Significance
In this study, we compare the differential impact of nonhomologous end-joining–deficient induced pluripotent stem cells (iPSCs) derived from patients with mutations in genes encoding DNA ligase 4, DNA-protein kinase catalytic subunit, or Artemis, on cellular reprogramming, DNA damage repair, and myeloid differentiation. We found a predominant role for ligase 4 in genomic stability and myeloid differentiation of iPSCs, but a minor one for Artemis. Disease modeling with patient-derived iPSCs may provide significant insights into phenotypic variability associated with mutations in various genes along the same pathway and may serve as a platform to explore tissue-specific consequences of single gene defects and development of therapeutic interventions.
Abstract
Nonhomologous end-joining (NHEJ) is a key pathway for efficient repair of DNA double-strand breaks (DSBs) and V(D)J recombination. NHEJ defects in humans cause immunodeficiency and increased cellular sensitivity to ionizing irradiation (IR) and are variably associated with growth retardation, microcephaly, and neurodevelopmental delay. Repair of DNA DSBs is important for reprogramming of somatic cells into induced pluripotent stem cells (iPSCs). To compare the specific contribution of DNA ligase 4 (LIG4), Artemis, and DNA-protein kinase catalytic subunit (PKcs) in this process and to gain insights into phenotypic variability associated with these disorders, we reprogrammed patient-derived fibroblast cell lines with NHEJ defects. Deficiencies of LIG4 and of DNA-PK catalytic activity, but not Artemis deficiency, were associated with markedly reduced reprogramming efficiency, which could be partially rescued by genetic complementation. Moreover, we identified increased genomic instability in LIG4-deficient iPSCs. Cell cycle synchronization revealed a severe defect of DNA repair and a G0/G1 cell cycle arrest, particularly in LIG4- and DNA-PK catalytically deficient iPSCs. Impaired myeloid differentiation was observed in LIG4-, but not Artemis- or DNA-PK–mutated iPSCs. These results indicate a critical importance of the NHEJ pathway for somatic cell reprogramming, with a major role for LIG4 and DNA-PKcs and a minor, if any, for Artemis.
DNA repair is a crucial process for cell integrity, and its failure may result in cell cycle arrest, apoptosis, senescence, and introduction of genomic abnormalities that may lead to neoplastic transformation (1). Cellular DNA damage occurs frequently and can be caused by exogenous factors, such as exposure to ionizing and UV radiation and chemical drugs, or may result from endogenous sources, in particular reactive oxygen species (ROS) and replication errors (2). Although these insults may lead to both DNA single-strand breaks (SSBs) and double-strand breaks (DSBs), the latter are more critical in terms of cell survival and mutation probability. Importantly, DNA DSBs are also physiologically introduced in the T-cell receptor (TCR) and Ig genes during V(D)J recombination and class switch recombination (3).
Homologous recombination (HR) and nonhomologous end joining (NHEJ) represent two major pathways of DNA DSB repair in mammalian cells (4). HR is a high fidelity mechanism that requires a sister chromatid as a template and therefore is restricted to late S and G2 phases of the cell cycle. NHEJ is the major repair pathway in mammalian somatic cells, operating in G0/G1 phases of the cell cycle and competing with HR in the late S and G2 phases (5). The Ku70/Ku80 heterodimer binds rapidly at DNA DSBs, resulting in recruitment of two DNA-dependent protein kinase catalytic subunit (DNA-PKcs) molecules and formation of the DNA-PK holoenzyme. In the classical NHEJ (C-NHEJ) pathway, DNA-PKcs activates the endonuclease Artemis, which processes the DNA ends with overhangs. Finally, the XRCC4-DNA ligase 4 (LIG4) complex is recruited and ligates the DNA strand with the help of the XRCC4-like factor (XLF) (2). In addition to XRCC4-LIG4–dependent C-NHEJ, at least one alternative end-joining (A-EJ) pathway exists, which involves microhomology (MH) and is mainly used in cells with defects affecting C-NHEJ (6).
Consistent with the key role played by NHEJ in V(D)J recombination, severe defects of NHEJ in humans result in severe combined immunodeficiency (SCID) with lack of T and B lymphocytes, as well as increased radiation sensitivity and a variable spectrum of extraimmune manifestations (7).
The majority of patients with radiation-sensitive SCID carry biallelic mutations in the DNA Cross-Link Repair 1C (DCLRE1C) gene encoding for Artemis. This condition is associated with mild cellular radiosensitivity but no evidence of bone marrow failure or symptoms of neurodevelopmental delay. By contrast, LIG4 deficiency results in a more complex syndrome with growth retardation, pancytopenia, microcephaly, developmental delay, and a tendency to develop bone marrow failure and malignancies (8). Patients with LIG4 deficiency carry a hypomorphic mutation on at least one of the two mutated alleles. Homozygous deletion of the Lig4 gene in mice is embryonically lethal due to elevated apoptosis in neuronal stem cells and progenitor cells (9). Deficiency of XLF causes combined immunodeficiency, associated with microcephaly and developmental delay (10). Finally, only two patients with mutations of the Protein Kinase, DNA-activated, Catalytic polypeptide (PRKDC) gene, encoding for DNA-PKcs, have been identified so far. The first patient carried a homozygous mutation that affected activation of Artemis protein, but left DNA-PK catalytic activity intact; this patient presented with an isolated T−B−NK+ SCID phenotype (11). More recently, a second patient has been reported, whose PRKDC mutations dramatically reduced DNA-PK catalytic activity. This patient presented with severe and progressive neurodevelopmental delay, microcephaly, and dysmorphisms, in addition to T−B−NK+ SCID (12). Overall, these data indicate that the clinical phenotype associated with defects of NHEJ in humans may vary, depending on the nature of the gene affected and the specific mutations.
