Significance
The mitochondrial uniporter is a highly selective calcium channel found in many diverse eukaryotes, but absent in the yeast Saccharomyces cerevisiae. Although the uniporter’s existence was recognized more than 50 y ago, its molecular components have been identified only recently. Here we use yeast as a facile reconstitution system to identify the minimal components sufficient for in vivo uniporter activity. We describe the simplified calcium uniporter of slime mold, consisting of one transmembrane component, DdMCU, which alone is sufficient for robust calcium uptake in yeast mitochondria. Intriguingly, the human uniporter requires two proteins, MCU and the animal-specific protein EMRE, that together are sufficient for uniporter activity. Our work provides a powerful reconstitution system for studying the evolution and function of this channel.
Abstract
The mitochondrial calcium uniporter is a highly selective calcium channel distributed broadly across eukaryotes but absent in the yeast Saccharomyces cerevisiae. The molecular components of the human uniporter holocomplex (uniplex) have been identified recently. The uniplex consists of three membrane-spanning subunits –mitochondrial calcium uniporter (MCU), its paralog MCUb, and essential MCU regulator (EMRE)– and two soluble regulatory components–MICU1 and its paralog MICU2. The minimal components sufficient for in vivo uniporter activity are unknown. Here we consider Dictyostelium discoideum (Dd), a member of the Amoebazoa outgroup of Metazoa and Fungi, and show that it has a highly simplified uniporter machinery. We show that D. discoideum mitochondria exhibit membrane potential-dependent calcium uptake compatible with uniporter activity, and also that expression of DdMCU complements the mitochondrial calcium uptake defect in human cells lacking MCU or EMRE. Moreover, expression of DdMCU in yeast alone is sufficient to reconstitute mitochondrial calcium uniporter activity. Having established yeast as an in vivo reconstitution system, we then reconstituted the human uniporter. We show that coexpression of MCU and EMRE is sufficient for uniporter activity, whereas expression of MCU alone is insufficient. Our work establishes yeast as a powerful in vivo reconstitution system for the uniporter. Using this system, we confirm that MCU is the pore-forming subunit, define the minimal genetic elements sufficient for metazoan and nonmetazoan uniporter activity, and provide valuable insight into the evolution of the uniporter machinery.
Mitochondria from all vertebrates tested possess a high-capacity calcium uptake mechanism called the uniporter (1, 2). The uniporter resides within the mitochondrial inner membrane and is classically defined based on its dependence on membrane potential and sensitivity to ruthenium red and Ru360. Whole mitoplast patch-clamp studies have indicated that the uniporter is a highly selective calcium channel that produces remarkably high current densities (3). Although its physiology has been studied extensively for over five decades, the uniporter’s molecular identity remained elusive until our group introduced a comparative genomics strategy to reveal its first molecular component (4). Key to identifying this component was the observation that uniporter activity is found across all vertebrate mitochondria, and is even documented in trypanosomes, yet is absent in the yeast Saccharomyces cerevisiae (5). We combined this taxonomic profile with comparative genomics (4) to search for mitochondrial proteins (6) that are conserved in vertebrates and kinetoplastids but absent in yeast. This approach led to the identification of mitochondrial calcium uptake 1 and 2 (MICU1/2), the calcium sensing regulatory components of the uniporter (4, 7–10), which then facilitated identification of the mitochondrial calcium uniporter (MCU), the putative pore-forming subunit (11–13).
Recent studies have begun to achieve a full molecular characterization of the uniplex, and the function of each component is emerging. We recently characterized the human uniporter holocomplex (uniplex) and reported that it consists of MCU, MCUb, MICU1/2, and a previously uncharacterized protein that we termed the essential MCU regulator (EMRE) (14). Electrophysiological characterization of HEK-293T cell mitochondria with human MCU (HsMCU) knockdown or ruthenium red-resistant MCU overexpression suggests that MCU is the pore-forming subunit (13). Although human MCU alone is apparently sufficient for conductance in lipid bilayers (12), in human mitoplasts, both HsMCU and HsEMRE are required for the electrophysiologically defined uniporter current, IMiCa (14). However, EMRE is metazoan-specific, without any obvious homologs in other organisms with uniporter activity encoding MCU (and MICU) homologs (14). Thus, the identity of the minimal components required for uniporter activity has remained a pressing issue in the field.
Because yeast does not have uniporter activity and its genome has no homologs of uniporter components, it represents an ideal in vivo reconstitution system for studying the uniporter in a physiologically relevant organellar membrane. Yeast has a highly reduced genome and facile genetics, and thus serves as a powerful system for systems and synthetic biology (15). In addition, it is a proven reconstitution system for pathways from other organisms, such as the RNAi machinery (16) and WT and mutant human metabolic enzymes (17). It also has been successfully used as a heterologous expression system for other mitochondrial transporters (18).
Here we report the establishment of yeast as a robust heterologous expression system for studying the uniporter. Because the human uniplex consists of multiple components, we first explored other eukaryotic organisms with simpler uniporter systems as candidates for in vivo reconstitution. Given that all animals appear to have the uniporter, and yeast do not, we considered Amoebazoa, the outgroup to animals and fungi. Here we demonstrate that Dictyostelium discoideum has mitochondrial calcium uptake machinery, and that the D. discoideum MCU homolog is able to rescue the mitochondrial calcium uptake defect in HEK-293T HsMCU or HsEMRE KO cells. Using this single defined genetic component, we were able to reconstitute uniporter activity in yeast mitochondria. Having established yeast as a reconstitution system for the uniporter, we asked which components of the human uniplex are required for in vivo reconstitution. We found that expression of HsMCU alone is insufficient and that joint expression of HsMCU and HsEMRE is necessary and sufficient for uniporter activity.
