Significance
African trypanosomes are a major plague in sub-Saharan Africa. They cause sleeping sickness in humans and nagana in cattle, preventing extensive cattle raising. They escape the immune system of their host through frequent changes in their major surface antigen, the variant surface glycoprotein (VSG). The control of VSG expression is poorly understood, and the level at which it takes place has been a matter of debate for the last 25 y. A better understanding of the mechanism by which it works would facilitate the identification of the proteins involved and would provide putative targets for drug design. We report data indicating an unusual control level, namely after transcription initiation, suggesting transcription elongation or RNA processing as a control step.
Keywords: antigenic variation, sleeping sickness, single cell RT-PCR
Abstract
African trypanosomes survive the immune defense of their hosts by regularly changing their antigenic coat made of variant surface glycoprotein (VSG). The Trypanosoma brucei genome contains more than 1,000 VSG genes. To be expressed, a given VSG gene must be located in one of 15 telomeric regions termed “VSG expression sites” (ESs), each of which contains a polycistronic transcription unit that includes ES-associated genes. Only one ES is fully active at a time, so only one VSG gene is transcribed per cell. Although this monoallelic expression is controlled at the transcriptional level, the precise molecular mechanism for this control is not understood. Here we report that in single cells transcription is initiated on several ESs simultaneously, indicating that the monoallelic control is not determined only at transcription initiation, but also at further control steps such as transcription elongation or RNA processing.
African trypanosomes are flagellated protozoan parasites transmitted by tsetse flies. The main population of these parasites lives as the procyclic form in the tsetse fly midgut. Once transmitted to their mammalian hosts, they multiply in the blood, causing a disease with both medical and veterinary consequences. Seventy million people are at risk for human African trypanosomiasis (HAT), and the World Health Organization reports that about 7,000 new cases appeared in 2010 (1). Nagana, the bovine form of the disease, prevents the development of efficient cattle raising in a large portion of sub-Saharan Africa. HAT is caused by the persistence of trypanosomes in the blood, sometimes for years, despite a specific immune response. This population survives by antigenic variation, a mechanism allowing trypanosomes regularly to change their antigenic coat made of variant surface glycoprotein (VSG) (2, 3). The change in VSG protein is driven by a change in the VSG gene expressed. More than 1,000 VSG genes and pseudogenes are packed as gene arrays or located at telomeres (3). To be expressed, a VSG gene must be located in one of approximately 15 telomeric expression sites (ESs). In a given cell only one ES is active at any time, and therefore only one VSG protein expressed (4). Two main mechanisms are used to change the expressed VSG gene and therefore perform antigenic variation: homologous recombination (gene conversion or reciprocal telomere exchange), which replaces the VSG gene in the active ES, and transcriptional switching between ESs (a process called “in situ activation”), which turns off the active ES and turns on a new one (2).
ES transcription results from the recruitment of RNA polymerase I (pol I) on a promoter of the ribosomal type, in a nonnucleolar nuclear structure termed the “ES body” (5). The mechanism controlling the selective activity of a single ES at a time is still unknown, although several factors have been found to be involved, such as differential chromatin/histone acetylation and methylation between active and inactive ESs (6–8); nucleosome depletion in the active ES (9, 10); and the influence of various negative transcription modulators such as a histone methylase, a high-mobility group protein, histone H1, NLP (a nucleoplasm-like protein), a FACT subunit (a chromatin remodeler), a SWI/SNF (a chromatin remodeling factor), ORC1 and MCM-BP (proteins involved in the initiation of DNA replication), RAP1 (a telomere-binding protein), histone chaperones, or a lamin-like protein (8, 11–21). However, no master ES regulator has been identified so far. Furthermore, the level at which the control takes place is unknown: Whether the transcription control takes place at the initiation or elongation level is debated. Data reporting low levels of transcripts from silent ESs upon RNAi-mediated knockdown of various factors (8, 11–21) cannot resolve this issue, because the data are compatible with both models. Previous experiments pointed to mixed conclusions. On the one hand, transcripts from inactive ESs could be detected in bloodstream clones and even in procyclic forms, which do not express VSG, suggesting that transcription initiation takes place in all ESs simultaneously (22–29). On the other hand, experiments tagging ESs with antibiotic-resistance genes showed that RNA expression can be detected from two ES, although the expression was unstable (30). This result gave rise to a model in which transcription is initiated on only two ESs, one being fully active and a second being preactive (moderately transcribed) (31). This model is equally able to account for the finding of transcripts from multiple ESs within a trypanosome population (29), provided the preactive ES differs among individual cells. Most experimental results can be accounted for by a multistep process in which at least two ESs are transcribed but the expression of only one is stabilized.
To decide if one, two, or multiple ES promoters can initiate transcription in a given cell, single-cell RT-PCR is required. Here we report results of single-cell RT-PCR analysis of ES transcription. The results showed that transcription initiation takes place even on silent ESs despite VSG monoallelic expression.
