 | 1.10 | 0.05 | 60 | 57 |
| 10 | 66M | 0.25 | 0.17 | −0.08 | 1.10 | 1.59 | 0.49 | 70 | 45 |
| 12 | 63F | 0.40 | 0.11 | −0.29 | 1.01 | 1.01 | 0.00 | 59 | 59 |
| 14 | 51F | 0.19 | 0.12 | −0.07 | 1.02 | 0.90 | −0.12 | 64 | 74 |
| 21 | 73M | 0.61 | 0.00 | −0.61 | 1.19 | 2.04 | 0.85 | 60 | 31 |
| KIM-1 Positive Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases # |
Age/ Gender |
Preoperative KIM-1 |
Postoperative KIM-1 |
Change in KIM-1 |
Preoperative sCr |
Postoperative sCr |
Change in sCr |
Preoperative CKD-EPI |
Postoperative CKD-EPI |
| 2 | 64F | 1.06 | 0.25 | −0.82 | 0.90 | 1.15 | 0.25 | 68 | 50 |
| 3 | 31F | 1.63 | 0.27 | −1.37 | 1.00 | 0.82 | −0.18 | 75 | 96 |
| 6 | 61M | 3.22 | 0.59 | −2.63 | 0.88 | 0.80 | −0.08 | 93 | 96 |
| 8 | 54M | 0.29 | 0.10 | −0.18 | 0.95 | 1.83 | 0.88 | 90 | 41 |
| 9 | 66M | 0.27 | 0.00 | −0.27 | 0.89 | 1.77 | 0.88 | 89 | 39 |
| 13 | 69M | 0.20 | 0.11 | −0.09 | 1.32 | 1.98 | 0.66 | 55 | 33 |
| 16 | 68F | 2.52 | 1.30 | −1.22 | 1.54 | 1.69 | 0.15 | 34 | 31 |
| 17 | 65F | 2.61 | 0.18 | −2.43 | 0.73 | 1.25 | 0.52 | 42 | 45 |
| 18 | 74M | 0.50 | 0.45 | −0.05 | 1.60 | 2.16 | 0.56 | 87 | 29 |
| 19 | 76M | 1.41 | 0.32 | −1.09 | 1.06 | 1.56 | 0.50 | 68 | 43 |
| 20 | 54M | 0.17 | 0.53 | 0.36 | 0.74 | 1.22 | 0.48 | 105 | 67 |
| 22 | 59M | 0.55 | 0.28 | −0.27 | 0.53 | 1.14 | 0.61 | 116 | 70 |
Figure 1.
Expression of KIM-1 and CD68 in chromophobe and papillary renal cell carcinoma. KIM-1 stained negatively in chromophobe renal cell carcinoma (A) but positive in papillary renal cell carcinoma (B). Similar to KIM-1, CD68 staining was negative in chromophobe renal cell carcinoma (C), but positive in papillary renal cell carcinoma (D). All magnifications are at × 400.
Table 2.
Procedure and Evaluation of Renal Lesions in Renal Negative and Positive KIM-1 cases
| KIM-1 Negative Stained Group | ||||||
|---|---|---|---|---|---|---|
| Cases | Pathological Type/Grade | Size of lesions | Stage | Lesion near collecting system | KIM-1 staining | CD68 staining |
| 4 | Ch-RCC,g2, | 7.1 cm | T2a | Yes | 0 | 0 |
| 5 | Ch-RCC, g2 | 10.2 cm | T3a | Yes | 0 | 0 |
| 7 | Benign complex cyst | 2.3 cm | NA | No | 0 | 0 |
| 10 | Benign complex cyst | 3.5 cm | NA | No | 0 | 0 |
| 12* | No lesion removed | See comment | NA | No | NA | NA |
| 14 | Benign complex cyst | 3.9 cm | NA | No | 0 | 0 |
| 21 | Hematoma | 4.5 cm | NA | No | 0 | 0 |
| KIM-1 Positive Stained Group | ||||||
|---|---|---|---|---|---|---|
| Cases | Pathological Type/Grade | Size of lesions | Stage | Lesion near collecting system | KIM-1 staining | CD68 staining |
| 2 | C-RCC, g3 | 6.5 cm | Tib** | No | 2+, 50% | 1+, 80% |
| 3 | C-RCC, g3 | 3.7 cm | T1a | No | 1+, 10% | 2+, 70% |
| 6 | C-RCC, g3 | 2.4 cm | T1a | No | 1+, 50% | 1+, 10% |
| 8 | C-RCC, g3 | 4.5 cm | T1b | No | 1+, 50% | 2+, 80% |
| 9 | C-RCC, g3 | 5.2 cm | T1b | Yes | 1+, 70% | 3+, 90% |
| 13 | P-RCC, g2 | 3.2 cm | T1a | No | 3+, 90% | 3+, 50% |
| 16 | P-RCC, g3 | 3.5 cm | T1a | No | 2+, 80% | 2+, 80% |