Results
D. discoideum Mitochondria Exhibit Uniporter Activity.
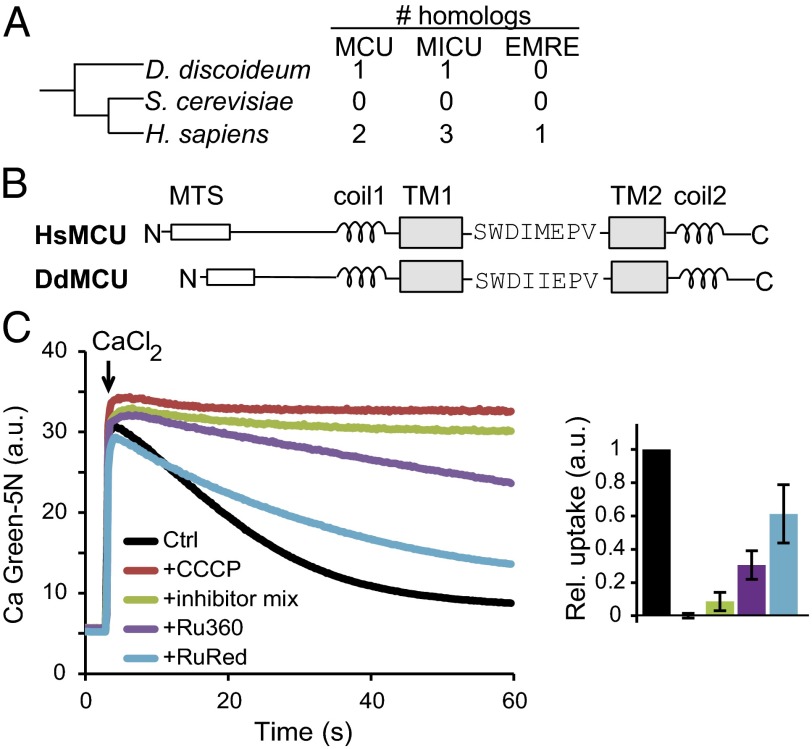
Classic studies of uniporter physiology have demonstrated that although uniporter activity is found within all animals tested, it is absent in the yeast S. cerevisiae. The evolutionary sister group of Opistokonta (a supergroup containing Metazoa and Fungi) is Amoebozoa, which includes the genetic model organism D. discoideum (19) (Fig. 1A). The D. discoideum genome has one homolog of MCU (which we term DdMCU) and one homolog of MICU1, and thus has a simpler predicted uniporter than that of humans, which has two MCU homologs (MCU and MCUb), three MICU1 homologs (MICU1, MICU2, and MICU3), and EMRE (Fig. 1A). Multiple sequence alignment indicates that DdMCU closely aligns with HsMCU, with a shared domain organization (Fig. 1B): a N-terminal mitochondrial targeting sequence, two coiled-coil domains surrounding two transmembrane domains that flank the “DIME motif,” which faces the intermembrane space and contains the conserved acidic residues essential for calcium transport (11).
Fig. 1.
The mitochondrial calcium uniporter in D. discoideum. (A) Taxonomic relationship of D. discoideum with respect to S. cerevisiae and H. sapiens, based on the NCBI taxonomy tree. Included are the number of uniplex homologs in each species. (B) Domain organization of the D. discoideum MCU homolog (DdMCU) compared with human MCU (HsMCU). MTS, mitochondrial targeting sequence; coil, coiled-coil domain; TM, transmembrane domain. (C) (Left) Representative traces of mitochondrial calcium uptake in permeabilized D. discoideum amoebae without added drugs (Ctrl), in the presence of uncoupler (CCCP), mitochondrial respiratory chain inhibitors (antimycin A, rotenone, and oligomycin), Ru360, or ruthenium red (RuRed) in response to 45 µM CaCl2. (Right) Histogram showing relative uptake rate of calcium, defined as the slope of linear fits between 20–30 s, normalized to Ctrl (mean ± SD); n ≥3.
Based on the presence of DdMCU in the D. discoideum genome and a previously reported mitochondrial inhibitor-sensitive calcium pool in D. discoideum (20, 21), we hypothesized that this organism has uniporter activity. To evaluate whether D. discoideum mitochondria are capable of uptaking calcium via a uniporter, we permeabilized the plasma membrane of vegetative D. discoideum amoebae and delivered a high pulse of calcium, which was rapidly taken up (Fig. 1C). The observed calcium uptake is membrane potential-dependent, and the uncoupler carbonyl cyanide 3-chlorophenylhydrazone (CCCP) disrupts it. Moreover, highly potent inhibitors of the mitochondrial respiratory chain also eliminate the observed uptake. Finally, the uptake is partially inhibited by the classical uniporter inhibitors ruthenium red and Ru360. Although D. discoideum has other calcium-buffering compartments (20, 21), the inhibition of calcium uptake by specific mitochondrial inhibitors and CCCP strongly suggests that other organelles are not involved in the measured activity. Collectively, these experiments provide strong evidence that D. discoideum has mitochondrial uniporter activity.
DdMCU Is the Functional Ortholog of Human MCU.