Results
The claim that RT-PCR products originate from a single cell requires stringent technical controls that were performed as prerequisites. Serial trypanosome dilutions were seeded in 96-well plates to achieve an average density of 0.33 cells per well (Fig. 1). In three experiments with MiTat 1.2 (Trypanosoma brucei strain 328-114) parasites, 27, 32, and 30 of the 96 wells in each experiment were found to contain growing cells. With AnTat 1.3 parasites from the EATRO1125 strain, the cloning efficiency was lower, with less than 10% positive wells per plate. Therefore, most of the work was performed with the MiTat 1.2 variant. To confirm the validity of the serial dilution, we generated 10 different T. brucei cell lines that could be discriminated by PCR based on the sequence or/and size of a transfected insert. Thus, 10 plasmids containing different inserts in the same p2T7177 plasmid backbone (32) were transfected separately in the 328-114 cell line. It should be noted that RNAi was not induced and that none of the 10 genes potentially targeted by RNAi are known to interfere with the RNA pol I transcription process. These cell lines were mixed together, and PCR was performed on DNA extracted from this mix using primers targeting sequences flanking the insert. This approach allowed us to amplify from the whole population a mix of 10 different fragments with the sizes expected from the combination of each insert and flanking sequences (Fig. S1). After serial dilution of this population, the genomic DNA was extracted and analyzed by PCR from positive wells. In the analysis of 27 samples, only one of the 10 possible inserts was found per well, except in one case where two inserts were present (Fig. S1A). Therefore, most populations growing in individual wells after serial dilution originated from a single cell.
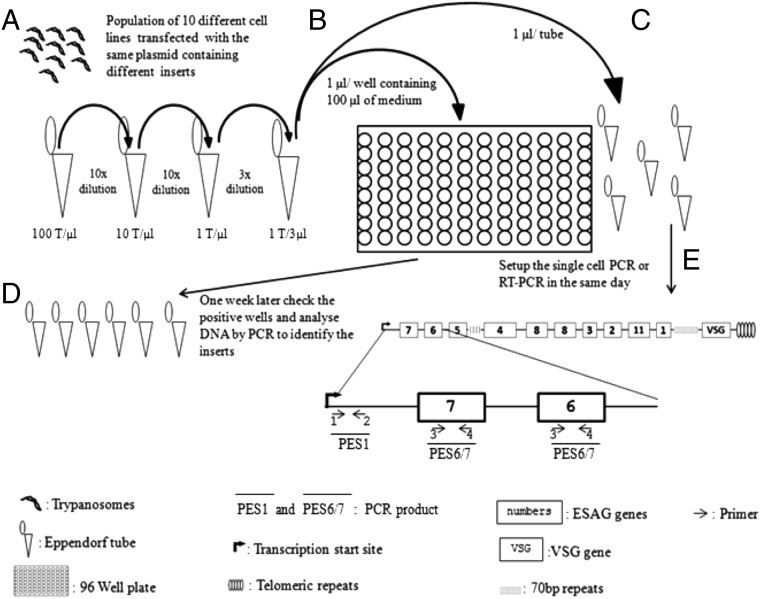
Fig. 1.
Schematic of the single-cell RT-PCR experiment. (A) Ten different bloodstream 328-114–derived cell lines transfected with the same plasmid (p2T7177) backbone containing different inserts were mixed. Serial dilution was performed on this population to obtain one cell (Trypanosome:T) per3 µL (suspension A). (B) One microliter of suspension A was added to each well of a 96-well plate. Each well contained 100 µL of culture medium. The suspension was cultivated for 7 d. (C) One microliter of suspension A also was added to each of five Eppendorf tubes, which were frozen and stored before the single-cell PCR or RT-PCR experiment was performed. (D) After 1 wk, all the positive wells were subjected to DNA extraction, and PCR was performed using primers binding to the plasmid backbone to amplify the different inserts. (E) After the validity of the serial dilution experiment in the 96-well plate (step D) was confirmed, RT-PCR was performed on the Eppendorf tubes containing an average of one trypanosome per three tubes using primers located (as shown at the beginning of a typical ES) and amplifying the beginning (nucleotide +1 to nucleotide +149) of the ES (PES1) or ESAG6/7 (PES6/7).