| 17 | C-RCC, g2 | 4.5 cm | T1b | Yes | 2+, 10% | 1+, 10% |
| 18 | C-RCC, g3 | 3.5 cm | T1a | No | 2+, 80% | 1+, 30% |
| 19 | P-RCC, g2 | 4.1 cm | T1b | No | 3+, 80% | 2+, 80% |
| 20 | C-RCC, g4 | 10.5 cm | T3b# | Yes | 1+, 5% | 1+, 50% |
| 22 | P-RCC, g2 | 6.5 cm | T1b | Yes | 2+, 90% | 3+, 90% |
C-RCC, clear renal cell carcinoma, P-RCC, papillary renal cell carcinoma, ch-RCC, chromophobe renal cell carcinoma, and BC - benign cyst, NA, no applicable.
The initial CT scan showed a 9 mm × 8 mm exophytic mass in the patient's left kidney. However, an intraoperative ultrasound failed to confirm the small mass in the kidney, thus no operation was performed,
- with renal capsular invasion, and # with renal vein invasion.
Pre-operative and post-operative urine KIM-1 levels are listed in Table 1. All of the KIM-1 negative cases had uKIM-1/uCr levels lower than 1.0 ng/mg urine creatinine. Six out of twelve (50%) KIM-1 positive staining cases had uKIM-1/uCr levels greater than 1.0 ng/mg Ucr (Table 1). In the KIM-1 negative group, uKIM-1/uCr did not significantly change before (0.367 ± 0.095 ng/mg Cr) and after nephrectomy (0.323 ± 0.014 ng/mg Cr) (Figure 2A). However, in the KIM-1 positive group, the elevated baseline uKIM-1/uCr at 1.200 ± 0.310 ng/mg Ucr was significantly reduced to 0.362 ± 0.099 ng/mg Ucr after the operation (Figure 2A) representing a 70% reduction in uKIM-1/uCr after total or partial nephrectomy in the KIM-1 positive group. Urine KIM-1 levels were not significantly associated with tumor grade, size or distance to collecting system.(Table 2). Pre-operative and post-operative serum creatinine levels are listed in Table 1. Mean serum creatinine levels and eGFR were increased to equivalent extents in KIM-1 negative and positive groups following the surgical procedures (Figure 2B–2C).
Figure 2.
Normalized urine KIM-1 (uKIM-1/uCr), serum creatinine levels and eGFR in KIM-1 negative and positive groups before and after surgery. Normalized uKIM-1 was significantly reduced in KIM-1 positive group but not in KIM-1 negative group (panel A). Serum creatinine levels (panel B) and eGFR (panel C) were significantly elevated in both groups. * p < 0.05 vs pre-operative value.
Observe expression of KIM-1 and CD68 in archival renal tumors
CD68 staining was negative in renal lesions negatively stained for KIM-1 but positive in RCC with positive KIM-1 staining (Table 2, Figure 1C and 1D). To evaluate whether this parallel relationship between KIM-1 and CD68 staining was seen in larger numbers of renal masses, we stained for both KIM1-1 and CD68 in a tissue microarray block containing various of renal tumors and found similar CD68 staining pattern to KIM-1 staining in 90 renal tumors of different types (Table 3). In renal tumors derived from proximal tubules (clear cell RCC and papillary RCC) most stained positively for both KIM-1 and CD68 while none of the tumors derived from distal nephron tubules (chromophobe RCC and oncocytoma) were positive for either marker.
Table 3.
