Significance
Here we show that both sialylated O-linked sugar T antigen (sTn) and its attached peptide are required for recognition of paired immunoglobulin-like type 2 receptor α (PILRα) by O-glycosylated mucin and related proteins. The crystal structure of PILRα complexed with the O-glycosylated peptide reasonably explains this simultaneous recognition of both the sTn O-glycan and the compact peptide structure, distinct from other sugar-recognition receptors. The structure provides a novel framework for the receptor-binding mode of O-glycosylated mucin proteins, which are relevant in immune responses and viral infection.
Keywords: paired receptor, O-glycosylated protein, X-ray crystallography, immune regulation
Abstract
Paired Ig-like type 2 receptor α (PILRα) recognizes a wide range of O-glycosylated mucin and related proteins to regulate broad immune responses. However, the molecular characteristics of these recognitions are largely unknown. Here we show that sialylated O-linked sugar T antigen (sTn) and its attached peptide region are both required for ligand recognition by PILRα. Furthermore, we determined the crystal structures of PILRα and its complex with an sTn and its attached peptide region. The structures show that PILRα exhibits large conformational change to recognize simultaneously both the sTn O-glycan and the compact peptide structure constrained by proline residues. Binding and functional assays support this binding mode. These findings provide significant insight into the binding motif and molecular mechanism (which is distinct from sugar-recognition receptors) by which O-glycosylated mucin proteins with sTn modifications are recognized in the immune system as well as during viral entry.
Mucin-type O-glycosylation is highly important in physiological events, but it is too complicated to be fully understood because the structures of modified O-linked sugars are basically heterogeneous and are extremely difficult to predict. However, this complexity is beneficial in discriminating cellular conditions. To date, a number of molecules have been shown to recognize O-glycans, which are important in many biological events, including tumorigenesis and immune evasion (1), as well as infectious diseases. Selectins recognize the O-glycan parts of mucin-type glycoproteins on target cell surfaces to regulate the rolling and attachment of lymphocytes in inflammation and in cancer metastasis. However, how O-glycosylated proteins are recognized by specific receptors has remained largely unclear.
To regulate function, the immune system has a fine-tuning mechanism using paired receptors families in which members show opposite (activating and inhibitory) signaling even though their cognate ligand recognitions are essentially similar. It is believed that activating members exhibit lower affinity to ligands than inhibitory members, possibly preventing undesired overactivation of immune responses (2–4) [e.g., killer cell Ig-like receptors, CD94/NKG2 heterodimers, leukocyte Ig-like receptors, and CD28/CTLA-4 (5)]. Paired Ig-like type 2 receptor (PILR) is a member of a paired receptor family comprising activating and inhibitory members designated “PILRβ” and “PILRα,” respectively. PILRs have one Ig-like domain in the extracellular region, which is responsible for the ligand recognition. Our previous reports showed that PILRs recognize the O-glycosylated mucin proteins, mouse CD99 (6), and PILR-associating neural protein (PANP) (7). A recent study identified two O-glycosylated ligands for PILRs, neuronal differentiation and proliferation factor-1 (NPDC1) and collectin-12 (COLEC12), expanding the PILR-recognizing O-glycosylated ligand family (8). All PILR ligands are mucin or have mucin-like domains in which the heavy O-glycosylation occurs and which are significantly important for a wide range of cell–cell interactions. Furthermore, PILRs show unique features for recognizing not only O-linked sugars but also the peptide sequences attached to the sugars.
PILRs are expressed broadly on immune-related cells, including monocytes/macrophages, dendritic cells, B cells, natural killer (NK) cells, and neutrophils, as well as on neurons (6). A recent study showed that PILRs are important for the control of acute Streptococcus aureus infection in the lung in vivo (9). Moreover, we recently demonstrated that in Pilra-deficient mice neutrophil infiltration was increased at the LPS-induced inflammatory sites by increased adhesion to the β2 integrin and that these mice were highly susceptible to endotoxin shock (10). These findings indicate that PILRα is relevant for the regulation of neutrophil recruitment in inflammatory responses, clearly demonstrating the importance of PILRα in innate immune responses. Mouse CD99 is broadly expressed on myeloid lineage cells, and it is believed that PILR recognition of CD99 regulates the immune response. Interestingly, activating PILRβ exhibits lower affinity for CD99 than inhibitory PILRα (11), likely to prevent overactivation of immune responses as suggested above. Similarly to CD99, COLEC12 is expressed on monocytes and vascular endothelial cells, likely involving the immune system. PILRs also bind to PANP and NPDC1, both of which are expressed on neural tissues, enabling PILR-expressing cells either to stimulate or to inhibit their function; this binding indicates that PILRs have a role in neural tissues.
On the other hand, paired receptor families often are targets for recognition by microorganism proteins or products that contribute to infection or to the suppression of the immune system (4), e.g., Ly49H for m157 of murine CMV (12), LILRB1 for human CMV UL18 (13), unknown ligands of Staphylococcus aureus (14), and KIR3DL2 for CpG DNA of bacteria (15), among others. We recently found that human PILRα is an entry receptor for an essential unit, glycoprotein B (gB) of HSV-1 (16, 17). Human PILRα is an inhibitory receptor, and thus its binding to HSV-1 gB could induce immune suppression and subsequently promote HSV-1 immune evasion, even though gB cannot bind to PILRβ at a level that is detectable in infection assays (17).