We then performed PCR on DNA extracted from a population of 5 × 106 trypanosomes to validate the primer pairs specifically targeting the ESs. First, we used primers designed to hybridize to many ESs and to generate a 149-bp PCR product corresponding to the sequence starting at position +1 bp downstream from the transcription initiation site in the FM162568 (BES3/TAR2) ES (see Fig. S2 for the nomenclature of ESs and PCR products). These primers already had been validated on several antigenic variants of the EATRO1125 strain (29). The PCR products were cloned in the TOPO vector, and DNA was isolated from individual clones and sequenced (Dataset S1). The 94 PCR products analyzed were found to be of 29 types generally present from one to seven times, except for G6, which was generated 30 times (Table 1, column 2). The sequences were numbered according to the nomenclature adopted in ref. 29 (Table 1 and Table S1, column 1), and correspondence was made to the nomenclature of Hertz-Fowler et al. (33) (Table S2). All sequences reported by Hertz-Fowler et al. (33) were recovered. Some sequences described in ref. 29 were not represented, possibly because different trypanosome strains were analyzed. As previously noticed (29, 33, 34) these sequences can be classified as evolving one from another. Not classified in these groups are 11 different sequences (11.7% of the total) generated by loner mutations that possibly could originate from sequencing/PCR mistakes (Table 1, column 2).
Table 1.
Analysis of PCR and RT-PCR products amplified from the beginning of the ES using primer pair 1 and 2 (product PES1 in Fig. 1) and ESAG 6 and 7 genes using primer pair 3 and 4 (product PES6/7 in Fig. 1)
| PES1 sequence name* | Number of PCR products of PES1 | Number of RT-PCR products of PES1 | ||||
| Genomic population | Genomic single cell | Single BF cell (experiment 1) | Single BF cell (experiment 2) | Single BF cell (experiment 3) | Single PF cell | |
| G1 | 6 | 6 | 30 | 40 | 37 | — |
| G2 | 2 | 3 | — | — | — | 1 |
| G3 | — | — | — | — | — | 2 |
| G6 | 30 | 17 | 3 | 5 | 3 | 14 |
| G7 | — | 4 | — | — | — | 4 |
| G8 | — | — | — | — | — | 1 |
| G9 | 5 | 8 | 4 | 3 | 2 | 11 |
| G11 | — | — | 5 | — | 4 | — |
| G14 | — | — | — | — | — | 5 |
| G16 | 3 | 6 | 9 | 6 | 7 | — |
| G17 | — | — | — | — | — | 15 |
| G18 | 3 | 6 | — | 1 | 3 | |
| G19 | 1 | 1 | — | 1 | — | — |
| G20 | — | — | 4 | — | — | — |
| G21 | — | 1 | — | — | 5 | 2 |
| G23 | 1 | 2 | 11 | 7 | — | 11 |
| G25 | 5 | 2 | 3 | 4 | — | 4 |
| G28 | 7 | 6 | 2 | 1 | 2 | 2 |
| G29 | — | — | — | 1 | — | — |
| G32 | 1 | 1 | — | 3 | — | 1 |
| E58 | 3 | — | — | — | — | — |
| E50/r50 | — | — | — | — | — | 4 |
| A1 | 3 | 5 | 2 | 3 | — | — |
| A2 | 3 | 3 | 1 | — | 4 | — |
| A3 | 2 | 4 | — | 4 | 3 | — |
| A4 | 5 | 1 | 3 | — | — | — |
| A5 | 1 | 1 | — | 1 | — | — |
| A6 | 2 | 1 | 1 | — | — | — |
| Av1 | — | 2 | — | — | — | — |
| Av2 | — | — | 2 | — | — | — |
| Av3 | — | — | — | — | 2 | — |
| Av4 | — | — | — | — | 2 | — |
| Av5 | — | — | 1 | — | 1 | — |
| Av6 | — | — | 1 | 1 | — | — |
| No. of additional unique sequences | 11 | 12 | 13 | 11 | 17 | 15 |
| Total no. | 94 | 92 | 95 | 92 | 90 | 90 |
PCR (genomic) and RT-PCR analyses were performed on the indicated materials. The PCR products were cloned, and individual inserts were sequenced. The total number of analyzed sequences is given at the bottom of each column. The number of times each sequence was recovered is indicated. Sequences were generated by PCR on DNA extracted from a 328-114 population, by PCR on DNA extracted from a single cell, by RT-PCR performed on RNA extracted from a BF single cell in three independent experiments, and by PCR on RNA extracted from a PF single cell. BF, T. Brucei bloodstream form; PF, T. Brucei procyclic form.
Sequences beginning with “G” and “E” are named according to Vanhamme et al. (29). Sequences beginning with “A” are described by Hertz-Fowler et al. (33) but were not found by Vanhamme et al. (29). Sequences beginning with “Av” have not been reported previously. Additional sequences that did not match any of the previously obtained sequences were observed.
As a further control, single-cell genomic DNA was analyzed by PCR. As expected, this analysis identified sequences that were identical to those found in genomic PCR products from the cell populations analyzed above and three previously unidentified sequences, namely G7, G21, and Av1 (Table 1, column 3). Twelve of these sequences (13% of the total) could not be classified in known groups. Only one sequence from the population (namely, E58) was not recovered in the single-cell PCR. This result could be linked to a limited coverage in the 92 PCR products analyzed here. Again, from one to eight copies of the different sequences were recovered, except for the G6 sequence, which was found in 17 copies.