Tissue Microarray Expression of KIM-1 and CD68 in 90 Various Renal Tumors
| Positive KIM-1 case # /total cases # (% of positivity) | Positive CD68 case #/total case # (% of positivity) | |
|---|---|---|
| Clear RCC | 18/22 (82 %) | 13/22 (59 %) |
| Papillary RCC | 18/20 (90 %) | 17/20 (85 %) |
| Chromophobe RCC | 0/25 (0 %) | 0/25 (0 %) |
| Oncocytoma of kidney | 0/23 (0 %) | 0/23 (0 %) |
RCC – renal cell carcinoma
Determine DNA sequence in archival renal tumors
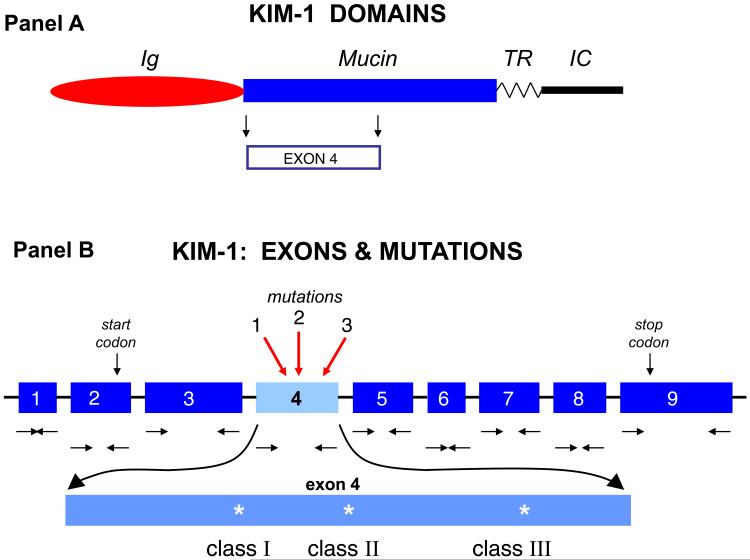
No differences in DNA sequence for exons 2, 3, 5, 6, 7, 8, and 9 were encountered among the four subtypes of renal tumors. However, a number of mutations were observed at three loci within exon 4, a region of the protein thought to be in KIM-1's mucin-like domain (Figure 3). The DNA sequence of exon 4 from papillary RCC agreed exactly with the published sequence of the protein (Table 4). The remaining three subtypes of renal tumors showed a 15 base deletion in exon 4, resulting in a deletion of 5 amino acids, MTTVP (Table 4). Additional mutations in exon 4 were observed: 1) a T to C mutation resulting in an amino acid change from Leu157 to Pro157; 2) a T to G silent mutation in both ChRCC and CRCC (Thr207); and 3) a double mutation with the same codon (T to G and A to G) in oncocytoma resulting in a change from Thr207 to Ala207. As only minor alterations were noted in the DNA sequence from all tumors, it seems unlikely that these minor mutations play a role in the regulation of KIM-1 expression, particularly exon 4 was not at the promoter region of KIM-1 (Figure 3).
Figure 3.
KIM-1 Domains (panel A). KIM-1 is a type one transmembrane protein. The larger extracellular portion is made up of an immunoglobulin-like (Ig) domain and a mucin-like (Mucin) domain, where Exon 4 is located. The transmembrane domain (TR) and the short, intracellular segment (IC) are also indicated (based on reference 2). KIM-1 Exons and Mutations (panel B). The nine exons are represented together with the positions of the nested primers (horizontal sets of arrows, → ←) that were used to amplify the genomic DNA sequence. An expanded view of exon 4 is shown to indicate the three mutations we detected: class 1, class 2, and class 3 (see deduced amino acid alterations from RCC in Table 4).
Table 4.
