Significance
Microscopic marine algae (phytoplankton) are responsible for much of Earth's photosynthesis, serving as the base of a massive food web supporting fisheries. Phytoplankton compete for limiting resources, with some species producing noxious compounds that kill competitors or inhibit their growth. The red-tide dinoflagellate Karenia brevis is one such allelopathic species, causing growth suppression of other phytoplankton and negatively impacting coastal ecosystems. Metabolomic and proteomic approaches were used to characterize the sublethal physiological impacts of K. brevis allelopathy on two competing phytoplankton, providing insights into the physiological mechanisms by which allelopathy occurs and the metabolic pathways that enable resistance in co-occurring competitors.
Keywords: chemical ecology, systems biology, mass spectrometry
Abstract
Competition is a major force structuring marine planktonic communities. The release of compounds that inhibit competitors, a process known as allelopathy, may play a role in the maintenance of large blooms of the red-tide dinoflagellate Karenia brevis, which produces potent neurotoxins that negatively impact coastal marine ecosystems. K. brevis is variably allelopathic to multiple competitors, typically causing sublethal suppression of growth. We used metabolomic and proteomic analyses to investigate the role of chemically mediated ecological interactions between K. brevis and two diatom competitors, Asterionellopsis glacialis and Thalassiosira pseudonana. The impact of K. brevis allelopathy on competitor physiology was reflected in the metabolomes and expressed proteomes of both diatoms, although the diatom that co-occurs with K. brevis blooms (A. glacialis) exhibited more robust metabolism in response to K. brevis. The observed partial resistance of A. glacialis to allelopathy may be a result of its frequent exposure to K. brevis blooms in the Gulf of Mexico. For the more sensitive diatom, T. pseudonana, which may not have had opportunity to evolve resistance to K. brevis, allelopathy disrupted energy metabolism and impeded cellular protection mechanisms including altered cell membrane components, inhibited osmoregulation, and increased oxidative stress. Allelopathic compounds appear to target multiple physiological pathways in sensitive competitors, demonstrating that chemical cues in the plankton have the potential to alter large-scale ecosystem processes including primary production and nutrient cycling.
Marine phytoplankton are responsible for ∼50% of global net primary production (1), which ultimately drives the global carbon cycle (2). This primary production is vital for higher trophic levels with interactions among species playing a critical role in controlling the flux of biomass and nutrients in the water column (3). Chemical cues and signals mediate many interactions among planktonic organisms, including competition (4, 5), defense against grazers (6, 7), predator detection (8), prey capture (9), and signaling between neighbor cells during bloom events (10). Therefore, chemical cues that affect ecological interactions between microalgae are hypothesized to alter these large-scale ecosystem processes.
Allelopathy, the production and release of chemical compounds to inhibit or kill competitor species, is a form of interference competition that alters community composition in terrestrial (11) and benthic aquatic communities (12, 13). In the plankton, allelopathy is noted as a strong structuring force that alters species succession (14, 15) and community composition (16, 17) by influencing a variety of species-specific physiological targets in competitors. Some allelopathic interactions result in massive mortality of competitors (18) or initiation of programmed cell death pathways (15). However, allelopathy is not always lethal as it can cause sublethal physiological responses in competitors, such as cyst formation (19), altered cell swimming behavior (5, 6), or reduced growth rate (e.g., refs. 20 and 21).
The red-tide dinoflagellate Karenia brevis is known for a suite of potent neurotoxins, brevetoxins, responsible for neurotoxic shellfish poisoning in humans as well as fish and marine mammal mortalities during bloom events in the Gulf of Mexico (22). Although brevetoxins have been found to lack allelopathic potency in most studies thus far (4, 23–25), K. brevis produces an additional group of unstable, polar compounds (24) that negatively influence several competing phytoplankton species (4, 25). As a relatively weak exploitation competitor (26), K. brevis may use allelopathy to maintain monospecific blooms in near-shore waters (4). Competitor susceptibility to K. brevis allelopathy is at least partly mediated by ecological context and not all competitors are equally affected (4). The presence of particular competitor species modulates allelopathic potency (27), and competitors in earliest growth stages are most susceptible to K. brevis allelopathy (23). Allelopathic compounds produced by K. brevis cause sublethal reductions in growth and photosynthetic efficiency as well as increased cell membrane permeability (25), although the exact cellular metabolic targets of K. brevis allelopathy are unknown.
Metabolomics and proteomics provide systems-level snapshots of the metabolism of a cell or organism at the time of harvest (28–30). Proteins are responsible for cellular signals, structural integrity, and catalysis of most biochemical reactions including the production and conversion of the vast array of metabolites required for cellular survival. Although metabolomics and proteomics have been used in the past to examine diatoms adapting to various stressors (31–34), the current work represents, to our knowledge, the first instance of metabolites and proteins measured simultaneously to understand the effects of allelopathy or any form of competition. To identify potential cellular targets of K. brevis allelopathy and better understand effects of sublethal chemical cues in marine ecosystems, we examined two diatom species exposed to K. brevis allelopathic compounds using whole-cell proteomics and metabolomics. This integrated systems biology approach provided multiple lines of evidence from which we distinguished metabolic responses of these two competitors to K. brevis allelopathy.
Results and Discussion
Allelopathy Impacts Competitor Metabolism.