PILRs can bind to above-mentioned O-glycosylated proteins, expanding PILR ligand family members. In recognizing O-glycan, PILR exhibits three unique characteristics: (i) the requirement for sialylation; (ii) core-1 structure specificity; and (iii) dependency on the peptide sequence attached to O-glycan (8, 18). Here we report that human PILRα recognizes both the sialylated O-linked sugar T antigen (sTn) and its attached peptide region (representatively selected from HSV-1 gB). Furthermore, we show that the O-glycosylated sTn peptide can be used to inhibit PILRα-mediated immune regulation and HSV-1 fusion. Furthermore, we have determined the crystal structures of both the free and complex (attached to the sTn peptide) forms of PILRα. The structures clearly reveal a previously unidentified mode of sugar–peptide–protein interaction that is dependent on both the O-linked sugar and, uniquely, on the peptide, thus simultaneously recognizing both the O-liked sugar and its attached peptide. Taken together, these results provide functional and structural insights on recognition modes to O-glycosylated mucin family including sTn modifications (well-known cancer targets), which are important in immune system as well as HSV-1 entry mechanism.
Results
sTn O-Glycosylated Peptide Is an Essential Unit for PILRα Recognition.
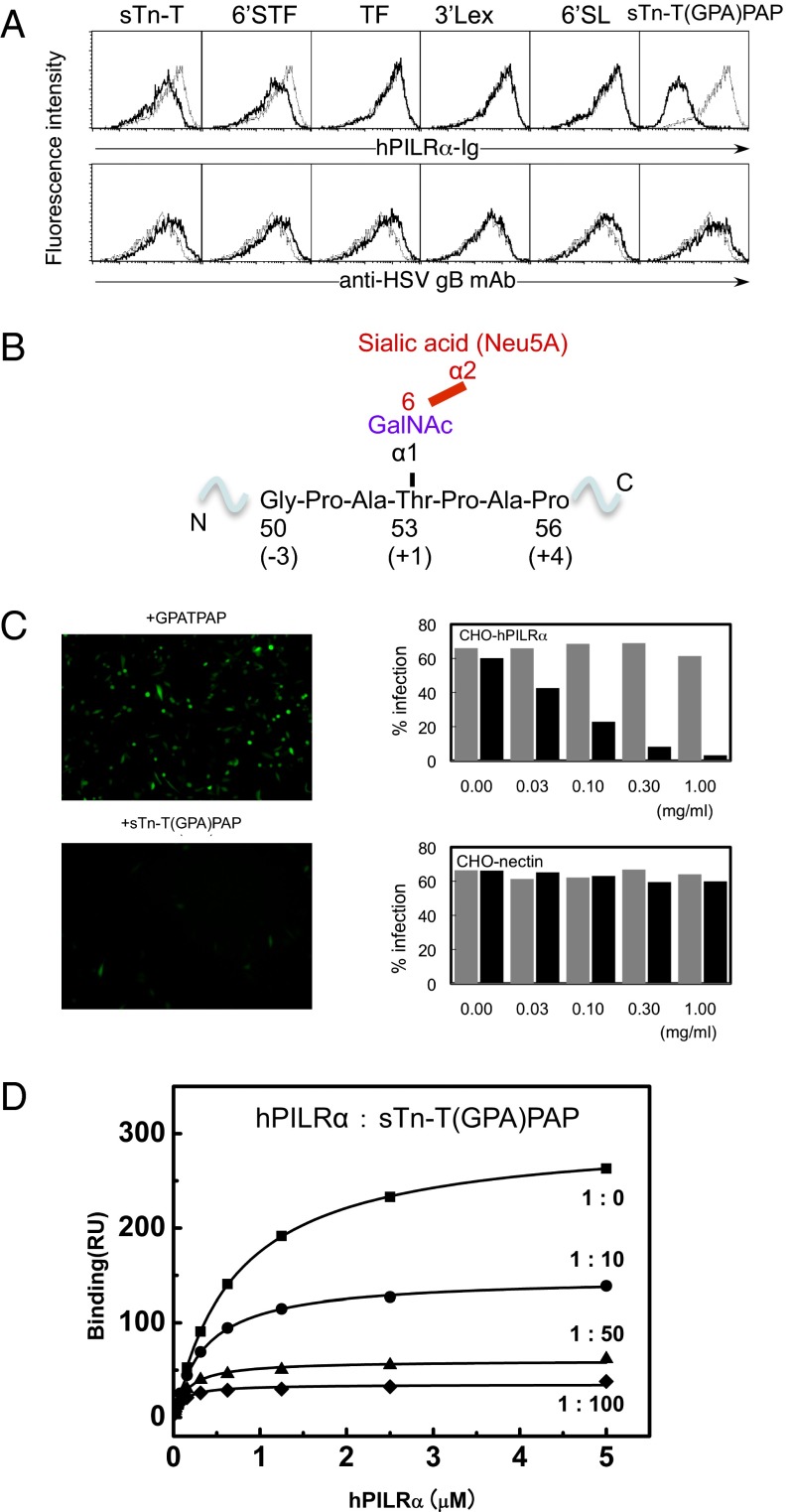
Our and other previous reports found that the O-linked sugar modifications on the ligand proteins are essential for PILRα recognition (6–8, 17). Here we examine the molecular mechanism of such recognition. First, we used the sugar array system of the Consortium for Functional Glycomics (CFG) to clarify the precise structure of the sugar ligand necessary for PILRα binding and clearly showed that sugars in general do not exhibit a positive response (www.functionalglycomics.org/glycomics/HServlet?operation=view&sideMenu=no&psId=primscreen_2977). In the present study, we selected HSV-1 gB as a representative ligand of PILRα. We performed a competition assay in which PILRα was allowed to bind to gB-expressing HEK293 cells in the presence of a set of sugar compounds. Fig. 1A shows that the core 1 O-linked sugar moiety, sTn-T (a sialyl T antigen, Neu5Acα2–6GalNAcα-O-Thr), and its derivative, 6′STF [Galβ1–3(Neu5Acα2–6)GalNAcα-O-Thr] competed slightly with PILRα binding, indicating that sTn is involved in PILRα binding (the galactose extension does not contribute to the binding). Sun et al. (8) also showed that only a very high concentration (125 mM) of 6′-sialyllactose containing sialic acid in α2–6 linkage could inhibit the PILRα–hNPDC1 interaction, but 3′-sialyllactose containing sialic acid in α2–3 linkage did not. These findings suggest that the PILRα binds to a complex structure involving both sTn and peptides.