Finally, a control for DNA contamination was performed before starting the experiments. For that purpose, we took advantage of the only gene in T. brucei documented to contain a unique intron, the gene coding for poly(A) polymerase (35). PCR primers were designed to anneal inside the intron and in exonic regions flanking this intron. PCR and RT-PCR analyses using different combinations of these primers allowed the detection of PCR products corresponding only to unspliced products from DNA analysis, to unspliced and spliced products from RNA untreated by DNase, only to spliced products from DNase-treated RNA, and to no product from RT-minus reactions performed on DNase-treated RNA (Fig. S3). The results indicated that DNase treatment was completely efficient.
Because all the control experiments gave satisfactory results, we assessed transcription at the beginning of ESs in single cells. Three independent RT-PCR experiments were performed on RNA extracted from single-cell samples generated by serial dilutions, each time analyzing five Eppendorf tubes. (Statistically, only one Eppendorf tube in three should contain one trypanosome.) The extracted RNA was split into two halves, one of which was analyzed with reverse transcriptase omitted. As shown in Fig. S1B, all reactions performed in absence of reverse transcriptase gave negative results, whereas three of the 15 tubes analyzed in presence of reverse transcriptase proved positive. The PCR products were cloned, and inserts from individual colonies were sequenced. A majority of the sequences obtained (78/95, 80/92, and 70/90; Table 1, columns 4–6) were identical to genomic sequences. Most sequences were present in 1–11 copies, except for G1, which was present in 30–40 copies. Because G1 is present in both FM162566 and FM162572 (33), and FM162566 is the active ES in this isolate, the relatively high proportion of this PCR product was in keeping with the higher transcription efficiency in the dominant ES (36) and with the more efficient processing of transcripts from the active ES (29). Here again, 11–17 outliers (12–19% of the total sequences) were generated. As further controls, we attempted to clone the hypothetical material coming from RT-PCR analyses performed in absence of reverse transcriptase and appearing negative by gel electrophoresis. Plasmids extracted from 300 bacterial colonies generated from three independent experiments were sequenced, and readable sequences were shown to contain short DNA inserts not matching any trypanosome sequences except PCR primer fragments (Dataset S2). According to Vanhamme et al. (29), the large transcript heterogeneity found in some antigenic variants at the beginning of the expression site was lost during downstream progression from the promoter. Notably, between 3 and 5 kbp downstream from the promoter the heterogeneity was only ∼2, corresponding to the transcripts from the two homologous genes, ESAG6 (expression site associated gene 6) and ESAG7 (expression site associated gene 7), located in tandem in all ESs except BES8/TAR64 (FM162575) (33, 34). Single-cell RT-PCR analysis of the ESAG6/7 region was performed in the MiTat isolate. First, a genomic control generated four sequences found by both Vanhamme et al. (29) and Hertz-Fowler et al. (33) and three sequences that were found only by Vanhamme et al. (29) (Table S1, column 2). After serial dilution, cloning and sequencing, single-cell RT-PCR generated a major product (77/90 sequences), a minor type (2/90 sequences), and 11 different outliers (Table S1, column 3). The major product exhibited the sequence found by Hertz-Fowler et al. (33) for both ESAG7 and ESAG6 in the active ES in this isolate. Therefore, at the ESAG7/6 level, 3–5 kbp downstream from the transcription initiation start site, only transcription in the active ES could be detected readily, along with weak transcription from another ES, confirming the data obtained by Vanhamme et al. (29) from a trypanosome population.
Then RT-PCR was performed to detect VSG transcripts at a single-cell level. From 152 sequences only one type of product was detected, corresponding to the MiTat 221 VSG.
In the last two cases (ESAG6/7 and VSG), the recovered sequences matched known/expected sequences in 100% of the cases.
Finally, we performed single-cell RT-PCR of the procyclic differentiation stage that does not express VSG. We analyzed single trypanosomes of the EATRO1125 strain, which had been analyzed as a population by Vanhamme et al. (29). We sequenced 92 products, including 14 different sequences present in 1–15 copies (Table 1, column 7). The most abundant product corresponded to a dominant ES in the AnTaR repertoire of this isolate (EATRO1125), suggesting that, as also noticed by Vanhamme et al. (29) and Rudenko et al. (23), this ES remained among the most actively transcribed ESs even in its procyclic form. The pattern of products matched that of Vanhamme et al. (29), confirming both the reliability of the method and the conclusion that in procyclic cells transcription is initiated in all ESs.