Although previous studies (e.g., refs. 4 and 25) have shown that K. brevis allelopathy reduces the growth of competitor species, the current study demonstrates the impact of allelopathy on global cellular function and indicates that, even in the case of modest growth inhibition, metabolism was altered. K. brevis allelopathy differentially affected growth of the two competitors (Fig. 1), reflected in both the metabolomes (Fig. 2 and Fig. S1) and proteomes (Table 1) of each competitor. In the presence of K. brevis, Thalassiosira pseudonana growth was reduced by 85% (Fig. 1B; n = 15; P < 0.0001), with growth suppression and subsequent population decline observed after 4 d of coculture with species physically separated by dialysis membrane. Analysis of the T. pseudonana metabolome (Table 2 and Fig. 2 A and B) and proteome (Table 1) revealed that K. brevis allelopathy impacts multiple metabolic pathways, with some cellular responses possibly used to compensate for negative effects of allelopathy. In contrast, the overall growth of Asterionellopsis glacialis was only reduced by 35% when exposed to K. brevis allelopathy, an effect that was not statistically significant (Fig. 1B; n = 15; P = 0.089), indicating, at most, a weak allelopathic effect of K. brevis on A. glacialis in this experiment. In the A. glacialis experiment, final K. brevis cell concentrations in flasks averaged 15.3 × 103 cells per mL, whereas in the T. pseudonana experiment, mean K. brevis concentrations were 8.3 × 103 cells per mL at the time of harvest (Fig. 1A). Nutrients were not limiting in these experiments (SI Materials and Methods), suggesting that the negative effects of K. brevis on competitor growth were mediated by allelopathy rather than exploitation competition. Overall, it is clear that, based on both the proteomic and metabolomic data, a sensitive competitor exposed to allelopathy operates in a functionally compromised metabolic state (Fig. 2 and Fig. S1).
Fig. 1.
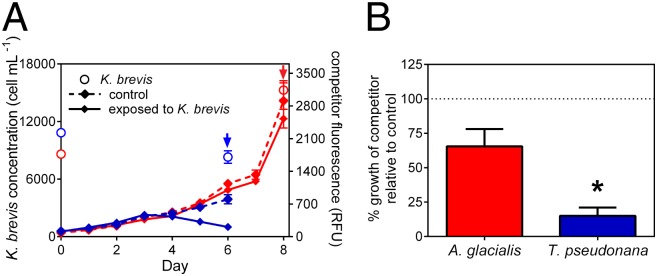
Effects of exposure to exudates of live K. brevis on the growth of A. glacialis and T. pseudonana. (A) A. glacialis (red) in vivo and T. pseudonana (blue) in vivo fluorescence (arrow indicates day of harvest for metabolomics and proteomics). The solid lines indicate fluorescence of diatom-only controls, and the dashed lines indicate fluorescence of diatoms exposed to K. brevis. Initial K. brevis (red open circles for A. glacialis experiment; blue open circle for T. pseudonana experiment) concentrations from cultures used to fill dialysis tubes (n = 1), final concentrations from experimental flasks at time of harvest (n = 15). (B) Calculated percentage growth of competitors A. glacialis (red) and T. pseudonana (blue) relative to their own controls after 8 and 6 d exposure to K. brevis, respectively. The dotted line indicates growth equivalent to control. n = 15. P < 0.0001 indicated by asterisk (*), unpaired t test. Error bars represent ±1 SEM.
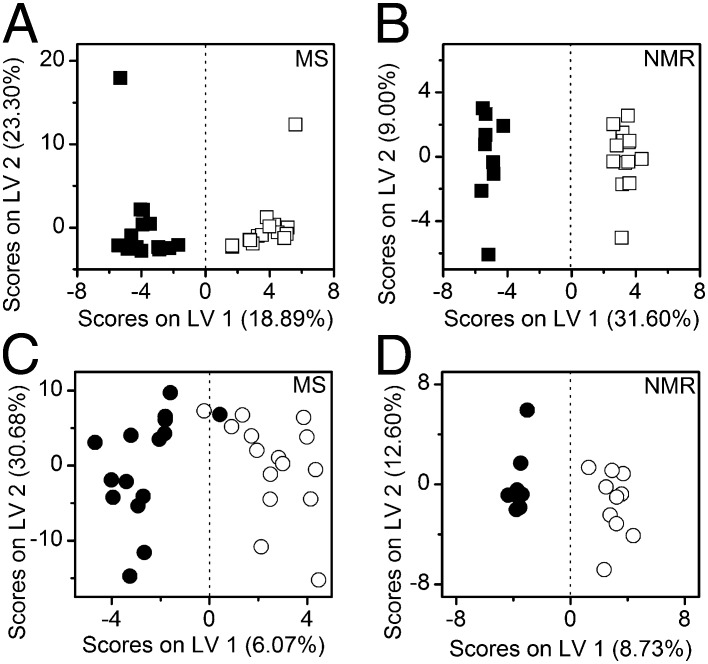
Fig. 2.
Orthogonal PLS-DA shows effects of K. brevis allelopathy on the metabolomes of competitor diatoms. PLS-DA scores plot of (A) UHPLC/MS metabolic features and (B) 1H NMR spectral data for T. pseudonana exposed to K. brevis (filled squares) or dilute media control (open squares) with cross-validated accuracies of 87% and 100%, respectively. PLS-DA scores plot of (C) UHPLC/MS metabolic features and (D) 1H NMR spectral data for A. glacialis exposed to live K. brevis (filled circles) or dilute media controls (open circles) with cross-validated accuracies of 57% and 63%, respectively.
Table 1.