Fig. 1.
Identification of the region in HSV-1 gB that is required for PILRα recognition. (A) Competition assay with various sugar compounds assessing the ability of the PILRα-Ig fusion protein to bind to HEK293 cells expressing the HSV-1 gB molecule. The black and gray lines indicate binding in the presence or absence of additional sugar compounds (final concentration, 0.5 mg/mL), respectively. sTn-T, Neu5Acα2–6GalNAcα-O-Thr; 6′STF, Galβ1–3[Neu5Acα2–6]GalNAcα-O-Thr; TF, Galβ1–3GalNAcα-O-Thr; 3′SLex, Neu5Acα2–6Galβ1–4[Fucα1–3]GalNAcβ-Sp; 6′SL, Neu5Acα2–6Galβ1–4Glcβ-Sp. (B) The detailed structure surrounding Thr53 of gB with an O-linked sugar and the region that represents the sTn-T(GPA)PAP peptide. (C) Effects of sTn-T(GPA)PAP or GPATPAP on HSV-1 infection. (Left) Representative images. (Right) Dose-dependent effects of sTn-T(GPA)PAP (black) or GPATPAP (gray) on the infection efficiency in CHO cells expressing PILRα or nectin-1. (D) SPR competition assay examining the ability of the sTn-T(GPA)PAP glycopeptide to inhibit PILRα binding to HSV-1 gB. Closed squares, PILRα only; closed circles, PILRα:sTn peptide, 1:10; closed triangles, PILRα:sTn peptide, 1:50; closed diamonds, PILRα:sTn peptide, 1:100.
The N-terminal region of gB including O-glycosylated Thr53, which is essential for PILRα binding, is rich in glycines, prolines, and serines and thus is a flexible domain [as indicated by this region’s not being visible in the crystal structure (19)]. Furthermore, the O-linked sugar modifications generally occur on flexible regions, e.g., of mucins and related molecules. Thus, we designed and attached a sialylated O-linked sugar to a specific gB peptide (residues 50–56), NH2-Gly-3Pro-2Ala-1Thr+1(sTn)Pro+2Ala+3Pro+4-CO2H [hereafter designated “sTn-T(GPA)PAP)”] (Fig. 1B). sTn-T(GPA)PAP competed with PILRα binding to gB-expressing HEK293 cells much more significantly than did either sTn-T or 6′STF (Fig. 1A) and inhibited HSV-1 infection of CHO cells expressing PILRα, whereas the GPATPAP peptide did not inhibit infection (Fig. 1C). This glycopeptide also inhibited PILRα binding to gB in an in vitro assay using surface plasmon resonance (SPR) (Fig. 1D). Therefore, the sTn-T(GPA)PAP glycopeptide represents the essential unit required for PILRα recognition, indicating that PILRα has the unique ability to see the molecular features of both the O-linked sugar and the surrounding peptide.
Structures of PILRα and Its Complex with sTn-Modified Peptide.
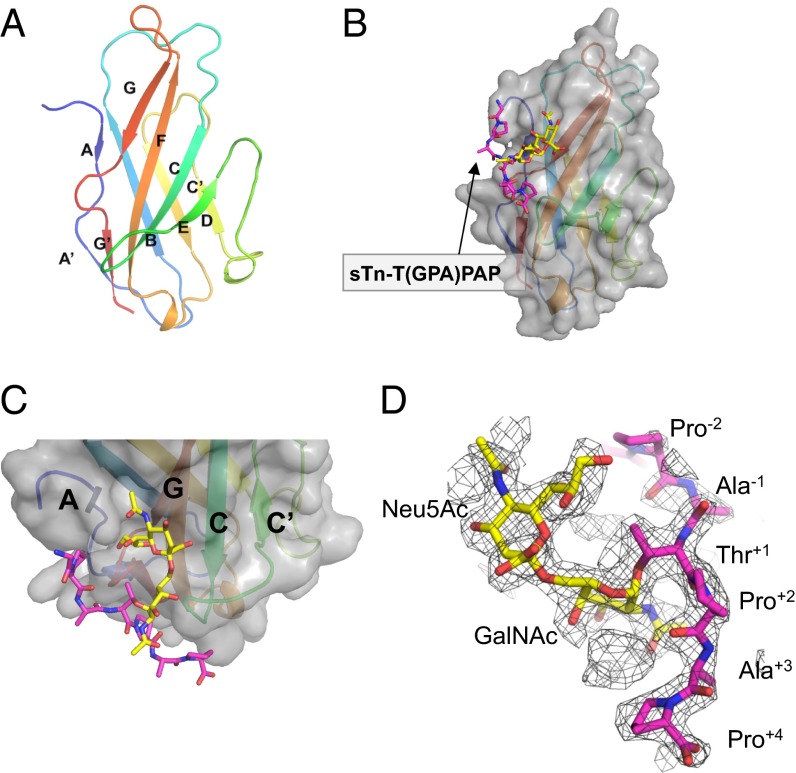
To understand the structural elements that are involved in the PILRα interaction with the sTn O-glycosylated peptide, we successfully produced and crystallized the free human PILRα protein comprising one Ig-like domain (residues 13−131) (20) (Fig. S1A). We could determine the crystal structure of free PILRα at 1.3-Å resolution by using the single anomalous dispersion (SAD) method of iodide derivate crystals (Fig. 2A; see also Table S1 and Fig. S2). The PILRα ectodomain harbored a V-set Ig-like β-sandwich fold (Fig. 2A). A search of the DALI database identified sialic acid-binding Ig-like lectin (Siglec)-1/sialoadhesin (21) as the structure closest to PILRα (rmsd = 1.6 Å for 97 Cα atoms; Protein Data Bank ID: 1URL). The structure and topology of PILRα were similar to structures reported for members of the Siglec family (Siglec-1, -5, and -7) (21–23), which consists of 14 Siglecs in human, even though the sequence similarity among them is very low (about 22%).
