Significance
Nucleosomes limit access to DNA, which antagonizes gene expression and prevents recruitment of transcription factors that cannot bind DNA wrapped around the histone octamer. Numerous studies using large cell populations determined that active genes promoters tend to be nucleosome-depleted. We developed a method to examine nucleosome positioning in single cells and revealed significant heterogeneity of nucleosome configurations within a population. In an inactive gene loaded with nucleosomes, a small subpopulation of nucleosome-depleted cells exists that were engaged in transcription. Single-cell mapping revealed that even in apparently nucleosome-free regions, some cells were occupied by nucleosomes. These data reveal an underlying complexity of nucleosome positioning and its role in regulating gene expression.
Keywords: chromatin structure, gene regulation
Abstract
Nucleosomes, the basic unit of chromatin, have a critical role in the control of gene expression. Nucleosome positions have generally been determined by examining bulk populations of cells and then correlated with overall gene expression. Here, we describe a technique to determine nucleosome positioning in single cells by virtue of the ability of the nucleosome to protect DNA from GpC methylation. In the acid phosphatase inducible PHO5 gene, we find that there is significant cell-to-cell variation in nucleosome positions and shifts in nucleosome positioning correlate with changes in gene expression. However, nucleosome positioning is not absolute, and even with major shifts in gene expression, some cells fail to change nucleosome configuration. Mutations of the PHO5 promoter that introduce a poly(dA:dT) tract-stimulated gene expression under nonpermissive conditions led to shifts of positioned nucleosomes similar to induction of PHO5. By contrast, mutations that altered AA/TT/AT periodicity reduced gene expression upon PHO5 induction and stabilized nucleosomes in most cells, suggesting that enhanced nucleosome affinity for DNA antagonizes chromatin remodelers. Finally, we determined nucleosome positioning in two regions described as “fuzzy” or nucleosome-free when examined in a bulk assay. These regions consisted of distinct nucleosomes with a larger footprint for potential location and an increase population of cells lacking a nucleosome altogether. These data indicate an underlying complexity of nucleosome positioning that may contribute to the flexibility and heterogeneity of gene expression.
The eukaryotic genome is packaged into chromatin, which consists of a basic repeating unit of nucleosomes arranged in regularly spaced arrays. A nucleosome is comprised of a histone octamer wrapped around 147 bp of DNA (1, 2). Nucleosomes coat much of genomic DNA, but specific functional regions of the DNA such as promoters, enhancers, and terminators are relatively depleted of nucleosomes (3–7). Although “pioneer” DNA-binding transcription factors can bind nucleosomal DNA (reviewed in ref. 8), many other factors compete with nucleosomes for DNA binding (9–13) and often cannot bind to DNA without the removal of nucleosomes (reviewed in ref. 14). For example, the TATA-binding protein cannot bind nucleosomal DNA, resulting in a failure of recruitment of RNA polymerase II and inhibition of transcription initiation (15). Conversely, a well-positioned nucleosome can promote transcription factor binding if it sits proximal to the binding site, thereby forcing the DNA to be accessible for factor binding. Thus, understanding how nucleosomes are naturally positioned and how such positioning changes under physiological and stress conditions can provide significant insight into predicting gene expression.
Histone octamers do not bind DNA randomly, instead, a “nucleosome code,” which consists of the primary DNA sequence (4) and the secondary DNA structures, helps dictate where nucleosomes form (16–18). Twisting of DNA and hence the ability of DNA to wrap around the histone octamer is facilitated by the periodicity of the bendable dinucleotide AA/TT/AT sequence, spaced every 10 bp to fit in the minor grove of the helix (4, 19, 20). By contrast, the homopolymeric sequences of poly(dA:dT) are inherently stiff (16, 21–23) and can create nucleosome-free regions (22, 24, 25). Sequence, however, is not the sole determinant of nucleosome positioning. Proteins compete with nucleosomes for binding to specific DNA sequence (26) and ATP-dependent chromatin remodelers actively displace nucleosomes (27). Barriers created by DNA sequence or protein factors that exclude nucleosomes may create a particularly well-positioned nucleosome adjacent to that barrier. As a result, subsequent nucleosomes may be constrained, and an array of positioned nucleosomes can thus be created (28). The relative contributions of DNA sequence, protein binding, and histone modifications to nucleosome positioning are still not fully elucidated.
One major drawback to the nucleosome mapping studies to date is that they are performed as bulk experiments. Much of our knowledge of nucleosome-positioning patterns relies only on gene-averaged events and even upon examination of a specific locus, nucleosome positioning is based on information averaged from millions of cells. Similarly, gene expression studies are generally performed on bulk populations, and gene expression may vary significantly from cell to cell. When single cells are examined, a more “digital” on or off state for gene expression may be observed. An illustrative example of the difference between a bulk and single-cell expression is the case of yeast cells that are grown in galactose, glucose, or galactose–glucose mixtures as their carbon source (29). The galactose inducible GAL genes are highly expressed when yeast are grown in galactose, and these genes are silenced when the organism is grown in glucose. When grown in a mixture of galactose and glucose, the population expresses the GAL genes at intermediate levels that depend quantitatively on the galactose/glucose ratio. Single-cell analysis, however, revealed that in a galactose–glucose mixture, intermediate gene expression does not result from each cell transcribing a moderate level of protein and each cell gradually changing its expression level with increasing galactose. Instead, each individual cell behaves in a binary fashion: either strongly expressing or strongly repressing the GAL genes. As the ratio of galactose/glucose increases, the probability of any given cell expressing a set level of GAL gene increases. Similarly, population studies of nucleosome positioning could be misleading. Indeed, recent work from the Boeger laboratory examines nucleosome positioning by electron microscopy and finds that numerous nucleosome configurations exist on the promoter, which include both nucleosome-rich and nucleosome-free architectures (30). Thus, examination of single cells at the level of nucleosome positioning may help explain the molecular basis of the cellular decision to activate a gene.
Here, we present a method for mapping nucleosome positioning at specific loci based upon the ability of a nucleosome to protect DNA from methylation by an exogenous enzyme. Using this method, we examined the cell-to-cell variation of nucleosome positioning in the acid phosphatase PHO5 promoter under nutrient-rich conditions associated with low basal level gene expression and upon phosphate starvation, which induces PHO5 expression (31). Furthermore, we examined the nucleosome landscape when the PHO5 promoter was mutated by introduction of a poly(dA:dT) tract, to antagonize nucleosomes, or by altering the AA/TT/AT periodicity to stabilize nucleosomes. These experiments provide direct evidence for a correlation of nucleosome positioning and gene expression. Finally, we used our method to deconvolute the nucleosome positioning of two loci that have previously been described as “fuzzy,” a term used to describe nucleosomes that are not well localized (3). These data demonstrate that considerable cell-to-cell variability of nucleosome positioning exists, and low basal levels of gene expression within a population are attributable to a subset of cells having permissive nucleosomal positioning. Furthermore, we found that even upon major shifts in gene expression not every cell of a population shifts positions in the same way or becomes nucleosome-depleted. This method allows for the survey of less well-positioned nucleosomes and determines the range of sites nucleosomes can associate, which will help us to further refine our understanding of the “nucleosome code.” Variation in nucleosome positioning may contribute to flexibility of gene expression in a cell population and cell-to-cell heterogeneity of gene expression.