Metabolic pathways and cellular functions from Biological Process and Molecular Function Gene Ontology categories for proteins in T. pseudonana whose concentrations increased (△) or decreased (▼) in response to K. brevis allelopathy
| Effect on concentration | Metabolic pathway/function | No. of proteins* | P value† |
| △ | Generation of precursor metabolites and energy | 25 | 8.9E-12 |
| △ | Metabolic process | 101 | 3.2E-10 |
| △ | Cellular carbohydrate metabolic process | 16 | 2.0E-08 |
| △ | Monosaccharide metabolic process (hexose and glucose) | 14 | 2.1E-08 |
| △ | Alcohol metabolic process | 15 | 3.6E-08 |
| △ | Energy coupled proton transport, down electrochemical gradient | 10 | 4.7E-07 |
| △ | ATP metabolic process | 11 | 1.7E-06 |
| △ | Oxidative phosphorylation | 10 | 3.8E-06 |
| △ | Nucleoside triphosphate metabolic process | 11 | 4.5E-06 |
| △ | Cellular metabolic process | 73 | 5.7E-06 |
| △ | Proton-transporting ATPase activity, rotational mechanism | 9 | 9.7E-10 |
| △ | H-ion transport ATP synthase activity, rotational mechanism | 8 | 2.2E-08 |
| △ | Catalytic activity | 96 | 4.2E-08 |
| △ | ATPase activity, coupled to transmembrane movement of ions | 10 | 9.3E-08 |
| △ | Hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | 11 | 8.2E-07 |
| △ | Lyase activity | 15 | 2.2E-06 |
| △ | Cofactor binding | 19 | 3.3E-06 |
| ▼ | Photosynthesis | 12 | 7.7E-11 |
| ▼ | Generation of precursor metabolites and energy | 13 | 1.3E-09 |
| ▼ | Chromatin assembly | 8 | 1.6E-09 |
| ▼ | Cellular macromolecular complex subunit organization | 8 | 3.5E-07 |
| ▼ | Cellular component biogenesis | 9 | 8.5E-07 |
| ▼ | Electron carrier activity | 6 | 1.3E-03 |
| ▼ | Iron ion binding | 6 | 2.8E-03 |
| ▼ | 4 Iron, 4 sulfur cluster binding | 3 | 3.0E-03 |
| ▼ | Metal cluster binding | 4 | 4.3E-03 |
Number of proteins represents the number of proteins that correlate to the annotation category.
P value is the modified Fisher exact P value for protein enrichment analysis.
Table 2.
Candidate metabolites tentatively identified by NMR spectra and UHPLC/MS to increase in concentration (△) or decrease in concentration (▼) in T. pseudonana exposed to K. brevis
| Effect on concentration | Candidate metabolite | Biological subcategory | Fold change* | Method of detection | P value† |
| Cell protection | |||||
| △ | Lipid (C31H42O9)‡ | Membrane constituent | +10.3 | UHPLC/MS | 6.9E-17 |
| △ | Phospholipid (C24H47O12P)‡ | Membrane constituent | +10.3 | UHPLC/MS | 6.9E-17 |
| △ | Sarcosine§ | Glycine metabolism/osmoregulation | +0.32 | 1H-HSQC NMR | 4.9E-1 |
| ▼ | Mannan¶|| | Cell wall constituent | −3.4 | UHPLC/MS | 9.8E-16 |
| ▼ | Phospholipid (C26H43O9P)** | Membrane constituent | −1.4 | UHPLC/MS | 2.5E-15 |
| ▼ | Lipid (C26H42O11)** | Membrane constituent | −1.4 | UHPLC/MS | 2.5E-15 |
| ▼ | Terpene glycoside | Membrane constituent | −1.2 | UHPLC/MS | 6.9E-15 |
| ▼ | Betaine | Osmoregulation | −0.58 | 1H-HSQC NMR | 2.9E-3 |
| ▼ | myo-Inositol†† | Osmoregulation | −0.41 | 1H-HSQC NMR | 6.2E-1 |
| ▼ | Homarine | Osmoregulation | −2.5 | 1H-HSQC NMR | 3.0E-5 |
| ▼ | Polyphenol¶ | Oxidative stress | −3.4 | UHPLC/MS | 9.8E-16 |
| ▼ | Taurine | Amino acid metabolism | N/A | 1H-HSQC NMR | N/A |
| Energy metabolism | |||||
| △ | Acetate | Carbon metabolism/glycolysis | +0.92 | 1H-HSQC NMR | 1.8E-2 |
| △ | Dimethylamine§ | Glycine metabolism/osmoregulation | +0.32 | 1H-HSQC NMR | 4.9E-1 |
| ▼ | Alanine | Amino acid metabolism | −0.48 | 1H-HSQC NMR | 1.1E-1 |
| ▼ | Stachyose¶|| | Carbon metabolism | −3.4 | UHPLC/MS | 9.8E-16 |
| ▼ | Cellotetraose¶|| | Carbon metabolism | −3.4 | UHPLC/MS | 9.8E-16 |
| ▼ | Maltopentaose‡‡ | Carbon metabolism | −3.2 | UHPLC/MS | 3.3E-20 |
| ▼ | Amylopectin‡‡ | Carbon metabolism | −3.2 | UHPLC/MS | 3.3E-20 |
| ▼ | Cellopentaose‡‡ | Carbon metabolism | −3.2 | UHPLC/MS | 3.3E-20 |
| ▼ | Verbascose‡‡ | Carbon metabolism | −3.2 | UHPLC/MS | 3.3E-20 |
| ▼ | Glucose†† | Carbon metabolism/glycolysis | −0.41 | 1H-HSQC NMR | 2.1E-1 |
| ▼ | Glycerate | Photorespiration | −0.024 | 1H-HSQC NMR | 9.2E-1 |
| ▼ | Dihydrouracil | Pyrimidine metabolism | −0.91 | 1H-HSQC NMR | 2.4E-2 |
| Cell protection/energy metabolism | |||||
| △ | Glutamate | Amino acid metabolism/osmoregulation | +1.22 | 1H-HSQC NMR | 1.4E-3 |
| △ | Proline | Amino acid metabolism/osmoregulation | +0.052 | 1H-HSQC NMR | 8.8E-1 |
Fold change was calculated as the base 2 logarithm of the average peak area ratios for exposed T. pseudonana samples and control samples.
P values calculated using unpaired, two-tailed t test.
Isobars.
Metabolites share observed chemical shifts.
Isobars.
Isomers.
Isobars.