Discussion
Antigenic variation in T. brucei relies on monoallelic gene expression. The precise underlying molecular mechanism is still not understood. Although several molecular players have been shown to be involved (8, 11–20), depletion of any of these factors had only a moderate effect on the repression of silent VSG genes. Therefore, either a master regulator is still to be discovered or, alternatively, the process is controlled by multiple collaborative factors ensuring a cumulative tight lock. Uncovering whether such a key player exists would be facilitated by delineating the step at which control is exerted. This control must happen at the level of transcription or posttranscriptional mRNA processing/stability, because we detected only one VSG mRNA per cell. Previously, it had been assumed that control occurred at the level of transcription initiation, in conformance with the common rule in eukaryotes and in the absence of any experiment properly addressing the point.
Several observations pointed to a control operating after the initiation step (reviewed in ref. 37). Thus, VSG ESs are active when transfected as episomes in cells, indicating that several promoters can be active simultaneously (38). This conclusion is also true in procyclic forms, where the ES promoters are normally not active (38). The very simple structure and ribosomal nature of the ES promoters (39, 40) is in keeping with the absence of regulatory sequences. In support of this notion, promoters from active or inactive ESs share the same sensitivity to DNase (24), and antigenic variation still occurs properly if the promoter of the active ES is replaced by the ribosomal promoter (41, 42). Other arguments include the detection of transcripts originating from inactive ESs such as RNA transcribed from the ES beginning in procyclic forms (29), RNA from antibiotic-resistance genes inserted in inactive ESs (25), or RNA from different ESAG6 from inactive ESs, accounting for up to 20% of the total ESAG6 transcripts (28). Taken together these observations support the notion of leaky ES transcription (27).
A different conclusion was reached when tagging ESs with antibiotic-resistance genes to force their expression. Under these conditions it was impossible to express more than two ESs at a time, and this expression was found to be unstable (31). This observation suggested an alternative model in which ES expression is controlled at the level of transcription initiation from one fully active and one preactive ES per cell (30). These two models are in agreement with a more consensual proposal that transcription is initiated in two or several ES and that expression is stabilized in only one of them.
RT-PCR allowing the detection of ES transcripts in various bloodstream clones revealed several products at the beginning of the ES, with heterogeneity decreasing with progression downstream from the promoter (29). These results did not allow discrimination between the models, because the heterogeneous transcripts could originate in each cell from transcription of either multiple or only two ESs (an active and a preactive one), provided that the preactive ES is different in different cells of the population. A way to distinguish clearly between these possibilities is single-cell RT-PCR. The study presented here unambiguously allowed the detection of transcripts originating from several ESs, definitely showing that transcription can be initiated simultaneously from these ESs. Although transcriptional leakage could be invoked as an alternative explanation accounting for the quantitative difference between RT-PCR products from different ESs, this explanation is not likely: We demonstrated earlier (29) that the relative difference in the levels of RT-PCR products between the active and “inactive” ESs was reduced or erased when analyzing native RNA transcription and cannot be ascribed fully to a difference between initiation rates but rather arises from differing rates of transcript processing, cytoplasmic export, and stability. Nevertheless, this observation does not exclude any difference in initiation rates among ESs. In agreement with that possibility, a basal class I transcription factor (CITFA) was shown to bind the ES promoter and to be essential for initiating RNA pol I transcription in vitro. It was present in the expression site body (ESB) in vivo. The occupancy by this factor and RNA pol I was several times higher in the first few hundred base pairs of the active ES than in the equivalent region of a silent ES. However, significant CITFA occupancy also was found on silent ESs (43, 44). Thus, these analyses, together with the results presented herein, allow us to claim that transcription initiation is not the determinant level of ES control. They are compatible with a short basal promoter having a permanent ability to recruit the transcription machinery, but with the major control being exerted at other levels.
Several observations argue for the reliability of the analysis including reverse transcriptase, PCR, and sequencing. First, a majority of the characterized sequences corresponded to sequences that already were known and characterized (Table S2) (3, 29, 33). Second, no single point mutation/mistake could be detected in the 152 RT-PCR products generated from the VSG transcripts. Third, 100% of the ESAG6/7 sequences obtained matched previously reported sequences and also confirmed the identity of ESAG6/7 in the active MiTat1.2 ES. Finally, the concern that the observed single-cell RT-PCR products could originate from contaminating genomic DNA was excluded thrice: by (i) negative results of reactions omitting RT; (ii) our inability to clone any relevant trypanosome DNA from RT-negative reactions already proven negative by gel electrophoresis; and (iii) the absence of PCR products corresponding to unspliced RNAs in our analysis of poly(A) polymerase transcripts, the only documented intron-containing gene in T. brucei (35).
However, some ES sequences had not been reported before. Among other possibilities, such as PCR/sequencing artifacts linked to some amplicons, these findings could result from incomplete genomic ES sequencing in current databases, for example on intermediate chromosomes that are known to contain ESs (45). Whatever the reason, the presence of these outliers (in the range of 10–20% of total sequences) cannot interfere significantly with our conclusions.
