Significance
Nicotinamide adenine dinucleotide (NAD+) is an endogenous small molecule that has effects on diverse processes, including obesity, lifespan, and cancer. A major goal is to identify the NAD+-regulated cellular pathways that may mediate these effects. In this study, we demonstrate that NAD+ regulates the microtubule cytoskeleton. We find that these effects are mediated by the mitochondrial sirtuin-3. Our findings have implications for many clinically used chemotherapeutics that target microtubules, as we demonstrate that high NAD+ levels can reduce sensitivity to these drugs. These results are also significant because they demonstrate for the first time that NAD+, a molecule regulated by age, diet, and disease state, can influence basic microtubule functions.
Abstract
Nicotinamide adenine dinucleotide (NAD+) is an endogenous enzyme cofactor and cosubstrate that has effects on diverse cellular and physiologic processes, including reactive oxygen species generation, mitochondrial function, apoptosis, and axonal degeneration. A major goal is to identify the NAD+-regulated cellular pathways that may mediate these effects. Here we show that the dynamic assembly and disassembly of microtubules is markedly altered by NAD+. Furthermore, we show that the disassembly of microtubule polymers elicited by microtubule depolymerizing agents is blocked by increasing intracellular NAD+ levels. We find that these effects of NAD+ are mediated by the activation of the mitochondrial sirtuin sirtuin-3 (SIRT3). Overexpression of SIRT3 prevents microtubule disassembly and apoptosis elicited by antimicrotubule agents and knockdown of SIRT3 prevents the protective effects of NAD+ on microtubule polymers. Taken together, these data demonstrate that NAD+ and SIRT3 regulate microtubule polymerization and the efficacy of antimicrotubule agents.
Nicotinamide adenine dinucleotide (NAD+) is an endogenous dinucleotide that is present in the cytosol, nucleus, and mitochondria. Athough it serves an important role as a redox cofactor in metabolism, NAD+ is also a substrate for several families of enzymes, including the poly(ADP ribose) polymerases and the sirtuin deacetylase enzymes (reviewed in refs. 1 and 2). The level of intracellular NAD+ is regulated by many factors, including diet and energy status (3), axonal injury (4), DNA damage (5), and certain disease states (6), suggesting that NAD+-dependent signaling is dynamically modulated in diverse contexts.
NAD+-dependent signaling can be induced by treatment of cells with exogenous NAD+, which increases intracellular NAD+ levels and results in diverse effects in cells and animals. These effects include enhanced oxygen consumption and ATP production (7), as well as protection from genotoxic stress and apoptosis (3). Mice treated with nicotinamide riboside, a NAD+ precursor that is metabolized into NAD+, have enhanced oxidative metabolism, increased insulin sensitivity, and protection from high-fat diet-induced obesity (8). These results demonstrate that NAD+-dependent pathways can enhance metabolic function and improve a variety of disease phenotypes.
An NAD+-regulated pathway also inhibits axonal degeneration elicited by axonal transection (4). Treatment of axons with 5–20 mM NAD+ markedly delays the axon degenerative process (9). Additionally, animals that express the Wallerian degeneration slow (WldS) protein, a fusion of the NAD+ biosynthetic enzyme Nicotinamide mononucleotide adenylyl transferase 1 and Ube4a, exhibit markedly delayed degeneration of the distal axonal fragment after axonal transection (10), and expression of WldS mitigates disease phenotypes in several neurodegenerative disease models (11–14). Thus, understanding the intracellular pathways regulated by NAD+ may be important for understanding the pathogenesis of numerous disorders.
Despite the diverse beneficial effects of genetically and pharmacologically augmenting NAD+ levels, the cellular processes that are affected by NAD+ treatment are incompletely understood. In this study, we show that microtubule dynamics and polymer stability are markedly influenced by NAD+ levels. We show that elevation of intracellular NAD+ levels markedly alters the stability of microtubule polymers in cells, and renders these polymers resistant to depolymerization elicited by antimicrotubule agents, such as vinblastine, nocodazole, and colchicine. We find that these effects are mediated by sirtuin-3 (SIRT3), a mitochondrial NAD+-dependent deacetylase, and that elevated SIRT3 levels also blocks the effects of antimicrotubule agents on the cytoskeleton. Furthermore, we find that both NAD+ and SIRT3 reduce the sensitivity of cells to the cytotoxic effects of vinblastine. Taken together, these data identify a new role for NAD+ and SIRT3 in regulating the effects of antimicrotubule agents, and link the actions of NAD+ to microtubule stabilization in cells.
Results
Screen to Identify Pathways Mediating Effects of NAD+.
To identify pathways that mediate the effects of NAD+, we set up an assay to detect NAD+-dependent responses in cells. We based this assay on the well-described ability of NAD+ to prevent axon degeneration induced by axonal transection (4, 9). This effect is readily detectable by phase microscopy and can be quantified using an automated algorithm (15). We reasoned that molecules that exhibit an NAD+-like effect in this assay may be acting through the same pathways regulated by NAD+. We therefore screened small molecules that have known effects on cellular pathways and identified compounds that mimicked the effect of NAD+ on axonal degeneration.
In this screen, we assayed axonal degeneration using rat embryonic day (E) 14.5 dorsal root ganglia (DRG) neurons. Dissociated neurons were cultured in microfluidic chambers for 2 d, at which point axons had extended through the 450-µm microgrooves into the axonal compartment (16). Axonal transection was elicited by flushing media through the cell body compartment, resulting in shearing of every axon (Fig. S1A). Following axonal transection, axons remained morphologically intact for the first 2 h (Fig. S1 B and C). However, after 4 h, axons exhibited characteristic fragmentation and blebbing, as is commonly seen using in vitro models of axon degeneration (reviewed in ref. 17). As expected, NAD+ treatment led to a marked delay in axonal degeneration after axonal transection (4) (Fig. S1 D and E).
In our screen, we searched for compounds that mimicked the protective effects of NAD+ on axons. One compound, the microtubule-stabilizing agent taxol (1 µM), substantially inhibited axonal degeneration at 4 h to a similar degree seen with NAD+ treatment (Fig. S1 B and C). Similar results were observed in DRG explant cultures, suggesting that these results are not unique to the microfluidic culture system (Fig. S1 F and G). Axon degeneration was not inhibited by jasplakinolide, which stabilizes actin polymers, indicating the effect is not a general result of altered cytoskeletal stability but specific to microtubule stability (Fig. S1 D and E). Because both NAD+ and taxol prevent axon degeneration, these results raised the possibility that NAD+ might mediate its effects by influencing the stability of microtubule polymers.
NAD+ Prevents Microtubule Polymer Disassembly Elicited by Vinblastine.
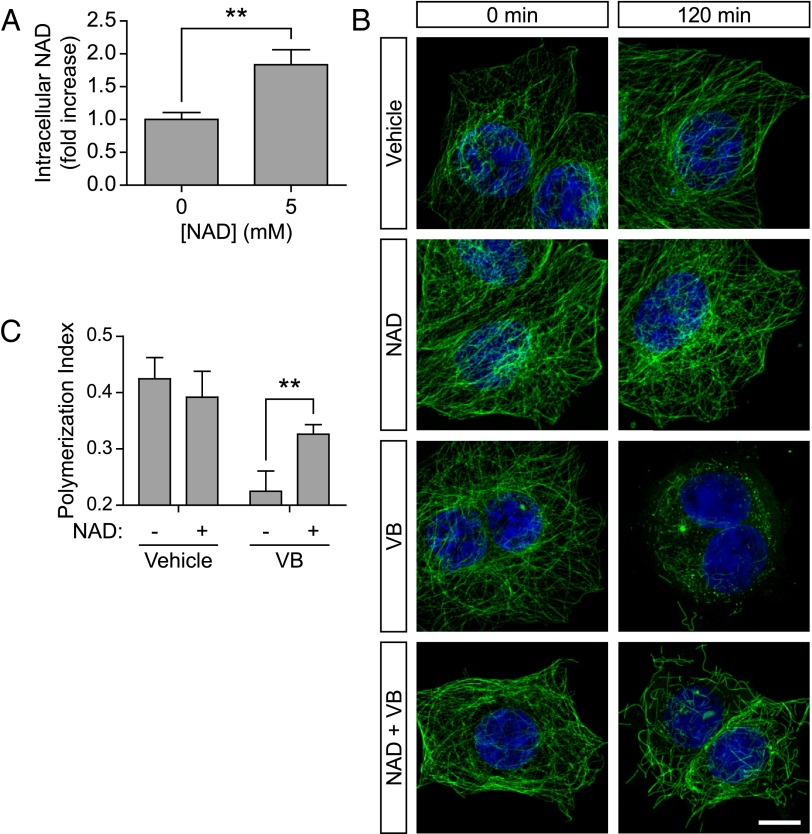
To address this possibility, we examined the effect of increasing intracellular NAD+ on the microtubule cytoskeleton in MCF-7 human breast adenocarcinoma cells, which have a large cytoplasm suitable for microtubule network visualization. We first sought to confirm that treatment of cells with NAD+ leads to an increase in intracellular NAD+ levels in this cell line. Measurement of intracellular NAD+ levels showed that application of 5 mM NAD+ for 2 h resulted in a nearly twofold increase in intracellular NAD+ concentration (Fig. 1A). These results confirmed studies in other cell lines (7) that exogenous application of NAD+, which is relatively cell impermeable, can increase intracellular NAD+ levels.