Deduced Amino Acid Alteration in KIM-1 from Renal Tumors
| TUMOR TYPE | Patient | Class I | Class II | Class III |
|---|---|---|---|---|
| CLEAR CELL RCC | A | NO deletion or Thr159_Thr160ins | NONE or Leu179Pro | NONE or 1154T>G (silent) |
| B | Met158_Pro162del* | Leu179Pro | 1154T>G (silent) | |
| C | Thr159_Thr160ins | Leu179Pro | 1154T>G (silent) | |
|
| ||||
| PAPILLARY RCC | D | Met158_Pro162del* | Leu179Pro | Thr207Ala |
| E | NO deletion | NONE | NONE | |
| F | Met158_Pro162del* | Leu179Pro | 1154T>G (silent) | |
|
| ||||
| CHROMOPHOBE RCC | G | Met158_Pro162del* | Leu179Pro | 1154T>G (silent) |
| H | NO deletion or Thr159_Thr160ins | NONE or Leu179Pro | NONE or 1154T>G (silent) | |
|
| ||||
| ONCOCYTOMA | I | Met158_Pro162del* | Leu179Pro | Thr207Ala |
| J | Met158_Pro162del* | Leu179Pro | 1154T>G (silent) | |
The 5 amino acids deleted are: Met-Thr-Thr-Val-Pro.
DISCUSSION
Although our previous study showed that uKIM-1/uCr was higher than in patients with RCC than normal healthy controls, there are only five post-operative follow-up cases available in that study [12] (Table 5). In the current KIM-1 negative group (Group 1), both prior and postoperative levels of normalized uKIM-1 remained low and none of this group had an uKIM-1/uCr level greater than 1.0 ng/mg Ucr. By contrast, 50 % of 12 KIM-1 positive cases (Group 2) had baseline uKIM-1/uCr higher than 1.0 ng/mg Ucr and after the operation uKIM-1/uCr was reduced significantly to the mean level of the KIM-1 negative group. If we use 1.0 ng/mg Ucr as a cuff off to detect RCC, the specificity at this cut-off level is 100% for both groups, although the sensitivity is relatively low at 50%. Thus the main findings of our current investigation are that RCCs that stained positively for KIM-1 demonstrated a marked reduction in uKIM-1/uCr after surgical resection. This finding demonstrates potentially significant clinical utility. In KIM-1 positively stained tumors, urine KIM-1 could potentially serve as a surrogate marker for disease preventing unnecessary radiological exposure, psychological pressure and too-late detection of progression. Moreover, however not evaluated in the current study, uKIM-1 may aid in timing of treatment of recurrent cancers earlier than normally occurs with radiological follow-up.
Table 5.
Summary of 3 Studies Using Urine KIM-1 (uKIM-1) to Detect Renal Cell Carcinoma (RCC)
| KIM-1 Antibodies used for ELISA | Retrospective Data | Prospective Data | Follow-up period | Major Findings | |
|---|---|---|---|---|---|
| Han et al, 2005 (reference 12) | Monoclonal KIM-1 antibody from Bonventre's lab | Yes Con n = 30 RCC n = 42 |
Yes Con n = NA RCC n = 5 |
4 –6 weeks | RCC cases had higher levels of uKIM-1 and 5 RCC cases had reduced uKIM-1 after nephrectomy |
| Morrissey et al 2011 (reference 14) | Polyclonal KIM-1 antibody, commercially available with R & D system | No | Yes Con n = 41 RCC n = 51 |
1 – 3 months | Higher levels of uKIM-1 in RCC cases were significantly reduced following nephrectomy |
| Zhang et al, current study | Monoclonal KIM-1 antibody from Bonventre's lab and commercially available with Bioassayworks LCC | No | Yes KIM-1 − n = 7 KIM-1 + n = 12 |
1 month | High levels of uKIM-1 in KIM-1 positive group were significantly reduced after nephrectomy |
In our previous study, the healthy normal controls had very low levels of uKIM-1/uCr (0.05 +/− 0.01 ng/mg Ucr) [12]. A recent study from another group also confirms low uKIM-1/uCr in their controls at 0.03 ng/mg Ucr [14]. As KIM-1 has been well known as a specific injury maker for proximal tubules [4–6] and any kidney injury may cause a rise in uKIM-1/uCr in clinical studies [11, 19, 20], we are not surprised that there were slightly higher uKIM-1/uCr levels in the KIM-1 negative group containing either KIM-1 negative chromophobe RCC or benign renal mass lesions. There is one issue we would like to point out in the recent study showing reduced uKIM-1/uCr in RCC patients after nephrectomy [14] (Table 5) - RCC should be highly suspected when high uKIM-1/uCr levels are found in patients who have normal renal function. In the current study, nephrectomy resulted in significant reduction in uKIM-1/uCr in the KIM-1 positive group with normal pre-operative serum creatinine in many cases. A reduction in uKIM-1/uCr levels was seen in all patients except patient #20. Even though patient #20 had the highest stage (T3b) and Fuhrman grade 4, the lack of response may be related to the percentage of cells that stained for KIM-1 (5%). Further studies are required to evaluate this finding. The second possibility in patient #20 without a reduction of urine KIM-1 after the nephrectomy is that the patient had residual tumor at the venous margin or metastatic disease. Retrospectively, the patient developed a pulmonary metastasis 5 months postoperatively. Currently there is no single urine or serum marker that can predict RCC in clinical practice and the surgery selection for renal cystic and solid lesions is chiefly determined by imaging studies. Although uKIM-1/uCr seems not perfect for all RCC cases in group 2, this non-invasive index worked for at least 50% of RCC in group 2 cases. Furthermore, all benign cases in group 1 had lower uKIM-1/uCr before and after surgery, imply that urologists may find the urine index very useful to determine benign versus malignant renal lesions taking together with the results of imaging studies.