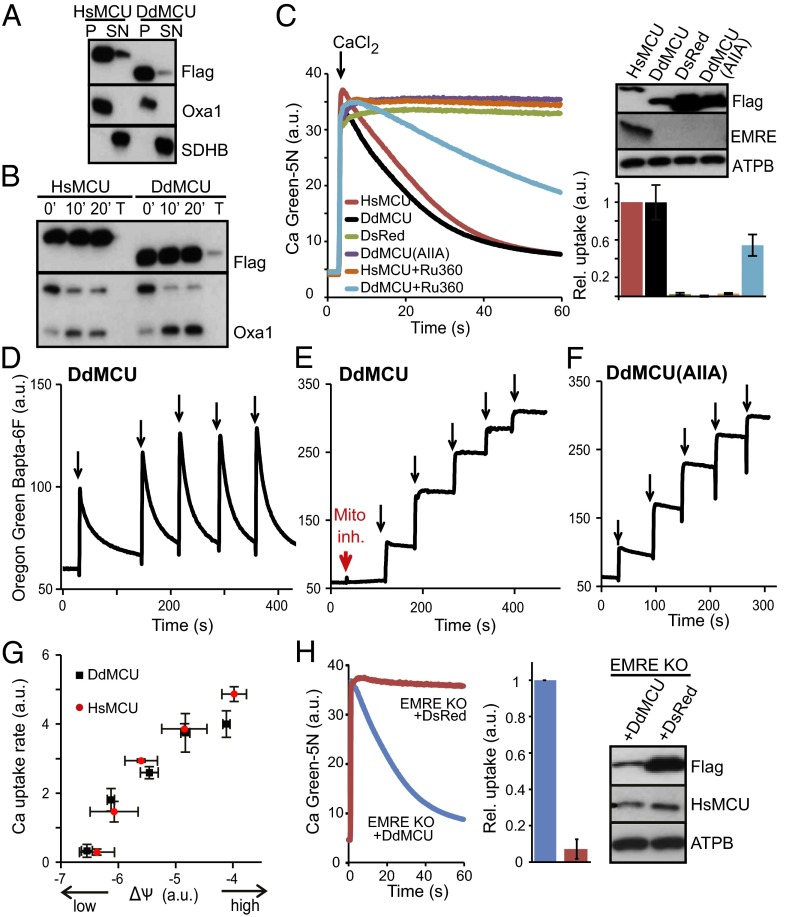
We next sought to determine whether DdMCU is indeed the functional ortholog of HsMCU. We stably expressed C-terminally FLAG-tagged DdMCU, HsMCU (positive control), and mitochondrial-targeted DsRed (red fluorescent protein; negative control) in MCU KO HEK-293T cell lines (14). Carbonate extraction and limited proteolysis experiments with proteinase K of isolated mitochondria confirmed mitochondrial inner membrane localization for both HsMCU and DdMCU, with the proper N-in, C-in topology (Fig. 2 A and B), similar to the HsMCU topology confirmed earlier by limited proteolysis experiments and high-resolution EM (11, 22). Although HsMCU KO cells expressing the control protein DsRed do not take up any calcium, expression of DdMCU fully rescues calcium uptake, with nearly identical uptake kinetics to HsMCU (Fig. 2C). DdMCU-mediated calcium uptake is sensitive to Ru360, with altered efficacy compared with Ru360 inhibition of HsMCU. Highly conserved acidic residues in the HsMCU “DIME motif” are essential for uniporter function (11, 12). Similarly, the DIME mutant of DdMCU [DdMCU(AIIA)] has no ability to conduct calcium when expressed in HsMCU KO cells (Fig. 2C), although the mutant protein is properly localized (Fig. S1). DdMCU expression is capable of restoring calcium uptake in response to repeated pulses of calcium, in a manner dependent on the respiratory chain and the DIME motif (Fig. 2 D–F). We also manipulated mitochondrial membrane potential levels with different concentrations of uncoupler in permeabilized cells, and found a similar voltage dependence of the initial uniporter calcium uptake rate in mitochondria expressing HsMCU and mitochondria expressing DdMCU (3) (Fig. 2G).
Fig. 2.
Functional complementation of the human uniporter by DdMCU in human cells. (A) Carbonate extraction of mitochondria isolated from HEK-293T MCU KO cells expressing HsMCU or DdMCU. P, pellet; SN, supernatant; Oxa1, mitochondrial inner membrane control; SDHB, mitochondrial matrix control. (B) Proteinase K treatment of mitoplasts from MCU KO cells expressing either HsMCU or DdMCU. T, pretreatment with 1% Triton X-100. 0’, 10’, and 20’ indicate the duration of incubation with proteinase K. (C) Representative traces of calcium uptake in digitonin-permeabilized MCU KO cells expressing the indicated protein and treated with Ru360 where indicated. Histogram shows relative rates of calcium uptake defined as the slope of linear fits between 20–30 s, normalized to HsMCU (mean ± SD); n ≥3. Immunoblot analysis shows protein expression in whole cell lysates. ATPB: complex V beta subunit. (D–F) Representative traces of calcium uptake during repeated additions of 5 µM CaCl2 (black arrows) to permeabilized MCU KO cells expressing DdMCU in the absence or presence of mitochondrial inhibitors (as in Fig. 1) (D and E) and permeabilized MCU KO cells expressing DdMCU(AIIA) mutant (F). (G) Calcium uptake rate as a function of mitochondrial membrane potential (ΔΨ) in permeabilized MCU KO cells expressing DdMCU or HsMCU. Mitochondrial membrane potential was varied using 0–500 nM CCCP. The rate of calcium uptake 15–20 s after CaCl2 injection is plotted as a function of initial membrane potential (mean ± SD); n = 3. (H) Representative traces of calcium uptake and Western blots of cell lysates of HEK-293T EMRE KO cells expressing DdMCU or DsRed, performed similarly as in C.
HsMCU KO cells do not express EMRE at a detectable level (14) (Fig. 2C). Interestingly, EMRE is not detected in the presence of DdMCU either—only HsMCU expression restores EMRE expression (Fig. 2C). To further confirm that DdMCU-mediated calcium uptake does not require EMRE, we expressed DdMCU in EMRE KO HEK-293T cells, which, like MCU KO cells, lack calcium uniporter activity (14), although MCU is expressed (Fig. 2H) . Intriguingly, DdMCU also rescues calcium current in EMRE KO cells (Fig. 2H). Collectively, these studies demonstrate that DdMCU is the bona fide functional ortholog of HsMCU, and that DdMCU does not need EMRE for uniporter current, in contrast to HsMCU.
Reconstitution of Uniporter Activity in Yeast with DdMCU.