Induced pluripotent stem cells (iPSCs) represent a novel and powerful platform for disease modeling. Somatic cells can be reprogrammed into iPSCs by overexpression of the four factors OCT4, SOX2, KLF4, and c-MYC (13). However, reprogramming imposes cellular stress by induction of significant changes of gene expression (14) and enhanced cell metabolism, leading to increased levels of ROS (15). Reprogramming-associated DNA DSBs trigger the DNA damage response (DDR) and up-regulate the p53 pathway, thereby promoting cell death and senescence and limiting the efficiency of cell reprogramming (16, 17). Consistent with this, impaired reprogramming efficiency has been reported for human Fanconi anemia (FA) cells (18), LIG4- and XLF-mutated human fibroblasts (19, 20), and Ataxia Telangiectasia Mutated (ATM)-deficient mouse embryonic fibroblasts (MEFs) (21).
In this study, we used fibroblasts and iPSCs from patients with different mutations in various NHEJ genes (DCLRE1C, LIG4, and PRKDC) to model heterogeneity of the disease phenotype and to compare the impact of naturally occurring mutations in these genes on somatic cell reprogramming, NHEJ function, DDR, and myeloid differentiation in vitro.
Results
Defects of LIG4- and DNA-PK Catalytic Activation, but Not Artemis, Severely Reduce Efficiency of Reprogramming.
To investigate the impact of NHEJ on the efficiency and quality of reprogramming, we took advantage of a series of fibroblast cell lines derived from patients carrying mutations in DCLRE1C (Artemis), PRKDC (DNA-PKcs), or LIG4 genes and presenting with a variable severity of the clinical phenotype (Table 1). For each of these lines, a typical profile of DDR was observed on exposure to ionizing irradiation (IR) and up to 48 h after irradiation (Fig. S1).
Table 1.
Cell lines and patient characteristics
| Patient code | Gene | Mutation (allele 1; 2) | Immunological phenotype | Extraimmune manifestations | Reference |
| F04415 | DCLRE1C | del exon 1–3; del exon 1–3 | T−B−NK+ SCID | None | |
| F96224 | DCLRE1C | G126D; L70del | TloBlo CID | None | (42) |
| ID177 | PRKDC | L3062R; L3062R | T−B−NK+ SCID | None | (12) |
| NM720 | PRKDC | A3574V; del exon 16 | T−B−NK+ SCID | Severe microcephaly, sensorineural deafness, visual impairment | (13) |
| HYGM022 | LIG4 | R814X; K449Q | Blo | Microcephaly | |
| F07614 | LIG4 | A3V, T9I, R278H; A3V, T9I, R278H | TloB− CID, hyper-IgM | Microcephaly, mild developmental delay | (26) |
| 411BR | LIG4 | A3V, T9I, R278H; A3V, T9I, R278H | T−BloNK+ CID | Microcephaly, developmental delay, non-EBV lymphoma | (8, 9) |
| SCID072 | LIG4 | Q558P; K424fs | TloBlo CID, pancytopenia | Microcephaly, IUGR, developmental delay |
IUGR, intrauterine growth retardation.
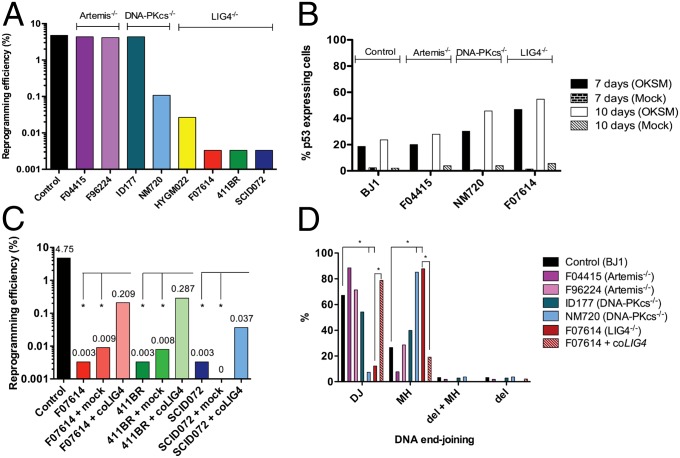
Fibroblast lines were transduced with a lentiviral vector containing the 4-in-1 transcription factor cassette (OKSM) and dTomato as a reporter (22). After transduction at an multiplicity of infection (MOI) of 1–2, we were able to obtain iPSC colonies from each fibroblast line. All iPSC lines tested expressed pluripotency markers as shown by immunofluorescence staining (Fig. S2A) and quantitative PCR (qPCR) (Fig. S2A) and were confirmed to carry the same mutations as in parental fibroblasts (Fig. S3). Artemis-deficient fibroblasts and the fibroblast cell line ID177, whose PRKDC mutation affects Artemis activation, showed reprogramming efficiencies similar to control iPSCs (4.75%) (Fig. 1A). By contrast, only one morphologically intact iPSC clone could be recovered after a prolonged period from each of the LIG4-deficient lines 411BR, F07614, and SCID072. Eight iPSC colonies were obtained from the LIG4-deficient line HYGM022, which was derived from a patient with a less severe phenotype (Table 1). The DNA-PK catalytically defective line NM720 was also reprogrammed with markedly reduced efficiency (0.1%), albeit higher than what observed for LIG4-deficient lines (Fig. 1A).