Fig. 1.
NAD+ stabilizes microtubule polymers. (A) Intracellular NAD+ concentration. To confirm that the intracellular concentration of NAD+ increases when cells are treated with exogenous NAD+, MCF-7 cells were treated with NAD+ (5 mM) for 2 h followed by NAD+ quantification. Consistent with our hypothesis, we find a nearly twofold increase in intracellular NAD+ concentration. These data are consistent with the idea that NAD+ acts intracellularly to affect the stability of microtubule polymers. Bar graph represents mean ± SEM; **P < 0.01 (unpaired, two-tailed Student t test). (B) NAD+ protects against the microtubule depolymerization effect of vinblastine. To test whether NAD+ exerts a microtubule-stabilizing effect, we treated MCF-7 cells with vinblastine (VB), an antimicrotubule agent that leads to depolymerization of the dynamic microtubule pool. Treatment with vinblastine (100 nM) results in a loss of microtubule polymers as measured by anti–Tyr-tubulin immunostaining (green) beginning at 30 min (Fig. S2). By 2 h, few microtubule polymers remain, and only a green haze is observed (from soluble tubulin dimers). Coapplication of NAD+ (5 mM) with vinblastine (100 nM) preserves a significant portion of the microtubule polymer mass, indicating that NAD+ can prevent microtubules from being depolymerized in vinblastine-treated cells. (Scale bar, 10 µm.) (C) Quantification of results in B. To quantify microtubule polymer mass in MCF-7 cells, polymerized microtubules were identified and binarized using the Tubeness plugin in Fiji, which identifies linear structures. The total area of the cell was calculated and the graph represents the ratio of microtubule polymers to cell area, which we refer to as the polymerization index. Microtubule mass was measured in ∼100 cells from n = 3 replicates. Bar graph represents mean ± SEM **P < 0.01 (unpaired, two-tailed Student t test).
To determine if elevated intracellular NAD+ has a microtubule polymer-stabilizing effect in these cells, we asked if NAD+ could interfere with microtubule destabilization elicited by antimicrotubule agents. Vinblastine induces depolymerization of dynamic microtubules by binding soluble microtubule dimers and preventing their incorporation into the microtubule polymer, as well as by binding the tips of microtubule polymers (reviewed in ref. 18). Application of 100 nM vinblastine to MCF-7 cells resulted in a loss of microtubule polymer networks in as little as 30 min, as measured by immunostaining for tyrosinated-α-tubulin (Tyr-tubulin) (Fig. S2A). Within 2 h, the majority of the microtubules were depolymerized, with tubulin dimers and oligomers diffusely localized throughout the cytosol (Fig. 1 B and C), consistent with the well-known effects of vinblastine (19). However, coapplication of 5 mM NAD+ with vinblastine markedly interfered with vinblastine-induced microtubule depolymerization (Fig. 1 B and C). We observed similar results in HEK293 cells, indicating our results are not unique to MCF-7 cells (Fig. S2B).
Importantly, the microtubule polymers seen after vinblastine treatment in NAD+-treated cells exhibited a slightly curved morphology toward the cell periphery, with distinct large microtubule fragments perinuclearly, rather than the more linear centrosomally nucleated microtubules seen in untreated cells. This result is reminiscent of microtubule fragments induced by tubulin depolymerizing drugs as a result of dissociation of the minus-ends of the microtubules from the centrosome (20). These results are also consistent with recent X-ray crystallographic analysis showing that vinblastine binds at the interface between two or more tubulin dimers, inducing a curved microtubule assembly (21, 22). Finally these results suggest that the protective effect of NAD+ on cellular microtubules does not involve the microtubule minus-end but rather affects the plus-end GTP-cap of microtubules.
To further assess the ability of NAD+ to affect microtubule polymer stability, we monitored microtubules by time-lapse imaging of live MCF-7 cells stably expressing GFP–α-tubulin (23). In these experiments, vinblastine treatment (25 nM) induced the depolymerization of microtubules, which was blocked by NAD+ treatment (Fig. S3 and Movies S1–S4). The effect of NAD+ was particularly evident at 75 min, when nearly all microtubules were completely depolymerized in vinblastine-treated cells. However, in the presence of NAD+, a significant number of dynamic microtubules remained in the cells. The effect of NAD+ treatment was more pronounced on microtubule preservation toward the cell membrane, where microtubule plus-ends are localized, in agreement with our results in Fig. 1. Taken together, these data indicate that NAD+ preserves microtubule polymers that would by otherwise depolymerized by vinblastine.
NAD+ Increases Dynamic Instability of Microtubule Polymers.
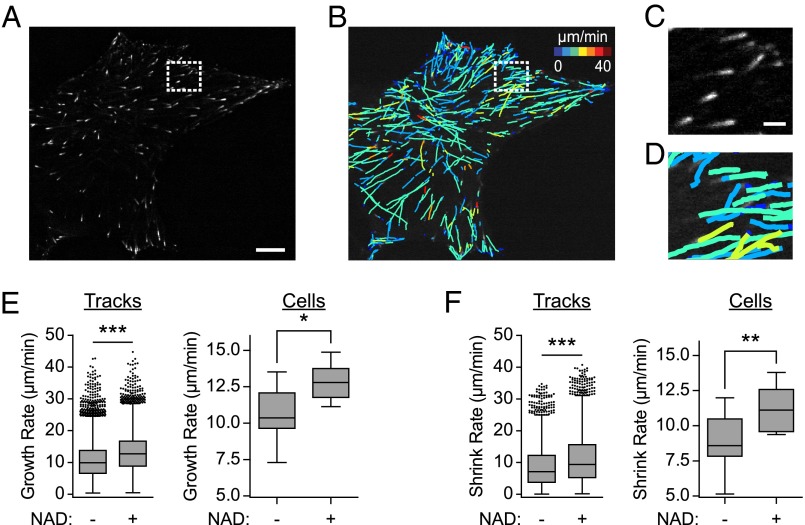
To assess how NAD+ affects microtubule stability, we examined microtubule dynamics in NAD+-treated cells. Studies of microtubule dynamics have shown that dynamic instability, the phenomenon of alternating microtubule assembly and disassembly at microtubule tips, is required for processes such as cellular growth and proliferation (19, 24, 25). Dynamic instability is markedly inhibited by antimicrotubule agents (26). Because of the density of microtubules in cells, microtubule dynamics can only be reliably measured in the cell periphery, which can bias measurements of microtubule dynamics and does not allow monitoring of microtubule behavior toward the center of the cell (27). To overcome this bias, recent studies have monitored microtubule dynamics by imaging fluorescently labeled microtubule plus-end tracking proteins, such as EB1ΔC (referred to throughout the manuscript as EB1, for simplicity) (27, 28). Imaging of EB1-GFP fluorescence typically results in the observation of “comets,” which reflects EB1-GFP binding to the rapidly growing plus ends of microtubule polymers. Microtubule polymers undergoing catastrophe or that have paused growth do not have EB1 associated with their plus-end tips. EB1-GFP imaging gives similar measurements of microtubule dynamics, as is seen in cells injected with small amounts of fluorescently labeled tubulin (29–31). Imaging EB1-GFP enables imaging of both microtubule growth rates as well as the duration of growth phases. The frequency and duration of both pause and shrink rates can also be inferred using this approach (27).
To assess the effects of NAD+ on microtubule dynamics, we monitored over 20,000 EB1-GFP comets in control and NAD+-treated cells (Fig. 2 A–D). The results indicated that NAD+ treatment significantly increased comet growth rate without altering the underlying rate distributions (Fig. 2E). Computational analysis further revealed that NAD+ treatment significantly increased microtubule shrink rate (Fig. 2F). We also found significant changes in the gap speed and probability that a shrink occurs (Table S1). Taken together, these data indicated that NAD+ increased the dynamic instability at microtubule tips. Thus, NAD+ appears to have the opposite effect on dynamic instability compared with microtubule-targeting drugs, which suppress dynamic instability at low-doses, consistent with the ability of NAD+ to counteract the effects of vinblastine.