In conclusion, the mechanism ensuring monoallelic ES expression does not happen primarily at transcription initiation but occurs at later steps of gene expression. For instance, the active expression site is uniquely devoid of nucleosomes (9, 10), probably favoring transcription elongation and correlating with a higher occupancy by RNA pol I and RNA pol I transcription factor at the beginning of the active ES (44). Also, rates of transcript processing and cytoplasmic export and stability are different in transcripts of the active and inactive ESs (29). Thus, the model that best explains VSG monoallelic expression at present involves multiple layers of control. These include factors repressing silent ES and specifically favoring expression of the active ES, including transcription factors, RNA pol I itself, chromatin remodeling factors, and elongation and RNA-processing factors. As already suggested (5), recruitment of these factors, including a functional RNA factory, could be possible only in the unique ES body, allowing full elongation of transcription, and hence VSG transcription, in a single ES at a time.
Materials and Methods
Trypanosomes and Cell Culture.
T. brucei procyclic forms were from the EATRO1125 (46) or Lister 427 (47) strains. They were grown at 27 °C with 5% CO2 in SDM79 medium (48) supplemented with 15% (vol/vol) FBS. Bloodstream forms were from the EATRO1125 strain of the AnTaR1 repertoire (46) or the 328-114 cell line from the Lister 427 strain of the MiTaR1 repertoire (49). They were grown at 37 °C with 5% CO2 in HMI-9 medium (50) supplemented with 10% (vol/vol) FBS (Sigma) and 10% (vol/vol) SERUM PLUS (SAFC Biosciences). Electroporation and selection procedures were conducted as described previously (51).
For control experiments, 328-114 cells were transfected separately with 10 plasmids constructed on a p2T7177 (32) background by insertion of gene fragments allowing expression of double-stranded RNAs targeting the following genes (the size of the amplified fragment is given in parentheses): Tb927.10.13320 (Rpb5: DNA-directed RNA polymerase II subunit) (485 bp); Tb927.11.7280 (Rpb9:DNA-directed RNA polymerase II, subunit 9) (413 bp); Tb927.9.11000 (RAB7:GTP- binding protein) (405 bp); Tb927.1.2980 (hypothetical protein) (590 bp); Tb927.10.16100 [FK506-binding protein (FKBP)-type peptidyl-prolyl isomerase] (two different regions, of 370 and 351 bp); Tb927.10.4620 (peptidyl-prolyl cis-trans isomerase) (493 bp); Tb927.1.2990 (hypothetical protein) (640 bp); Tb927.10.8400 (legume-like lectin) (469 bp); and Tb927.7.3420 (peptidyl-prolyl cis-trans isomerase) (1,065 bp).
PCR and RT-PCR.
Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen). RNA was extracted using the TRIzol reagent (Invitrogen). RT-PCR was performed using the OneStep RT-PCR Kit (Qiagen). PCR was performed using Phusion High-Fidelity DNA Polymerase FINNZYMES. PCR and RT-PCR conditions were per the manufacturer’s guidelines with a variation in the number of cycles.
Serial Dilutions, Single-Cell Pcr, and Single-Cell RT-PCR.
Trypanosomes were washed with PBS, counted, and diluted to reach a density of one trypanosome per 3 µL of PBS (suspension A). Then 1 µL of suspension A was dispensed per well already containing 100 µL of culture medium in 96-well plates. As expected, after 7 d of incubation approximately one third of the wells contained parasites (27/96, 32/96 and 30/96 wells in three independent experiments). Suspension A was dispensed simultaneously in Eppendorf tubes at 1 µL per tube (statistically, only one in three Eppendorf tubes should contain one trypanosome) and was frozen immediately in liquid nitrogen. After validation of the serial dilution procedure, these tubes were subjected to five freeze/thaw cycles (1 min in liquid nitrogen/1 min at 37 °C). Then 1 µL of TURBO DNase buffer and 1 µL of RNase-free TURBO DNase (Ambion) were added to each tube, and digestion was allowed to proceed for 15 min at 37 °C, followed by incubation for 15 min at 65 °C. The content of each tube then was split into two PCR tubes. RT-PCR was performed for 45 cycles using the Qiagen OneStep RT-PCR Kit according to the manufacturer’s instructions, omitting reverse transcriptase in one of the tubes. RT-PCR products then were cloned in the TOPO plasmid according to the Zero Blunt TOPO PCR Cloning Kit procedure and were electroporated in Escherichia coli. Plasmids were prepared from individual colonies and sequenced. PCR amplification of single-cell genomic DNA was performed using high-fidelity Phusion DNA polymerase (Thermo Scientific) in Eppendorf tubes coming from the same serial dilution procedure and submitted to the same five freeze-thaw cycles.