Results
Cell-to-Cell Diversity of Nucleosome Positioning.
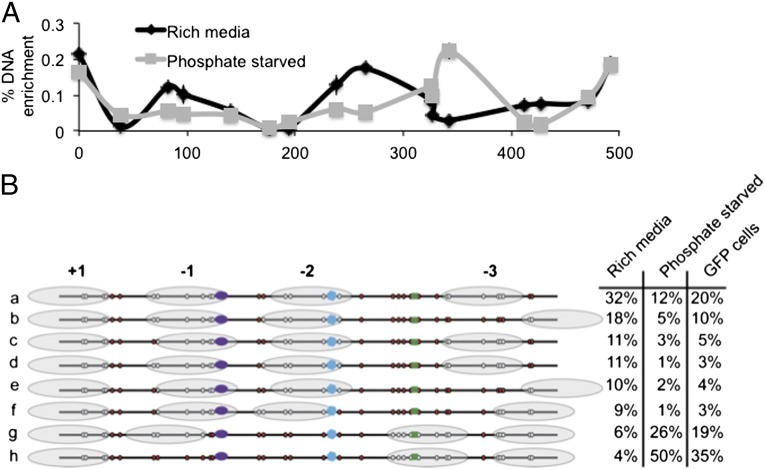
As a model for determining cell-to-cell diversity of nucleosome positioning, we used the PHO5 promoter as the position of its nucleosomes was well characterized in bulk studies (canonical nucleosome positioning depicted in Fig. 1A). PCR-based nucleosome-scanning assay (Fig. 1B) and genome-wide micrococcal nuclease (MNase-seq) were performed (Fig. 1C), confirming previously reported nucleosome positioning on the PHO5 promoter for positions of the +1 through −3 nucleosomes of the PHO5 promoter under phosphate-rich conditions, which is nonpermissive for gene expression (30, 32–35).
Fig. 1.
Nucleosome architecture of the PHO5 promoter can be ascertained in individual cells and demonstrates heterogeneity. (A) Canonical position of nucleosomes (gray ovals) in the PHO5 promoter. White circles indicate the locations of cytosines that are part of GC dinucleotides. (B) Mapping of mononucleosomal DNA of cells grown in rich media using a nucleosome-scanning assay. Chromatin was digested with MNase, mononucleosomal DNA was purified, and MNase protection was determined by quantitative PCR. Enrichment of mononucleosomal DNA (y axis) is indicated by the midpoints of each amplicon (x axis), and error bars represent 1 SD from two independent biological replicates. (C) MNase-seq track of the PHO5 promoter from cells grown in rich media. (D) Nucleosome architecture of 806 cells from three bulk populations revealed eight conformations (a to h). Nucleosomes are depicted as gray ovals. Red circles indicate methylated cytosines, and white circles indicate unmethylated cytosines that are part of GC dinucleotides. The fraction of total cells that demonstrated each protection pattern is indicated on the right. The SDs for the three experiments were all less than 1.5%.
To map nucleosomes in individual cells, we modified a method developed in the Kladde and Jones laboratories based upon the ability of nucleosomes to block the GpC-specific DNA methyltransferase M.CviPI from modifying DNA wrapped around a histone octamer (34, 36, 37). In comparison with other nucleosome-mapping methods, this method is less destructive than MNase for mapping the DNA at the ends of the nucleosome or chemical mapping, which cleaves the DNA at the center of the nucleosome (19). MNase cleavage also has sequence specificity, which can bias the results of nucleosome mapping (38–41). Although chemical mapping nucleosomes by incorporation of a modified histone circumvents the sequence specificity, it is not a very efficient reaction and thus can only map a fraction of the nucleosomes within a cell (19, 41).
To map nucleosomes in single cells, yeast are converted to permeable spheroplasts and treated with M.CviPI, which methylates the cytosines of GpC dinucleotide that are not protected by DNA-bound proteins including nucleosomes. Single cells were isolated by serial dilution until there was, on average, zero or one cell per well. DNA from single cells was extracted, digested, and treated with sodium bisulfite to convert unmethylated cytosines to uracil. Each cellular equivalent of DNA was subjected to PCR amplification using primers complimentary to the promoter of interest, and DNA amplicons were sequenced. The pattern of GpC methylation within the promoter of each single cell was compared with all possible GpC sites in the promoter to infer the locations of nucleosomes (Fig. 1D).
It is important to note that this method allows for the inference of nucleosome location by protection patterns of the GpCs and therefore does not reveal information regarding nucleosomes being smaller or larger than 147 bp. Moreover, the methyltransferase activity and the bisulfite conversion needed to be as close to 100% efficient as possible to accurately map nucleosomes in individual cells. If any cytosine that should have been methylated or bisulfite-converted were not modified, this would give an inaccurate result. To measure enzymatic efficiency, a time course was performed with M.CviPI followed by bisulfite sequencing to determine the incubation time necessary to achieve methylation of all accessible GpCs (Fig. S1).
In our pilot study of 235 cells, 212 did not have a nucleosome over the low-affinity upstream activating sequence (UAS), a pattern further supported in our final data, explained in detail below (Fig. 1D, conformations a to f). Of the pilot set, only six cells had unprotected GpCs flanking the low-affinity UAS, suggesting transcription factor binding. Two other cells had a single unprotected GpC among the seven GpCs between the −2 and −3 nucleosomes. These single unprotected GpCs could be attributable to an inability of the methyltransferase to access this region of DNA because of a protein factor or secondary nucleic acid structure or inefficient methyltransferase or bisulfite conversion (i.e., less than 100% efficiency); however, taken together, the data from the pilot experiment strongly indicated that methylation and bisulfite conversion are extremely effective and thus can be used for single-cell nucleosome mapping.
We further note that as a quality control for each single-cell nucleosome-mapping experiment, bulk genomic DNA was treated with M.CviPI followed by bisulfite conversion and cloning; 25 colonies were sequenced to confirm that all cytosines within GpCs were methylated and all free cytosines were bisulfite-converted. Moreover, to corroborate that PCR products of single-cell experiments indeed represented DNA from a single cell, several PCR products from a single cell were cloned from each experiment and a minimum of 10 colonies were sequenced and shown to be exactly the same. Together, these controls demonstrate that GpC methyltransferase activity and bisulfite conversion are ∼99% efficient and PCR amplicons do represent single cells. Thus, we concluded that the technique described above could be used to determine GpC protection patterns of individual cells.