Fig. 2.
NAD+ increases microtubule dynamics. (A) Example of EB1 comets in an MCF-7 cell. To monitor microtubule dynamics, we used MCF-7 cells expressing EB1-GFP, a microtubule end-binding protein that tracks the tips of growing microtubules. A representative image is shown to demonstrate the robustness of this approach in tracking microtubule +tips. This image is a snapshot from a 1-min time-lapse recording. Boxed region is enlarged in C. (Scale bar, 2.5 µm.) (B) Example of EB1 comet track overlays in an MCF-7 cell. To quantify microtubule dynamics based on the movement of EB1-GFP comets, we used an image analysis algorithm described previously (27). Briefly, this algorithm detects EB1-GFP comets and calculates the microtubule growth rates, shown here as heat-mapped lines representing growth rate and trajectory. Using spatiotemporal clustering of EB1-GFP growth tracks, we can infer microtubule behaviors during phases of pause and shortening. See Materials and Methods for a more thorough explanation. Lines represent overall EB1-GFP comet movement for a 1-min time-lapse recording. Boxed region is enlarged in D. (C and D) Enlarged images of EB1-GFP comets. Selected regions from A and B were enlarged to more clearly display EB1-GFP comets (C) and microtubule tracks (D). (Scale bar, 0.5 µm.) (E) NAD+ increases microtubule polymerization rate. To monitor microtubule polymerization rate, we used MCF-7 cells expressing EB1-GFP, a microtubule end-binding protein that tracks the tips of growing microtubules. The cells were treated with NAD+ (5 mM) for 2 h and time-lapse images were acquired every 0.5 s for 1 min. Microtubule growth rates were calculated by directly observing the EB1-GFP comets. In cells treated with NAD+ (5 mM), we observed a significant increase in the growth rate of all tracked EB1-GFP comets (left; 10.5 ± 0.04 µm/min vs. 13.0 ± 0.04 µm/min) and the per cell means (Right; 10.53 ± 0.51 vs. 12.88 ± 0.35 µm/min). These data indicate that NAD+ treatment is sufficient to increase microtubule polymerization rates. Box plots indicate the 25th percentile (bottom boundary), median (middle line), 75th percentile (top boundary), and nearest observations within 1.5-times the interquartile range (whiskers). n = 11 cells per condition; *P < 0.05; ***P < 0.00005 (permutation t test). (F) NAD+ increases microtubule shrink rate. To determine microtubule shrink rate, we used MCF-7 cells expressing EB1-GFP, a microtubule end-binding protein that tracks the tips of growing microtubules. In contrast to the directly observed growth rates (E), microtubule shrink rates were inferred computationally, as described previously (27). The cells were treated with NAD+ (5 mM) for 2 h, and time-lapse images were acquired every 0.5 s for 1 min. In cells treated with NAD+ (5 mM), we observe a significant increase in both the shrink rate of all tracked EB1-GFP comets (Left; 9.01 ± 0.16 µm/min vs. 11.45 ± 0.16 µm/min) and the per cell means (Right; 8.86 ± 0.61 vs. 11.16 ± 0.47 µm/min). These data indicate that NAD+ alone can increase microtubule shrink rate. NAD+ treatment does not alter the underlying distribution of microtubule growth or shrink rates (n > 20,000 and n > 2,000, respectively), as determined by a two-sample Kolmogorov–Smirnov test (P > 0.05). However, we find a significant increase in the mean microtubule growth (E) and shrink rates (F). These data indicate that NAD+ alone can increase microtubule dynamics, which are an indicator of healthy cells (reviewed in ref. 76). Box plots indicate the 25th percentile (bottom boundary), median (middle line), 75th percentile (top boundary), and nearest observations within 1.5-times the interquartile range (whiskers). Points represent outliers. n > 20,000 comets and n > 2,000 inferred shrink rates per condition; **P < 0.01; ***P < 0.00005 (permutation t test).
To further explore the effect of NAD+ on dynamic instability, we also measured microtubule dynamics in DRG neurons. In these experiments, we monitored microtubule dynamics using EB3-GFP, a neuronal microtubule plus-end tracking protein (31). In neurons, the microtubule growth rate was significantly lower than that measured in MCF-7 cells, consistent with previous studies showing that neuronal microtubules have reduced growth rates and dynamic instability (31). Following axon transection, however, we observed a significant decrease in the microtubule growth rate (Fig. S4 A–F). This alteration in microtubule dynamics was prevented by treatment with NAD+ before transection (Fig. S4G). Although we did not observe a significant increase in dynamics with NAD+ treatment alone, this is not entirely unexpected because of the highly stable nature of axonal microtubules. Taken together, these results demonstrate that NAD+ can counteract microtubule depolymerization caused by diverse insults, as well as in diverse cell types.
NAD+ Acts Intracellularly to Influence Microtubule Atability.
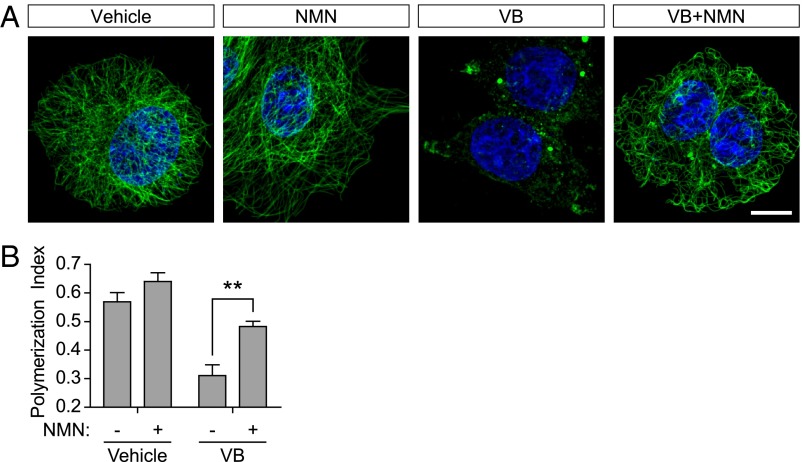
As an additional control, we sought to confirm that the effects of NAD+ were mediated by NAD+ acting inside the cell, rather than through the activation of a cell-surface receptor. To test this, we used nicotinamide mononucleotide (NMN), which is converted intracellularly to NAD+ (32). As with NAD+ treatment, NMN also impaired the microtubule depolymerizing effects of vinblastine (Fig. 3). These data suggest that NAD+ treatment leads to increased intracellular NAD+ levels, which affect intracellular microtubule stability and dynamics.
Fig. 3.
NAD+ acts intracellularly to stabilize microtubules. (A) NMN also exhibits a microtubule-protective effect. We wondered if NAD+ exerts its effects on microtubules by binding and activating a cell-surface receptor, or if NAD+ acts intracellularly. To address this question, we used NMN, an NAD+ precursor that is metabolically adenylated to form NAD+ in the cell. As seen in Fig. 1 A and B, vinblastine (VB; 100 nM) treatment for 2 h resulted in a loss of the majority of microtubule polymers in MCF-7 cells as measured by anti–Tyr-tubulin immunostaining (green). Like NAD+, NMN (5 mM) also protects the microtubule cytoskeleton from the depolymerizing effects of vinblastine (100 nM). These data suggest that intracellular NAD+ is required for its microtubule protective effects. This conclusion is supported by subsequent experiments that identify SIRT3 as a mediator of the effects of NAD+. (Scale bar, 10 µm.) (B) Quantification of results in A. To quantify microtubule polymer mass in MCF-7 cells, polymerized microtubules were identified and binarized using the Tubeness plugin in Fiji, which identifies linear structures. The total area of the cell was calculated, and the graph represents the ratio of microtubule polymers to cell area, which we refer to as the polymerization index. We measured ∼50 cells from n = 3 replicates. Bar graph represents mean ± SEM; **P < 0.01 (unpaired, two-tailed Student t test).
NAD+ Does Not Directly Interact with Microtubules.