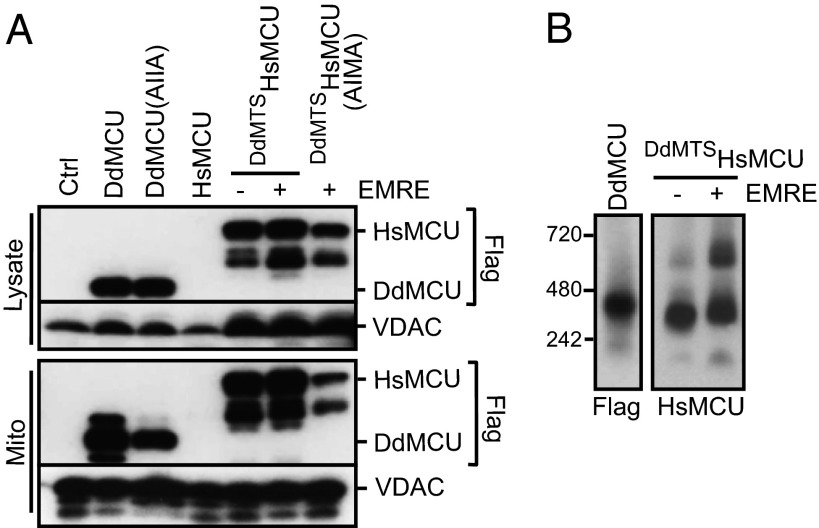
Because yeast mitochondria do not have a uniporter (23), we next explored whether DdMCU could be used to reconstitute uniporter activity in vivo in yeast. We expressed the full-length DdMCU with a C-terminal FLAG tag in the yeast S. cerevisiae. DdMCU expresses and localizes to yeast mitochondria (Fig. 3A). Moreover, DdMCU evidently forms a higher-molecular-weight oligomer (Fig. 3B). Mitochondria purified from yeast expressing the empty vector control (Ctrl), DdMCU, or DdMCU(AIIA) all become energized (i.e., increased membrane potential; Fig. S2 A–C) after the addition of fuel substrates. Furthermore, they undergo robust membrane potential transitions in response to added ADP (the initial decrease in membrane potential recovers after ADP depletion), indicating that oxidative phosphorylation in these mitochondria is intact (Fig. S2 A–C).
Fig. 3.
Heterologous expression of DdMCU, HsMCU, and EMRE in yeast mitochondria. (A) Immunoblot analysis of cell lysates and mitochondria from yeast cells expressing the indicated proteins or the empty vector control (Ctrl) using VDAC as a loading control. Lower molecular weight bands in DdMCU or DdMTSHsMCU expressing cells are most likely degradation products of the proteins. (B) BN-PAGE of digitonin-solubilized mitochondria isolated from yeast cells expressing the indicated proteins. Standard molecular weight markers are shown on the left.
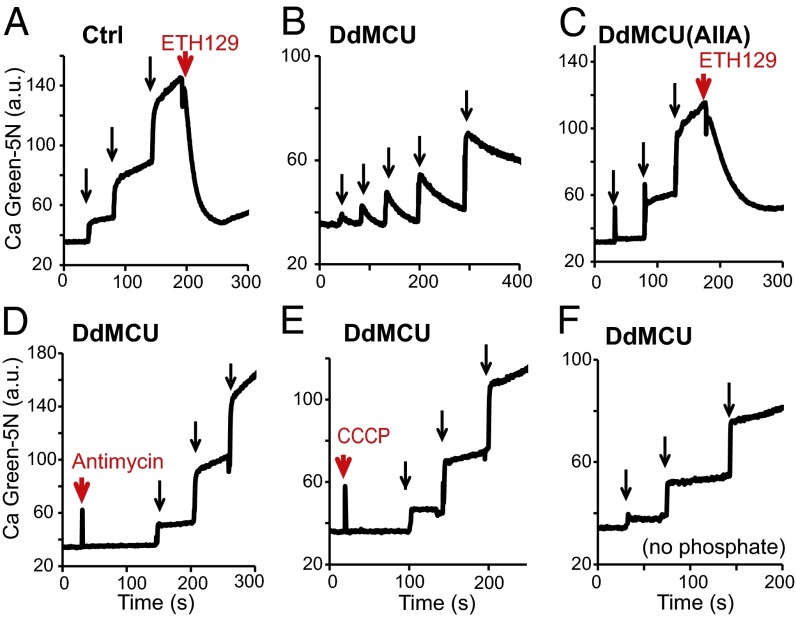
Although the control yeast mitochondria generate a robust membrane potential in response to fuel substrates, they are incapable of calcium uptake (Fig. 4A). They do, however, exhibit robust calcium uptake activity after addition of the membrane potential-dependent calcium ionophore ETH129, indicating that the mitochondria have the necessary membrane potential and buffering capacity within the matrix for calcium transport (24).
Fig. 4.
Reconstitution of mitochondrial calcium uniporter activity in yeast with DdMCU, a single defined genetic component. (A–C) Representative traces of repeated 20 µM CaCl2 additions (black arrows) to energized mitochondria isolated from empty vector control (Ctrl) (A) and from DdMCU-expressing (B) and DdMCU(AIIA)-expressing (C) yeast cells, using the calcium ionophore ETH129 as a control, monitoring extramitochondrial calcium levels with Calcium Green-5N. (D–F) Representative traces of calcium uptake during repeated additions of 20 µM CaCl2 (black arrows) to DdMCU expressing yeast mitochondria preincubated with 1 µM respiratory chain complex III inhibitor antimycin A (D) or 1 µM uncoupler CCCP (E), or in a buffer containing no phosphate (F).