Using single-cell mapping, we examined nucleosome occupancy in three biological replicate mapping experiments, including the pilot examination of 235 cells, for a total of 806 single cells (Fig. 1D). We observed that in a population of cells grown under nutrient-rich, nonpermissive conditions for PHO5, the majority of individual cells demonstrated nucleosome architecture that resembles the canonical nucleosome map (compare Fig. 1 A and D). Specifically, 32% of cells had promoters in the same nucleosome configuration that fits the canonical nucleosome map (Fig. 1D, conformation a). An additional 11% of the cells demonstrate only a slight difference of one GpC at the 3′ end of the −1 nucleosome (compare conformations a and c in Fig. 1D), which may be attributable to a small shift in the nucleosome location. It is also possible that there is a difference in the ability of the methyltransferase to modify that particular GpC because there is a single base that separates the two GpCs. Approximately 91% of all cells had a nucleosome-rich promoter with nucleosomes occluding both the TATA box and high-affinity UAS and a nucleosome-free low-affinity UAS (Fig. 1D, conformations a to f). These data suggest that generally only small variations in nucleosome positions exist in the population.
Under nutrient-rich conditions, the PHO5 promoter displayed four well-positioned nucleosomes (Fig. 1D). The +1 nucleosome appears to shift only slightly, if at all, in any of the cells, which is in agreement with previous reports indicating that nucleosome positioning is strongest at the +1 position (3, 42). The −1 nucleosome that occludes the TATA box appears less well positioned by MNase-seq (Fig. 1C); however, upon closer investigation, there are still about 500 DNA sequence reads that span the 162-bp region (Fig. 1C), which is in agreement with the nucleosome-scanning assay (Fig. 1B).
The single-cell data further reveal more differences in positioning of the −1 nucleosome compared with the +1 nucleosome because four configurations (including the absence of the −1 nucleosome) were identified. The vast majority of the cells (Fig. 1D, conformations a to f) demonstrated nucleosome occupancy at the −1 position in two different protection patterns: ∼6% of the cells had a nucleosome shifted away from the TATA box, and only ∼4% of the cells did not show the presence of a nucleosome at the −1 position. The −2 nucleosome was also well positioned with only small cell-to-cell variations. The −2 nucleosome was easier to map accurately compared with the −1 based on GCs located on each end of the nucleosome, and tight positioning correlated with the relatively sharp peak in the MNase-seq data (Fig. 1C). The majority of these cells had a −2 nucleosome occluding the high-affinity UAS, as previously shown by other methods (30, 32–35). However, the single-cell analysis showed that about 10% of cells do not have a nucleosome over the high-affinity UAS (Fig. 1D, conformations g and h) but instead have a nucleosome shifted 3′ to the −2 nucleosome that occludes the low-affinity UAS, a result that could not be ascertained from the bulk MNase-seq or nucleosome-scanning methods.
The −3 nucleosome demonstrated the most variation in positioning, which is in agreement with the MNase-seq track and nucleosome-scanning assay. The two peaks in the MNase-seq are in very close proximity in a region too small to accommodate two nucleosomes, so the exact position of these nucleosomes is more difficult to ascertain from bulk methods. The single-cell data indicated that the majority of cells occupy one of two conformations at the −3 position (Fig. 1D, confirmations a to e), but almost 20% of the population has a nucleosome that spans the region between the two conformations (Fig. 1D, confirmations f to h). Taken together with the data above, this single-cell analysis illustrates that although many cells conform to the general pattern of the nucleosome architecture defined by assay of a bulk population, at the single-cell level, considerable variability of nucleosome positioning exists.
Single-Cell Mapping Deconvolutes Fuzzy Nucleosomes.
Although the PHO5 promoter was chosen as one model because of its well-positioned nucleosomes, our single-cell analysis revealed distinct cell-to-cell variation in exact nucleosome positions. Nucleosome positioning can range from almost perfect positioning (e.g., the +1 nucleosome of PHO5), with no variability, to undefined positioning, where a nucleosome can be located at many sites within a region of DNA. MNase-seq and other nucleosome-mapping techniques can yield fuzzy nucleosomes in which a broad region without a peak demonstrates partial protection from digestion. Only single-cell analysis can determine whether this “fuzziness” is caused by the nucleosome forming along a continuous stretch of DNA at numerous positions or is attributable to a smaller set of cells that contain discrete positions assumed by the nucleosome.
We examined two loci: the 5′ end of CHA1 and the promoter of CYS3, both of which display poorly characterized nucleosomes by MNase-seq (Fig. 2). In the case of CHA1, the nucleosome-scanning assay and MNase-seq revealed two potential nucleosomes at the 5′ end of the gene followed by a fuzzy, or nucleosome-free, region depicted by a low amount of enrichment of mononucleosomal DNA in the nucleosome-scanning assay (Fig. 2A) and the broad flat region in the MNase-seq track (Fig. 2B). We analyzed a total of 481 cells and identified 68 different nucleosome conformations of four potential nucleosomes in a 575-bp region of CHA1. The seven most common conformations were seen in 31% of cells (Fig. 2C, conformations a to g). However, a great many more conformations exist, some of which were identified in only one cell (Fig. S2). It is highly likely that even more variability exists in this region because we are limited by our ability to only detect differences in positioning via GpC protection. Although nucleosome scanning and MNase-seq confirmed DNA sequences were protected from digestion, specific locations for nucleosomes are unclear. Single-cell analysis revealed that in the +3 and +4 nucleosome region, only 17% of cells were actually nucleosome-free and an additional 31% contained only one nucleosome in this region.
Fig. 2.
Single-cell analysis of nucleosome architecture reveals distinct positions of nucleosomes previously described as fuzzy. (A) Nucleosome-scanning assay of the 5′ region of CHA1 from cells grown in rich media. The experiment was performed as described in Fig. 1. (B) MNase-seq track of CHA1 from cells grown in rich media. (C) Nucleosome architecture of 481 cells from two bulk experiments revealed 68 conformations of nucleosomes in the 5′ region of CHA1 (Fig. S2). An overlay of all possible nucleosome positions is depicted followed by the top seven conformations that represent 31% of the total population. (D) Nucleosome-scanning assay of the CYS3 promoter from cells grown in rich media. The experiment was performed as described in Fig. 1. (E) MNase-seq track of CYS3 promoter from cells grown in rich media. (F) Nucleosome architecture of 550 cells from two bulk experiments revealed 20 conformations of nucleosomes in the CYS3 promoter (Fig. S3). An overlay of all possible nucleosome positions is depicted, followed by the top six conformations that represent 31% of the total population.
Examination of the CYS3 promoter also revealed a collection of nucleosome conformations (Fig. 2F and Fig. S3). We analyzed 550 cells, and overall we found three nucleosomes positioned in 20 conformations spanning 540 bp in the promoter and 5′ end of CYS3. Nucleosome scanning and MNase-seq demonstrated a very well-defined nucleosome in the −2 and +1 positions of CYS3 (Fig. 2 D and E). We only detected one conformation for the nucleosome at the −2 position and four conformations at the +1 nucleosome that span a region of 166 bp (Fig. 2F and Fig. S3). By contrast, we detected five positions for the −1 nucleosome spanning a larger region of 184 bp, and in some cells, there was complete absence of a nucleosome in this region. In the majority of cases, a nucleosome is present in one of four conformations at the −1 position, but 36% of cells entirely lack a nucleosome in the promoter. These results demonstrate that our method can detect distinct nucleosome locations that previously were described as fuzzy or even nucleosome-free.
Nucleosome Remodeling Is Observed upon Phosphate Starvation.