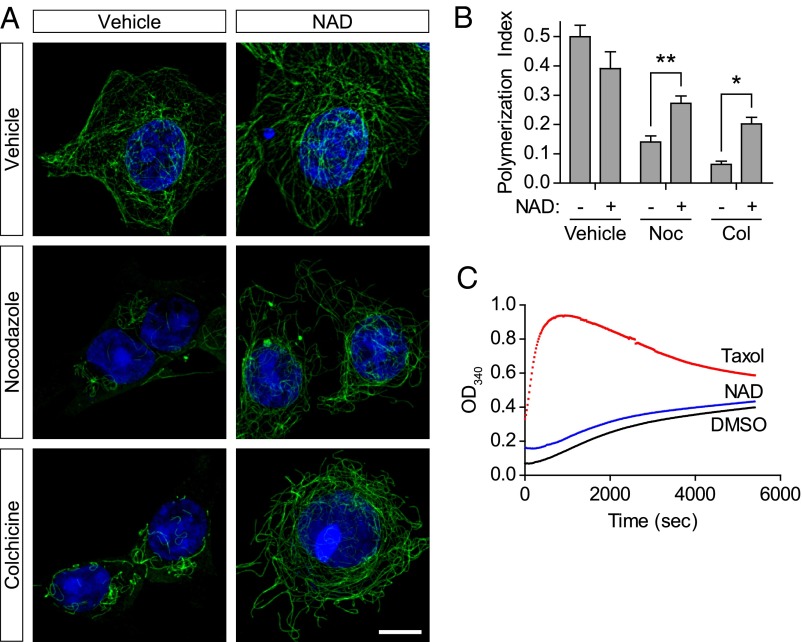
We next sought to understand the mechanism by which NAD+ affects microtubule polymer stability. We first considered the possibility that NAD+ directly binds microtubules and competes with vinblastine for binding to microtubules. To test this theory, we asked if NAD+ prevents microtubule depolymerization induced by colchicine, which binds at a distinct site on microtubules (21). Additionally, we tested whether NAD+ prevents microtubule depolymerization elicited by nocodazole, which is a structurally distinct microtubule-depolymerizing agent that binds to the vinca alkaloid site on β-tubulin (22, 33). As seen with vinblastine treatment, NAD+ treatment markedly reduced microtubule depolymerization induced by either colchicine or nocodazole (Fig. 4 A and B). As with vinblastine-treated cells, the microtubule polymer network exhibited increased curvature at the cell periphery (Fig. 4 A and B). These data using distinct microtubule depolymerizing agents argue against direct competition of NAD+ and vinblastine for binding to microtubules.
Fig. 4.
NAD+ indirectly stabilizes microtubules. (A) NAD+ prevents microtubule depolymerization elicited by diverse antimicrotubule agents. We wondered if NAD+ competes with vinblastine in binding to the vinblastine-binding site on microtubules. In this case, NAD+ would not be able to prevent the depolymerizing effects of colchicine, a structurally distinct antimicrotubule agent, which binds to a site on microtubules that does not overlap with vinblastine (21, 33). MCF-7 cells were treated with either vehicle, 1 µM nocodazole (Noc; a structurally distinct drug that also binds at the vinblastine-binding site), or 15 µM colchicine (Col) for 15 min or 30 min, respectively. In the absence of NAD+, microtubule polymer mass was noticeably reduced, as measured by anti–Tyr-tubulin immunostaining (green). However, coapplication of 5 mM NAD+ markedly preserved the microtubule mass in cells treated with either microtubule-depolymerizing drug. These data suggest that NAD+ does not prevent the actions of vinblastine by completing for its binding to microtubules. (Scale bar, 10 μm.) (B) Quantification of results in A. To quantify microtubule polymer mass in MCF-7 cells, polymerized microtubules were identified and binarized using the Tubeness plugin in Fiji, which identifies linear structures. The total area of the cell was calculated, and the graph represents the ratio of microtubule polymers to cell area, which we refer to as the polymerization index. Microtubule mass was measured in ∼100 cells from n = 3 replicates. Bar graph represents mean ± SEM *P < 0.05; **P < 0.01 (one-way ANOVA with Tukey’s post hoc test). (C) NAD+ does not affect the polymerization of purified microtubules. To obtain further evidence that NAD+ does not bind tubulin directly, we compared the effect of NAD+ and taxol on microtubule polymerization. Purified bovine brain tubulin was incubated in polymerization buffer (80 mM Pipes pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 3% glycerol), and the formation of microtubule polymers was detected by diffraction absorption spectrometry. Taxol (10 µM) elicited rapid induction of microtubule polymer formation at stoichiometric concentrations with tubulin, as is expected for a drug that directly binds and stabilizes tubulin; However, even a high concentration of NAD+ (10 mM) had no significant effect on the rate of microtubule polymer formation. Graph displays representative data from n = 3 independent experiments.
To further assess whether the microtubule-stabilizing effects of NAD+ are caused by direct binding to microtubules, we monitored the effect of NAD+ on in vitro polymerization of purified tubulin. Addition of taxol, a microtubule-stabilizing agent that binds directly to tubulin, enhances polymerization of purified tubulin in vitro (Fig. 4C). However, addition of NAD+ to purified tubulin did not affect the polymerization rate (Fig. 4C). Thus, the effects of NAD+ are likely to be a result of the activation of an intracellular NAD+-dependent signaling pathway rather than by direct binding to microtubule polymers.
SIRT3 Expression Stabilizes Microtubule Polymers.
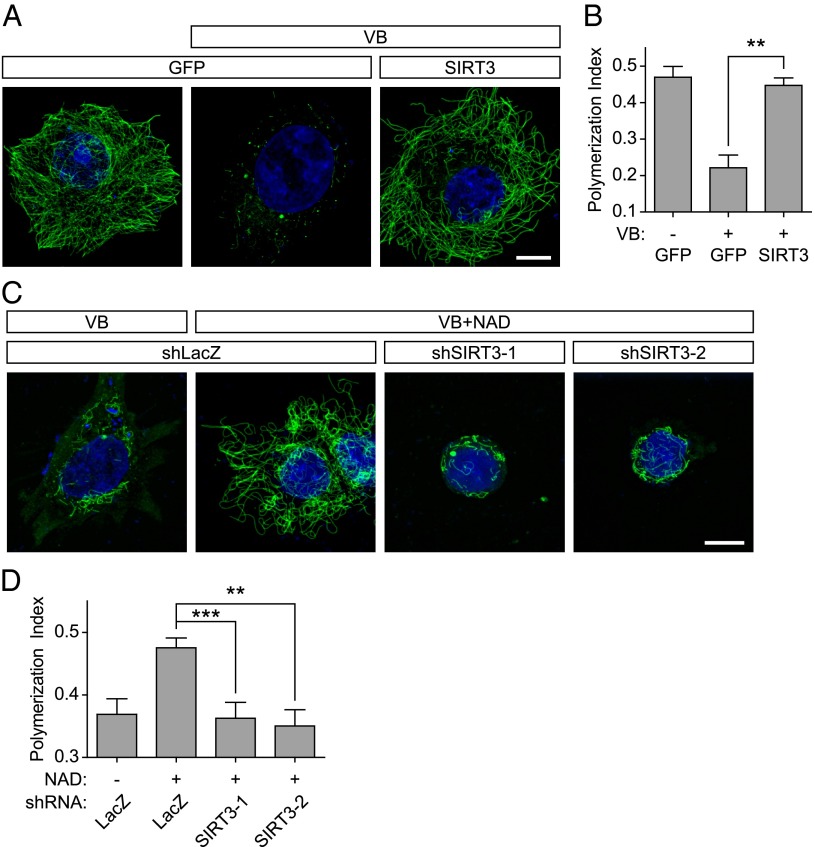
We next asked if the microtubule-stabilizing effects of NAD+ are mediated by sirtuins, a family of enzymes that catalyze NAD+-dependent deacetylation or deacylation of proteins (reviewed in refs. 34 and 35). To explore this idea, we overexpressed each sirtuin to determine if any could recapitulate the effect of NAD+ treatment on microtubule stability. In these experiments, SIRT1–7 were expressed with a C-terminal GFP tag in MCF-7 cells, and then subsequently treated with 100 nM vinblastine for 2 h to depolymerize the microtubule cytoskeleton. In each case, sirtuin overexpression had a minimal effect on vinblastine-induced microtubule depolymerization, except for cells expressing SIRT3. In SIRT3-expressing cells, vinblastine-induced microtubule depolymerization was markedly inhibited (Fig. 5 A and B and Fig. S5 A and B). The residual microtubules exhibited a similar curved morphology as seen in NAD+-treated cells following vinblastine treated cells (Fig. 1). These data suggest that among the sirtuins, SIRT3 has the most pronounced influence on microtubule polymer stability.
Fig. 5.