Remarkably, heterologous expression of DdMCU alone endows yeast mitochondria with the ability to uptake calcium (Fig. 4B). Expression of this single protein mimics the effects of the calcium ionophore ETH129. The DdMCU(AIIA) mutant is incapable of transporting calcium (Fig. 4C), despite being expressed and localized to mitochondria (Fig. 3A); however, mitochondria expressing the DdMCU(AIIA) mutant show calcium uptake in the presence of the ionophore (Fig. 4C), indicating that they have the appropriate driving force for calcium uptake. Moreover, mitochondrial calcium uptake by DdMCU-expressing yeast mitochondria is dependent on the respiratory chain and membrane potential and requires incubation with inorganic phosphate (Fig. 4 D–F), all established properties of the uniporter.
All of the foregoing observations are consistent with mediation of calcium uptake by the uniporter. The requirement for a functional respiratory chain for calcium uptake further indicates that the observed calcium uptake is mitochondrial and not related to possible contamination of other organelles (e.g., vacuoles). Collectively, these studies demonstrate the successful in vivo reconstitution of uniporter activity in yeast using a single defined genetic component, DdMCU.
Reconstitution of Human Uniporter Activity with HsMCU and HsEMRE.
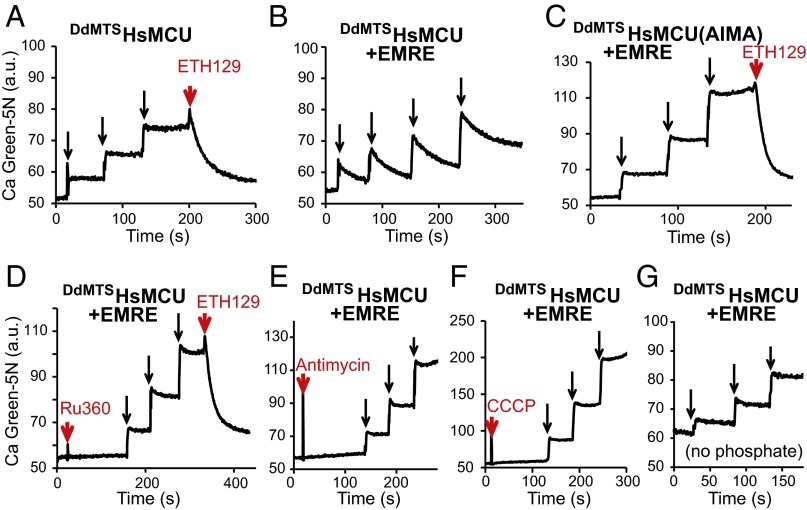
Having established a yeast reconstitution system with DdMCU, we next considered the more challenging problem of reconstituting the human uniporter to explore the minimal essential uniporter components in humans. Initial attempts to express HsMCU in yeast mitochondria led to little or no protein expression (Fig. 3A); therefore, we created a chimeric protein, consisting of the DdMCU mitochondrial targeting sequence, fused to the mature form of HsMCU (DdMTSHsMCU), which led to the successful expression of HsMCU in the mitochondrial membrane (Fig. 3A and Fig. S3B). The protein assembles into a complex of a similar molecular weight observed in HsEMRE KO HEK-293T cells (Fig. 3B) (14). Although these mitochondria are capable of generating membrane potential with substrate addition and undergo standard membrane polarization and depolarization during respiratory state transitions (Fig. S2D), they are unable to take up calcium unless the calcium ionophore ETH129 is added (Fig. 5A). Thus, HsMCU on its own, even though properly assembled into a complex within the yeast mitochondrial membrane, is not sufficient to reconstitute a calcium current.
Fig. 5.
Reconstitution of the human mitochondrial calcium uniporter activity in yeast. (A–C) Representative traces of repeated 20 µM CaCl2 additions (black arrows) to energized mitochondria isolated from yeast cells expressing DdMTSHsMCU (A), DdMTSHsMCU + EMRE (B), and DdMTSHsMCU(AIMA) + EMRE (C), using the calcium ionophore ETH129 as a control. (D–G) Representative traces of calcium uptake during repeated additions of 20 µM CaCl2 (black arrows) to DdMTSHsMCU + EMRE expressing yeast mitochondria preincubated with 2 µM Ru360 (D), 1 µM respiratory chain complex III inhibitor antimycin A (E), or 10 µM uncoupler CCCP (F), or in a buffer containing no phosphate (G).
Biochemical characterization of the human uniplex and loss-of-function studies have revealed that HsMCU and HsEMRE are both essential for the in vivo uniporter current (13, 14). Therefore, we attempted to coexpress HsEMRE with DdMTSHsMCU (Fig. 3A). Expression of HsEMRE could be detected only after the sample was enriched for HsEMRE by coimmunoprecipitation of FLAG-tagged DdMTSHsMCU, likely owing to the low affinity of the HsEMRE antibody (Fig. S3A). DdMTSHsMCU and HsEMRE form a slightly higher-molecular-weight complex compared with DdMTSHsMCU alone on native gels (Fig. 3B) (14). A second, higher-molecular-weight complex becomes more pronounced when HsEMRE is coexpressed (Fig. 3B). Remarkably, DdMTSHsMCU- and HsEMRE-expressing mitochondria are capable of calcium uptake (Fig. 5B) that meets all of the criteria for uniporter activity; the calcium uptake is sensitive to Ru360, dependent on the membrane potential and the respiratory chain, and requires inorganic phosphate (Fig. 5 D–G). Mitochondrial expression of the DIME mutant DdMTSHsMCU [DdMTSHsMCU(AIMA)] (Fig. 3A) in combination with HsEMRE is incapable of transmitting calcium (Fig. 5C); however, these mitochondria have intact oxidative phosphorylation (Fig. S2F) and are able to take up calcium after addition of the ionophore (Fig. 5C). Thus, coexpression of HsMCU and HsEMRE is sufficient for full reconstitution of human uniporter activity.
Discussion
In this study, we have (i) established yeast as an in vivo reconstitution system for the mitochondrial calcium uniporter; (ii) demonstrated that the nonmetazoan D. discoideum uniporter can be reconstituted using a single genetic component, DdMCU, thereby confirming that MCU is the pore-forming subunit; and (iii) shown that HsMCU and HsEMRE are the minimal genetic elements sufficient for human uniporter activity in vivo.