Fig. 1.
Reprogramming efficiency of NHEJ-deficient fibroblasts. (A) Human Artemis-, DNA-PKcs–, and LIG4-deficient fibroblasts were reprogrammed to iPSCs by lentiviral-mediated vector transduction. Reprogramming efficiency was calculated as the percentage of morphologically intact iPSC colonies obtained per total number of fibroblasts transduced. (B) Fibroblasts from given cell lines were either OKSM- or mock-transduced and sorted for dTomato expression after 48 h. Percentage of p53-expressing cells was assessed by immunofluorescence after 7 and 10 d of culture. (C) LIG4-deficient fibroblasts transduced with either human codon optimized LIG4 cDNA (coLIG4)-expressing vectors or with mock vectors were reprogrammed to iPSCs. Complementation of LIG4 partially rescued the reprogramming efficiency obtained in the previous experiments (*P ≤ 0.05). (D) The percentages of plasmids repaired by direct joining (DJ), microhomology-mediated joining (MH), combinations of MH and deletions (MH + del), or deletions (del) not associated with MH or insertion were analyzed in iPSCs transfected with the linearized pDVG94 construct and are reported for given cell lines (*P ≤ 0.05).
Because activation of the p53 pathway by DSBs is known to account for impaired survival of LIG4-deficient mice (9) and constrains efficiency of somatic cell reprogramming (16), we analyzed whether the reduced reprogramming efficiency of LIG4-mutated and of DNA-PK catalytically defective cells was due to increased and prolonged induction of p53. To test this, Artemis-, DNA-PKcs-, LIG4-deficient, and control fibroblasts were transduced with the OKSM-containing reprogramming vector and, in parallel, with a mock vector lacking the 4-in-1 transcription factor cassette. Cells were sorted for dTomato expression 48 h after transduction and analyzed for p53 expression by immunofluorescence staining after 7 and 10 d of culture. Marked up-regulation of p53 expression was observed after 10 d of culture in DNA-PKcs– (NM720) and LIG4-mutated (F07614) lines, (45% and 55%, respectively) compared with control fibroblasts (23%) and Artemis-deficient cells (28%) (Fig. 2B). By contrast, no significant induction of p53 was observed on transduction with the mock vector in any of the lines tested. These data rule out that the abnormal response of F07614 and NM720 lines could reflect DNA damage following lentivirus integration and rather indicate a critical role for epigenetic modifications and cell stress in inducing p53 expression and constraining efficiency of somatic cell reprogramming (23).
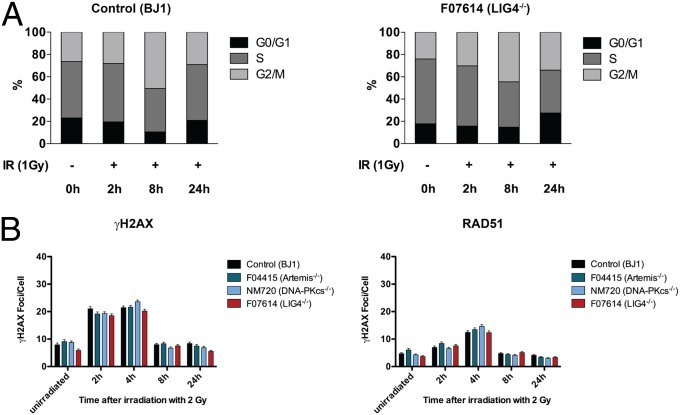
Fig. 2.
Cell cycle and DNA damage response in nonsynchronized iPSCs. (A) The percentages of cells in G0/G1, S, and G2/M phases of the cell cycle are shown for both unirrradiated cells and for cells exposed to 1-Gy IR at 2, 8, and 24 h of culture. (B) Nonsynchronized iPSCs from indicated cell lines were irradiated (0.5 Gy) and fixed at given time points. Nuclear γH2AX and RAD51 foci formation were analyzed by coimmunostaining and confocal microscopy. Results were obtained in multiple sets of experiments and are expressed as mean numbers of foci per cell + SEM.
LIG4-Deficient Cells Tend to Acquire Clonal Karyotypic Abnormalities During Culture and/or Reprogramming.
Karyotype analysis was performed on two to eight clones of each iPSC line, as well as on the parental fibroblasts. Two of four LIG4-deficient iPSC lines showed clonal karyotypic abnormalities; in some, chimerism was documented in parental fibroblasts, whereas others were apparently acquired de novo during reprogramming and/or expansion of iPSCs (Table S1). By contrast, no karyotypic abnormalities were identified in any of the other iPSC lines tested. These findings of increased genomic instability in LIG4 deficiency are consistent with a recent report (21).