NAD+ exerts its stabilizing effects on microtubules through SIRT3. (A) SIRT3 overexpression inhibits vinblastine-mediated microtubule depolymerization. To determine if a sirtuin mediates the effects of NAD+ on microtubules, we tested each sirtuin for its effects on microtubule stability. In these experiments, MCF-7 cells were transfected with C-terminal GFP-tagged versions of SIRT1–7, and microtubule depolymerization was elicited by treatment of cells with vinblastine (VB; 100 nM) for 2 h. Microtubules were visualized by anti–Tyr-tubulin immunostaining (green). In control-transfected cells, minimal microtubule polymer mass remained after vinblastine treatment. However, cells expressing SIRT3-GFP exhibited marked protection of microtubules from vinblastine treatment. (Scale bar, 10 µm.) (B) Quantification of results in A. To quantify microtubule polymer mass in MCF-7 cells, polymerized microtubules were identified and binarized using the Tubeness plugin in Fiji, which identifies linear structures. The total area of the cell was calculated, and the graph represents the ratio of microtubule polymers to cell area, which we refer to as the polymerization index. We measured ∼100 cells from n = 3 replicates. Bar graph represents mean ± SEM; **P < 0.01 (one-way ANOVA with Tukey’s post hoc test). (C) SIRT3 is necessary for the effects of NAD+ on microtubule depolymerization. To determine if the effects of NAD+ on microtubule stability are mediated through SIRT3, we monitored the effects of SIRT3 knockdown on NAD+-induced stabilization of microtubules. MCF-7 cells were treated with 100 nM vinblastine for 2 h, which results in both loss of the microtubule cytoskeleton and subsequent cell shrinkage. Microtubules were visualized by anti–Tyr-tubulin immunostaining (green). Treatment of these cells with 1 mM NAD+ prevents the loss of the microtubule cytoskeleton and plasma membrane collapse. However, this effect of NAD+ is lost in cells expressing either of two SIRT3-specific shRNA. These cells also display more membrane collapse than control cells, suggesting that the loss of SIRT3 may make the cells more sensitive to vinblastine. These data indicate that SIRT3 is required for the protective effects of NAD+ on the microtubule cytoskeleton. (Scale bar, 10 µm.) (D) Quantification of results in C. Quantification was performed as described in B. We measured ∼100 cells from n = 3 replicates. Bar graph represents mean ± SEM; **P < 0.01, ***P < 0.001 (one-way ANOVA with Tukey’s post hoc test).
We observed a slight but nonsignificant protection of microtubules from vinblastine when we overexpressed SIRT1 in MCF-7 cells (Fig. S5 A and B). This moderate effect may reflect the ability of SIRT1 to induce the transcriptional up-regulation of SIRT3 via peroxisome proliferator-activated receptor-γ coactivator-α (PGC-1α) (36, 37). Notably, we did not see an effect of SIRT2 on microtubule polymer stability. SIRT2 can deacetylate lysine-40 in α-tubulin (38). However, in MCF-7 cells treated with NAD+, we found no NAD+-dependent decrease in acetyl-tubulin levels, as measured by Western blotting (Fig. S5C). Thus, the effects of NAD+ appear unlikely to be mediated by SIRT2-dependent deacetylation of tubulin.
SIRT3 Mediates the Stabilizing Effect of NAD+ on Microtubule Polymers.
We next asked if SIRT3 is required for the effects of NAD+ on microtubule polymer stability. In MCF-7 cells expressing control shRNA, vinblastine treatment (100 nM) for 2 h resulted in loss of microtubule polymers and contraction of the plasma membrane. This effect was blocked by treatment of cells with 5 mM NAD+ (Fig. 5 C and D). However, in cells expressing either of two SIRT3-specific shRNA (Fig. S5D), NAD+ treatment did not block vinblastine-induced microtubule depolymerization and contraction of the cellular membrane (Fig. 5 C and D). These data indicate that SIRT3 is required for the microtubule-stabilizing effects of NAD+.
One of the best described functions of SIRT3 is to suppress the production of reactive oxygen species (ROS) in mitochondria (reviewed in ref. 39). The physiological targets of SIRT3 include over 100 mitochondrial proteins, including diverse proteins that suppress ROS generation (40). Acetylation of these proteins typically leads to increased ROS production, whereas SIRT3-mediated deacetylation leads to decreased ROS levels (41–43). Indeed, treatment of MCF-7 cells with NAD+ reduced the increase in ROS induced by vinblastine (Fig. S6A). Reduction of ROS levels using either α-tocopherol (44) or ascorbic acid resulted in reduced vinblastine-induced microtubule depolymerization, similar to the effect seen with NAD+ treatment or SIRT3 overexpression (Fig. S6 B–D). Because both antioxidants and NAD+ appear to exhibit similar effects on microtubule polymer stability, the mechanism of NAD+ treatment and SIRT3 overexpression may relate to suppression of ROS that contribute to the microtubule-destabilizing effect of vinblastine.
NAD+ Reduces the Cytotoxic Effects of Vinblastine.
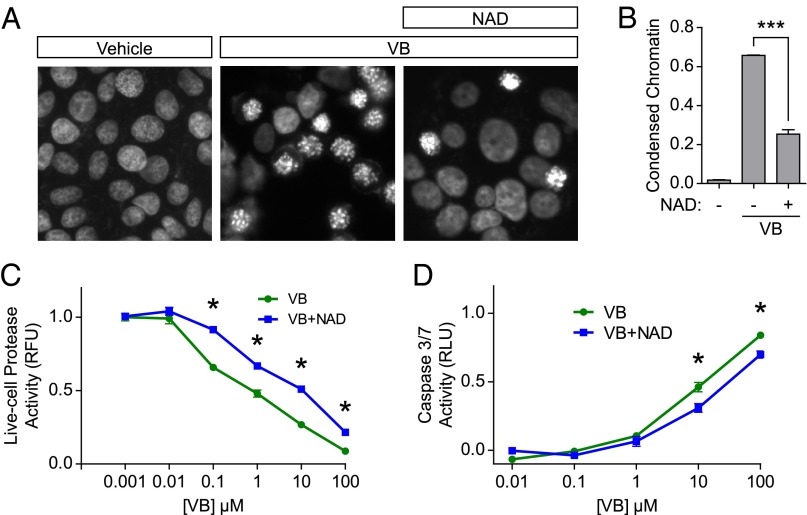
Vinblastine is one of many microtubule-targeting drugs commonly used in cancer chemotherapy. Because augmented NAD+ or SIRT3 can prevent the destabilizing effects of vinblastine on the microtubule cytoskeleton, we wondered if NAD+ would also alter the ability of vinblastine to induce cell death. To test this idea we asked if NAD+ treatment affects the sensitivity of cells to vinblastine-induced cell death. In these experiments, cells were treated with vinblastine (100 nM) for 24 h. Cell death was assayed by monitoring nuclear fragmentation using DAPI staining, which provides assessment of cell death induced by diverse pathways. In vinblastine-treated cells, over 60% of the cells exhibited nuclei with condensed chromatin, representing mitotic (prometaphase) arrest, which is a hallmark of vinblastine toxicity. However, when cells received NAD+ treatment along with vinblastine, we observed a significant reduction in the number of cells arrested in mitosis (Fig. 6 A and B). This result suggests that increasing NAD+ levels reduces the sensitivity of cells to vinblastine, likely because of the diminished antimicrotubule effects elicited by the drug in the presence of NAD+.
Fig. 6.
NAD+ alters the chemotherapeutic sensitivity of vinblastine. (A) Increased NAD+ levels inhibit growth arrest because of vinblastine. To determine if NAD+ can antagonize the chemotherapeutic effects of vinblastine, MCF-7 cells were treated with vinblastine (100 nM) and nuclei were visualized with DAPI. After 24 h of vinblastine treatment, greater than 60% of the cells exhibited nuclei with condensed chromatin, representing mitotic arrest. When cells received coapplication of NAD+ (5 mM), however, there was a significant reduction in the number of cells arrested in mitosis. These results suggest that NAD+ can inhibit the cytotoxic effects of vinblastine. (Magnification: A, 20×.) (B) Quantification of results in A. To quantify the percent of fragmented nuclei, cells were stained with DAPI and the number of nuclei exhibiting condensed, fragmented nuclei were counted. The results were expressed as the ratio of fragmented nuclei to total nuclei. We counted ∼400 cells per condition from n = 3 replicates. Bar graph represents mean ± SEM; ***P < 0.001 (unpaired, two-tailed Student t test). (C) Increased NAD+ levels reduce the sensitivity of cells to vinblastine. MCF-7 cells were treated with increasing concentrations of vinblastine for 24 h and cell viability was measured by a live-cell protease activity in which a compound is cleaved into a fluorescent molecule by a protease only active in intact cells. Vinblastine-induced a dose-dependent decrease in viability, consistent with its known effects. When cells were treated with NAD+ (5 mM) along with vinblastine, however, there was a 10-fold decrease in vinblastine sensitivity. These data demonstrate that increased NAD+ levels can reduce the sensitivity of cells to vinblastine. We measured ∼10,000 cells per well in a 96-well plate from n = 3 replicates. Each point represents the mean ± SEM normalized to vehicle or NAD+ control. *P < 0.05 (unpaired, two-tailed Student t test). (D) Increased NAD+ levels reduce vinblastine-induced apoptosis. We measured the effect of NAD+ on vinblastine-induced cell death using an alternate assay for cell viability. In these experiments, we measured apoptosis using a caspase 3/7 activity assay. Vinblastine treatment induced a dose-dependent increase in apoptosis, as expected. When cells were also treated with NAD+ (5 mM), however, there was a significant reduction in the levels of apoptosis. These data demonstrate that increased NAD+ levels can reduce the sensitivity of cells to vinblastine-induced apoptosis. We measured ∼10,000 cells per well in a 96-well plate from n = 3 replicates. Each point represents the mean ± SEM normalized to vehicle or NAD+ control. *P < 0.05 (unpaired, two-tailed Student t test).