Using our method for determining nucleosome positioning in single cells, we next directly tested the correlation between nucleosome positioning and gene expression through the study of the yeast PHO5 gene. When grown in nutrient-rich media, PHO5 expression is low to absent in yeast. Most of the yeast cells grown under these conditions displayed a nucleosome over the TATA box and high-affinity UAS (Fig. 1D, −1 and −2 nucleosomes), presumably blocking transcription factor accessibility and leading to gene silencing. Nevertheless, even under these nonpermissive conditions, about 10% of cells lacked a nucleosome over the high-affinity UAS site and the TATA box (Fig. 1D, conformations g and h).
PHO5 expression is induced upon phosphate starvation (ref. 31 and Fig. S4), and the nucleosomes of this gene were shown to undergo significant rearrangement upon phosphate starvation, as determined by electron microscopy (30) and methyltransferase accessibility (34). In agreement with previous studies (35, 43, 44), PCR-based nucleosome-scanning assay showed marked shifts in chromatin structure (Fig. 3A). The +1 nucleosome was positioned and present in the presence or absence of phosphate, but upon phosphate starvation, the −1 nucleosome was, on average, threefold less likely to be occupied, and the position of the −2 nucleosome was significantly shifted downstream (Fig. 3A). Indeed, previous predictions and experimental work suggested that the −2 nucleosome needed to be removed in order for the −1 nucleosome to remodel (30, 34). Single-cell nucleosome mapping showed that upon phosphate starvation and activation of the PHO5 gene, 76% of cells now had a nucleosome-depleted architecture, compared with only 10% when cells were grown in phosphate-rich media (Fig. 3B, conformations g and h). The +1 nucleosome was present in all cells regardless of nutrient status, but 50% of cells starved of phosphate lacked the −1 nucleosome (Fig. 3B, conformation h), and an additional 26% of the cells had the −1 nucleosome shifted away from the TATA box, allowing it to be accessible to protein factors (Fig. 3B, conformation g). Although these data confirmed patterns of nucleosome rearrangement upon phosphate starvation mapped in the bulk population of yeast, they also demonstrate that ∼25% of cells failed to undergo a remodeling of the nucleosome architecture and thus remained in a nucleosome-rich confirmation (Fig. 3B, conformations a to h), demonstrating a unique subpopulation of cells.
Fig. 3.
Remodeling of nucleosomes upon phosphate starvation correlates with an increase in gene expression. (A) Nucleosome-scanning assay of the PHO5 promoter from cells grown in rich media (black) and shifted to media lacking phosphate (gray) for 3 h. The experiment was performed as described in Fig. 1. (B) The nucleosome architecture of cells grown in rich media (column 1 in the table to the right) is from the experiments in Fig. 1. Nucleosome architecture of 465 cells grown in rich media and shifted to media lacking phosphate (column 2, Phosphate starved) from three bulk experiments revealed the same eight conformations of nucleosomes for the PHO5 promoter, but there is a shift in a greater population of more nucleosome-free conformations (g and h). Nucleosome architecture of 638 cells expressing PHO5 from two bulk experiments (column 3, GFP cells). Cells that contain an EGFP tag on the C terminus of PHO5 were grown in rich media and sorted for GFP-positive cells, which represents ∼1% of the total population.
PHO5 expression is very low under phosphate-rich conditions. Theoretically, this level of expression could arise from low-level transcription from all cells or could result from gene silencing in most cells and transcription from a small minority of cells. A yeast strain in which the PHO5 gene contains a C-terminal EGFP tag was grown under nutrient-rich, nonpermissive conditions. Less than 1% of the cells were GFP-positive, which indicates that a low basal level of transcription was occurring at the PHO5 locus. These GFP-positive cells were sorted by FACS and processed for single-cell mapping. The sorted population showed the same eight conformations of nucleosomes that were observed in the unsorted population, but there was a significant shift to more cells displaying an open chromatin structure (Fig. 3B, conformations g and h, and Fig. S5). Specifically, 54% of the GFP-positive cells showed a loss of nucleosomes over the UAS and a loss or shift away of a nucleosome over the TATA box, compared with only 10% of cells in an unsorted population.
Furthermore, we found that when cells were grown in the permissive, low-phosphate conditions, GFP-negative cells demonstrated the same distribution of nucleosome architectures as GFP-negative cells grown in rich media (Fig. S5), specifically these cells had a strong tendency to have the −1 and −2 nucleosome sites occupied. These data strongly suggest that the small population of cells that remain in a relatively nucleosome-free configuration under nonpermissive conditions account for “leaky,” low-level basal gene expression. The shift from nucleosome-rich to nucleosome-free promoter configurations correlates directly with gene expression indicating that nucleosome remodeling is a barrier to regulate gene expression.
Altering DNA Sequence Shifts Nucleosome-Positioning Preferences.
DNA sequence itself plays a role in nucleosome positioning because nucleosome formation is affected by the overall ability for a 147-bp sequence to bend around the histone octamer (16). The homopolymeric sequence poly(dA:dT) is intrinsically stiff and strongly inhibitory to nucleosome formation (16, 21–23). Poly(dA:dT) tracts of only 12–15 bp result in nucleosome-free regions of more than 150 bp (3, 45). Thus, we asked how the introduction of a poly(dA:dT) tract in the PHO5 promoter would affect nucleosome organization at the single-cell level.
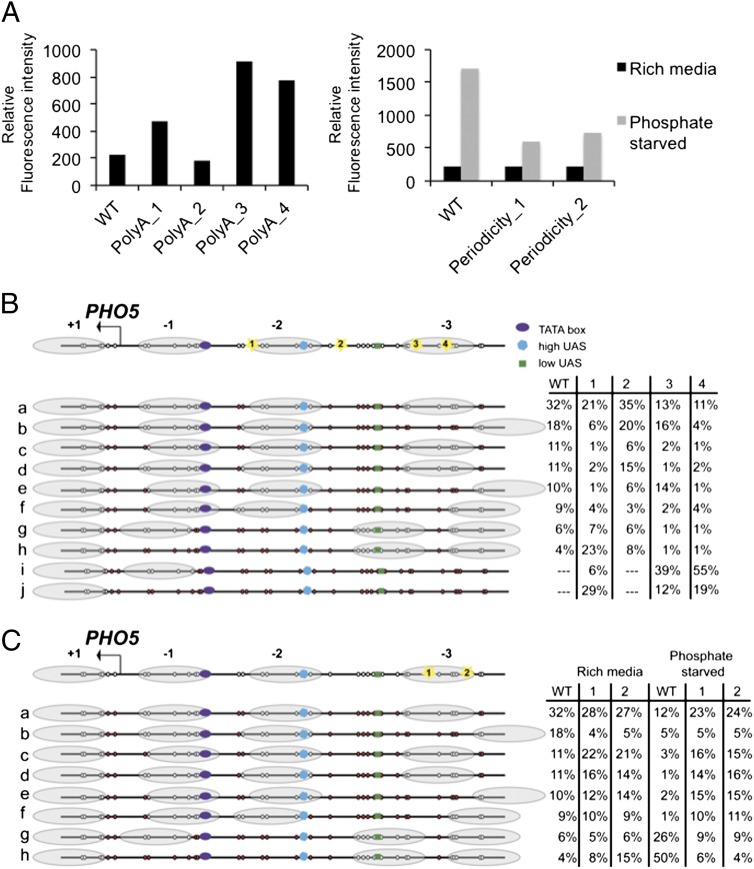
We tested four mutations: two of which were tracts comprised of a mixture of As and Ts (PolyA_1 and PolyA_2), and two of which were poly(dA:dT) tracts (PolyA_3 and PolyA_4). These mutations all changed base composition but did not change the length of the promoter sequence and were not in regions believed to contain transcription factor binding sites. All but one of the mutations led to an increase in PHO5-GFP expression, as determined by flow cytometry (Fig. 4A). In the mixed A and T tract, PolyA_2 did not alter gene expression (Fig. 4A), nor did it significantly alter the nucleosome architecture, because the distribution of nucleosome locations was similar to that observed in wild-type cells (Fig. 4B, mutation 2). One possibility for the lack of change may be that the mixed A and T tract did not sufficiently stiffen the DNA to antagonize nucleosome formation. However, another mixed A and T tract, PolyA_1, demonstrated significant alterations in its nucleosome locations that were similar to the poly(dA:dT) tract mutants PolyA_3 and PolyA_4. These mutations all demonstrated the eight conformations previously seen in the wild type cells (Fig. 4B, conformations a to h), as well as two additional conformations that are nucleosome-free (Fig. 4B, conformations i and j).