Lentivirus-Mediated Complementation of LIG4-Deficient Fibroblasts Restores Cellular Radiosensitivity and NHEJ Activity and Partially Rescues Reprogramming Efficiency.
LIG4-deficient fibroblast lines were transduced with a lentiviral vector expressing human codon-optimized LIG4 (coLIG4) cDNA, along with cDNAs of EGFP and blasticidin resistance (BSD), or with a lentiviral vector expressing EGFP and BSD only (mock). GFP-expressing fibroblasts were sorted and grown in blasticidin-containing medium. Both coLIG4- and mock-transduced fibroblasts were then tested for γH2AX nuclear foci formation in response to IR with 5 Gy to confirm complementation of LIG4 deficiency, which rescued the radiation sensitivity phenotype (Fig. S4). Subsequently, coLIG4- and mock-complemented 411BR, F07614, SCID072, and HYGM022 fibroblast cell lines were reprogrammed for iPSC generation. Although reprogramming efficiency of mock-transduced lines remained very low, partial rescue of reprogramming efficiency was observed for the coLIG4-complemented lines (Fig. 1C).
Real-time PCR confirmed that the coLIG4-complemented iPSC lines expressed pluripotency markers, as shown in Fig. S5 for iPSCs obtained from the F07614 line. However, karyotypic analysis revealed chromosomal abnormalities (Table S1). Some of these were acquired during the process of fibroblast complementation or mock transduction and others during reprogramming.
LIG4 and DNA-PKcs, but Not Artemis, Are Critical for NHEJ Activity in Pluripotent Stem Cells.
We tested iPSC lines for their ability and efficiency to perform NHEJ using a plasmid ligation assay (24). A linearized plasmid with blunt ends harboring a 6-bp sequence homology was transfected into cells, and plasmid DNA was recovered after 24 h. When repair junctions from joined plasmids were sequenced, limited use of direct joining was observed in LIG4-deficient and in the DNA-PK catalytically defective iPSCs, which mainly used MH-mediated joining. This pattern was reversed toward WT in coLIG4-complemented F07614 iPSCs (Fig. 1D). By contrast, the pattern of end-joining was similar in control iPSCs and in both the Artemis-deficient lines F04415 and F96224, as well as in the DNA-PK-mutated line ID177, in which Artemis activation is affected, but DNA-PK catalytic activity is intact (Fig. 1D).
Nonsynchronized LIG4-Deficient iPSCs Do Not Show Enhanced Abnormalities of DDR on Exposure to Ionizing Irradiation.
It has been reported that embryonic stem (ES) cells and iPSCs are highly susceptible to IR with increased apoptosis and cell cycle arrest in G2/M (25, 26). Consistent with this, when we exposed control and patient-derived iPSCs to IR, even at low dosage (0.5–1 Gy), we observed massive apoptosis and severe disruption of colonies (Fig. S6).
To test DDR in NHEJ-deficient iPSCs, initially we evaluated cell cycle progression in control and LIG4-deficient F07614 nonsynchronized, unirradiated cells and at 2, 8, and 24 h after irradiation (1 Gy). No significant differences were detected in the cell cycle distribution of control vs. LIG4-deficient iPSCs at any time point before or after IR (Fig. 2A and Fig. S7). We hypothesized that the DNA DSBs repair in nonsynchronized control and NHEJ-deficient iPSC lines occurred predominantly by HR rather than NHEJ. To test for this, we analyzed RAD51 together with γH2AX nuclear foci formation. The number of γH2AX foci increased early (30 min) after IR and declined to baseline levels after 8 h. As previously described (27), RAD51 foci formation peaked at 2 h and returned to baseline levels by 8 h after IR (Fig. 2B). Around 60–80% of iPSCs showed colocalization of RAD51 and γH2AX nuclear foci at all of the time points, suggesting a frequent use of HR for DNA DSB repair in iPSCs. Furthermore, we observed frequent colocalization of γH2AX and RAD51 foci also in unirradiated iPSCs, consistent with their high replicative activity.
On Cell Cycle Synchronization and Exposure to IR and LIG4 and DNA-PK Catalytically Deficient iPSCs, Arrest in G0/G1 and NHEJ-Deficient iPSCs Reveal a DNA Repair Defect.