Fig. 2.
Structures of PILRα and its sTn peptide complex. (A) Cartoon representation of the overall structure of the free human PILRα protein. The secondary structural elements are rainbow colored with the N terminus in blue and the C terminus in red. (B) Surface representation of the PILRα-sTn peptide complex. The sialylated O-linked sugar and surrounding peptide are shown as sticks and are colored yellow and magenta, respectively. The same presentation is used in C and D. (C) Close-up view of the sTn-binding region. PILRα is shown as a cartoon model with the surface presentation and coloring as in A. (D) Composite omit map around the sTn peptide (mesh in 1.2σ cutoff).
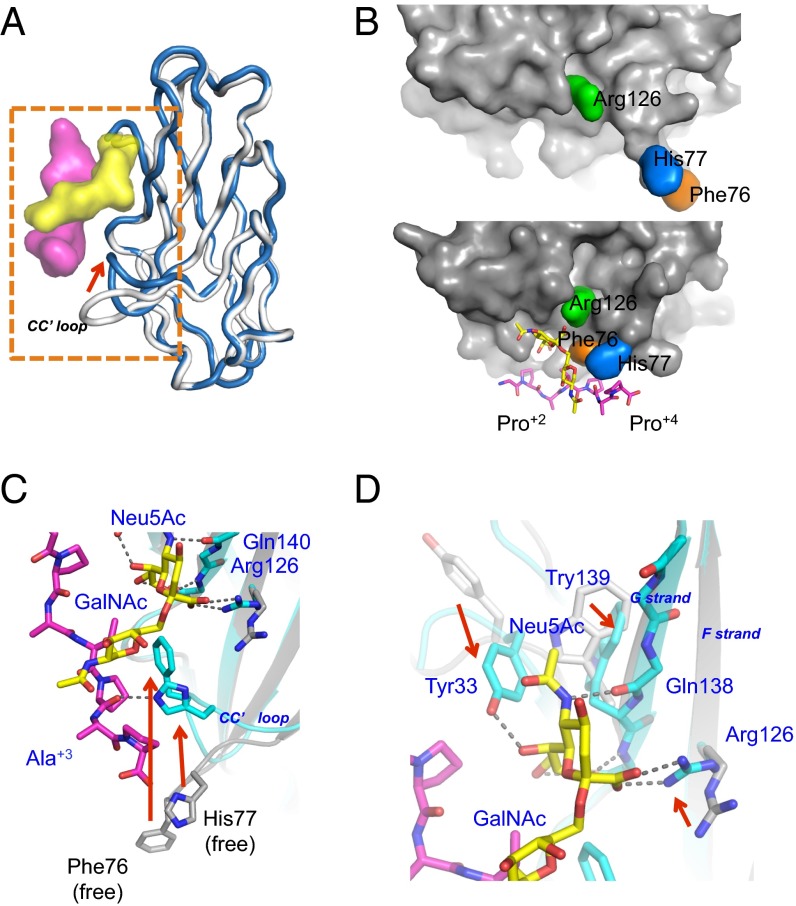
Next, we successfully crystallized and determined the structure of PILRα complexed with sTn-T(GPA)PAP (Figs. 2 B and C and 3, Figs. S3 and S4, and Table S1). The overall structure of the complex was similar to the free form (Fig. 3A), and the composite omit map of electron densities for the sTn peptide was clearly identified (Fig. 2D). However, PILRα unexpectedly underwent large structural rearrangements to bind both the sugar moiety and amino acids simultaneously within this sTn–O-glycosylated peptide (Fig. 3 and Fig. S4A). The buried surface upon the complex formation was 1,940 Å2, indicating a relatively large binding interface despite the low molecular weight of the compound. The O-glycosylated peptide was positioned on the AGF sheet as well as at the CC′ loop (Fig. 2C). The N-terminal peptide region (GPA) was not largely involved in this interaction, but the C-terminal site (PAP) wrapped around the CC′ loop, and Pro+2 had an essential role in this interaction (Figs. 3B and 4). This conformation is consistent with the observation that the amino acid sequences of C-terminal residues (sTn-TPxP or sTn-TPxxP) are conserved among the determined or predicted recognition sites of PILRα ligands (Table 1).
Fig. 3.
Detailed views and mutagenesis studies of the PILRα–sTn peptide interaction. (A) Superposition of the PILRα–sTn peptide complex (dark blue) with the free form of PILRα (gray). The red arrows indicate the structural rearrangement of the CC′ loop upon complex formation. The sTn-T(GPA)PAP peptide is shown as a surface model colored as in Fig. 2. (B) Surface presentation around the binding interface of the PILRα protein in the complexed (Upper) and free (Lower) structures. (C and D) Detailed views of the large structural rearrangements in the free (gray, stick and cartoon) and sTn peptide-complexed (cyan, stick and cartoon) forms of PILRα. The glycopeptides are colored yellow (sugar) and magenta (peptide), respectively.
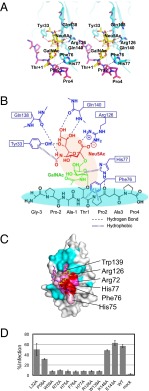
Fig. 4.