Fig. 4.
Changes in gene expression correlate with changes in nucleosome positioning. (A) PHO5 gene expression was determined from wild-type (WT) and PHO5 mutants containing a C-terminal EGFP tag. Gene expression was measured by the relative fluorescence intensity by flow cytometry. In the left graph, gene expression is shown for wild-type and polyA mutant cells grown in rich media. In the right graph, gene expression is shown for wild-type and periodicity mutant cells grown in rich media (black) or shifted to phosphate starvation media for three hours (gray). (B, Upper) The canonical nucleosome architecture of PHO5 as described in Fig. 1. The location of each of the polyA mutation is indicated by a number in a yellow star. (B, Lower) The ten nucleosome conformations (a to j) determined from analysis of WT and polyA mutants grown in rich media. Two bulk experiments were performed and the number of cells analyzed for each mutant was as follows: PolyA_1, 385 cells; PolyA_2, 412 cells; PolyA_3, 368 cells; and PolyA_4, 404 cells. The same eight conformations (a to h) were observed as in Fig. 1, along with two additional conformations (i and j). The table to the right shows each cell type and the percentage of cells that were determined to be in each conformation. Data for WT were shown in Fig. 1. (C, Upper) The canonical nucleosome architecture of PHO5 shown as described in Fig. 1. The location of each periodicity mutant is indicated by a number in a yellow star. (C, Lower) The eight nucleosome conformations (a to h) of WT and periodicity mutants are shown. Two bulk experiments were performed, and the number of cells analyzed for each mutant was as follows: Periodicity_1, 443 cells in rich media and 612 cells in phosphate starvation; and Periodicity_2, 461 cells in rich media and 602 cells in phosphate starvation. The table to the right shows each cell type under “Rich media” and “Phosphate starvation” conditions, and the percentage of cells that were determined to be in each conformation. In the table, the data shown for WT, Rich media is from Fig. 1 and WT, Phosphate starvation is from Fig. 3.
Whereas the wild-type PHO5 gene had a nucleosome in the −2 position in 90% of cells, poly A_1 contains a poly(dA:dT) tract located at the edge of the −2 nucleosome, which led to loss of the −2 nucleosome in 65% of cells. These cells were also missing a −1 nucleosome or the −1 nucleosome was shifted away from the TATA box (Fig. 4B, conformations g to j). Furthermore, introduction of a poly(dA:dT) tract in the −3 nucleosome (Fig. 4B, mutants 3 and 4) led to an increase in cells lacking both the −2 and −3 nucleosomes (Fig. 4B, configurations i and j), suggesting that changes in primary DNA sequence can alter the phasing of a set of nucleosomes. In the previously unidentified conformations i and j (Fig. 4B), the low- and high-affinity UAS sites and TATA box are devoid of nucleosomes. This change in nucleosome conformation strongly suggests that the introduction of the poly(dA:dT) tract directly antagonizes nucleosome formation and affects gene expression. It is notable, however, that the change of nucleosome positions is not uniform throughout the population of yeast cells. Despite the unfavorable thermodynamics of DNA bending, some cells harboring the poly(dA:dT) tracts maintained the canonical basal nucleosomal configuration of the PHO5 promoter (Fig. 4B, configuration a). These data indicate that although the DNA sequence of a promoter is one factor that plays a role in positioning nucleosomes, other factors such as the position of sequence-specific binding proteins play an important role. Single-cell analysis suggests that changes in DNA sequence alters the probability that a nucleosome may occupy a particular site and contributes to the potential of a gene to be activated or repressed.
Nucleosome formation is favored by the presence of stretches of dinucleotides AA/TT/TA on the face of the helical repeat that can directly interact with the histone (4, 19, 20). Nucleosome occupancy has been observed to increase dramatically when a DNA sequence is altered to fit 5-bp intervals of AA/TT/TA dinucleotides with GC dinucleotides (46). Therefore, we determined how altering the periodicity would change nucleosome positioning of the PHO5 promoter. We examined two mutations predicted to enhance the periodicity (Periodicity_1 and Periodicity_2). Under nutrient-rich conditions, these mutants exhibited a stabilized −3 nucleosome. Specifically, there was an increase in the proportion of cells in configurations c and d and decrease in configuration b in both mutants compared with wild type (Fig. 4C). Not surprisingly, neither mutation had an effect on gene expression when cells were grown under nutrient-rich conditions, because these mutations did not lead to changes in the −2 nucleosome, which occludes the high-affinity UAS.
Upon phosphate starvation, the periodicity mutants demonstrated decreased expression compared with the wild-type PHO5 gene (Fig. 4A, Right). We reasoned that these mutations stabilize the nucleosomes in a manner that antagonizes remodeling upon phosphate starvation. Single-cell mapping of nucleosomes of these mutants upon phosphate starvation revealed similar nucleosome positioning to that of the cells when grown in nutrient-rich media (Fig. 4C). Whereas 76% of cells lose nucleosomes in the −2 position and the −1 nucleosome shifts away from the TATA box under phosphate-starvation conditions in the wild-type cells (Fig. 4C, conformations g and h), the periodicity mutants demonstrated no increase in the number of cells in nucleosome-depleted conformations g and h. These data indicate that the mutations that enhance the AA/TT/TA periodicity stabilize the nucleosomes in a manner that antagonizes chromatin remodelers.