The following PCR primers were used in this study:
Primer pair 1 and 2: ACGGTGTGGCTGACGTCTCGAACCG (primer 1) and CCACGGGATAAAGCGTAGATGAG (primer 2), generating an amplified sequence of 149 bp starting at +1 bp downstream from the transcription initiation site of the BES3/TAR2 VSG ESs (3)
Primer pair 3 and 4: ACATTCCAGCAGGAGTTGGAGGAAATGAGG (primer 3) and CACGTGAATCCTCAATTTTGTAAAGGGTTTCAGGTCC (primer 4), generating an amplified sequence of 171 bp shared between ESAG 6 and ESAG7 (starting at 330 bp and ending at 501 of the ORF) (3)
SL (splice leader): TAGAACAGTTTCTGTACTATATTG and UTR: TTTTTTTTTTASGTGTTAAAATATATC, amplifying the whole VSG sequence
TiTA FORW (X): CCGCTCTAGAACTAGTGGA and HYG1 REV (Y): CGCGGTGAGTTCAGGC, used to check the different RNAi target genes in P2T7177 plasmid
Primer A: PAP5′EXONFOW: AGTCCCAATCCGGTTTTAACGC
Primer B: PAP5′INTRONFOW: GCATCTCCCCTCTTCTTCC
Primer C: PAP3′INTROREV: CGACGACGAACAGACTCACATC
Primer D: PAP3′EXONREV: CAGAGTTATATAACAACCTCGGAAACTCATCG.
Primers A and D anneal respectively to the exon sequences flanking PAP intron at 5′ and 3′. Primers B and C anneal to the PAP intron sequence.
Supplementary Material
Acknowledgments
We thank Pierrick Uzureau, Laurence Lecordier, and Fadi Abdel-Sater for advice and technical help and Sylvie Dickens, Cyril Gueydan, and Oberdan Leo for comments on the manuscript. This work was supported by the Interuniversity Attraction Pole Program of the Belgian Science Policy Office, a grant from the Belgian National Fund for Scientific Research (FRS-FNRS) (to L.V.), and the Brachet Foundation. A.K. was the recipient of a Fonds pour la formation à la Recherche dans l'Industrie et l'Agriculture Fellowship and now is the recipient of a fellowship from the association “Les Amis de l'Institut Pasteur à Bruxelles.” L.V. is Director of Research at the FRS-FNRS.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404873111/-/DCSupplemental.
References
- 1.Leung E, Weil DE, Raviglione M, Nakatani H. World Health Organization World Health Day Antimicrobial Resistance Technical Working Group The WHO policy package to combat antimicrobial resistance. Bull World Health Organ. 2011;89(5):390–392. doi: 10.2471/BLT.11.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry JD, McCulloch R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 3.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 4.Glover L, et al. Antigenic variation in African trypanosomes: The importance of chromosomal and nuclear context in VSG expression control. Cell Microbiol. 2013;15(12):1984–1993. doi: 10.1111/cmi.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414(6865):759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 6.Kawahara T, et al. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol Microbiol. 2008;69(4):1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel TN, et al. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol Microbiol. 2008;67(4):762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6(7):e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo LM, Cross GA. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9(1):148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanne TM, Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot Cell. 2010;9(1):136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narayanan MS, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41(5):2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei. PLoS Pathog. 2012;8(11):e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan MS, et al. NLP is a novel transcription regulator involved in VSG expression site control in Trypanosoma brucei. Nucleic Acids Res. 2011;39(6):2018–2031. doi: 10.1093/nar/gkq950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denninger V, Fullbrook A, Bessat M, Ersfeld K, Rudenko G. The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei. Mol Microbiol. 2010;78(2):459–474. doi: 10.1111/j.1365-2958.2010.07350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes K, et al. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26(9):2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benmerzouga I, et al. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol Microbiol. 2013;87(1):196–210. doi: 10.1111/mmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137(1):99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiengwe C, et al. Identification of ORC1/CDC6-interacting factors in Trypanosoma brucei reveals critical features of origin recognition complex architecture. PLoS ONE. 2012;7(3):e32674. doi: 10.1371/journal.pone.0032674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsford S, Horn D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012;40(20):10150–10160. doi: 10.1093/nar/gks813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DuBois KN, et al. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10(3):e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HS, Park SH, Günzl A, Cross GA. MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS ONE. 2013;8(2):e57001. doi: 10.1371/journal.pone.0057001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pays E, Coquelet H, Pays A, Tebabi P, Steinert M. Trypanosoma brucei: Posttranscriptional control of the variable surface glycoprotein gene expression site. Mol Cell Biol. 1989;9(9):4018–4021. doi: 10.1128/mcb.9.