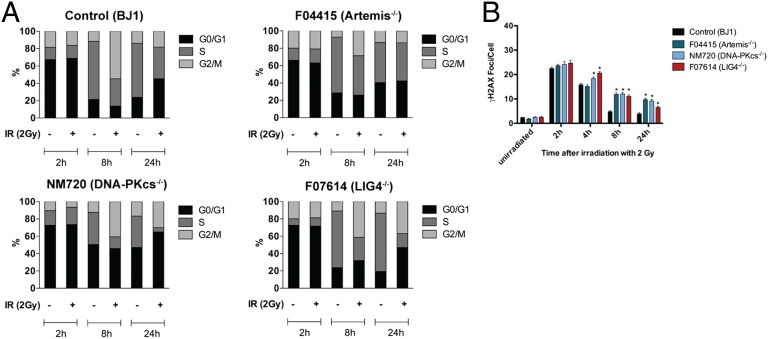
Because the high replicative activity of iPSCs could confound interpretation of γH2AX foci formation data, we synchronized cell cycles with nocodazole for 16 h (28). Two hours after release from synchronization, about 70% of iPSCs were in G0/G1 phase. In the absence of irradiation, control and NHEJ-deficient iPSCs resumed high proliferative activity. On irradiation with 2 Gy, significant differences in cell cycle progression were observed among iPSC lines. In particular, compared with control and Artemis-deficient lines, the NM720 and F07614 iPSC lines showed fewer cells in S phase at 8 and 24 h, and a large fraction (45–60%) of cells were in G0/G1 at 24 h after exposure to IR (Fig. 3A).
Fig. 3.
Cell cycle and DNA damage response in synchronized iPSCs. (A) Indicated cell lines were synchronized and irradiated with 2 Gy, and cell cycle was analyzed by flow cytometry. Diagrams display the percentages of live cells in G0/G1, S, and G2/M cell cycle phases at 2, 8, and 24 h under nonirradiated and irradiated (2 Gy) conditions. Data shown are representative for results from three independent experiments. (B) Mean numbers of γH2AX foci per cell + SEM are shown for indicated iPSC lines at all time points tested (*P ≤ 0.05).
Next, we analyzed γH2AX foci at 2, 4, 8, and 24 h after 2-Gy irradiation of control and NHEJ-deficient synchronized iPSCs. To distinguish between cells in G0/G1 and G2/M, cells were stained for the centromere protein CENP-F (29). Compared with control irradiated lines, the mean numbers of γH2AX foci per cell declined with slower kinetics in all NHEJ-deficient iPSC lines that were in G0/G1 (Fig. 3B and Fig. S8).
LIG4-Deficient iPSCs Have Reduced Potential for Hematopoietic Differentiation Toward the Myeloid Lineage.
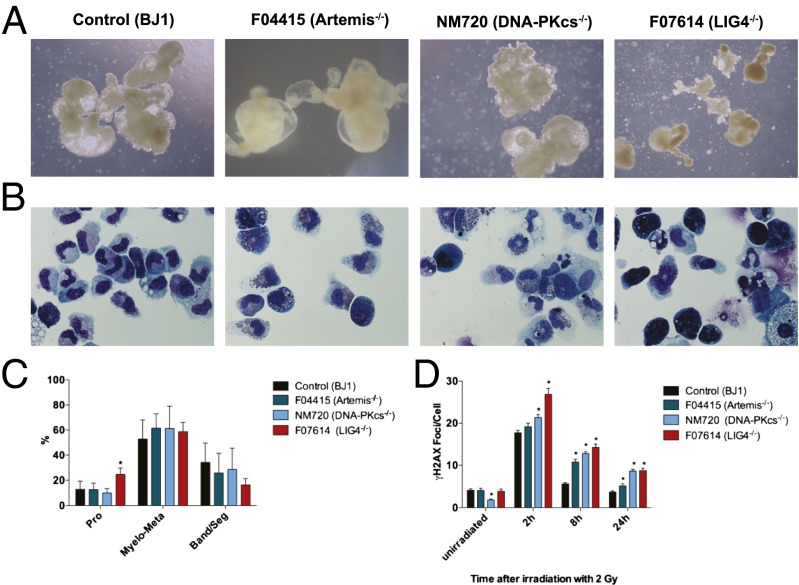
LIG4 deficiency is associated with an increased risk of bone marrow failure (8). Progressive loss of hematopoietic stem and progenitor cells has been demonstrated in a mouse model of the disease (30). We tested the ability of F07614 LIG4-mutated iPSCs to expand and generate embryoid bodies (EBs). In multiple experiments, we observed that F07614 LIG4-mutated iPSCs yielded poorly developed EBs and embryoid sacs, with features of apoptosis (Fig. 4A). To test and compare the potential of NHEJ-deficient iPSCs for hematopoietic differentiation, we performed a colony forming unit (CFU) assay. The total numbers of CFUs obtained from F07614 EBs were reduced to 52–54% compared with control iPSCs, and morphological analysis of granulocyte maturation revealed skewing to more immature stages, with higher frequency of promyelocytes in LIG4-deficient compared with control iPSCs (Fig. 4 B and C). By contrast, the efficiency of myeloid differentiation was not altered in Artemis (F04415)- and DNA-PKcs (NM720)–deficient cell lines. However, myeloid cells derived from all NHEJ-deficient iPSCs showed increased and persistent formation of γH2AX foci on irradiation with 2 Gy (Fig. 4D), recapitulating what was observed in parental fibroblasts and synchronized iPSCs.
Fig. 4.
Differentiation efficiency and DNA damage response of myeloid cells derived from NHEJ-deficient iPSCs. (A) EBs were developed from human control, Artemis-, catalytically DNA-PK– and LIG4-deficient iPSCs. One representative experiment of five is shown. (B) EBs from the indicated cell lines were cultured in methylcellulose and granulocyte colonies (CFU-G) were picked after 14 d. (C) Granulocyte maturation was assessed based on nuclear and cytoplasmic morphology and divided into groups of promyelocytes, myelocytes/metamyelocytes, and bands/segmented cells (*P ≤ 0.05). (D) Myeloid cells (CFU-G, CFU-M, and CFU-GM) of indicated cell lines were irradiated with 2 Gy, and γH2AX foci formation is expressed as mean foci numbers per cell + SEM (*P ≤ 0.05).