Discussion
Nucleosome positioning can control the accessibility of promoters to transcription factors and RNA polymerase and thereby plays a critical role in the control of gene expression. Here, we present a method for mapping nucleosome positioning at specific loci in individual cells. Before this work, much of our knowledge of nucleosome positioning was built on studies of large populations of cells that give an approximation of nucleosome positions based upon averages with the exception of two single-molecule experiments. One set of experiments used a methyltransferase assay followed by bisulfite conversion that monitored the accessibility of a site in the center of the nucleosome by sequencing clones from a bulk population. This work revealed minimal changes in nucleosome positioning under nonpermissive conditions for PHO5 but considerable heterogeneity upon phosphate starvation (34). Because this technique monitored changes in a methylation site only at the center of the nucleosome, the resolution only allowed for determining the presence or absence of a nucleosome. In agreement, recent electron microscopy also demonstrates significant heterogeneity in the nucleosomal configuration of the activated PHO5 gene (30). Here, we demonstrate significant heterogeneity in the PHO5 promoter in both activated and repressed conditions, albeit there is a shift in populations depending upon PHO5 expression. This heterogeneity is even more apparent in the case of fuzzy nucleosomes, because such regions frequently lack a nucleosome or form a nucleosome at a larger number of discrete positions. We also demonstrate a distinct correlation between nucleosome rearrangements and gene expression as cells expressing PHO5 frequently display chromatin in a more open and partially nucleosome-free structure. Furthermore, DNA mutations predicted to alter nucleosome formation greatly affect the number of cells with more open chromatin states and result in changes in gene expression.
Cellular Diversity in Nucleosome Positioning.
Single-cell nucleosome mapping demonstrates that even for promoters that display relatively well-positioned nucleosomes by cell population assays, significant cell-to-cell variation still exists. For example, under nutrient-rich conditions, a small fraction of cells were in an open chromatin state in the PHO5 promoter, and this small population of cells likely accounts for a low level of gene expression in the bulk population of cells (Figs. 1 and 3). By contrast, under conditions in which the PHO5 promoter was fully induced, a fraction of cells did not undergo chromatin shifts, and presumably gene expression was silenced (Fig. 3 and Fig. S5). Previous experiments performed in bulk yeast cultures showed that at maximal PHO5 expression, the TATA box is accessible in a fraction of the cells (47). This is consistent with our data showing that under permissive conditions for PHO5 expression, half of the cell population has no nucleosome near the PHO5 TATA box, about 25% of the population has a nucleosome shifted away from the TATA box, and the remaining 25% of the population has a nucleosome occluding the TATA box.
Our method also allows for the determination of the status of nucleosomes that cannot be mapped by bulk methods. Indeed, we detected 68 and 20 combinations of nucleosome positioning in the 5′ region of CHA1 and the promoter of CYS3, respectively (compared with only eight conformations in the PHO5 region). It is highly likely that significantly more nucleosome conformations exist that cannot be detected by our method, which is dependent upon the location of GpC dinucleotides. In both of these examples, the broad, flat track of MNase-seq of the bulk cell population correlated with nucleosomes occupying a number of different configurations but did not indicate that nucleosomes were necessarily absent. Single-cell analysis demonstrated that fuzzy nucleosomes are a composite of multiple different discrete binding configurations such that the average footprint spans ∼180 bp, similar to previous reports where the fuzzy nucleosomes spanned 156–174 bp (48). This larger span stands in contrast to the footprint of a well-positioned nucleosome, such as the −2 position in PHO5 that spans just 152 bp.
Because of the lack of firm positioning of a nucleosome, such regions of chromatin are more likely to be accessible to the actions of sequence-specific transcription factors that could activate gene expression. Intriguingly, fuzzy regions do not necessarily occupy significantly more conformations than well-positioned nucleosomes, because the −1 nucleosome in CYS3 occupies four positions or can be absent (Fig. 2F and Fig. S3) compared with the −2 nucleosome in PHO5 promoter, which occupies three conformations or can be absent (Fig. 1D). The main differences between a fuzzy nucleosome and a well-positioned nucleosome appear to be a larger footprint, meaning the ability of a nucleosome to form over a wider range of DNA sequence and the fact that by bulk methods, a greater percentage of the population lacks the nucleosome entirely. For example, in the PHO5 promoter, the −2 nucleosome is absent in only 10% of the cells (Fig. 1D), whereas in the CYS3 promoter the nucleosome is absent in 36% of the cells (Fig. 2F and Fig. S3). Additionally, CHA1 has a fuzzy region that can contain two nucleosomes (+3 and +4), but in 17% of the population, there are no nucleosomes and an additional 31% of the population only has one nucleosome in that region (Fig. 2C and Fig. S2). Although there are many conformations that the +1 and +2 nucleosomes of CHA1 can occupy, only 5% of the population lacks any nucleosomes in that region and an additional 11% contain only one nucleosome. The region of a fuzzy nucleosome offers opportunities for tuning of gene expression through dynamic positioning of nucleosomes. A population of cells displaying a fuzzy nucleosome may show cell-to-cell diversity in gene expression depending on the exact placement of the nucleosome in a given cell.
Intriguingly, a very well-positioned nucleosome was found at the −2 position in the promoter of CYS3 and seems to create a barrier that the fuzzy nucleosome of CYS3 cannot cross (Fig. 2F). A barrier also exists in the CHA1 locus between the +2 nucleosome and the two potential nucleosomes downstream of it (Fig. 2C). These barriers are in agreement with an analysis of 26 genome-wide maps in Saccharomyces cerevisiae that found nucleosomes are arranged around a well-defined center, preventing them from invading distinct ranges of other nucleosomes (48). One reason for the barrier could be that the fuzzy region of both loci is very AT-rich, and numerous studies in yeast have revealed that GC-rich sequences enhance nucleosome formation (reviewed in ref. 49). Thus, a relatively well-positioned nucleosome in a GC-rich stretch of DNA cannot move beyond the confines of this sequence-specific region.
DNA Sequence Can Alter Nucleosome Occupancy.
The importance of DNA sequence in dictating nucleosome positioning has been debated (16). Although the histone octamer’s affinity for a given 147-bp DNA sequence varies over three orders of magnitude (50), nucleosome remodelers can override intrinsic DNA sequence preferences (16). We tested the two major sequence determinants that affect bending of DNA and can influence nucleosome formation: poly(dA:dT) tracts and periodicity. As predicted, the poly(dA:dT) tracts that exhibited an increase in gene expression had a more nucleosome-depleted architecture (Fig. 4). These results were also in agreement with the Nucleosome Positioning Prediction Engine (NuPoP) (51). As predicted by NuPoP, PolyA_1, PolyA_3, and PolyA_4 all had changes in nucleosome positioning compared with the wild-type sequence. Specifically, the probability of the −3 nucleosome occupancy was reduced in the PolyA_1 mutation compared with the wild-type sequence. The probability of a −3 nucleosome in the PolyA_3 and PolyA_4 mutations was even further reduced to less than 0.05. NuPoP predicted a change of positioning in the −2 nucleosome for the PolyA_1 mutation, and accordingly we found that at least 65% of the harboring this mutant promoter cells lacked this nucleosome, whereas only 10% of the wild-type cells lacked this nucleosome. PolyA_2 was predicted by NuPoP to have only a modest change in −2 nucleosome positioning.
