Significance
The ever-increasing demand for transportation fuels, the decrease in global petroleum reserves, and the negative impact of greenhouse gases resulting from burning petroleum make renewable and sustainable biofuels an imperative for the future. First-generation biofuels produced from food crops are limited by cost and competition with food supply. Considerable effort has been made to produce fuels from lignocellulosic biomass, but the need for chemical and enzymatic pretreatment to solubilize the biomass prior to microbial bioconversion is a major economic barrier to the development of an industrial process. Here we report the metabolic engineering of a bacterium, Caldicellulosiruptor bescii, that is capable of using unprocessed switchgrass, an abundant, environmentally desirable, and economically sustainable lignocellulosic plant biomass, as feedstock to produce ethanol.
Keywords: metabolic engineering, bioenergy and thermophiles
Abstract
Ethanol is the most widely used renewable transportation biofuel in the United States, with the production of 13.3 billion gallons in 2012 [John UM (2013) Contribution of the Ethanol Industry to the Economy of the United States]. Despite considerable effort to produce fuels from lignocellulosic biomass, chemical pretreatment and the addition of saccharolytic enzymes before microbial bioconversion remain economic barriers to industrial deployment [Lynd LR, et al. (2008) Nat Biotechnol 26(2):169–172]. We began with the thermophilic, anaerobic, cellulolytic bacterium Caldicellulosiruptor bescii, which efficiently uses unpretreated biomass, and engineered it to produce ethanol. Here we report the direct conversion of switchgrass, a nonfood, renewable feedstock, to ethanol without conventional pretreatment of the biomass. This process was accomplished by deletion of lactate dehydrogenase and heterologous expression of a Clostridium thermocellum bifunctional acetaldehyde/alcohol dehydrogenase. Whereas wild-type C. bescii lacks the ability to make ethanol, 70% of the fermentation products in the engineered strain were ethanol [12.8 mM ethanol directly from 2% (wt/vol) switchgrass, a real-world substrate] with decreased production of acetate by 38% compared with wild-type. Direct conversion of biomass to ethanol represents a new paradigm for consolidated bioprocessing, offering the potential for carbon neutral, cost-effective, sustainable fuel production.
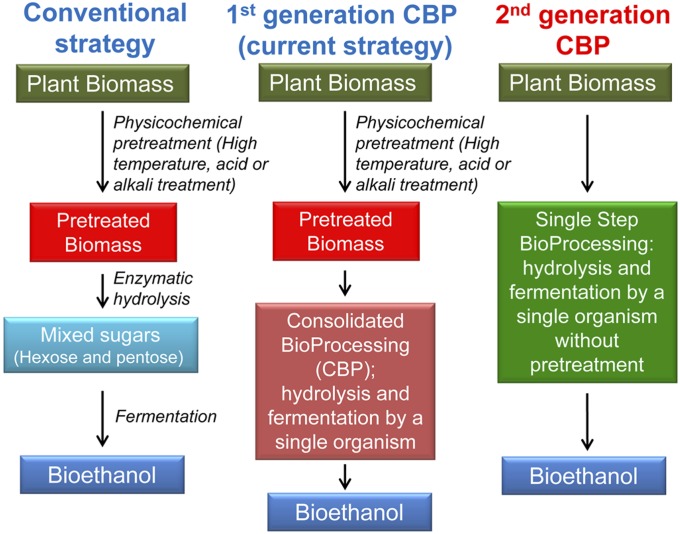
Increasing demand for fuels, geopolitical instability, the limitation of global petroleum reserves, and the impact on climate change induced by greenhouse gases have increased the need for renewable and sustainable biofuels (1–5). First-generation biofuels produced from food crops, such as corn, are limited by cost and competition with food supply (6, 7). Switchgrass is a perennial grass native to North America, and its high productivity on marginal farmlands and low agricultural input requirements make it an attractive feedstock for the production of biofuels and biochemicals (8). A yield of 36.7 Mg⋅ha−1 was achieved in field trials in Oklahoma (9), and switchgrass has the potential to produce 500% or more energy than is used for its cultivation (10). The use of abundant lignocellulosic plant biomass as feedstock is environmentally desirable and economically essential for enabling a viable biofuels industry (11). Current strategies for bioethanol production from lignocellulosic feedstocks require three major operational steps: physicochemical pretreatment, enzymatic saccharification, and fermentation (Fig. 1) (6, 12). Pretreatment and enzymatic hydrolysis represent substantial cost and it is estimated that the use of cellulolytic microbes for consolidated bioprocessing and eliminating pretreatment would reduce bioprocessing costs by 40% (2). Considerable effort has been made to develop single microbes capable of both saccharification and fermentation to avoid the substantial expense of using saccharolytic enzyme mixtures (13). Heterologous expression of saccharolytic enzymes has been demonstrated in a number of organisms, including Saccharomyces cerevisiae, Zymomonas mobilis, Escherichia coli, and Bacillus subtilis to ferment various model cellulosic and hemicellulosic substrates (13–15). Although these approaches have resulted in progress in cellulose utilization, the overall enzyme activity is still very low compared with that of naturally cellulolytic organisms and the rates of hydrolysis are not sufficient for an industrial process (13).