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudenko G, Blundell PA, Taylor MC, Kieft R, Borst P. VSG gene expression site control in insect form Trypanosoma brucei. EMBO J. 1994;13(22):5470–5482. doi: 10.1002/j.1460-2075.1994.tb06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanhamme L, Berberof M, Le Ray D, Pays E. Stimuli of differentiation regulate RNA elongation in the transcription units for the major stage-specific antigens of Trypanosoma brucei. Nucleic Acids Res. 1995;23(11):1862–1869. doi: 10.1093/nar/23.11.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarro M, Cross GA. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol Cell Biol. 1996;16(7):3615–3625. doi: 10.1128/mcb.16.7.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro M, Cross GA. In situ analysis of a variant surface glycoprotein expression-site promoter region in Trypanosoma brucei. Mol Biochem Parasitol. 1998;94(1):53–66. doi: 10.1016/s0166-6851(98)00049-8. [DOI] [PubMed] [Google Scholar]
- 27.Alarcon CM, Pedram M, Donelson JE. Leaky transcription of variant surface glycoprotein gene expression sites in bloodstream african trypanosomes. J Biol Chem. 1999;274(24):16884–16893. doi: 10.1074/jbc.274.24.16884. [DOI] [PubMed] [Google Scholar]
- 28.Ansorge I, Steverding D, Melville S, Hartmann C, Clayton C. Transcription of ‘inactive’ expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol Biochem Parasitol. 1999;101(1-2):81–94. doi: 10.1016/s0166-6851(99)00060-2. [DOI] [PubMed] [Google Scholar]
- 29.Vanhamme L, et al. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36(2):328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 30.Chaves I, et al. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95(21):12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18(17):4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125(1-2):211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 33.Hertz-Fowler C, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE. 2008;3(10):e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young R, et al. Isolation and analysis of the genetic diversity of repertoires of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum. BMC Genomics. 2008;9:385. doi: 10.1186/1471-2164-9-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mair G, et al. A new twist in trypanosome RNA metabolism: Cis-splicing of pre-mRNA. RNA. 2000;6(2):163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zomerdijk JC, Kieft R, Duyndam M, Shiels PG, Borst P. Antigenic variation in Trypanosoma brucei: A telomeric expression site for variant-specific surface glycoprotein genes with novel features. Nucleic Acids Res. 1991;19(7):1359–1368. doi: 10.1093/nar/19.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhamme L, Lecordier L, Pays E. Control and function of the bloodstream variant surface glycoprotein expression sites in Trypanosoma brucei. Int J Parasitol. 2001;31(5-6):523–531. doi: 10.1016/s0020-7519(01)00143-6. [DOI] [PubMed] [Google Scholar]
- 38.Jefferies D, Tebabi P, Pays E. Transient activity assays of the Trypanosoma brucei variant surface glycoprotein gene promoter: Control of gene expression at the posttranscriptional level. Mol Cell Biol. 1991;11(1):338–343. doi: 10.1128/mcb.11.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zomerdijk JC, et al. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 1990;9(9):2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pays E, et al. Trypanosoma brucei: Constitutive activity of the VSG and procyclin gene promoters. EMBO J. 1990;9(10):3145–3151. doi: 10.1002/j.1460-2075.1990.tb07512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanhamme L, Pays A, Tebabi P, Alexandre S, Pays E. Specific binding of proteins to the noncoding strand of a crucial element of the variant surface glycoprotein, procyclin, and ribosomal promoters of trypanosoma brucei. Mol Cell Biol. 1995;15(10):5598–5606. doi: 10.1128/mcb.15.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83(4):547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 43.Brandenburg J, et al. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007;26(23):4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen TN, Müller LS, Park SH, Siegel TN, Günzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res. 2014;42(5):3164–3176. doi: 10.1093/nar/gkt1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berriman M, et al. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol. 2002;122(2):131–140. doi: 10.1016/s0166-6851(02)00092-0. [DOI] [PubMed] [Google Scholar]
- 46.Bajyana Songa E, Van Meirvenne N, Murray M. [Characterization of the AnTAR 2 antigen of Trypanosoma brucei brucei] Ann Soc Belg Med Trop. 1984;64(1):13–20. [PubMed] [Google Scholar]
- 47.Cross GA, Manning JC. Cultivation of Trypanosoma brucei sspp. in semi-defined and defined media. Parasitology. 1973;67(3):315–331. doi: 10.1017/s0031182000046540. [DOI] [PubMed] [Google Scholar]
- 48.Brun R, Jenni L. A new semi-defined medium for Trypanosoma brucei sspp. Acta Trop. 1977;34(1):21–33. [PubMed] [Google Scholar]
- 49.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 50.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75(6):985–989. [PubMed] [Google Scholar]
- 51.Devaux S, et al. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol Biochem Parasitol. 2006;148(1):60–68. doi: 10.1016/j.molbiopara.2006.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.