Metabolites share multiple observed chemical shifts.
Isomers.
A Co-occurring Competitor Has a Robust Metabolism in Response to Allelopathy.
To date, metabolomics and proteomics are underused tools for investigating the impacts of ecological interactions on organism physiology. This study revealed that metabolomic and proteomic analyses of competitors can demonstrate the species-specific effects of K. brevis allelopathy. The diatom A. glacialis appeared to maintain a relatively robust metabolism in response to K. brevis, which resulted in mild allelopathic effects on growth (Figs. 1 and 2 C and D). Using multivariate analysis [orthogonal partial least-squares discriminant analysis (PLS-DA)], we identified groups of metabolites whose covarying concentrations allowed us to distinguish metabolomes of K. brevis-exposed and control cultures of competitors (SI Materials and Methods). Although concentrations of these compounds may not have significantly differed between treatments and controls when each compound was considered alone, these compounds displayed explanatory power when part of a discriminating panel (35). Orthogonal PLS-DA analysis suggested that concentrations of only a few aliphatic metabolites were enhanced in A. glacialis by exposure to allelopathy (i.e., glycerophosphocholine and lactose or polysaccharides, as well as two unidentified compounds; Table S1). However, only 6–9% (Fig. 2 C and D) of the observed among-culture variation in the A. glacialis metabolome was due to K. brevis exposure, indicating that this species maintains relatively normal functioning in response to K. brevis. Additionally, only one protein increased in abundance when exposed to K. brevis according to differential expression protein analyses (ATP synthase), whereas five proteins were significantly suppressed (e.g., cytochrome b559, photosystem II chlorophyll A antenna apoprotein; Dataset S1). The strain of A. glacialis used in this experiment was originally isolated from the Gulf of Mexico, where K. brevis blooms regularly occur (22). Thus, A. glacialis may have evolved partial resistance to K. brevis allelopathy due to its potential for frequent exposure to K. brevis blooms (36). T. pseudonana used in the current study, however, was from the North Atlantic Ocean where K. brevis does not occur. It is possible that the robust metabolome of A. glacialis has resulted from multiple metabolic adaptations that mitigate allelopathic impacts, allowing A. glacialis cells to function when challenged by K. brevis allelopathy.
A Sensitive Competitor Experiences Altered Energy Metabolism and Increased Cell Stress in Response to Allelopathy.
Because of its sensitivity to K. brevis allelopathy (Fig. 1), T. pseudonana serves as a suitable model for exploring mechanisms by which allelopathy affects competitor physiology. From our integrated systems biology approach, we found T. pseudonana energy metabolism to be greatly impacted by K. brevis allelopathy (Fig. 3). An observed significant increase of acetate concentrations (+0.92-fold), as well as decreased myo-inositol and/or glucose concentrations, as distinguished by orthogonal PLS-DA (Table 2, Table S2, and Fig. S2), with concomitant increased concentrations of glycolysis enzymes (e.g., pyruvate kinase +3.0-fold; Table 1 and Dataset S2), suggests that glycolysis was enhanced in T. pseudonana. Starch stores were depleted, as indicated by significantly lowered concentration of maltopentose, amylopectin, cellopentose, and/or verbascose (−3.2-fold) (Table 2 and Table S3), whereas concentrations of small (e.g., C8-C10) fatty acids increased (Table S2 and Fig. S2). Several fatty acid biosynthesis and metabolism enzymes (Table 1) were more abundant in T. pseudonana in response to K. brevis allelopathy, including acetyl CoA carboxylase (+2.3) and acyl CoA dehydrogenase (+3.0-fold) (Dataset S2). Together, this suggests that allelopathy caused enhanced glycolysis in T. pseudonana, while also stimulating synthesis and β-oxidation of fatty acids. Allelopathy may have altered the photosynthetic capability of T. pseudonana, as evidenced by the decreased abundance of 12 photosynthetic proteins, even though the abundance of RuBisCO, the key enzyme responsible for carbon fixation, remained unchanged (Table 1 and Dataset S2). The reduction in photosystem complexes and surge in fatty acid β-oxidation may indicate that cells experiencing allelopathy became carbon limited.
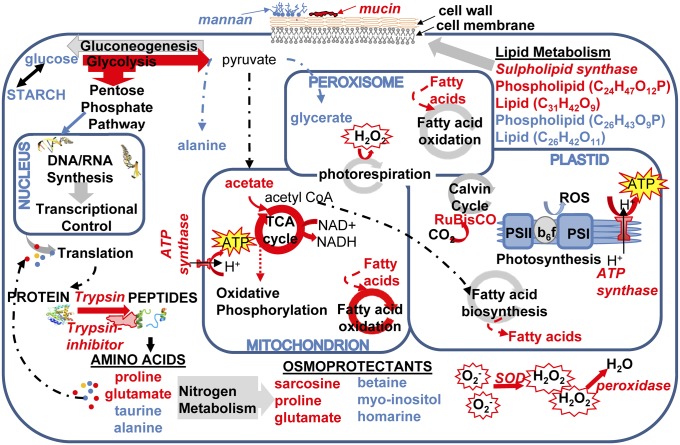
Fig. 3.
Network of cellular pathways, enzymes, and metabolites in the diatom T. pseudonana impacted by exposure to K. brevis allelopathy, derived from NMR and MS metabolomics and proteomics. Pathways and metabolites enhanced by allelopathy are indicated by red arrows and compound names, respectively. The blue arrows and compound names denote pathways and metabolites that were suppressed by allelopathy.
Pathways Involved in Cell Protection Are Impacted by K. brevis Allelopathy.