Discussion
Cellular reprogramming is associated with DNA and histone modifications and massive induction of gene expression. Although the integration of viral vectors delivering reprogramming transgenes requires DNA DSBs repair (31), it has been demonstrated that DNA DSBs are also generated when nonintegrating systems are used for somatic cell reprogramming (23). This likely reflects significant cellular and genotoxic stress, which leads to up-regulation of the p53 pathway (16) and induction of cell death and senescence (32) and thus somatic cell reprogramming efficiency.
By comparing side-by-side human cell lines with different mutations in a broader panel of NHEJ genes, our data demonstrate for the first time important differences in the requirement for specific components of NHEJ and of specific mutations in these genes during somatic cell reprogramming, DDR, and in vitro differentiation of iPSCs. In particular, we observed increased expression of p53 and severe impairment of somatic cell reprogramming on transduction of LIG4- and DNA-PK catalytically deficient fibroblasts with the lentiviral reprogramming vector. By contrast, normal reprogramming efficiency and levels of p53 expression comparable to control were observed for Artemis-deficient fibroblasts and for the line ID177, whose PRKDC homozygous mutation affects Artemis activation, but leaves DNA-PK catalytic activity intact (12). Altogether, these data demonstrate a critical role for the NHEJ machinery for DNA repair and survival in cells undergoing reprogramming.
It has been reported that the spectrum of chromosomal instability is different and less severe in Artemis-deficient cells compared with LIG4- or DNA-PKcs–deficient lymphocytes (33). Artemis plays a role only in a subset of NHEJ events, in particular processing DNA ends with overhangs or opening the hairpin (34), and it is involved in an ATM-dependent NHEJ pathway repairing with slower kinetics (35). Our data extend these observations by demonstrating that Artemis is dispensable for somatic cell reprogramming and genomic stability, although its role in NHEJ can be appreciated in synchronized iPSCs at later time points (8–24 h) after IR.
In the absence of C-NHEJ, DNA DSBs may be joined by the A-EJ pathway (6). This process facilitates chromosomal translocations with MH joins at the junctions (36). We observed increased genomic instability, including emergence of structural chromosomal abnormalities, in iPSC lines derived from LIG4-deficient fibroblasts, and similar results have been recently reported by others (19). In particular, several chromosomal abnormalities have been identified in LIG4-mutated iPSCs that were not present in parental fibroblasts. These chromosomal abnormalities may have occurred at various stages during iPSC generation and culture, including fibroblast transduction, reprogramming, and/or iPSC expansion. We used fluorescent in situ hybridization to analyze the parental fibroblast lines for any translocations that had been observed in the derived iPSCs (Table S1). However, because of the limited number of cells analyzed, chromosomal abnormalities possibly present at very low frequency in the parental fibroblasts may have been missed. Although genomic instability and mutagenesis have been reported in Prkdc−/− mouse models (37, 38), no chromosomal abnormalities were detected in iPSCs deficient in DNA-PK catalytic activity. Lack of chromosomal abnormalities in DNA-PK catalytically deficient fibroblasts and iPSCs studied here may be due to the low level of spontaneous genomic instability or to tissue specificity, as suggested by the observation that p53−/− scid mice predominantly develop lymphoma from erroneously repaired programmed DNA breaks (39).
Using a plasmid ligation assay, we were able to confirm defective C-NHEJ activity and preferential use of the A-EJ pathway with a higher proportion of MH joins in LIG4-deficient iPSCs, thus recapitulating a hallmark of this disease (6). Increased proportion of MH joins was observed also in NM720 DNA-PK catalytically deficient iPSCs. Large deletions are also expected to occur during joining of DNA DSBs in DNA-PKcs–deficient cells; however, the plasmid ligation assay does not permit to properly identify large deletions and score their relative frequency.
Pluripotent stem cells are characterized by high proliferative activity with a large proportion of cells in S phase of the cell cycle and shortened G1 and G2 phases, which is important for maintaining pluripotency (40) and favors use of HR for DNA DSB repair (27). Consistent with these observations, we found around 50% of unirradiated nonsynchronized control- and NHEJ-deficient iPSCs in S phase and observed no significant difference among nonsynchronized NHEJ-deficient and control iPSCs in both cell cycle and DDR on irradiation. However, it has been shown on synchronized human ES cells (ESCs) irradiated with UV C (UVC) (28) and on human nonsynchronized iPSCs in response to IR (27) that pluripotent stem cells are capable to interrupt cell cycle progression in response to DNA damage. To investigate more properly the DDR of NHEJ-defective cells, we performed cell cycle synchronization, and exposed iPSCs to IR. Under these conditions, a more severe phenotype (cell cycle arrest at the G0/G1 checkpoint) was observed in iPSCs with LIG4 deficiency or impaired DNA-PK catalytic activity but not in Artemis-deficient iPSCs.