After the identification of MCU, the first known transmembrane subunit of the uniporter (11, 12), multiple studies pointed to its role as the channel-forming subunit, but the important question of whether it is sufficient for uniporter activity remained. Our previous in vivo whole mitoplast patch-clamp studies (13), using an MCU point mutant resistant to ruthenium red, already suggested that MCU is the pore-forming subunit. In addition, a previous study (12) performed in vitro reconstitution of purified human MCU in planar lipid bilayers and observed a calcium current. Nevertheless, given the discrepancies between results obtained from planar lipid bilayers and mitoplast patch-clamp studies, the question arose as to whether MCU can function autonomously to achieve uniporter activity in vivo. In a subsequent study, we identified EMRE as a second component required for uniporter conductance in human cells, suggesting that human MCU alone is insufficient to achieve uniporter activity in vivo (14). In the present study, we have resolved the foregoing paradox by establishing yeast as an in vivo reconstitution system and have defined the minimal genetic elements sufficient for uniporter current, which, interestingly, differ across evolutionary space.
In our yeast reconstitution system, DdMCU alone is sufficient to achieve uniporter activity, confirming that MCU is indeed the channel-forming subunit responsible for calcium uptake. In contrast, human cells require both HsMCU and EMRE, a protein that is present only in Metazoa. At present, why EMRE is required for HsMCU but not for DdMCU activity is unclear. In a previous loss-of-function study, we found that EMRE has at least two roles (14); it is required for the conductance of MCU and bridges MCU with its regulators MICU1/2. Although the overall domain architecture of DdMCU and HsMCU are the same, there are sequence divergences. Which of these differences may be responsible for allowing DdMCU to function independent of EMRE is unclear. Despite its lack of conductance in the absence of EMRE, HsMCU is still expressed in the mitochondrial membrane and forms a high-molecular-weight complex both in HEK-293T cells (14) and in yeast (Fig. 3B). Thus, EMRE seems to be intimately involved in the function of the channel rather than functioning as an assembly factor. Assembly factors of OXPHOS complexes are not a part of the mature complex, but rather are present only in intermediate assembly complexes (25). In contrast, EMRE is part of the mature uniplex and serves as a critical accessory subunit. One possibility is that in the absence of EMRE, MCU assembles but is in a closed conformation, and the presence of EMRE is required to maintain the channel in an open form. Establishment of yeast as a reconstitution system will make it possible to test this intriguing hypothesis in the future.
This study significantly contributes to our understanding of uniporter evolution. Studies dating back to the 1960s established that vertebrates (5, 26), Drosophila (27), plants (28), and trypanosomes (29, 30) harbor uniporter activity, but yeast does not (5). Our previous phylogenomic analysis of MCU and MICU1 homologs (31) revealed the presence of homologs in most eukaryotic branches, including those in which uniporter activity has already been reported, and suggested that certain fungal lineages, notably yeast, had lost the genes coding for uniporter machinery during evolution. We studied D. discoideum, the nearest outgroup of Metazoa and Fungi, and demonstrated that it has uniporter activity, and that DdMCU alone is sufficient for reconstituting uniporter current.
Our present work provides compelling experimental evidence that the uniporter was a feature of unikont mitochondria and was lost during fungal evolution. Notably, in contrast to the human complex, D. discoideum only needs DdMCU for uniporter activity and lacks EMRE homologs, indicating that EMRE is a more recent innovation specifically required in animals. Establishment of yeast as an in vivo reconstitution system should allow us to better understand the evolution of the uniporter and elucidate in detail its molecular mechanisms.
Materials and Methods
Sequence Analysis.
Multiple sequence alignment was performed with ClustalW. Mitochondrial targeting sequences and coiled-coil and transmembrane domains were predicted by MitoProt II version 1.101, COILS, and TMHMM version 2.0, respectively.
D. discoideum and Human Cell Lines.
D. discoideum strain AX3 was grown in HL5 medium (ForMedium) supplemented with ampicillin (100 µg/mL) and streptomycin sulfate (300 µg/mL) at 21 °C and harvested in the midlog phase of growth (1–5 × 106 cells/mL).
HEK-293T cells were grown as described previously (14). MCU and EMRE KO HEK-293T cell lines were generated by TALE nuclease (TALEN) (14). To generate DdMCU-, HsMCU-, and DsRed-expressing stable cell lines, full-length DdMCU (XM_632658 codon optimized for human expression by Genewiz), HsMCU (NM_138357.1), and DsRed (BAE53441.1) were cloned into a lentiviral mammalian expression vector with a C-terminal FLAG tag. DsRed also contained the mitochondrial targeting sequence of cytochrome c oxidase subunit 8 (amino acids 1–29). DdMCU(AIIA) mutant (D192A and E195A) and HsMCU(AIMA) mutant (D261A and E264A) were created using the Stratagene QuikChange mutagenesis kit. MCU KO or EMRE KO HEK-293T cells were infected with lentivirus and selected with 2 µg/mL puromycin (Life Technologies). Cell lysates were prepared as described previously (14). Samples were analyzed on SDS/PAGE by Western blot analysis against the FLAG tag (Cell Signaling; 2368), HsMCU (Sigma-Aldrich; HPA016480), EMRE (Santa Cruz Biotechnology; sc-86337), or ATPB (Abcam; ab14730), using rabbit or mouse secondary antibodies (GE Healthcare).
Yeast Strains.