In previous work, K. brevis exudates were found to increase cell membrane permeability of several competitors, via unknown mechanisms (25). In the current study, metabolic and biosynthetic activities associated with cell membranes were altered in T. pseudonana in response to K. brevis allelopathy, as evidenced by decreased mannan and C26 phospholipid or C31 lipid concentrations, as well as an increase in a C24 phospholipid or C26 lipid (Table 2 and Table S3). This is suggestive of membrane restructuring and a possible increase in permeability because mannans and phospholipids are primary membrane components of T. pseudonana (37) and other photosynthetic organisms (38). These changes could be the result of membrane instability and the subsequent attempt of T. pseudonana to alter membrane phospholipid content via increased biosynthesis. Recently, Martin et al. (39) revealed that phosphorus-deprived T. pseudonana substitute betaine lipids and sulfolipids for phospholipids in cellular membranes. In addition, Riekhof et al. (40) demonstrated that betaine lipid synthesis is associated with the degradation of membrane phospholipids. These coordinated pathways provide a possible explanation to the significantly reduced betaine concentrations (−0.58-fold) because betaine would be consumed to build betaine-based lipids (Table 2 and Table S2). Enzymes involved in sulfolipid biosynthesis were present in higher concentrations in T. pseudonana responding to K. brevis (e.g., sulfolipid biosynthesis protein; Dataset S2), suggesting a modification of T. pseudonana’s cell membrane content via an increase in sulfolipids. Although phosphate was not limiting to T. pseudonana in this experiment (SI Materials and Methods), it is possible that K. brevis allelopathy reduced the capacity of T. pseudonana to use nutrients present in the media. Plankton lipids mediate a number of important ecological interactions, including grazing (41), susceptibility to viral infection (42), and toxin sensitivity (43). Thus, future experiments testing how phytoplankton allelopathy alters lipid biochemistry and membrane stability could lead to discovery of cascading ecological effects on other types of interactions.
Another critical aspect of membrane structure and function is the ability of a cell to control solute osmosis. From the current study, it is apparent that osmotic regulation was impacted by K. brevis allelopathy: concentrations of homarine (−2.5-fold) and betaine were suppressed when T. pseudonana was exposed to K. brevis (Table 2 and Table S2) and are recognized osmolytes in marine planktonic organisms (44, 45). T. pseudonana may respond to K. brevis allelopathy by compensating for impeded osmotic regulation by enhancing or maintaining concentrations of both proline and dimethylamine (33, 46) (Table 2).
When T. pseudonana was exposed to K. brevis allelopathy, multiple enzymes involved in critical pathways related to oxidative stress were enhanced (Table 1). In addition, several enzymes responsible for mitigating high oxidative stress were more abundant. Fifteen oxidoreductases increased in abundance, including manganese superoxide dismutase and ascorbate peroxidase (Dataset S2). Our PLS-DA model suggests glycerate concentrations were reduced, as part of a group of compounds whose concentrations covaried, in T. pseudonana exposed to K. brevis indicating that photorespiration was disrupted (Table 2 and Fig. S2), which would also likely increase oxidative stress (47, 48). Recently, Schlegel et al. (49) proposed that an increase in oxidative stress may force cells to reduce the abundance of photosynthetic and electron carrier enzymes. Consistent with that prediction, in the current study proteins involved in electron carrier activity were suppressed, whereas proton transport activity was enhanced, suggesting potential compensation within T. pseudonana cells (Table 1 and Dataset S2). Seven proteins involved in the pentose phosphate pathway, a side step to glycolysis that yields reducing equivalents of NADPH (31), yielded heightened abundances (Table 1 and Dataset S2). Recent findings also show that diatoms experiencing oxidative stress lose their ability to assimilate nitrogen, due to either altered redox states of critical nitrogen assimilation enzymes, or the loss of reducing equivalents (e.g., NADPH) (50). This suggests that, in the current study, oxidatively stressed T. pseudonana may have been forced to recycle internal nitrogen stores as a downstream effect of exposure to K. brevis allelopathy. We also observed alterations in the concentrations of several amino acids (increased concentrations of glutamate and proline; decreased concentrations of taurine; Table 2 and Table S2), suggesting a reshuffling and possible recycling of internal nitrogen stores. Together, this suggests that T. pseudonana was responding to increased oxidative stress as a result of K. brevis allelopathy.
Cells in Crisis.
Overall, it appears that T. pseudonana cells exposed to K. brevis allelopathy experienced substantially disrupted metabolic processes indicative of heightened stress (Fig. 3). In addition to the metabolic and proteomic changes described above, we observed a fourfold increase in the abundance of the diatom’s trypsin with a concomitant increase in the abundance of an anti-trypsin protease inhibitor as well as reduced concentrations of a serine protease inhibitor (Dataset S2). For trypsin annotation, a total of 19 unique peptides were identified, none of which resembled the pure porcine trypsin used for digestion during sample preparation. These findings suggest that protein degradation pathways were induced, which may approach conditions typically observed for programmed cell death response in diatoms (51). In addition, concentrations of several DNA replication enzymes, as well as eight chromatin assembly proteins were suppressed (e.g., histone-3: −3.4-fold; Table 1 and Dataset S2), consistent with a strong negative impact of allelopathy on T. pseudonana growth (Fig. 1).
Mucins, gel-like glycoproteins, were in higher abundance in T. pseudonana exposed to allelopathy (Dataset S2). Although largely unexplored for phytoplankton, in other organisms mucins have been found to reduce adherence of bacteria (52). If diatom mucins prevent colonization by harmful bacteria or encourage the aggregation of diatom cells, this may represent a valuable defense mechanism or a response to stress. Furthermore, if mucins are shown to be involved in diatom aggregation, then allelopathy may directly influence the flux of particulate carbon from the water column to depth in the ocean. Mucins have also been found to demonstrate antiapoptopic activity (53, 54). Previous studies on T. pseudonana have found ascorbate peroxidase genes to be up-regulated in iron-limited situations, whereas mucin genes were down-regulated, demonstrating a balance between reactive oxygen species reducing mechanisms and the initiation of programmed cell death pathways in stressful situations (55). This balancing act ultimately allows acclimation to stress, which may be disrupted by allelopathy.