A detailed view of simultaneous recognition by PILRα of the O-glycan and peptide region of the sTn peptide and functional assays with the mutant PILRα proteins. (A) Stereoview of the interactions between PILRα and sTn peptide. (B) Schematic representation of the simultaneous recognition by PILRα of the O-glycan and the attached peptide of the sTn peptide. Sialic acid (red), GalNAc (green), the peptide region (black), and PILRα residues (blue) are shown. (C) gB binding with wild-type and mutant PILRα proteins. In the SPR study, the ligand was gB, and the analytes were the PILRα proteins. Significant residues that showed no (red) or reduced (pink) binding affinity based on the structure of the PILRα-sTn peptide complex are mapped. Cyan indicates residues in which mutations did not affect gB binding. (D) The bar graph shows the infectious activity of HSV-1 in cells expressing wild-type or mutant PILRα.
Table 1.
Comparison of the peptide sequence around the O-glycosylated Thr+1 of PILRα ligands
| Amino acid position |
||||||||||
| Ligand | −3 | −2 | −1 | +1 | +2 | +3 | +4 | +5 | +6 | Kd, μM |
| HSV-1 gB (53-61) | G | P | A | T | P | A | P | P | A | 2.3* |
| mCD99 Thr45 (42-50) | M | K | P | T | P | K | A | P | T | 2.8† |
| mCD99 Thr50 (47-55) | K | A | P | T | P | K | K | P | S | 3.3† |
| PAMP§ | G | L | A | T | P | H | P | N | S | Not measured |
| NPDC1§ | L | P | S | T | P | G | T | P | T | 0.049‡ |
| COLEC12§ | N | E | P | T | P | A | P | E | D | 1.1‡ |
The N-terminal region (30–108) of gB was used for the binding study.
The single Ala mutation (T45 or T50) did not cause any reduction of PILRα binding compared with mCD99 wild type (Kd = 2.2 μM) (11).
These affinity contained the avidity effect of Fc-fused dimerization (8).
These are the predicted O-glycosylation sites of each PILRα ligand.
Recognition of O-glycosylated sTn Peptide.
The sialylation of the O-linked sugar attached to the PILRα ligands is necessary for PILRα binding and HSV-1 infection (8, 17, 24), and the designed sialylated O-glycosylated peptide was sufficient for the binding, as described above. The carboxyl group of the sialic acid (Neu5Ac) residue interacted directly with Arg126 (Figs. 3 B–D and 4A, Fig. S4C, and Table S2). This interaction resembled the sialic acid recognition of a conserved arginine, called “essential R,” of the Siglec family (21–23), even though these proteins have low amino acid sequence conservation. On the other hand, there is a hydrogen-bonding network among Neu5Ac and Tyr33, Gln138, and Gln140 (Figs. 3 C and D and 4A and Table S2). Within the site that binds to the hydrophobic face of the ring and the glycerol portion of the Neu5Ac residue, the aromatic rings of Tyr33 and Trp139 were largely flipped (Fig. 3D). An Arg126Ala or Trp139Ala mutation significantly reduced binding (Fig. 4C and Table S3) and infection (Fig. 4D and Fig. S5). Sun et al. (8) also showed that the Arg126Ala mutant failed to interact with hNPDC1, mCD99, hCOLEC12, and HSV-1 gB. Furthermore, Trp139Leu mutation significantly impaired cell-fusion activity for HSV-1 and binding to gB (25). PILRα has Leu139 and does not act as a HSV-1 gB receptor. Taken together, these interactions with the Neu5Ac residue, distinct from Siglec family members (Fig. S4B), contribute significantly to the overall binding affinity.
Recognition of the Peptide Region.
Unexpectedly, the complex formation resulted in remarkable structural rearrangements within a wide area of the sTn peptide-binding interface. The CC′ loop as well as the continuous C′ strand (residues 74−80) in PILRα were largely changed as the result of direct interactions with both the GalNAc residue and the C-terminal P+2A+3P+4 peptide region (Figs. 3 B–D and 4A), whereas the conformation of the CC′ loop in the free form seemed to be somewhat stabilized by crystal packing. The GalNAc residue alone made a hydrophobic interaction with the aromatic ring of His77 in this CC′ loop (Fig. S4C), as is typical in protein–sugar interactions (26, 27).
Although the N-terminal side of the sTn peptide interacted with PILRα only minimally [only the pyrolidine ring of Pro−2 (sTn) is located close to Met31 and Ile142], the bound C-terminal region of the main-chain structure had a compact configuration. Phe76 and His77 had van der Waals contacts with the hydrophobic pyrolidine rings of two proline residues in the C-terminal part of the sTn peptide (Figs. 3B and 4A, Fig. S4C, and Table S2). His77 also formed a hydrogen bond with the carboxyl group of Pro+2 within the peptide, supporting the kinked main-chain orientation in the P+2A+3P+4 region. Furthermore, the connection between the sTn sugar moiety and the peptide region also was kinked to form the compact structure. This kinking was underpinned by (i) a hydrophobic contact between the aromatic ring of Phe76 and the hydrophobic faces of Thr+1 and Pro+2 (sTn) and (ii) interactions between the N atom of the N-acetyl group of the GalNAc residue and two backbone carboxyl groups of Thr+1 and Pro+2 in the peptide. Notably, one of the latter interactions, GalNAc(N)-Thr+1(O), also was observed in the preferential state of the GalNAc–O-glycosylated IgA hinge peptide by NMR (28) and further facilitated the trans configuration of the adjacent proline residue corresponding to Pro+2 of the sTn peptide (Fig. S4C). Therefore, PILRα selectively captures the major conformation of the free form of the sTn peptide. A PILRα mutant harboring Phe76Ala or His77Ala mutations completely lost the ability to bind to gB (Fig. 4C, Fig. S1C, and Table S3) and no longer facilitated HSV-1 infection (Fig. 4D and Fig. S5). These mutations had remarkable effects that were comparable to that of Arg126, a determinant of sialic acid recognition, confirming that the PILRα–sTn–peptide interaction requires the recognition of both sugar and peptide as well as the proper configuration.