The amount of KIM-1 present in urine may be related to several factors such as productivity of KIM-1 by individual tumor cells, tumor size and pathways to dump KIM-1 into urine. There are only two ways that KIM-1 from RCC could be detected in urine samples, one by glomerular filtration and the other by tumor invasion to urinary collecting system. Theoretically, shed soluble KIM-1 protein at 90kD (medium size of protein) can be partially filtered through glomerular filtration membranes, but some of the filtered KIM-1 would be reabsorbed back by proximal tubules depend on the amount filtered when renal function is normal. Microscopic evidence of invasion to benign renal tubules and collecting duct system was not present in all RCC cases except venous invasion in case #20. Thus it is still puzzling to us why some RCC had higher uKIM-1 than those with similar size and stage of RCC with low uKIM-1. Developing a test to detect serum KIM-1 can be an important direction to resolve at least part of the mystery.
Our study demonstrated that except for minor changes in the mucin-binding domain, the DNA sequence of KIM-1 in different types of RCC was essentially intact, implying that KIM-1 proteins in RCC are relatively intact (the data were only published in an abstract form [21]) and may have a function in RCC. KIM-1 staining using immunohistochemical method is usually negative in chromophobe RCC, but small amount of KIM-1 can be detected with more sensitive techniques such as PCR in the current study and, western blot or mRNA analysis in our previous studies [12, 13]. The ELISA technique may detect only small amount of uKIM-1 from chromophobe RCC, although our current two cases with chromophobe RCC were divided into KIM-1 negative group based on their negative stains for KIM-1 using immunohistochemical method.
To further assess why KIM-1 is upregulated in proximal tubule-derived RCC, we examined the relationship between CD 68, a 110-kDa glycoprotein which is expressed by macrophages [17, 18], and KIM-1, known to be involved in phagocytic activity of injured proximal tubules [7, 22]. We then examined both KIM-1 and CD68 expression using tissue microarrays containing 90 renal tumors. CD68 is upregulated in the cytoplasm of injured proximal tubules [23, 24]. We observed a parallel expression between KIM-1 and CD68 in injured proximal tubules (unpublished data). One study reports upregulated CD68 in three out of five RCC (60 %) when the authors examine a large scale of different lymphomas, sarcomas and carcinoma [25]. In the current tissue microarray, CD68 was upregulated in the majority of RCC expressed KIM-1 whereas the KIM-1 negative renal tumors, such as chromophobe RCC, did not express CD68 (Table 3). In addition, CD68 expression paralleled KIM-1 staining in group 1 and 2 lesions; CD68 was negative in KIM-1 negative renal lesions in group 1 but was positive in KIM-1 positive RCC (group 2). As phagocytosis is a part of lysosome-autophagy system involved in scavenging role and cell survival in normal and tumor cells [26, 27], we speculate that both KIM-1 and CD68 may contribute to cellular scavenging in RCC. Additional studies are needed to confirm this possibility and investigate whether KIM-1 and CD68 play important roles in the pathobiology of RCC.