Conclusions
Whole-cell metabolomic and proteomic analyses revealed differing responses of planktonic competitors to K. brevis allelopathy, suggesting that co-occurring species may have evolved partial resistance to allelopathy via robust metabolic pathways. In contrast, a “naïve” competitor, T. pseudonana, which does not co-occur with K. brevis blooms, suffered greater metabolic disruption and growth suppression when exposed to K. brevis allelopathy. Critical metabolic processes including glycolysis, photosynthesis, cell membrane maintenance, osmoregulation, and responses to oxidative stress were all impacted in this sensitive competitor. If other sensitive competitors are also found to undergo similarly altered metabolic pathways, then allelopathy among phytoplankton may affect ecosystem-level processes even more than previously believed due to the importance of phytoplankton on carbon fixation and nutrient dynamics in the world’s oceans. This interdisciplinary systems biology approach provides an unbiased opportunity to establish novel, testable hypotheses toward understanding the mechanisms by which allelopathy alters species composition in plankton communities, and thus the roles of chemical cues in mediating important ecological interactions.
Materials and Methods
Phytoplankton Culturing.
Cultures of the diatoms Asterionellopsis glacialis strain CCMP 137 and Thalassiosira pseudonana strain CCMP 1335 were grown in artificial seawater (Instant Ocean, 35 ppt) amended with L1 plus Si media (56). Cultures were haphazardly arranged in a Percival incubator set to a 12:12 light/dark cycle with irradiance of 100–145 μmol⋅m−2⋅s−1 (Biospherical Instrument QSL2100), at 21 °C. Cultures of Karenia brevis strain CCMP 2228 were grown in L1 media-amended artificial seawater in conditions mentioned above. Populations of diatoms were quantified using either visual counts or in vivo fluorescence as a proxy for cell concentration; K. brevis cell concentrations were determined with FlowCAM (SI Materials and Methods).
Experimental Design.
A coculture design was used to investigate antagonistic interactions between K. brevis and each of two competitor species. The diatoms A. glacialis and T. pseudonana were each cocultured with the dinoflagellate K. brevis (n = 15) with the latter placed within permeable dialysis membrane tubing (e.g., ref. 57). This design allowed for ongoing exudation of allelopathic compounds from K. brevis through the dialysis membrane over the course of the experiment, but prevented cell contact between species. For controls (n = 15 for each experiment), competitors were exposed to dialysis tubes filled with L1 media diluted to 65% (vol/vol) of full L1 nitrate, vitamins, and trace metal concentrations, and 90% (vol/vol) of full L1 phosphate concentrations to mimic the nutritional environment of exponentially growing K. brevis. After a period of coculture with live K. brevis (treatments) or dilute media (controls), competitor cells were harvested, extracted, and their polar metabolite profiles compared using 1H NMR spectroscopy and ultrahigh performance liquid chromatography–mass spectrometry (UHPLC/MS). Cell pellets of diatoms were also harvested for proteomics and analyzed via LC/MS. (See SI Materials and Methods for sample preparation, procedures, and analytical method parameters.)
Metabolite Analysis and Annotation.
Before multivariate analysis, spectral data (NMR and MS) were preprocessed (SI Materials and Methods). Principal-component analysis and orthogonal PLS-DA were used to investigate alterations in the metabolomes of K. brevis-exposed diatoms in comparison with media-exposed diatoms of the same species (MATLAB, version 7.13.0, with PLS_Toolbox, version 6.71; Eigenvector Research). Those metabolites with discriminatory power were tentatively annotated using the Metlin database (58), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (59), LIPID Metabolites and Pathways Strategy (LIPID MAPS) database (60), the Madison–Qingdao Metabolomics Consortium Database (MMCD) (61), the Human Metabolome Database (62), and Chenomx Profiler. Representative extracts from each experiment were used to collect 2D NMR spectral data [standard heteronuclear single quantum coherence (HSQC) and total correlation spectroscopy experiments, 500 MHz] to aid in annotation.
Proteomics.