C-terminally FLAG-tagged DdMCU, HsMCU, and DdMTSHsMCU chimera [mitochondrial targeting sequence of DdMCU (amino acids 1–30) and the mature part of HsMCU (amino acids 58–351)] were cloned into the HindIII sites of pACT2 (lacking the GAL4-HA activation domain) using the In-Fusion Cloning Kit (Clontech). The GAL4 activation domain was retained as a negative control in the empty vector. HsEMRE (NP_201575) was cloned between the XmaI and BamHI sites of the pRS426 vector containing a CIT1 promoter between the HindIII and EcoRI sites. Plasmids were introduced into the yeast strain W303 (MATa ade2-1 his3-11,15 leu2-3,112 ura3, trp1-1, can1-100) using the lithium acetate transformation method (32). Cells were grown in synthetic dropout medium lacking leucine (and uracil in the presence of HsEMRE) as a selection marker(s). To test protein expression, cells were vortexed for 10 min at 4 °C in the presence of acid-washed glass beads (Sigma-Aldrich) in 50 mM Tris pH 8.0, 1 mM EDTA, 3 mM DTT, and cOmplete protease inhibitor mixture (Roche). Samples were pelleted, and supernatant was analyzed by Western blot analysis with FLAG tag and VDAC (Abcam; ab110326) antibodies.
Permeabilized Cell Calcium Uptake Assays and Membrane Potential Measurements.
Calcium uptake measurements of permeabilized HEK-293T cells were performed essentially as described previously (33). In brief, cells were permeabilized in KCl buffer (125 mM KCl, 2 mM K2HPO4, 1 mM MgCl2, 10 µM EGTA, and 20 mM Hepes, pH 7.2) supplemented with 0.005% digitonin (Sigma-Aldrich), 5 mM glutamate, 5 mM malate, and 1 µM cell-impermeable Calcium Green-5N (Life Technologies). D. discoideum cells were treated similarly, except that the pH was set to pH 7.0, and the medium contained 0.01% digitonin, 3 mM glutamate, 3 mM malate, and 3 mM succinate. Fluorescence was monitored with a PerkinElmer Envision plate reader before and after injection of 45 μM CaCl2 using FITC filter sets (excitation/emission, 485 nm/535 nm) every 0.2 s at room temperature. The following reagents were used where indicated: 1 µM CCCP (Sigma-Aldrich), 1 µM antimycin A (Sigma-Aldrich), 1 µM rotenone (Sigma-Aldrich), 1 µM oligomycin, 3 µM Ru360 (Calbiochem), and 3 µM ruthenium red (Sigma-Aldrich).
Calcium uptake during repeated additions of 5 μM CaCl2 was measured with a PerkinElmer LS55 luminescence spectrometer every 0.2 s at room temperature [excitation/emission: 494 nm/524 nm with Oregon Green 488 Bapta-6F (Life Technologies); slit length, 10 nm/2.5 nm]. For measurements of membrane potential and calcium uptake in parallel, 1 million MCU KO HEK-293T cells stably expressing DdMCU or HsMCU were permeabilized in 150 µL of KCl buffer, supplemented with 0.005% digitonin, 5 mM glutamate, 5 mM malate, and 1.3 μM tetramethylrhodamine (TMRM; Life Technologies) (34), with or without 1 μM Calcium Green-5N and 0–500 nM CCCP. Fluorescence measurements were performed with a PerkinElmer Envision plate reader (excitation/emission, 485 nm/535 nm).
Isolation and Analysis of Crude HEK-293T Mitochondria.
Crude mitochondria isolation was performed as described previously (14), except that cells were lysed at 800 psi for 15 min in a Parr bomb, followed by five strokes with a Potter–Elvehjem Teflon homogenizer.
To prepare mitoplasts, mitochondria were incubated in ice-cold water for 10 min. After incubation, mitoplasts were pelleted and resuspended in mitochondria isolation buffer (14). Protein topology was determined using 100 µg of mitoplasts for each condition. The mitoplasts were treated with 40 µg/mL proteinase K (Sigma-Aldrich) for 0, 10, or 20 min at 4 °C (35). The reaction was stopped by incubation with 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich) for 5 min on ice. In the 0 min samples, the addition of PMSF was followed by the addition of protease. Triton X-100 (1%) treated samples were incubated with proteinase K for 20 min. After the reaction, samples were pelleted and resuspended in SDS/PAGE sample buffer. Oxa1 (mitochondrial inner membrane control) undergoes cleavage of its intermembrane space-facing domain by proteinase K, thereby giving rise to a shift in its molecular weight.
To determine membrane localization, 100 µg of crude mitochondria was treated with 100 mM Na2CO3 for 30 min on ice (35). Samples were pelleted (100,000 rpm for 30 min, in a TLA100 rotor), and the supernatant was subjected to TCA precipitation (20%) and an acetone wash. Samples were resuspended in SDS/PAGE sample buffer and analyzed on SDS/PAGE by Western blot analysis against the following antibodies: FLAG, Oxa1 (BD Biosciences; 611980), succinate dehydrogenase subunit B (SDHB; Abcam; ab14714), cytochrome c oxidase subunit II (MTCO2; Abcam; ab110258), Tomm20 (Santa Cruz Biotechnology; sc17764), Timm23 (BD Biosciences; 611222), and Hsp60 (Abcam; ab3080).
Isolation and Analysis of Yeast Mitochondria.
Yeast crude mitochondria were isolated following the protocol of Yaffe (36), except that the mitochondrial isolation buffer was supplemented with cOmplete protease inhibitor mixture tablets, 0.5 mg/mL BSA, and 1 mM EDTA, and no density gradient centrifugation was applied. For spheroplasting, Zymolyase 20T was purchased from Amsbio.
For determining membrane localization, carbonate extraction was performed similarly to that for HEK-293T cells using FLAG, VDAC, and cytochrome c (Abcam; AB110325) antibodies. For blue native PAGE (BN-PAGE) analysis, 5 µg of crude mitochondria was solubilized in 1% digitonin (11, 37), and BN-PAGE was performed according to the manufacturer’s instructions on a NativePAGE Novex 3–12% Bis-Tris gel (Life Technologies) using NativeMark unstained standards (Life Technologies). Coimmunoprecipitation was performed using mitochondria under the conditions described previously (14).