Fig. 1.
Comparison of bioethanol production strategies and a predicted fermentative pathway in C. bescii. Depiction of “single step bioprocessing” accomplished by engineered C. bescii. CBP, consolidated bioprocessing.
High-temperature fermentations facilitate biomass deconstruction and may reduce contamination and volatilize toxic products, such as alcohols. Clostridium thermocellum and Thermoanaerobacterium saccharolyticum have been used in mixed culture fermentations successfully for laboratory-scale demonstration of first-generation consolidated bioprocessing (13, 16) (Fig. 1). C. thermocellum is one promising candidate for consolidated bioprocessing because it is naturally cellulolytic, able to hydrolyze cellulose at 2.5 g⋅L−1⋅h−1, and produces ethanol as one fermentation product, but it has not yet been engineered to produce ethanol at high yield and lacks the ability to ferment hemicellulosic sugars (13, 17). Caldicellulosiruptor bescii, on the other hand, is the most thermophilic cellulolytic bacterium so far described, growing optimally at ∼80 °C with the ability to use a wide range of substrates, such as cellulose, hemicellulose, and lignocellulosic plant biomass without harsh and expensive chemical pretreatment (17, 18), efficiently fermenting both C5 and C6 sugars derived from plant biomass (17, 18). C. bescii uses the Embden–Meyerhof–Parnas pathway for conversion of glucose to pyruvate, and the predominant end-products are acetate, lactate, and hydrogen (Fig. 2) (18). A mutant strain of C. bescii (JWCB018) was recently isolated in which the lactate dehydrogenase gene (ldh) was disrupted spontaneously via insertion of a native transposon (19, 20). A complete deletion of ldh was also engineered (21), and this strain no longer produced lactate, instead diverting metabolic flux to additional acetate and H2, demonstrating the utility of the newly developed tools to provide a platform for further strain engineering. The recent development of genetic methods for the manipulation of this organism (19, 21, 22) opens the door for metabolic engineering for the direct conversion of unpretreated plant biomass to liquid fuels, such as ethanol, via “single step bioprocessing” (Fig. 1).
Fig. 2.
Overview of C. bescii fermentative pathways for bioconversion of hexose sugars. Pathway 1 (blue) results in 2 mol of acetic acid and 4 mol of H2 per mole of glucose. Pathway 2 (green) produces 2 mol of lactic acid per mole of glucose. Pathway 3 (red) is a new pathway resulting from heterologous expression of the C. thermocellum adhE gene to synthesize 2 mol of ethanol per mole of glucose.
Results and Discussion
Although many mixed-acid fermentation organisms use a bifunctional acetaldehyde/alcohol dehydrogenase (AdhE) to reduce acetyl-CoA into acetaldehyde and then into ethanol, bioinformatic analysis indicates that the C. bescii genome does not encode an obvious AdhE or acetaldehyde dehydrogenase (AldH) (23) (Fig. 2), and indeed no ethanol production is detected from this strain (18). The phylogenetically related, thermophilic Firmicute C. thermocellum, however, encodes an NADH-dependent AdhE (Cthe0423), which is one of the most highly expressed genes (24) and abundant proteins (25) in C. thermocellum and is the key enzyme in ethanol production in this organism. Based on its known thermostability, coenzyme specificity (NADH-dependent), similarity in codon use, and favorable catalytic stoichiometry (26), this adhE was a promising candidate to generate an ethanol production pathway in C. bescii. The adhE gene from C. thermocellum was expressed in the C. bescii ldh mutant strain (JWCB018), resulting in strain C. bescii JWCB032 (Table 1).
Table 1.
Strains and plasmids used in this study
| Strains | Strain and genotype/phenotype | Source |
| JWCB001 | C. bescii DSMZ6725 wild-type/(ura+/5-FOAS) | DSMZ† |
| JWCB018 | ΔpyrFA ldh::ISCbe4 Δcbe1/(ura−/5-FOAR) | (19, 20) |
| JWCB032 | ΔpyrFA ldh::ISCbe4 Δcbe1::PS-layer Cthe-adhE2/(ura/5-FOAR) | Present study |
| JWCB033 | ΔpyrFA ldh::ISCbe4 Δcbe1::PS-layer Cthe-adhE*(EA)3/(ura/5-FOAR) | Present study |
| pDCW88 | cbeI deletion vector (ApramycinR) | (19) |
| pDCW139 | Intermediate vector 1 (ApramycinR) | Present study |
| pDCW140 | Intermediate vector 2 (ApramycinR) | Present study |
| pDCW142 | Intermediate vector 3 (ApramycinR) | Present study |
| pDCW144 | Integrational vector containing the PS-layer Cthe-adhE‡ | Present study |
| pDCW145 | Integrational vector containing the PS-layer Cthe-adhE*§ | Present study |
German Collection of Microorganisms and Cell Cultures.