CONCLUSION
This prospective study revealed that preoperative high levels of uKIM-1/uCr in patients with KIM-1 positive RCC was significantly reduced to the baseline level of KIM-1 negative control cases. uKIM-1/uCr appeared useful in detecting 50% of KIM-1 positive renal tumors when it had a limitation for differentiating chromophobe RCC from benign renal lesions such as renal cysts. While in general, both clear cell and papillary RCC represent more than 90% of all renal malignant tumors, our current preliminary findings suggests that uKIM-1/uCr may be a non-invasive surrogate marker for clear cell and papillary RCC.
What's known on the subject? And what does the study add?
Currently, monitoring kidney cancers requires expensive and frequent imaging procedures, making a reliable urinalysis test an ideal tool to develop for disease management. Kidney injury molecule-1 (KIM-1) is a marker found via histological testing to be upregulated in proximal tubule-derived RCC including clear cell (C-RCC) and papillary (P-RCC) but not in chromophobe RCC (distal tubular tumor). KIM-1, when produced, can be found in a urinalysis, making it a possible marker for at least two types of common RCC.
This study was designed to prospectively examine urine KIM-1 levels before and one month after removal of renal tumors to evaluate its usefulness as an adjuvant test for certain RCC cases, in additional to imaging studies. Two groups of patients, both KIM-1 negative and KIM-1 positively staining cases, were evaluated for pre- and post-nephrectomy urine KIM-1 (uKIM-1) normalized to urine creatinine levels (uCr). Upon completion of the study, KIM-1 positive cases had significantly reduced uKIM-1 levels post-nephrectomy, suggesting a possible use as a monitoring system for KIM-1 positive RCC cases.
ACKNOWLEDGEMENT
Authors thank Beaumont Health System and Geisinger Health System for funding the internal grants and National Institutes of Health awards DK 39773, DK 72381, and DK 38452 (to J.V.B.). Authors appreciate excellent assistance from Ms. Sharon K. Hick.
Footnotes
DISCLOSURES J.V.B is a co-inventor of KIM-1 patents that are assigned to Partners HealthCare and licensed by Partners to Johnson & Johnson, Sekisui Medical, Biogen Idec, and a number of research reagent companies. J.V.B. is a consultant for Sekisui Medical.
REFERENCE
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273(7):4135–42. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 3.Ichimura T, Hung CC, Yang SA, Stevens JL, Bonventre JV. Kidney injury molecule-1: a tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am J Physiol Renal Physiol. 2004;286(3):F552–63. doi: 10.1152/ajprenal.00285.2002. [DOI] [PubMed] [Google Scholar]
- 4.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62(1):237–44. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 5.van Timmeren MM, van den Heuvel MC, Bailly V, Bakker SJ, van Goor H, Stegeman CA. Tubular kidney injury molecule-1 (KIM-1) in human renal disease. J Pathol. 2007;212(2):209–17. doi: 10.1002/path.2175. [DOI] [PubMed] [Google Scholar]
- 6.Zhang PL, Rothblum LI, Han WK, Blasick TM, Potdar S, Bonventre JV. Kidney injury molecule-1 expression in transplant biopsies is a sensitive measure of cell injury. Kidney Int. 2008;73(5):608–14. doi: 10.1038/sj.ki.5002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–68. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Borst MH, van Timmeren MM, Vaidya VS, de Boer RA, van Dalen MB, Kramer AB, et al. Induction of kidney injury molecule-1 in homozygous Ren2 rats is attenuated by blockade of the renin-angiotensin system or p38 MAP kinase. Am J Physiol Renal Physiol. 2007;292(1):F313–20. doi: 10.1152/ajprenal.00180.2006. [DOI] [PubMed] [Google Scholar]
- 9.Kramer AB, van Timmeren MM, Schuurs TA, Vaidya VS, Bonventre JV, van Goor H, et al. Reduction of proteinuria in adriamycin-induced nephropathy is associated with reduction of renal kidney injury molecule (Kim-1) over time. Am J Physiol Renal Physiol. 2009;296(5):F1136–45. doi: 10.1152/ajprenal.00541.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaidya VS, Ramirez V, Ichimura T, Bobadilla NA, Bonventre JV. Urinary kidney injury molecule-1: a sensitive quantitative biomarker for early detection of kidney tubular injury. Am J Physiol Renal Physiol. 2006;290(2):F517–29. doi: 10.1152/ajprenal.00291.2005. [DOI] [PubMed] [Google Scholar]