Discussion
Our structural analysis reveals the molecular details of the recognition not simply of O-glycan sugars but also of the O-glycosylated part of proteins. The PILRα residues that contact the sTn, representative of cancer targets, as well as the Pro-rich peptide are essential to form the characteristic compact conformation of the sTn peptide. Importantly, the ligand specificity of PILRα appeared to be determined by the amino acid sequence. The N terminus seemed to be hardly involved in the structure, but the C-terminal parts, Pro+2 and Pro+4 and/or Pro+5, are conserved among the known PILRα ligands (Table 1). Further binding and structural analyses using variants of sTn O-glycosylated peptides are required to evaluate the consensus motifs, including the adjacent residues of conserved proline residues for PILRα binding. As described above, PILRα adopts a binding structure for this preferential conformation of the O-glycosylated peptide. The sequence similarity between PILRα and Siglec family members is rather low; however, their recognition of the sialic acid is similar. The distinct feature of PILRα is the large structural change of the CC′ loop upon the ligand binding; the CC′ loops of Siglec members use the additional sugar interaction instead (Fig. S4B). This unique peptide recognition determines the preference for expanding mucin-type PILR ligands. PILR finely senses the cellular condition by checking the combination of sugar modification and peptide sequence.
Somers et al. (29) reported the crystal structure of P-selectin complexed with P-selectin glycoprotein ligand 1 (PSGL-1), a mucin-like homodimeric glycoprotein. The structure revealed that P-selectin recognizes both the sialyl Lewisx (SLex) attached to Thr57 and the tyrosine sulfate residues (Tyr48 and Tyr51) of PSGL-1 (29), but the interaction is distinct from that of the PILRα–glycopeptide complex. C-type lectin-like receptor-2 (CLEC-2) similarly showed peptide-dependent binding to the sialylated O-glycosylated region of the platelet aggregation-inducing factor, the mucin-type sialoglycoprotein podoplanin, which contributes to the regulation of tumor cell-induced platelet aggregation and tumor metastasis (30, 31). Future study of the mechanisms by which CLEC-2 recognizes sialylated O-glycopeptides might show which structures of the O-glycosylated targets are important for receptor recognition.
The number of PILR ligand members is expanding. PILRα also can bind to human peripheral blood mononuclear cells, although the ligands have not been not identified (8). Furthermore, Pilrb-knockout mice and mice administered the agonist anti-PILRα mAb exhibited resistance to S. aureus infection, whereas the wild-type mice could not clear the infection (9). Although PILRα/β ligands on S. aureus are not yet known, some O-linked sugar-modified peptide/proteins or related molecules might be ligands. As described above, the in vivo importance of PILR is reported in integrin-mediated inflammation and in viral and bacterial infection (10). It is likely that unknown self and nonself ligands form a large PILR-recognizing ligand family. Future experiments involving ligand fishing and posttranslational modification (O-glycosylation) based on the peptide motif of PILR ligands revealed by the present structural data will provide further insights on a wide range of physical regulations involving PILR. In turn, the inhibitors of PILR likely can be used for the diagnosis and medical treatment of both immune-related and infectious diseases.
HSV-1 is extremely difficult to eliminate completely, and dampened immune responses can induce herpes recurrence (32). HSV-1 evolved to select PILRα as a gB receptor, contributing to viral entry, as well as to the suppression of broad immune responses. The gB-PILRα association is important for HSV-1 to infect PILR-expressing cells, such as monocytes, directly and is required for efficient pathological infection (17, 33, 34). Importantly, the present data show that the N-terminal flexible site is a prerequisite for entry and that PILRα interacts with both the sialylated O-glycan and the surrounding protein structure. As described above, the sTn peptide definitely inhibited the HSV-1 entry and thus can serve an initial frame for a potential therapeutic compound. The targeting host receptor is beneficial for preventing mutation risk, unlike the targeted molecules of pathogens that can be mutated to evade immune responses. The short-term administration of an inhibitory compound for PILRα likely will have only a small effect on the immune system, because the Pilr-knockout mice demonstrate some effect on neutrophil function but do not exhibit any harmful effects. Therefore, the present structure may contribute to the rational design of antiviral drugs.
Materials and Methods
Sample Preparation and Crystallization.
Sample preparation for the Ig V-set domain in the extracellular region of human PILRα was described previously (11). Briefly, the gene encoding the Ig V-set domain of PILRα (residues 13–131) was cloned into the plasmid vector pGMT7 (11). For the mutagenesis studies, alanine mutations were introduced by PCR. The resulting plasmid was transformed into Escherichia coli Rosetta (DE3), and the protein was overexpressed as inclusion bodies. Then, the inclusion bodies were solubilized and refolded by a standard dilution method. The refolded PILRα was purified by gel filtration (Fig. S1) followed by Resource S column chromatography (GE Healthcare). The sTn-attached HSV-1 gB peptide, NH2-GPAT(sTn)PAP-CO2H, was synthesized chemically by Sussex Research.
As described previously (20), crystals of the free form of PILRα were grown by the sitting-drop method at 20 °C by mixing 6 mg/mL of the PILRα protein with Classics Suite (QIAGEN) No. 7 [0.1 M tri-Na citrate (pH 5.6), 20% isopropanol, 20% PEG 4000]. For the PILRα–sTn peptide complex, initial crystallization trials were performed with Crystal Screens 1 and 2 (Hampton Research) and the Classics Suite on Intelli-Plates using the automatic Mosquito crystallization-setup robot (TTP Labtech). The drop was a mixture of 0.2 μL of a 1:5 PILRα:sTn peptide complex solution [20 mM succinate (pH 5.0), 100 mM NaCl] and 0.2 μL of reservoir solution, and the crystallization plates included 90 μL of reservoir buffer at 293 K. Crystals of the PILRα complex were obtained using Crystal Screen 1 solution No.30 (0.2 M ammonium sulfate, 30% wt/vol, PEG 8,000).