Cells were lysed and then digested with trypsin in an acid-cleavable detergent following manufacturer’s guidelines (SI Materials and Methods). Mass spectrometry was performed on a Thermo Fisher QExactive, and all results were searched and interpreted with SEQUEST (PVM, version 27 20070905). Further information on proteomic sample preparation and MS analyses are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful for the insights provided by Drs. M. Zhou and M. E. Monge for UHPLC/MS method development and analysis, respectively. We thank T. Alexander, E. McMillan, R. Poulin, and Dr. M. Teasdale for assistance with culturing experiments; J. Gonzales, S. Weber, and Dr. J. Montoya for nutrient analyses; Drs. L. Gelbaum and J. Leisen for assistance with NMR spectroscopy; Drs. J. Byrne and R. Davidson for assistance with NMR metabolomic analysis, supported by the UK Natural Environment Research Council's (NERC) Biomolecular Analysis Facility at the University of Birmingham (R8-H10-61). We acknowledge support from National Science Foundation Grant OCE-1060300 and support in part by the University of Washington Proteomics Resource Bioinformatics Team (UWPR95794).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402130111/-/DCSupplemental.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281(5374):200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 3.Strom SL. Microbial ecology of ocean biogeochemistry: A community perspective. Science. 2008;320(5879):1043–1045. doi: 10.1126/science.1153527. [DOI] [PubMed] [Google Scholar]
- 4.Kubanek J, Hicks MK, Naar J, Villareal TA. Does the red tide dinoflagellate Karenia brevis use allelopathy to outcompete other phytoplankton? Limnol Oceanogr. 2005;50(3):883–895. [Google Scholar]
- 5.Tillmann U, John U, Cembella A. On the allelochemical potency of the marine dinoflagellate Alexandrium ostenfeldii against heterotrophic and autotrophic protists. J Plankton Res. 2007;29(6):527–543. [Google Scholar]
- 6.Tillmann U, John U. Toxic effects of Alexandrium spp. on heterotrophic dinoflagellates: An allelochemical defence mechanism independent of PSP-toxin content. Mar Ecol Prog Ser. 2002;230:47–58. [Google Scholar]
- 7.Teegarden GJ, et al. Copepod feeding response to varying Alexandrium spp. cellular toxicity and cell concentration among natural plankton samples. Harmful Algae. 2008;7(1):33–44. [Google Scholar]
- 8.Selander E, Thor P, Toth G, Pavia H. Copepods induce paralytic shellfish toxin production in marine dinoflagellates. Proc Biol Sci. 2006;273(1594):1673–1680. doi: 10.1098/rspb.2006.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng J, Malkiel E, Katz J, Adolf JE, Place AR. A dinoflagellate exploits toxins to immobilize prey prior to ingestion. Proc Natl Acad Sci USA. 2010;107(5):2082–2087. doi: 10.1073/pnas.0912254107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vardi A, et al. A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol. 2008;18(12):895–899. doi: 10.1016/j.cub.2008.05.037. [DOI] [PubMed] [Google Scholar]
- 11.Inderjit, Wardle DA, Karban R, Callaway RM. The ecosystem and evolutionary contexts of allelopathy. Trends Ecol Evol. 2011;26(12):655–662. doi: 10.1016/j.tree.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Thacker RW, Becerro MA, Lumbang WA, Paul VJ. Allelopathic interactions between sponges on a tropical reef. Ecology. 1998;79(5):1740–1750. [Google Scholar]
- 13.Rasher DB, Hay ME. Chemically rich seaweeds poison corals when not controlled by herbivores. Proc Natl Acad Sci USA. 2010;107(21):9683–9688. doi: 10.1073/pnas.0912095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keating KI. Allelopathic influence on blue-green bloom sequence in a eutrophic lake. Science. 1977;196(4292):885–887. doi: 10.1126/science.196.4292.885. [DOI] [PubMed] [Google Scholar]
- 15.Vardi A, et al. Dinoflagellate-cyanobacterium communication may determine the composition of phytoplankton assemblage in a mesotrophic lake. Curr Biol. 2002;12(20):1767–1772. doi: 10.1016/s0960-9822(02)01217-4. [DOI] [PubMed] [Google Scholar]
- 16.Fistarol GO, Legrand C, Graneli E. Allelopathic effect of Prymnesium parvum on a natural plankton community. Mar Ecol Prog Ser. 2003;255:115–125. [Google Scholar]
- 17.Uronen P, Kuuppo P, Legrand C, Tamminen T. Allelopathic effects of toxic haptophyte Prymnesium parvum lead to release of dissolved organic carbon and increase in bacterial biomass. Microb Ecol. 2007;54(1):183–193. doi: 10.1007/s00248-006-9188-8. [DOI] [PubMed] [Google Scholar]
- 18.Ma H, Krock B, Tillmann U, Cembella A. Preliminary characterization of extracellular allelochemicals of the toxic marine dinoflagellate Alexandrium tamarense using a Rhodomonas salina bioassay. Mar Drugs. 2009;7(4):497–522. doi: 10.3390/md7040497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fistarol GO, Legrand C, Rengefors K, Granéli E. Temporary cyst formation in phytoplankton: A response to allelopathic competitors? Environ Microbiol. 2004;6(8):791–798. doi: 10.1111/j.1462-2920.2004.00609.x. [DOI] [PubMed] [Google Scholar]
- 20.Suikkanen S, Engstrom-Ost J, Jokela J, Sivonen K, Viitasalo M. Allelopathy of Baltic Sea cyanobacteria: No evidence for the role of nodularin. J Plankton Res. 2006;28(6):543–550. [Google Scholar]
- 21.Ribalet F, Berges JA, Ianora A, Casotti R. Growth inhibition of cultured marine phytoplankton by toxic algal-derived polyunsaturated aldehydes. Aquat Toxicol. 2007;85(3):219–227. doi: 10.1016/j.aquatox.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Landsberg JH, Flewelling LJ, Naar J. Karenia brevis red tides, brevetoxins in the food web, and impacts on natural resources: Decadal advancements. Harmful Algae. 2009;8(4):598–607. [Google Scholar]
- 23.Poulson KL, Sieg RD, Prince EK, Kubanek J. Allelopathic compounds of a red tide dinoflagellate have species-specific and context-dependent impacts on phytoplankton. Mar Ecol Prog Ser. 2010;416:69–78. [Google Scholar]
- 24.Prince EK, Poulson KL, Myers TL, Sieg RD, Kubanek J. Characterization of allelopathic compounds from the red tide dinoflagellate Karenia brevis. Harmful Algae. 2010;10(1):39–48. [Google Scholar]
- 25.Prince EK, Myers TL, Kubanek J. Effects of harmful algal blooms on competitors: Allelopathic mechanisms of the red tide dinoflagellate Karenia brevis. Limnol Oceanogr. 2008;53(2):531–541. [Google Scholar]