To further examine the effect of NAD+ on vinblastine-induced cell death, we treated cells with increasing concentrations of vinblastine and measured cell viability. We observed a dose-dependent decrease in viability with vinblastine treatment (Fig. 6C), consistent with its known cytotoxic effects. When cells were treated with NAD+ at the same time as vinblastine treatment, we found a 10-fold decrease in the sensitivity to vinblastine (Fig. 6C). We found similar results using an MTT cell viability assay (Fig. S7A). We also observed an NAD+-dependent reduction in sensitivity to vinblastine in HEK and HeLa cells (Fig. S7 B and C), indicating that our results are not specific to MCF-7 cells.
As a final measure of vinblastine cytotoxicity, we measured apoptosis rather than viability. In these experiments, we measured apoptosis by monitoring caspase-3/7 activity, and we observed a dose-dependent increase in apoptosis following vinblastine treatment (Fig. 6D), again consistent with the known effects of vinblastine. Following addition of NAD+, we found an ∼twofold shift in the sensitivity of cells vinblastine-mediated apoptosis.
Finally, we asked if NAD+ alters the sensitivity to other microtubule depolymerizing drugs. As with vinblastine, NAD+ reduced the sensitivity of MCF-7 cells to vincristine-, vindesine-, and vinorelbine-induced cytotoxicity, as measured using the MTT cell viability assay (Fig. S7A). Taken together, these results suggest that NAD+ can reduce the cytotoxic effect of antimicrotubule drugs.
Discussion
Although NAD+ has pronounced effects on diverse cellular and physiologic processes, the specific cellular alterations induced by NAD+ are not fully understood. Our results demonstrate a striking effect of increased intracellular NAD+ on microtubule polymer stability. We show that NAD+ alters microtubule dynamic instability and markedly inhibits microtubule depolymerization induced by vinblastine and related microtubule-depolymerizing agents. Furthermore, we find that increasing NAD+ levels can reduce the cell death induced by antimicrotubule agents. We show that these effects of NAD+ require SIRT3, a mitochondrial NAD+-dependent protein deacetylase. Our data demonstrate that NAD+ and SIRT3 are novel regulators of the microtubule cytoskeleton and suggest that alterations in intracellular NAD+ levels can affect the cytotoxicity of anti-microtubule agents.
Our studies suggest that NAD+ alters—but does inhibit—the effect of vinblastine. Although cells treated with NAD+ retain microtubule polymers after treatment with diverse microtubule depolymerizing agents, the microtubule polymers exhibit altered morphology. These polymers exhibit increased curvature, particularly near the cell periphery. Microtubule curvature is influenced by microtubule-associated proteins, such as adenomatous polyposis coli protein among other +TiP proteins (45). Thus, the effects of NAD+ may reflect an alteration in microtubule-bound proteins, especially those that bind the plus-end of microtubules.
Our study identifies SIRT3 as the major mediator of the microtubule effects of NAD+. The importance of SIRT3 was determined using both knockdown and an overexpression approach to screen different sirtuins for their ability to phenocopy the effect of NAD+ treatment. Despite the effectiveness of SIRT3 in this overexpression assay, the other mitochondrial sirtuins, SIRT4 and SIRT5, were much less effective. Recent studies suggest that these sirtuins remove acyl modifications in proteins or deacetylate only a small subset of proteins in mitochondria (46–48). This finding contrasts with SIRT3, which is the major protein deacetylase in mitochondria (49), affecting the acetylation status of ∼100 proteins in mitochondria (40). Among the nonmitochondrial sirtuins, overexpression of SIRT1 showed a detectable, although statistically nonsignificant, inhibition of vinblastine-induced microtubule depolymerization. This finding may reflect a SIRT1-dependent activation of SIRT3 via PGC-1α, as previously reported (36, 37).
How might SIRT3 influence microtubule function? A simple explanation would be direct deacetylation of tubulin by SIRT3; however, studies suggest that SIRT3 is exclusively mitochondrial (50, 51). Furthermore, proteomics studies have not identified tubulin as a target for SIRT3 (40, 49, 52). Instead, our studies raise the possibility that the effects of SIRT3 may relate to its well-characterized ability to reduce the mitochondrial production of ROS (reviewed in ref. 39). ROS affects diverse intracellular signaling proteins, such as phosphatases (53). By changing the activity of these proteins, suppression of ROS could affect pathways that control microtubule polymer stability. Indeed, ROS have been shown to affect microtubule dynamics (54). However, it should be noted that some ROS scavengers, such as ascorbic acid, have diverse effects in cells, making it difficult to definitively determine if their effects are a result of reducing ROS levels. Thus, the specific mechanisms by which SIRT3 influences microtubule polymer stability remain to be established.
Our impetus for exploring the link between NAD+ and microtubule dynamics was based on a screen for inhibitors of axon degeneration. Several groups have shown that NAD+ can delay axon degeneration (4, 9). Although the precise mechanisms by which NAD+ delays axon degeneration are not known, a recent study suggests a possible role for SIRT3 (55). This earlier study showed that overexpression of SIRT3 delayed axon degeneration in an excitotoxic model. Future studies will be needed to determine if the axon-protective effects of NAD+ and SIRT3 overexpression are mediated by their microtubule-stabilizing effects. It is interesting to note that taxol promotes regeneration after axon injury (56, 57) and prevents the formation of retraction bulbs (58). Thus, microtubule stabilization through either taxol or NAD+, albeit by distinct mechanisms, may be valuable for minimizing pathologic damage after axon injury.
NAD+ levels might also influence, at least in part, the clinical activity of the microtubule-depolymerizing drugs used in cancer chemotherapy. Our data indicate that increased intracellular NAD+ can impair the effects of the vinca alkaloids and other microtubule depolymerizing agents. Although many of our experiments involved application of exogenous NAD+ to cells, in a physiologic setting NAD+ levels are increased by complex pathways, such as circadian rhythm (59). These physiological processes are associated with ∼twofold changes in NAD+, similar to the increases seen with exogenous application of NAD+.
Based on our finding that NAD+ can prevent vinblastine-induced microtubule depolymerization, it is conceivable that physiological or disease states associated with increased or decreased NAD levels will be more resistant or more responsive, respectively, to the effects of vinblastine and related antimicrotubule agents. For example, aging and obesity are associated with decreased biosynthesis of NAD+ (60–63). In contrast, calorie restriction is associated with increased NAD+ (64). Interestingly, several cancers are known to exhibit up-regulation of NAD+ biosynthetic enzymes (6, 65). Up-regulation of these enzymes may make these cancers less susceptible to microtubule depolymerizing agents or may contribute to resistance to these compounds. Thus, examination of expression levels or genetic alterations of these enzymes might be useful as predictive biomarkers of antimicrotubule therapy. It will be important to determine if these diverse mechanisms that affect NAD+ levels also influence the clinical outcome of antimicrotubule agent therapy to move toward chemotherapy customization.
It is plausible that reducing NAD+ levels may be useful for increasing sensitivity to antimicrotubule agents and to achieve more effective cancer cell death. Indeed, FK866 and CHS828, small-molecule inhibitors of NAD+ synthesis, are already in clinical trials for use in cancer treatment in combination with standard chemotherapeutic agents (66). The effects of NAD+ inhibitors may be particularly useful when combined with antimicrotubule agents, dependent on the integrity of the NAD+ biosynthesis pathway in each tumor type.