Yeast Mitochondrial Membrane Potential and Calcium Uptake Measurements.
Mitochondrial membrane potential was monitored in a custom spectrophotometer. For this, 270 µg of crude mitochondria was added to 500 µL of mitochondrial experimental buffer (0.6 M mannitol, 20 mM Hepes-KOH pH 7.4, 2 mM MgCl2, 1 mM EDTA, 10 mM KH2PO4, and 5 mg/mL BSA), supplemented with 1.25 μM TMRM (excitation/emission: 530 nm/580–600 nm). Calcium uptake measurements were performed similarly as described by Jung et al. (24), except that 3 mM glutamate, 3 mM malate, 3 mM succinate, 10 µM EGTA, and 1 μM Calcium Green-5N were present in the medium. Repeated additions of 20 μM CaCl2 were monitored in a PerkinElmer LS55 luminescence spectrometer (excitation/emission, 506 nm/532 nm); 5 μM ETH129 (Sigma-Aldrich; Calcium Ionophore II) was added as a positive control.
Supplementary Material
Acknowledgments
We thank Abbas Yaseen for technical guidance, Sarah Calvo for bioinformatics assistance, and Zenon Grabarek for helpful discussions. K.J.K. was supported by a graduate research fellowship from the National Science Foundation. Y.S. received support from Helen Hay Whitney Foundation. This work was supported by National Institutes of Health Grant R01 DK080261 (to V.K.M.). V.K.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400514111/-/DCSupplemental.
References
- 1.Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA. 1961;47(13885269):1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasington FD, Murphy JV. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237(13925019):2670–2677. [PubMed] [Google Scholar]
- 3.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427(6972):360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 4.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467(7313):291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carafoli E, Lehninger AL. A survey of the interaction of calcium ions with mitochondria from different tissues and species. Biochem J. 1971;122(5):681–690. doi: 10.1042/bj1220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csordás G, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca²⁺ uniporter. Cell Metab. 2013;17(6):976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallilankaraman K, et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012;151(3):630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plovanich M, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013;8(2):e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 2014;15(3):299–307. doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri D, Sancak Y, Mootha VK, Clapham DE. MCU encodes the pore conducting mitochondrial calcium currents. eLife. 2013;2(2):e00704. doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancak Y, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342(6164):1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornmann B, et al. An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drinnenberg IA, et al. RNAi in budding yeast. Science. 2009;326(5952):544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marini NJ, et al. The prevalence of folate-remedial MTHFR enzyme variants in humans. Proc Natl Acad Sci USA. 2008;105(23):8055–8060. doi: 10.1073/pnas.0802813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang CY, Hagen T, Mootha VK, Slieker LJ, Lowell BB. Assessment of uncoupling activity of uncoupling protein 3 using a yeast heterologous expression system. FEBS Lett. 1999;449(2-3):129–134. doi: 10.1016/s0014-5793(99)00441-x. [DOI] [PubMed] [Google Scholar]
- 19.Paps J, Medina-Chacón LA, Marshall W, Suga H, Ruiz-Trillo I. Molecular phylogeny of unikonts: New insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist. 2013;164(1):2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Europe-Finner GN, Newell PC. Inositol 1,4,5-triphosphate induces calcium release from a non-mitochondrial pool in amoebae of Dictyostelium. Biochim Biophys Acta. 1986;887(3):335–340. doi: 10.1016/0167-4889(86)90163-1. [DOI] [PubMed] [Google Scholar]
- 21.Milne JL, Coukell MB. Identification of a high-affinity Ca2+ pump associated with endocytotic vesicles in Dictyostelium discoideum. Exp Cell Res. 1989;185(1):21–32. doi: 10.1016/0014-4827(89)90033-5. [DOI] [PubMed] [Google Scholar]
- 22.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30(11):1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carafoli E, Balcavage WX, Lehninger AL, Mattoon JR. Ca2+ metabolism in yeast cells and mitochondria. Biochim Biophys Acta. 1970;205(1):18–26. doi: 10.1016/0005-2728(70)90057-5. [DOI] [PubMed] [Google Scholar]
- 24.Jung DW, Bradshaw PC, Pfeiffer DR. Properties of a cyclosporin-insensitive permeability transition pore in yeast mitochondria. J Biol Chem. 1997;272(34):21104–21112. doi: 10.1074/jbc.272.34.21104. [DOI] [PubMed] [Google Scholar]
- 25.Mckenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62(7):497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 26.Chance B. The energy-linked reaction of calcium with mitochondria. J Biol Chem. 1965;240:2729–2748. [PubMed] [Google Scholar]
- 27.Fieni F, Lee SB, Jan YN, Kirichok Y. Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nat Commun. 2012;3(1317):1317. doi: 10.1038/ncomms2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CH, Lehninger AL. Ca2+ transport activity in mitochondria from some plant tissues. Arch Biochem Biophys. 1973;157(1):183–196. doi: 10.1016/0003-9861(73)90404-9. [DOI] [PubMed] [Google Scholar]
- 29.Docampo R, Vercesi AE. Ca2+ transport by coupled Trypanosoma cruzi mitochondria in situ. J Biol Chem. 1989;264(1):108–111. [PubMed] [Google Scholar]
- 30.Huang G, Vercesi AE, Docampo R. Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat Commun. 2013;4:2865. doi: 10.1038/ncomms3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886–886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 33.Murphy AN, Bredesen DE, Cortopassi G, Wang E, Fiskum G. Bcl-2 potentiates the maximal calcium uptake capacity of neural cell mitochondria. Proc Natl Acad Sci USA. 1996;93(18):9893–9898. doi: 10.1073/pnas.93.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scaduto RC, Jr, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76(1 Pt 1):469–477. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- 36.Yaffe MP. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 37.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.