Defective EB generation and impaired myeloid differentiation was observed for F07614 LIG4-mutated iPSCs. Along with similar data generated in mice (30), these observations indicate that impaired proliferation of hematopoietic stem and progenitor cells is responsible for the increased risk of bone marrow failure in LIG4 deficiency. The decreased numbers of in vitro-differentiated myeloid cells we observed is likely due to the loss of progenitor cells as evidenced by the smaller and fewer EBs generated. By contrast, no defects of EB generation and myeloid differentiation were observed in Artemis- and DNA-PK catalytically deficient iPSCs. The relative increase in the percentage of promyelocytes among differentiated myeloid cells may be due to the proliferative defect we see in EB development, with an attrition of more mature myeloid forms.
In summary, we demonstrated that NHEJ is critical for somatic cell reprogramming and maintenance of genomic stability in iPSCs and that use of HR may obscure defects of DNA DSB repair in nonsynchronized NHEJ-deficient iPSCs. Importantly, we showed that important differences exist in the requirement for individual NHEJ factors in somatic cell reprogramming, as well as in iPSC differentiation and DDR. These gene- and mutation-specific differences of the biological behavior of NHEJ-defective iPSCs model distinctive features of the clinical phenotype observed in patients with these disorders, including bone marrow failure and susceptibility to malignancies. Differentiation of NHEJ-deficient iPSC lines to other lineages, such as neurons, may help model additional phenotypic differences observed in vivo in patients.
Materials and Methods
Cell Lines and Cell Culture.
Fibroblast cell lines were grown from punch skin biopsies, collected on informed consent under protocol 0409113R approved by Children's Hospital Institutional Review Board (IRB) or under IRB-approved protocols at other collaborating institutions and made available to L.D.N. under protocol S09-08-0435. Culture conditions for fibroblasts and iPSCs have been previously described (41).
Vector Cloning, Lentiviral Vector Production, and Gene Complementation.
Human fibroblasts were reprogrammed by transduction with a self-inactivating lentiviral vector containing the four transcription factors OCT4, SOX2, KLF4, and c-MYC (OKSM) and dTomato cDNA (22). Cloning of the complementation vector for LIG4 deficiency, lentiviral vector production, and genetic complementation of LIG4-deficient fibroblasts are described in SI Materials and Methods.
Reprogramming, iPSC Generation, and Assessment of Reprogramming Efficiency.
Viral transduction was performed in 3 × 104 fibroblasts at MOIs of 1 for the original fibroblast lines and 2 for the complemented lines as described. Reprogramming efficiencies were calculated by dividing the number of morphologically intact iPSC colonies obtained after 21 d by the number of fibroblasts transduced.
Plasmid Ligation NHEJ Assay.
The NHEJ assay was performed as previously described (24). The linearized plasmid pDVG94 with defined, blunt DNA ends with 6-bp direct repeats (ATCAGC) was transiently transfected into single cell suspensions of iPSCs by nucleofection (Amaxa Nucleofector and Cell Line Nucleofector Kit V; Lonza). Plasmid DNA was recovered after 24 h, and the recombination junctions were PCR amplified using primers FM30 and DAR5 (24). The resulting PCR products were gel purified, cloned into pGEM-T vectors, and sequenced by Sanger sequencing.
Synchronization of iPSCs and Cell Cycle Analysis.
iPSCs grown under feeder-free conditions were synchronized by treatment with 200 ng/mL nocodazole in culture media for 16 h, after which medium was replaced by mTESR without nocodazole, and cells were immediately irradiated with 2 Gy. For cell cycle analysis (Apoptosis, Cell Damage and Proliferation Kit; BD Biosciences), cells were pulsed with 10 µM BrdU for 45 min at 37 °C, enzymatically detached with Accutase (STEMCELL), and analyzed by flow cytometry at indicated time points. Nonirradiated cells fixed at same time points served as controls.
Irradiation, Immunocytochemistry, and Confocal Microscopy.
Irradiated fibroblasts (5 Gy) and iPSCs (0.5–2 Gy) were stained for anti-phospho Ser-139 Histone H2AX and RAD51 or CENP-F, respectively, and subsequently with Alexa fluor anti-mouse/anti-rabbit 488/594 antibodies, as described in SI Materials and Methods. Confocal imaging was performed on a Fluoview FV 1000 microscope (UplanApo 10× and 60×/1.2-NA water lenses; Olympus). The numbers of nuclear foci per cell were counted manually or with Volocity software (PerkinElmer) after careful setup.
To assess p53 expression in reprogramming fibroblasts, OKSM- and mock-transduced cells were sorted for dTomato expression and stained with anti-p53 (mouse monoclonal, clone DO-1, sc-126; Santa Cruz) and Alexa fluor anti-mouse 647 (Invitrogen) antibodies after 7 and 10 d of culture. Imaging was performed on a BD Pathway 435 bioimager (20× magnification).
Hematopoietic Differentiation of iPSCs Toward the Myeloid Lineage.