The periodicity mutations we assayed were located in the −3 nucleosome of the PHO5 promoter. These mutations increased the probability of a nucleosome at the −3 position and did not affect the other nucleosomes. Indeed, the NuPoP nucleosome occupancy score for the −2, −1, and +1 nucleosomes were about 1.0 in wild-type. The NuPoP algorithm however yielded only a 0.4 probability of nucleosome occupancy for the −3 nucleosome in the presence of the periodicity mutation, which is only a modest increase from 0.3 in the wild-type sequence. In wild-type cells, the −3 position is occupied in 72% of cells in nutrient-rich media, decreasing to 21% upon gene induction. By contrast, the periodicity mutants, while also occupied by a nucleosome under nutrient rich conditions in ∼70% of cells, failed to induce gene expression and ∼60% of cells continued to show occupancy of this position upon shift to low phosphate media. Our results indicate that although DNA sequence plays an important role in nucleosome positioning, its is not the only factor and interplay with other components of the transcriptional machinery clearly must play an important role in the placement of nucleosome during gene induction.
Nucleosome Positioning Directly Affects Gene Expression.
Numerous studies demonstrated that transcriptional noise generates significant variability in gene expression in a homogenous population of cells (52–55). Nucleosome positioning may be one cause of such fluctuations by contributing to variability of gene expression (30, 31, 56). Generally, under phosphate-rich conditions, PHO5 gene expression is very low, and the promoter is highly occupied by nucleosomes (Fig. 1). Upon phosphate starvation, there is a shift to a more nucleosome-free architecture (Fig. 3). However, the small fraction of cells that express PHO5 under phosphate-rich conditions also exhibits this nucleosome-free architecture (Fig. 3 and Fig. S5). These data strongly suggest that nucleosomes play a direct role in antagonizing gene expression. Why this small fraction of cells lose nucleosomes remains to be explained but could be attributable to nucleosome “breathing” (reviewed in ref. 57), which refers to the fact that nucleosomes are dynamic, and spontaneous unwrapping at the ends could allow for access to sequence-specific transcription factors to bind the DNA and interfere with nucleosome positioning. Furthermore, our data support a model where fluctuations in promoter chromatin structure could trigger pulses of transcriptional activity (30, 44, 58, 59).
The transition from inactive to active chromatin states of the PHO5 promoter in bulk population has been associated with the retention of an average of 1.1 nucleosomes on the promoter (35), which can attributable to static retention of the nucleosome or by dynamic changes in nucleosomes with nucleosome sliding and disassembly accompanied by reassembly (44). Although we observe more than one nucleosome in the active state (conformations g and h), several aspects of our data support nucleosome sliding and are consistent with a certain amount of randomness in positioning where mapping occupancy is a snapshot of nucleosomes assembling, disassembling, and sliding (30). Nucleosome-scanning assays in bulk population show the appearance of a nucleosome at a position between nucleosomes −2 and −3 upon phosphate starvation, whereas the −1 and −2 nucleosomes become indistinct (Fig. 3A). Single-cell mapping indicates that the −1 nucleosome either shifts toward the start site to reveal the TATA box or disassembles, and the −2 and −3 nucleosomes shift away from the start site (Fig. 3B, configurations g and h). Quantitative measurements made by linking number differences and limited nuclease digestion assays of chromatin circles excised from the PHO5 promoter in vivo revealed that when PHO5 is activated, nucleosomes that were retained remained in similar positions to their locations in the repressed state. Specifically, Kornberg and colleagues found the probability of a nucleosome at the −1 position was 0.6, 0.2 at the −2 nucleosome, and 0.3 at the −3 nucleosome (35). We found in the activated nucleosome states of PHO5 induced by phosphate starvation (Fig. 3B, states g and h), ∼50% of cells entirely lose −1 nucleosome, and in 25% of cells, −1 nucleosome shifts away from the TATA box (Fig. 3B). In the activated state, the −2 nucleosome shifts upstream from the start site and thereby shifts the −3 nucleosome as well. These data are consistent with the disassembly of one nucleosome and sliding of the two others. Furthermore, when a poly(dA:dT) mutation is introduced, a subset of promoters becomes completely devoid of nucleosomes (Fig. 4B, configuration j), indicating that although, on average, the PHO5 gene retains a nucleosome when activated, this retention is not required for transcriptional activation.
The addition of the poly(dA:dT) sequence, which makes bending of DNA thermodynamically unfavorable, opposes the formation of a nucleosome and can increase expression in a manner similar to increasing the affinity of a transcription factor binding site (24). Indeed, the introduction of poly(dA:dT) tracts led to a loss of positioned nucleosomes and correlated with increased gene expression (Fig. 4). By contrast, altering the sequence to enhance the periodicity of the AA/TT/AT motif of the −3 nucleosome of PHO5 prevented increase in gene expression upon phosphate starvation and led to nucleosomes not being remodeled (Fig. 4). These data strongly suggest that the stabilization of nucleosomes prevents the ability of chromatin remodelers to shift these nucleosomes. Although significant evidence exists for nucleosomes antagonizing gene expression, these results demonstrate directly that individual cells shift to a nucleosome-depleted promoter when a gene is expressed. Furthermore, these data provide direct evidence for sequence preferences for nucleosome positioning as mutations, which are only base alterations and do not add or delete sequence, can influence nucleosome occupancy and ultimately affect gene expression.
Generally, promoters of active chromatin are relatively nucleosome-free (16), but as we have demonstrated, there is significant heterogeneity within a cell population. This heterogeneity may explain differential gene expression that previously could not be understood by population nucleosome mapping. Indeed, recent work from the Fraser laboratory demonstrated that individual immune T-helper cells had variability within their chromatin structure because some cells had many interchromosomal interactions, whereas other cells had almost exclusively intrachromosomal interactions (60). In addition, they found that in individual cells, domains enriched for active chromatin interact with other active domains and the inactive domains interact with other inactive domains. These cell-to-cell differences also appear to apply to nucleosome positioning and differences in positioning, as well as the presence of a nucleosome, could explain differential gene expression.
Our work demonstrates a method for mapping nucleosomes at the single-cell level. We are now able to identify the heterogeneity within cells at positions that previously could only be described as fuzzy. Specifically, we found that fuzziness may be, in part, attributable to more conformations of nucleosomes but also a combination of conformations over a larger region, as well as a considerable population entirely lacking a nucleosome in a particular region. We have also provided strong, direct evidence for a correlation between nucleosome occupancy and gene expression. Understanding nucleosome shifts in individual cells will likely provide fruitful information because epigenetic changes are recognized as causes for cancer and other diseases. Indeed, our method provides further insight into nucleosome locations, and likely changes in nucleosome occupancy in a specific population of cells could result in detrimental changes in gene expression that previously could not be determined.
Materials and Methods
PHO5 Mutant Strain Construction.
yECS9 containing EGFP-tagged PHO5 was made by integrative transformation of BY4741 with DNA amplified from pYM28 (61) using primers specific for C-terminal tagging of PHO5 listed in Table S1. The PHO5 gene, along with 2,000 bp upstream and 1,000 bp downstream of the gene, was cloned into pRS316 (pECS24) by introducing SacII and XhoI sites and amplifying genomic DNA from yECS9. Mutations in the PHO5 promoter were introduced into pECS24 using the QuikChange Mutagenesis Kit (Stratagene). To incorporate these mutations into yeast, the plasmids were digested with SacII and XhoI (New England Biolabs), and the regions of interest were gel-purified and transformed into BY4741, creating PolyA_1 (ECS54), PolyA_2 (ECS45), PolyA_3 (ECS62), PolyA_4 (ECS68), Periodicity_1 (ECS53), and Periodicity_4 (ECS56).