Data Collection and Structure Determination.
The crystals of the free form of PILRα belonged to the space group P212121 with unit cell dimensions of a = 40.33 Å, b = 44.94 Å, c = 56.87 Å and contained one molecule per asymmetric unit. A diffraction data set was collected at 100 K, at beamline BL41XU of SPring8 (λ = 0.9 Å). To determine the experimental phases, the iodide-anion derivative crystals were made by soaking in cryoprotectant solution including 1 M KI for 30 s (20). The 1.8-Å SAD dataset could be collected at 100 K at beamline BL17A of the Photon Factory. The diffraction data were processed and scaled using the HKL2000 program package (35). Five iodine sites in the asymmetric unit were assigned and used for the phase calculation with the autoSHARP package (36), resulting in the initial mean figure-of-merit of 0.574 for acentric reflections and of 0.213 for centric reflections. We improved the phases using the program DM (37). The initial model was built by ARP/wARP (38), and refinement was done by Refmac5 (37). The final model showed an Rfree factor of 17.7% and an R factor of 14.4%. Detailed statistics are summarized in Table S1.
We also prepared a solution of the complex of PILRα with the synthetic O-linked sugar-connecting peptide ligand, NH2-GPAT(sTn)PAP-CO2H. The crystals of the PILRα-sTn peptide complex belonged to the space group C2221 with unit cell dimensions of a = 50.27 Å, b = 155.80 Å, and c = 67.31 Å and contained two complexes per asymmetric unit. The structure was phased by molecular replacement using Molrep in the CCP4 package (37) with the free PILRα structure as a search probe. The two PILRα molecules (chains A and B) were found by using reflections in the range of 50.0–2.3 Å. Further crystallographic refinement was carried out with Refmac and Crystallography and NMR System (39) alternated with manual rebuilding in the interactive graphics program COOT. The final model, which comprises two complexes, was refined to an Rfree factor of 29.9% and an R factor of 23.1%. Detailed crystallographic statistics are shown in Table S1. A Ramachandran plot of backbone angles gave 94.7% and 5.3% in the most favored and additionally allowed regions, respectively. The Ramachandran plot was calculated by RAMPAGE (40). Figures were generated using PyMOL (http://pymol.sourceforge.net).
Production of Soluble HSV-1 gB-Ig Fusion Proteins.
The DNA encoding either the N-terminal (30–108) or C-terminal (429–528) region of the HSV-gB ectodomain was ligated using the pME18S expression vector containing the Fc fragment of human IgG1 at the C terminus, as described previously (16). The vector DNA was transfected into HEK293T cells using polyethyleneimine for transient expression. The supernatants were collected 4 d after the transfection, and the Ig-fused gB was purified with a protein A affinity column. Human PILRα-Ig fusion protein was prepared as described previously (17).
Binding Analysis Using SPR.
The wild-type and alanine-scanning mutants of PILRα were dissolved in HBS-EP buffer [10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% Surfactant P20]. SPR experiments were performed with a Biacore 3000 system (GE Healthcare). The Ig-fused N-terminal region of gB was immobilized on the CM5 sensor chip (GE Healthcare) by amine-coupling using the standard amine coupling kit (about 3,000 response units). BSA was used as a negative control protein. All PILRα samples were injected at a flow rate of 10 μL/min. The data were analyzed using BIA evaluation version 4.1 (GE Healthcare) and ORIGIN version 7 software. Affinity constants (Kd) were derived by nonlinear curve fitting of the standard Langmuir binding isotherm.
For the competition assay, a range of concentrations of PILRα (5 μM, 2.5 μM, 1.25 μM, 0.63 μM, 0.31 μM, 0.16 μM, 0.08 μM, 0.04 μM, 0.02 μM, and 0.01 μM) mixed with the sTn peptide as indicated in the inset (Fig. 1D) was injected over the Ig-fused gB.
Binding Assays.
pPEP98-gB (16) was transiently transfected into HEK293 cells. Then 48 h after transfection, cells were stained by PILRα-Ig fusion protein or by anti-HSV gB mAb (1105) (final concentration 5 μg/mL) with or without sugar compounds (final concentration 0.5 mg/mL), and the transfectants were analyzed by flow cytometry.
Infection Assays.
CHO cells stably expressing PILRα or human nectin-1 (16) were infected with HSV-GFP at a multiplicity of infection (MOI) of 10, and the infected cells were analyzed by flow cytometry or fluorescence microscopy (Carl Zeiss) after 12 h. In some experiments, CHO cells were transiently transfected with wild-type or mutant PILRα. The transfectants were infected with HSV-GFP at an MOI of 5 48 h after transfection. Infected cells were analyzed by flow cytometry 20 h after infection. The percentage of GFP-positive cells represented the percentage of infection.
Reagents and Antibodies.
Mouse monoclonal antibody against human PILRα was prepared as described previously (16). Mouse monoclonal antibody against gB (1105) was purchased from Goodwin Institute. The peptide GPATPAP was synthesized by GeneDesign. Neu5Aca2-6GalNAca-PAA-biotin was purchased from GlycoTech. The other carbohydrate complexes were provided by the Consortium for Functional Glycomics (www.functionalglycomics.org).