It is important to note that NAD+ has pleiotropic effects. Thus, the reduced cell death seen in NAD+-treated cells might be because of a combination of many factors, only one of which might be altered microtubule dynamics. For example, NAD+ can inhibit cytochrome C release and can affect poly(ADP) ribose polymerases, all of which can influence cell death pathways (5). However, the pronounced inhibitory effect of NAD+ on vinblastine-induced microtubule depolymerization suggests that its antiapoptotic effects are likely to be mediated, at least in part, by its ability to stabilize microtubules.
In most of our experiments, we monitored the effects of exogenous application of NAD+. However, physiologic levels of NAD+ are not regulated by extracellular NAD+ levels. The levels of NAD+ are instead determined by intracellular biosynthetic pathways. The ∼twofold increase in intracellular NAD levels that we observed following application of NAD is consistent with the types of changes in intracellular NAD+ levels caused by dietary restriction or circadian rhythms (3, 59). However, it is more challenging to reduce intracellular NAD+. Although NAD synthesis inhibitors have been developed (67), these drugs only partially reduce NAD+ levels (68). Thus, the development of highly effective NAD+ biosynthesis inhibitors will be useful to more fully define the role of endogenous NAD+ levels in regulating microtubule-dependent pathways.
Materials and Methods
Reagents and Materials.
The following assays and reagents were used throughout this manuscript. Unless otherwise indicated, assays were performed according to the manufacturer’s instructions. To induce microtubule depolymerization, we used the following reagents: vinblastine (Sigma), colchicine (Sigma), and nocodazole (Sigma). To increase intracellular NAD levels, we used the following reagents: NAD+ (Affymetrix) and NMN (Sigma). To study cell death, we used the following assays: Multitox Fluor Cytotoxicity Assay (live-cell protease activity; Promega), Caspase-Glo 3/7 Assay (apoptosis assay; Promega); MTT assay (cell viability; Roche). To study in vitro tubulin polymerization, we used the Tubulin Polymerization Assay Kit (Cytoskeleton). To study ROS generation, cells were treated with α-tocopherol (Sigma) or ascorbic acid (Sigma), and ROS levels were measured using CellROX Deep Red reagent (Life Technologies). For additional details about the assays and reagents, please refer to SI Materials and Methods.
Immunofluorescence.
Cells were cultured on glass coverslips and fixed using PHEMO buffer (80 mM Pipes, 25 mM Hepes, 15 mM EGTA, 3 mM MgCl2, 10% (vol/vol) DMSO, pH 6.8) containing 3.7% (vol/vol) formaldehyde, 0.05% gluteraldehyde, and 0.5% Triton X-100 to preserve microtubules for visualization. Microtubules were immunolabeled with antityrosinated-tubulin antibody (1:1,000; Novus Biologicals) overnight and Alexa Fluor 488 conjugated secondary antibody (1:1,000; Life Technologies) for 30 min. Coverslips were mounted on a glass microscope slide (VWR) using Prolong Gold with DAPI (Invitrogen). See SI Materials and Methods for details about image acquisition.
Quantification of Microtubule Polymerization.
Microtubule polymerization was quantified using Fiji (69). First, polymerized tubulin was identified using the “Tubeness” plugin on a 3D image stack. This process determines how tube-like each pixel is and returns a mask of all tube-like structures. This algorithm was robust in detecting polymerized microtubules. Total microtubule mass was measured based on this binary mask. Then an Alexa Fluor 568 Phalloidin counterstain (Life Technologies) was used to generate a mask representing the total cellular area. The ratio of polymerized tubulin mass to cellular area was used as a measure for microtubule polymerization, which we refer to as the polymerization index.
Live Cell Microtubule Depolymerization Assay.
MCF-7 cells stably expressing GFP–α-tubulin were plated on 35-mm glass bottom No. 1.5 dishes (MatTek). Cells were pretreated with NAD+ for 2 h at 37 °C before exposure to vinblastine. Following drug exposure, live-cell confocal microscopy was performed as described in SI Materials and Methods. Time-lapse movies were generated by recording every 15 s for a total of 90 min to observe gross changes in microtubule structure.
EB Comet Imaging Assays and Analysis.
MCF-7 cells were plated on 35-mm glass bottom No. 1.5 dishes (MatTek) and infected with GFP-EB1ΔC (referred to as EB1 for simplicity) lentiviral particles (70). The cells were imaged 2 d postinfection to measure the dynamicity of microtubules ends labeled with GFP-EB1ΔC. To image dynamics in axons, dissociated DRG neurons (E14.5) were cultured in microfluidic devices on glass-bottom dishes. The neurons were infected with EB3-GFP (31) lentiviral particles and imaged on days in vitro (DIV) 4–5. Cells were pretreated with vehicle (water) or 10 mM NAD+ for 2 h at 37 °C before imaging. Images were acquired every 0.5 s for a total of 1 min for each time-lapse movie. All image analysis was performed using ClusterTrack, an algorithm that has been described in detail previously (27, 71). See SI Materials and Methods for additional details about image acquisition and analysis.
Axon Degeneration Analysis.
Dissociated DRG neurons (E14.5) were prepared as described previously (72) and cultured in polydimethylsiloxane microfluidic devices (73). On DIV5, treatments were added to the axonal compartment, and cells were transected by aspirating the cell bodies from their compartment. Phase-contrast images (20×) of the distal axonal compartment were acquired at the indicated time points. Axon degeneration was analyzed by calculating the degeneration index as previously described (15). See SI Materials and Methods for additional details.
NAD+ Quantification.
MCF-7 cells were treated with 5 mM NAD+ for 2 h. Cells were then trypsinized, pelleted, and washed three times with PBS. A small aliquot of cells was saved for protein quantification via bicinchoninic acid (BCA) assay. The remaining cells were pelleted and resuspended in 7% (vol/vol) perchloric acid. Samples were vortexed and sonicated, and any remaining insoluble debris was pelleted and discarded. The supernatant was neutralized (pH 7) with NaOH and 1 M phosphate buffer. Sample was added to a 96-well plate and mixed with cycling assay buffer (5 mM Tris, 5 mM MgCl2, 50 mM KCl, 54 M resazurin, 2.25 mM lactate, 0.4 U/mL lactate dehydrogenase). Finally, 0.7 U diaphorase was added to each well, and the production of resorufin was recorded (530-nm excitation, 580-nm emission) every 30 s for 15 min. NAD+ concentration was calculated based on a standard curve, and the results were expressed as NAD+ concentration per milligram of protein. Three technical replicates were performed for n = 3 independent experiments.
Preparation of Lentiviruses.
Lentiviruses were prepared using the third-generation lentiviral system (74). Four plasmids [transfer vector containing the transgene and cis-acting sequences for genomic RNA production and packaging, and three plasmids (pLP1, pLP2, and pLP/VSV-G) encoding transacting factors (Gag-Pol, Rev, and VSV-G) required for packaging] were cotransfected in HEK293T cells using CaPO4 precipitation (CalPhos; Clontech). Supernatant containing viral particles was collected and concentrated by ultracentrifugation (∼60,000 × g, 2 h, 22 °C; SW41 Ti rotor) and titred using HEK cells. The viral pellet was resuspended in PBS (pH 7.4) containing 1% BSA and stored at −80 °C.
Generation and Validation of the shSIRT3 Constructs.
Two shRNA constructs targeting distinct regions of SIRT3 were generated and cloned into the lentiviral expression system under the control of the H1 promoter. An shRNA targeting LacZ served as a negative control. Coexpression of mCherry served as a marker for infection. Sequences for the shRNA constructs are as follows: shLacZ, GACTACACAAATCAGCGATT; shSIRT3-1, GTGCGTGCTTCAAGTGTTGT; shSIRT3-2, GCCCAACGTCACTCACTACT; Human MCF-7 cells were infected with SIRT3 shRNA or control shRNA virus. Five days postinfection, cells were lysed and Western blotting was performed as described in SI Materials and Methods. SIRT3 expression (anti-Sirt3, 1:2,000; Cell Signaling) was normalized to actin (anti–β-actin, 1:4,000; GenScript) and knockdowns were compared with the shLacZ control. Results from at least three independent experiments were used to quantify the amount of SIRT3 knockdown.
Statistical Analysis.
Statistical analysis was performed using MATLAB (MathWorks) or GraphPad Prism v5.0 (GraphPad Software). All statistical tests are described in their respective figure legends. Briefly, for analyzing degeneration index and polymerization index, when comparing two groups, we used a two-tailed, unpaired Student t test. When comparing three or more groups, we used a one-way ANOVA with Tukey’s post hoc test. We used a permutation t test to analyze microtubule dynamics (75). This test compares the bootstrapped distributions of parameters in two samples. It does not make any assumption about the distribution of the samples.