- 26.Brand LE, Campbell L, Bresnan E. Karenia: The biology and ecology of a toxic genus. Harmful Algae. 2012;14:156–178. doi: 10.1016/j.hal.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince EK, Myers TL, Naar J, Kubanek J. Competing phytoplankton undermines allelopathy of a bloom-forming dinoflagellate. Proc Biol Sci. 2008;275(1652):2733–2741. doi: 10.1098/rspb.2008.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viant MR, Rosenblum ES, Tieerdema RS. NMR-based metabolomics: A powerful approach for characterizing the effects of environmental stressors on organism health. Environ Sci Technol. 2003;37(21):4982–4989. doi: 10.1021/es034281x. [DOI] [PubMed] [Google Scholar]
- 29.Sardans J, Penuelas J, Rivas-Ubach A. Ecological metabolomics: Overview of current developments and future challenges. Chemoecology. 2011;21(4):191–225. [Google Scholar]
- 30.Nunn BL, Timperman AT. Marine proteomics. Mar Ecol Prog Ser. 2007;332:281–289. [Google Scholar]
- 31.Nunn BL, et al. Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation. PLoS One. 2013;8(10):e75653. doi: 10.1371/journal.pone.0075653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho RN, Lettieri T. Proteomic analysis of the marine diatom Thalassiosira pseudonana upon exposure to benzo(a)pyrene. BMC Genomics. 2011;12:159. doi: 10.1186/1471-2164-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bromke MA, Giavalisco P, Willmitzer L, Hesse H. Metabolic analysis of adaptation to short-term changes in culture conditions of the marine diatom Thalassiosira pseudonana. PLoS One. 2013;8(6):e67340. doi: 10.1371/journal.pone.0067340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prince EK, Irmer F, Pohnert G. Domoic acid improves the competitive ability of Pseudo-nitzschia delicatissima against the diatom Skeletonema marinoi. Mar Drugs. 2013;11(7):2398–2412. doi: 10.3390/md11072398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson DG, Reily MD, Baker JD. Metabonomics in pharmaceutical discovery and development. J Proteome Res. 2007;6(2):526–539. doi: 10.1021/pr060535c. [DOI] [PubMed] [Google Scholar]
- 36.Badylak S, Phlips EJ, Baker P, Fajans J, Boler R. Distributions of phytoplankton in Tampa Bay Estuary, USA 2002-2003. Bull Mar Sci. 2007;80(2):295–317. [Google Scholar]
- 37.Chiovitti A, et al. Variations in the substituted 3-linked mannans closely associated with the silicified walls of diatoms. J Phycol. 2005;41(6):1154–1161. [Google Scholar]
- 38.Popper ZA, et al. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu Rev Plant Biol. 2011;62(1):567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- 39.Martin P, Van Mooy BA, Heithoff A, Dyhrman ST. Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J. 2011;5(6):1057–1060. doi: 10.1038/ismej.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riekhof WR, Andre C, Benning C. Two enzymes, BtaA and BtaB, are sufficient for betaine lipid biosynthesis in bacteria. Arch Biochem Biophys. 2005;441(1):96–105. doi: 10.1016/j.abb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Miralto A, et al. The insidious effect of diatoms on copepod reproduction. Nature. 1999;402(6758):173–176. [Google Scholar]
- 42.Vardi A, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326(5954):861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 43.Deeds JR, Place AR. Sterol-specific membrane interactions with the toxins from Karlodinium micrum (Dinophyceae)—a strategy for self-protection? Afr J Mar Sci. 2006;28(2):421–425. [Google Scholar]
- 44.Dickson DMJ, Kirst GO. The role of β-dimethylsulphoniopropionate, glycine betaine and homarine in the osmoacclimation of Platymonas subcordiformis. Planta. 1986;167(4):536–543. doi: 10.1007/BF00391230. [DOI] [PubMed] [Google Scholar]
- 45.Keller MD, Kiene RP, Matrai PA, Bellows WK. Production of glycine betaine and dimethylsulfoniopropionate in marine phytoplankton. I. Batch cultures. Mar Biol. 1999;135(2):237–248. [Google Scholar]
- 46.Dickson DMJ, Kirst GO. Osmotic adjustment in marine eukaryotic algae: The role of inorganic ions, quaternary ammonium, tertiary sulphonium and carbohydrate solutes. New Phytol. 1987;106(4):645–655. doi: 10.1111/j.1469-8137.1987.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 47.Allen AE, et al. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc Natl Acad Sci USA. 2008;105(30):10438–10443. doi: 10.1073/pnas.0711370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroth PG, et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One. 2008;3(1):e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlegel K, Welte C, Deppenmeier U, Müller V. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 2012;279(24):4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 50.Rosenwasser S, et al. Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc Natl Acad Sci USA. 2014;111(7):2740–2745. doi: 10.1073/pnas.1319773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bidle KD, Bender SJ. Iron starvation and culture age activate metacaspases and programmed cell death in the marine diatom Thalassiosira pseudonana. Eukaryot Cell. 2008;7(2):223–236. doi: 10.1128/EC.00296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caldara M, et al. Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr Biol. 2012;22(24):2325–2330. doi: 10.1016/j.cub.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Workman HC, Sweeney C, Carraway KL., 3rd The membrane mucin Muc4 inhibits apoptosis induced by multiple insults via ErbB2-dependent and ErbB2-independent mechanisms. Cancer Res. 2009;69(7):2845–2852. doi: 10.1158/0008-5472.CAN-08-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thamatrakoln K, Korenovska O, Niheu AK, Bidle KD. Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ Microbiol. 2012;14(1):67–81. doi: 10.1111/j.1462-2920.2011.02468.x. [DOI] [PubMed] [Google Scholar]
- 56.Guillard RRL, Hargraves PE. Stichochrysis-immobilis is a diatom, not a chyrsophyte. Phycologia. 1993;32(3):234–236. [Google Scholar]
- 57.Paul C, Barofsky A, Vidoudez C, Pohnert G. Diatom exudates influence metabolism and cell growth of co-cultured diatom species. Mar Ecol Prog Ser. 2009;389:61–70. [Google Scholar]
- 58.Smith CA, et al. METLIN: A metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 59.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sud M, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35(Database issue) Suppl 1:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui Q, et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26(2):162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 62.Wishart DS, et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue) Suppl 1:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.