Yeast Growth.
For rich media growth, cells were grown in YPD (at 30 °C to an OD of ∼0.3). For phosphate starvation growth, the cells were washed with PBS and transferred to complete synthetic dropout media without potassium phosphate for an additional three hours. Cells were counted using a hemocytometer. GFP-positive cells were sorted using a FACSAria II. Gene expression of poly(dA:dT) and periodicity mutants was determined by flow cytometric analysis (BD LSR II).
MNase Digestion and Library Preparation.
Mononucleosomal DNA was prepared as described previously from spheroplasted yeast grown in rich media (4). Purified DNA was prepared for Illumina sequencing using NEB Next DNA Library Prep Reagent Set for Illumina and NEB Next Singleplex Oligos for Illumina. To determine the center-weighted nucleosome occupancy score, we followed our previously published method (19). Briefly, for each given sequence of length between 100–210 bp, we assigned a weight of to a position d bp away from the center of the sequence for . The center-weighted occupancy score at any given position is the aggregated weight from all sequences.
Nucleosome-Scanning Assay.
Purified mononucleosomal DNA was purified as described above. The assay was performed as described previously (62) using primers listed in Table S1. Quantitative RT-PCR was performed using a LightCycler 480II (Roche). The relative occupancy was determined by the comparative Ct method.
Methylation and Bisulfite Conversion of DNA.
Production of spheroplasts was performed as previously described (63). Briefly, cells were treated with 0.1 mg of lyticase per 100 mL of culture in 1 M sorbitol and 5 mM 2-mercaptoethanol for 15 min at room temperature. Spheroplasts were washed with 1 M sorbitol three times before treatment with GpC methyltransferase and treated with 30 units of M.CviP (NEB) supplemented with 160 µM S-adenosylmethionine (NEB) at 37 °C for 45 min. The reaction was stopped with 10 mM Tris (pH 7.9), 300 mM NaCl, 0.5% SDS, and 5 mM EDTA. Spheroplasts were then diluted based on original cell count to 0.6 cells per microliter. For diluted cells, 500 ng of ssDNA was added as carrier. DNA was phenol chloroform-extracted, followed by ethanol precipitation. DNA was digested with 0.1 units of HindIII (NEB). For bulk cells, 1µg of DNA was used for bisulfite conversion, and for diluted cells, the entire eluted DNA was used for conversion. Bisulfite conversion was performed using the EZ DNA Methylation-Lightening Kit (Zymo Research).
Amplification and Sequencing of DNA.
Primers for amplification were designed to amplify regions of interest and did not contain GCs. The primers used are listed in Table S1. DNA was amplified using ZymoTaq (Zymo Research). PCRs were purified using the PCR Purification Clean-up Kit (Qiagen) or the TOPO-TA Cloning Kit (Life Technologies), and colonies or purified DNA were Sanger-sequenced. Raw sequences available in Dataset S1.
Supplementary Material
Acknowledgments
We thank Eran Segal, Rob Phillips, Robert Holmgren, and members of the J.D.L. laboratory for critical reading of the manuscript. This work was inspired and began in the J.W. laboratory. This work was supported by Ruth Kirschstein National Research Service Award F32GM086948 (to E.C.S.) and Physical Sciences Oncology Center Grant U54CA143869 (to J.D.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400517111/-/DCSupplemental.
References
- 1.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 4.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39(10):1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 6.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10(10):R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLOS Comput Biol. 2008;4(11):e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaret KS, Carroll JS. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ransom M, et al. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem. 2009;284(35):23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258(5):800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 11.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254(2):130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 12.Floer M, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141(3):407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ertel F, et al. In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo. Mol Cell Biol. 2010;30(16):4060–4076. doi: 10.1128/MCB.01399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20(15):2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 15.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 16.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20(3):267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13(7):436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer VR. Nucleosome positioning: Bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22(5):250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486(7404):496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 21.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 22.Segal E, Widom J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suter B, Schnappauf G, Thoma F. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 2000;28(21):4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44(7):743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 25.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14(11):2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasson T, Hartemink AJ. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19(11):2101–2112. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: Nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16(14A):6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20(12):3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown CR, Mao C, Falkovskaia E, Jurica MS, Boeger H. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 2013;11(8):e1001621. doi: 10.1371/journal.pbio.1001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinke H, Hörz W. Anatomy of a hypersensitive site. Biochim Biophys Acta. 2004;1677(1-3):24–29. doi: 10.1016/j.bbaexp.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Reinke H, Hörz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11(6):1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 34.Jessen WJ, Hoose SA, Kilgore JA, Kladde MP. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat Struct Mol Biol. 2006;13(3):256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- 35.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11(6):1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 36.Fatemi M, et al. Footprinting of mammalian promoters: Use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33(20):e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuwaki M, Verreault A. Maintenance DNA methylation of nucleosome core particles. J Biol Chem. 2004;279(4):2904–2912. doi: 10.1074/jbc.M310111200. [DOI] [PubMed] [Google Scholar]
- 38.Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hörz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung HR, et al. The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS ONE. 2010;5(12):e15754. doi: 10.1371/journal.pone.0015754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyle-Heyrman G, et al. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc Natl Acad Sci USA. 2013;110(50):20158–20163. doi: 10.1073/pnas.1315809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18(7):1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao C, et al. Quantitative analysis of the transcription control mechanism. Mol Syst Biol. 2010;6:431. doi: 10.1038/msb.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133(4):716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippert MJ, et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci USA. 2011;108(2):698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HD, O’Shea EK. A quantitative model of transcription factor-activated gene expression. Nat Struct Mol Biol. 2008;15(11):1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belch Y, et al. Weakly positioned nucleosomes enhance the transcriptional competency of chromatin. PLoS ONE. 2010;5(9):e12984. doi: 10.1371/journal.pone.0012984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen A, Verstrepen KJ. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2011;75(2):301–320. doi: 10.1128/MMBR.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thåström A, et al. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288(2):213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 51.Xi L, et al. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics. 2010;11:346. doi: 10.1186/1471-2105-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blake WJ, KÆrn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 53.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 54.Maheshri N, O’Shea EK. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 55.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301(5636):1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 57.Blossey R, Schiessel H. The dynamics of the nucleosome: Thermal effects, external forces and ATP. FEBS J. 2011;278(19):3619–3632. doi: 10.1111/j.1742-4658.2011.08283.x. [DOI] [PubMed] [Google Scholar]
- 58.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dadiani M, et al. Two DNA-encoded strategies for increasing expression with opposing effects on promoter dynamics and transcriptional noise. Genome Res. 2013;23(6):966–976. doi: 10.1101/gr.149096.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11):947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 62.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18(6):735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Kent NA, Mellor J. Chromatin structure snap-shots: Rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23(18):3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.