Development of EBs and myeloid differentiation were performed as described in SI Materials and Methods. For morphological analysis of granulocyte maturation, CFU-G colonies were selectively harvested, and cells were spun on slides and stained with May-Grunwald Giemsa. Random fields were acquired and analyzed on an Eclipse E800 microscope (Nikon) with bright field illumination (20×), and at least 1,000 cells were counted per cell line and experiment. DNA repair efficiency was analyzed on myeloid cells irradiated with 2 Gy in methylcellulose cultures and harvested on slides at the indicated time points. Assessment of γH2AX foci formation was performed as described above.
Statistical Analysis.
For analysis of DDR, the mean and SEM of γH2AX foci per cell obtained from multiple experiments were calculated. Relative quantification and SEs of qPCR data were calculated based on triplicate samples. The P values were calculated based on a two-tailed unpaired Student t test, and statistical significance was determined at P < 0.05.
Supplementary Material
Acknowledgments
We thank the patients and their families for collaboration and Dr. Penny Jeggo for supplying the cell line NM720 and providing helpful comments on the manuscript. We thank Ms. Giulia Notarangelo for technical help. This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases Grants 1R01AI100887 and 5P01AI076210-04 (to L.D.N.). K.F. was supported by the German Research Foundation (FE1253/1-1), A.S. was supported by networks of Bundesministerium für Bildung und Forschung (PidNet) and European Union (Cellpid), Q.P.-H. was supported by the Swedish Research Council, European Research Council (242551-ImmunoSwitch), S.G. was partially supported by the University of Brescia and Fondazione A. Nocivelli, and L.D.N. was partially supported by the Manton Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323649111/-/DCSupplemental.
References
- 1.d’Adda di Fagagna F. Living on a break: Cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8(7):512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 4.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat Res. 2012;751(2):158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 7.de Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 8.O’Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8(6):1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 9.Frank KM, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5(6):993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 10.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124(2):287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 11.van der Burg M, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119(1):91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodbine L, et al. PRKDC mutations in a SCID patient with profound neurological abnormalities. J Clin Invest. 2013;123(7):2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151(7):1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berniakovich I, Laricchia-Robbio L, Izpisua Belmonte JC. N-acetylcysteine protects induced pluripotent stem cells from in vitro stress: Impact on differentiation outcome. Int J Dev Biol. 2012;56(9):729–735. doi: 10.1387/ijdb.120070ji. [DOI] [PubMed] [Google Scholar]
- 16.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilgner K, et al. A human iPSC model of Ligase IV deficiency reveals an important role for NHEJ-mediated-DSB repair in the survival and genomic stability of induced pluripotent stem cells and emerging haematopoietic progenitors. Cell Death Differ. 2013;20(8):1089–1100. doi: 10.1038/cdd.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilgner K, et al. Brief report: A human induced pluripotent stem cell model of cernunnos deficiency reveals an important role for XLF in the survival of the primitive hematopoietic progenitors. Stem Cells. 2013;31(9):2015–2023. doi: 10.1002/stem.1456. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, et al. Ataxia-telangiectasia mutated (ATM) deficiency decreases reprogramming efficiency and leads to genomic instability in iPS cells. Biochem Biophys Res Commun. 2011;407(2):321–326. doi: 10.1016/j.bbrc.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Warlich E, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Molec Therap. 2011;19(4):782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González F, et al. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Reports. 2013;3(3):651–660. doi: 10.1016/j.celrep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verkaik NS, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol. 2002;32(3):701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Fan J, et al. Human induced pluripotent cells resemble embryonic stem cells demonstrating enhanced levels of DNA repair and efficacy of nonhomologous end-joining. Mutat Res. 2011;713(1-2):8–17. doi: 10.1016/j.mrfmmm.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Momcilović O, et al. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 2009;27(8):1822–1835. doi: 10.1002/stem.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momcilovic O, et al. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS ONE. 2010;5(10):e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bárta T, et al. Human embryonic stem cells are capable of executing G1/S checkpoint activation. Stem Cells. 2010;28(7):1143–1152. doi: 10.1002/stem.451. [DOI] [PubMed] [Google Scholar]
- 29.Beucher A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28(21):3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 31.Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12(Suppl 1):971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 32.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23(18):2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney S, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci USA. 2004;101(8):2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108(6):781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 35.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37(Pt 3):569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109(7):811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 37.Jhappan C, Morse HC, 3rd, Fleischmann RD, Gottesman MM, Merlino G. DNA-PKcs: A T-cell tumour suppressor encoded at the mouse scid locus. Nat Genet. 1997;17(4):483–486. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 38.Gilley D, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci USA. 2001;98(26):15084–15088. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nacht M, et al. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 1996;10(16):2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 40.Tsubouchi T, Fisher AG. Reprogramming and the pluripotent stem cell cycle. Curr Top Dev Biol. 2013;104:223–241. doi: 10.1016/B978-0-12-416027-9.00007-3. [DOI] [PubMed] [Google Scholar]
- 41.Pessach IM, et al. Induced pluripotent stem cells: A novel frontier in the study of human primary immunodeficiencies. J Allergy Clin Immunol. 2011;127(6):1400–1407. doi: 10.1016/j.jaci.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans PM, et al. Radiation-induced delayed cell death in a hypomorphic Artemis cell line. Hum Mol Genet. 2006;15(8):1303–1311. doi: 10.1093/hmg/ddl050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.