- 11.Han WK, Waikar SS, Johnson A, Betensky RA, Dent CL, Devarajan P, et al. Urinary biomarkers in the early diagnosis of acute kidney injury. Kidney Int. 2008;73(7):863–9. doi: 10.1038/sj.ki.5002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han WK, Alinani A, Wu CL, Michaelson D, Loda M, McGovern FJ, et al. Human kidney injury molecule-1 is a tissue and urinary tumor marker of renal cell carcinoma. J Am Soc Nephrol. 2005;16(4):1126–34. doi: 10.1681/ASN.2004070530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin F, Zhang PL, Yang XJ, Shi J, Blasick T, Han WK, et al. Human kidney injury molecule-1 (hKIM-1): a useful immunohistochemical marker for diagnosing renal cell carcinoma and ovarian clear cell carcinoma. Am J Surg Pathol. 2007;31(3):371–81. doi: 10.1097/01.pas.0000213353.95508.67. [DOI] [PubMed] [Google Scholar]
- 14.Morrissey JJ, London AN, Lambert MC, Kharasch ED. Sensitivity and specificity of urinary neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 for the diagnosis of renal cell carcinoma. Am J Nephrol. 2011;34(5):391–8. doi: 10.1159/000330851. [DOI] [PubMed] [Google Scholar]
- 15.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, 3rd, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endre ZH, Pickering JW, Walker RJ, Devarajan P, Edelstein CL, Bonventre JV, et al. Improved performance of urinary biomarkers of acute kidney injury in the critically ill by stratification for injury duration and baseline renal function. Kidney Int. 2011;79(10):1119–30. doi: 10.1038/ki.2010.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81(6):1607–13. [PubMed] [Google Scholar]
- 18.Saito N, Pulford KA, Breton-Gorius J, Masse JM, Mason DY, Cramer EM. Ultrastructural localization of the CD68 macrophage-associated antigen in human blood neutrophils and monocytes. Am J Pathol. 1991;139(5):1053–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya VS, Ozer JS, Dieterle F, Collings FB, Ramirez V, Troth S, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28(5):478–85. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liangos O, Perianayagam MC, Vaidya VS, Han WK, Wald R, Tighiouart H, et al. Urinary N-acetyl-beta-(D)-glucosaminidase activity and kidney injury molecule-1 level are associated with adverse outcomes in acute renal failure. J Am Soc Nephrol. 2007;18(3):904–12. doi: 10.1681/ASN.2006030221. [DOI] [PubMed] [Google Scholar]
- 21.Schworer CM, Zhang PL, Gerhard GS, Lin F. Mutations in the mucin binding domain of the human kidney injury molecule-1 (KIM-1) gene are present in renal epithelial neoplasms. Mod Pathol. 2009;22(Suppl 1):192A. [Google Scholar]
- 22.Ichimura T, Brooks CR, Bonventre JV. Kim-1/Tim-1 and immune cells: shifting sands. Kidney Int. 2012;81(9):809–11. doi: 10.1038/ki.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Suzuki Y, Shirato I, Takahara H, Shibata T, Sugaya T, et al. Tubular epithelial cells have the capacity to transdifferentiate into CD68-positive macrophage-like cells by oxidative stress. Inflamm Res. 2008;57(12):593–600. doi: 10.1007/s00011-008-7171-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhu L, Yang X, Ji Y, Chen W, Guan W, Zhou SF, et al. Up-regulated renal expression of TNF-alpha signalling adapter proteins in lupus glomerulonephritis. Lupus. 2009;18(2):116–27. doi: 10.1177/0961203308094764. [DOI] [PubMed] [Google Scholar]
- 25.Gloghini A, Rizzo A, Zanette I, Canal B, Rupolo G, Bassi P, et al. KP1/CD68 expression in malignant neoplasms including lymphomas, sarcomas, and carcinomas. Am J Clin Pathol. 1995;103(4):425–31. doi: 10.1093/ajcp/103.4.425. [DOI] [PubMed] [Google Scholar]
- 26.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226(2):255–73. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox TM, Cachon-Gonzalez MB. The cellular pathology of lysosomal diseases. J Pathol. 2012;226(2):241–54. doi: 10.1002/path.3021. [DOI] [PubMed] [Google Scholar]