Supplementary Material
Acknowledgments
We thank A. Sauve (Weill Cornell Medical College) for sirtuin constructs and helpful comments; A. Deglincerti (Weill Cornell Medical College) for imaging advice; S. Shenoy (Weill Cornell Medical College) for assistance with data analysis; T. Wittmann (University of California at San Francisco) for the EB1ΔC construct and valuable discussions; and Noo Li Jeon (Seoul National University) for assistance with microfluidic chambers. This work was supported by the New York State Spinal Cord Injury Research Trust Fund Department of Health Contract C023832 and National Institutes of Health Grants R21CA176638 and R01 NS56306 (to S.R.J.); National Insitutes of Health Grants U54 CA143876 and R01 CA137020 (to P.G.); and the Anne Moore Breast Cancer Fund (P.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404269111/-/DCSupplemental.
References
- 1.Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267(6):1550–1564. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 2.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305(5686):1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 5.Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J Biol Chem. 2004;279(18):18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- 6.Olesen UH, Hastrup N, Sehested M. Expression patterns of nicotinamide phosphoribosyltransferase and nicotinic acid phosphoribosyltransferase in human malignant lymphomas. APMIS. 2011;119(4-5):296–303. doi: 10.1111/j.1600-0463.2011.02733.x. [DOI] [PubMed] [Google Scholar]
- 7.Pittelli M, et al. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol. 2011;80(6):1136–1146. doi: 10.1124/mol.111.073916. [DOI] [PubMed] [Google Scholar]
- 8.Cantó C, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15(6):838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, et al. A local mechanism mediates NAD-dependent protection of axon degeneration. J Cell Biol. 2005;170(3):349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack TG, et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4(12):1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 11.Fischer LR, et al. The WldS gene modestly prolongs survival in the SOD1G93A fALS mouse. Neurobiol Dis. 2005;19(1-2):293–300. doi: 10.1016/j.nbd.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko S, et al. Protecting axonal degeneration by increasing nicotinamide adenine dinucleotide levels in experimental autoimmune encephalomyelitis models. J Neurosci. 2006;26(38):9794–9804. doi: 10.1523/JNEUROSCI.2116-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beirowski B, Babetto E, Coleman MP, Martin KR. The WldS gene delays axonal but not somatic degeneration in a rat glaucoma model. Eur J Neurosci. 2008;28(6):1166–1179. doi: 10.1111/j.1460-9568.2008.06426.x. [DOI] [PubMed] [Google Scholar]
- 14.Meyer zu Horste G, et al. The Wlds transgene reduces axon loss in a Charcot-Marie-Tooth disease 1A rat model and nicotinamide delays post-traumatic axonal degeneration. Neurobiol Dis. 2011;42(1):1–8. doi: 10.1016/j.nbd.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J Neurosci. 2009;29(17):5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11(8):1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JT, Medress ZA, Barres BA. Axon degeneration: Molecular mechanisms of a self-destruction pathway. J Cell Biol. 2012;196(1):7–18. doi: 10.1083/jcb.201108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 19.Jordan MA, Thrower D, Wilson L. Mechanism of inhibition of cell proliferation by Vinca alkaloids. Cancer Res. 1991;51(8):2212–2222. [PubMed] [Google Scholar]
- 20.Yang H, Ganguly A, Cabral F. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J Biol Chem. 2010;285(42):32242–32250. doi: 10.1074/jbc.M110.160820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gigant B, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435(7041):519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 22.Gigant B, Cormier A, Dorléans A, Ravelli RBG, Knossow M. Microtubule-destabilizing agents: structural and mechanistic insights from the interaction of colchicine and vinblastine with tubulin. Top Curr Chem. 2009;286:259–278. doi: 10.1007/128_2008_11. [DOI] [PubMed] [Google Scholar]
- 23.Marcus AI, et al. The synergistic combination of the farnesyl transferase inhibitor lonafarnib and paclitaxel enhances tubulin acetylation and requires a functional tubulin deacetylase. Cancer Res. 2005;65(9):3883–3893. doi: 10.1158/0008-5472.CAN-04-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci USA. 1993;90(20):9552–9556. doi: 10.1073/pnas.90.20.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhamodharan R, Jordan MA, Thrower D, Wilson L, Wadsworth P. Vinblastine suppresses dynamics of individual microtubules in living interphase cells. Mol Biol Cell. 1995;6(9):1215–1229. doi: 10.1091/mbc.6.9.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan MA. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem Anticancer Agents. 2002;2(1):1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 27.Matov A, et al. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7(9):761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long JB, et al. Multiparametric analysis of CLASP-interacting protein functions during interphase microtubule dynamics. Mol Cell Biol. 2013;33(8):1528–1545. doi: 10.1128/MCB.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komarova YA, Vorobjev IA, Borisy GG. Life cycle of MTs: Persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci. 2002;115(Pt 17):3527–3539. doi: 10.1242/jcs.115.17.3527. [DOI] [PubMed] [Google Scholar]
- 30.Piehl M, Cassimeris L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol Biol Cell. 2003;14(3):916–925. doi: 10.1091/mbc.E02-09-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepanova T, et al. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23(7):2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emanuelli M, et al. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem. 2001;276(1):406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 33.Xu K, Schwarz PM, Luduena RF. Interaction of nocodazole with tubulin isotypes. Drug Dev Res. 2002;55(2):91–96. [Google Scholar]
- 34.Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: Regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23(9):467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 37.Kong X, et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 39.Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Mol Cell. 2011;42(5):561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebert AS, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49(1):186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Bell EL, Emerling BM, Ricoult SJH, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahoney JR, Graf E. Role of alpha-tocopherol, ascorbic acid, citric acid and EDTA as oxidants in model systems. J Food Sci. 1986;51(5):1293–1296. [Google Scholar]
- 45.Reilein A, Nelson WJ. APC is a component of an organizing template for cortical microtubule networks. Nat Cell Biol. 2005;7(5):463–473. doi: 10.1038/ncb1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 47.Ahuja N, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282(46):33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 48.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411(2):279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 51.Cooper HM, Huang J-Y, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS ONE. 2009;4(3):e4986. doi: 10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sol EM, et al. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS ONE. 2012;7(12):e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banan A, et al. Activation of delta-isoform of protein kinase C is required for oxidant-induced disruption of both the microtubule cytoskeleton and permeability barrier of intestinal epithelia. J Pharmacol Exp Ther. 2002;303(1):17–28. doi: 10.1124/jpet.102.037218. [DOI] [PubMed] [Google Scholar]
- 55.Magnifico S, et al. NAD+ acts on mitochondrial SirT3 to prevent axonal caspase activation and axonal degeneration. FASEB J. 2013;27(12):4712–4722. doi: 10.1096/fj.13-229781. [DOI] [PubMed] [Google Scholar]
- 56.Hellal F, et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 2011;331(6019):928–931. doi: 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sengottuvel V, Leibinger M, Pfreimer M, Andreadaki A, Fischer D. Taxol facilitates axon regeneration in the mature CNS. J Neurosci. 2011;31(7):2688–2699. doi: 10.1523/JNEUROSCI.4885-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ertürk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27(34):9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H-J, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10(2):722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 61.Virág L, et al. Garcia Soriano F Diabetic endothelial dysfunction: The role of poly(ADP-ribose) polymerase activation. Nat Med. 2001;7(1):108–113. doi: 10.1038/83241. [DOI] [PubMed] [Google Scholar]
- 62.Charron MJ, Bonner-Weir S. Implicating PARP and NAD+ depletion in type I diabetes. Nat Med. 1999;5(3):269–270. doi: 10.1038/6479. [DOI] [PubMed] [Google Scholar]
- 63.Mouchiroud L, et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 65.Wang B, et al. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30(8):907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 66.von Heideman A, Berglund A, Larsson R, Nygren P. Safety and efficacy of NAD depleting cancer drugs: Results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother Pharmacol. 2010;65(6):1165–1172. doi: 10.1007/s00280-009-1125-3. [DOI] [PubMed] [Google Scholar]
- 67.Galli U, et al. Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J Med Chem. 2013;56(16):6279–6296. doi: 10.1021/jm4001049. [DOI] [PubMed] [Google Scholar]
- 68.Pittelli M, et al. Inhibition of nicotinamide phosphoribosyltransferase: Cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;285(44):34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gierke S, Wittmann T. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr Biol. 2012;22(9):753–762. doi: 10.1016/j.cub.2012.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thoma CR, et al. Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J Cell Biol. 2010;190(6):991–1003. doi: 10.1083/jcb.201006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu KY, et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor AM, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2(8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dull T, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hesterberg T, Monaghan S, Moore D. Bootstrap methods and permutation tests. Introd to Pract Stat. 2005;5:1–70. [Google Scholar]
- 76.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.