Supplementary Material
Acknowledgments
We thank the beamline staff at Photon Factory and SPring8 for technical help during data collection and Y. Kawaguchi, H. Aramaki, K. Tokunaga, I. Tanaka, K. Inaba, T. Oka, and K. Mihara for helpful discussions. This work was supported in part by Platform for Drug Discovery, Informatics, and Structural Life Science and other grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan. K.K. and J.W. are supported by Japan Society for the Promotion of Science Research Fellowships for Young Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3WUZ and 3WV0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324105111/-/DCSupplemental.
References
- 1.Tarp MA, Clausen H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta. 2008;1780(3):546–563. doi: 10.1016/j.bbagen.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290(5489):84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 3.Daëron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: A quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 4.Kuroki K, Furukawa A, Maenaka K. Molecular recognition of paired receptors in the immune system. Front Microbiol. 2012;3:429. doi: 10.3389/fmicb.2012.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanier LL. Face off—the interplay between activating and inhibitory immune receptors. Curr Opin Immunol. 2001;13(3):326–331. doi: 10.1016/s0952-7915(00)00222-3. [DOI] [PubMed] [Google Scholar]
- 6.Shiratori I, Ogasawara K, Saito T, Lanier LL, Arase H. Activation of natural killer cells and dendritic cells upon recognition of a novel CD99-like ligand by paired immunoglobulin-like type 2 receptor. J Exp Med. 2004;199(4):525–533. doi: 10.1084/jem.20031885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogure A, Shiratori I, Wang J, Lanier LL, Arase H. PANP is a novel O-glycosylated PILRα ligand expressed in neural tissues. Biochem Biophys Res Commun. 2011;405(3):428–433. doi: 10.1016/j.bbrc.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, et al. Evolutionarily conserved paired immunoglobulin-like receptor α (PILRα) domain mediates its interaction with diverse sialylated ligands. J Biol Chem. 2012;287(19):15837–15850. doi: 10.1074/jbc.M111.286633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee A, et al. Modulation of paired immunoglobulin-like type 2 receptor signaling alters the host response to Staphylococcus aureus-induced pneumonia. Infect Immun. 2010;78(3):1353–1363. doi: 10.1128/IAI.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Shiratori I, Uehori J, Ikawa M, Arase H. Neutrophil infiltration during inflammation is regulated by PILRα via modulation of integrin activation. Nat Immunol. 2013;14(1):34–40. doi: 10.1038/ni.2456. [DOI] [PubMed] [Google Scholar]
- 11.Tabata S, et al. Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J Biol Chem. 2008;283(14):8893–8901. doi: 10.1074/jbc.M709793200. [DOI] [PubMed] [Google Scholar]
- 12.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 13.Cosman D, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7(2):273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama M, et al. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178(7):4250–4259. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- 15.Sivori S, et al. A novel KIR-associated function: Evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. 2010;116(10):1637–1647. doi: 10.1182/blood-2009-12-256586. [DOI] [PubMed] [Google Scholar]
- 16.Satoh T, et al. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132(6):935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. Binding of herpes simplex virus glycoprotein B (gB) to paired immunoglobulin-like type 2 receptor α depends on specific sialylated O-linked glycans on gB. J Virol. 2009;83(24):13042–13045. doi: 10.1128/JVI.00792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Shiratori I, Satoh T, Lanier LL, Arase H. An essential role of sialylated O-linked sugar chains in the recognition of mouse CD99 by paired Ig-like type 2 receptor (PILR) J Immunol. 2008;180(3):1686–1693. doi: 10.4049/jimmunol.180.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313(5784):217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 20.Tabata S, et al. Expression, crystallization and preliminary X-ray diffraction analysis of human paired Ig-like type 2 receptor α (PILRα) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 1):44–46. doi: 10.1107/S1744309107065384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May AP, Robinson RC, Vinson M, Crocker PR, Jones EY. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol Cell. 1998;1(5):719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 22.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446(7139):1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 23.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 24.Teuton JR, Brandt CR. Sialic acid on herpes simplex virus type 1 envelope glycoproteins is required for efficient infection of cells. J Virol. 2007;81(8):3731–3739. doi: 10.1128/JVI.02250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Q, Longnecker R. The Ig-like v-type domain of paired Ig-like type 2 receptor alpha is critical for herpes simplex virus type 1-mediated membrane fusion. J Virol. 2010;84(17):8664–8672. doi: 10.1128/JVI.01039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quiocho FA. Carbohydrate-binding proteins: Tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- 27.Quiocho FA. Molecular features and basic understanding of protein-carbohydrate interactions: The arabinose-binding protein-sugar complex. Curr Top Microbiol Immunol. 1988;139:135–148. doi: 10.1007/978-3-642-46641-0_5. [DOI] [PubMed] [Google Scholar]
- 28.Narimatsu Y, et al. Effect of glycosylation on cis/trans isomerization of prolines in IgA1-hinge peptide. J Am Chem Soc. 2010;132(16):5548–5549. doi: 10.1021/ja9106429. [DOI] [PubMed] [Google Scholar]
- 29.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103(3):467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki-Inoue K, et al. Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. J Biol Chem. 2007;282(36):25993–26001. doi: 10.1074/jbc.M702327200. [DOI] [PubMed] [Google Scholar]
- 31.Kato Y, et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci. 2008;99(1):54–61. doi: 10.1111/j.1349-7006.2007.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet. 2001;357(9267):1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- 33.Arii J, et al. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor α. J Virol. 2009;83(9):4520–4527. doi: 10.1128/JVI.02601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arii J, et al. A single-amino-acid substitution in herpes simplex virus 1 envelope glycoprotein B at a site required for binding to the paired immunoglobulin-like type 2 receptor alpha (PILRalpha) abrogates PILRα-dependent viral entry and reduces pathogenesis. J Virol. 2010;84(20):10773–10783. doi: 10.1128/JVI.01166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in Oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12):2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, et al. Crystallography & NMR System (CNS), a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Lovell SC, et al. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.