Cthe-adhE (Cthe0423; Bifunctional acetaldehyde/alcohol dehydrogenase derived from Clostridium thermocellum ATCC27405).
Cthe-adhE*(EA) (Cthe0423-P704L,H734R: Bifunctional acetaldehyde-CoA/alcohol dehydrogenase derived from ethanol tolerant Clostridium thermocellum strain EA) (26).
To determine the concentration at which growth of C. bescii is inhibited by ethanol, wild-type C. bescii was grown in low osmolarity-defined (LOD) medium (27) with 1% cellobiose as the sole carbon source, and subjected to different levels of added ethanol at both 65 °C and 75 °C (Fig. S1). The doubling time at 65 °C and 75 °C was ∼4 h and ∼2.7 h, respectively, in the absence of ethanol (Table S1). Growth of C. bescii in the presence of 300 mM added ethanol (13.8 g⋅L−1) was comparable to that with no added ethanol reaching a maximum OD of ∼0.6 at both 65 °C (in 15–20 h) and 75 °C (in 10–15 h). The addition of 400 mM ethanol (18.4 g⋅L−1) had a modest effect on growth at 65 °C (a doubling time of ∼4.5 h and a maximum OD of 0.44), but it substantially inhibited growth at 75 °C (a doubling time of ∼5.2 h and a maximum OD of 0.34). Growth was severely affected at 500 mM added ethanol (23 g⋅L−1) at 65 °C and 450 mM added ethanol (20.7 g⋅L−1) at 75 °C. No growth was seen at higher ethanol concentrations (>600 mM) (Table S1). This level of ethanol tolerance in C. bescii is comparable to that observed in wild-type C. thermocellum (28) and suggests that C. bescii can be engineered to produce ethanol at high titer.
The C. thermocellum adhE gene (Cthe0423) was amplified from C. thermocellum chromosomal DNA and cloned into pDCW144 (Fig. S2) under the transcriptional control of the C. bescii S-layer protein (Cbes2303) promoter (PS-layer). RNA profiling has shown that S-layer protein RNA is abundant throughout growth, suggesting that the promoter may be strong and constitutive (29). A rho-independent transcription terminator derived from a region immediately downstream of Cbes2303 was fused to the end of the adhE gene (Fig. 3A and Fig. S2) and the vector adds a C-terminal His-tag to the AdhE protein. This PS-layer-adhE construct was flanked by two 1-kb DNA regions of homology from the intergenic region between Cbes0863 and Cbes0864 (Fig. 3A and Fig. S2) to allow targeted integration into the C. bescii chromosome. The vector contains a pyrF cassette as both a positive and counter-selectable marker (30) and does not contain an origin of replication for C. bescii. Thus, selection for uracil prototrophy requires plasmid integration into the chromosome. This plasmid was transformed into the C. bescii ldh mutant strain JWCB018 [ΔpyrFA ldh::ISCbe4 Δcbe1 (Table 1); herein referred to as ldh−] that is a uracil auxotroph and contains a deletion of the CbeI restriction enzyme to simplify genetic manipulation (19, 20). Counter-selection with 5-fluoroorotic acid (5-FOA) selected for segregation of the merodiploid as previously described (19), depicted in Fig. 3A. Two of the 40 5-FOA–resistant colonies analyzed by PCR amplification using primers DC477 and DC478 (Fig. 3B) contained segregated insertions of the PS-layer -adhE cassette at the targeted chromosome site, resulting in strain JWCB032 (ΔpyrFA ldh::ISCbe4 Δcbe1 PS-layer-adhE+; herein referred to as ldh− adhE+). As shown in Fig. 3B, the parent strain, JWCB018, produced the expected wild-type ∼2.44-kb band, whereas amplification from JWCB032 (ldh− adhE+) produced an ∼5.04-kb band indicating a knock-in of the expression cassette within this region.
Fig. 3.
Targeted insertion and expression of the C. thermocellum ATCC27405 adhE gene (Cthe0423) in C. bescii. (A) A diagram of the integration vector pDCW144 (Fig. S2), which contains the PS-layer-Cthe-adhE expression cassette and pyrF cassette (30) for selection of transformants. Homologous recombination can occur at the upstream- or downstream-targeted chromosomal regions, integrating the plasmid into the genome and generating a strain that is a uracil prototroph. Counter selection with 5-FOA selects for loss of the plasmid sequences but not the adhE expression cassette. Bent arrows depict primers used for verification of the integrated expression cassette (2.6 kb). ApR, apramycin resistance gene cassette. (B) Gel depicting PCR products amplified from the targeted chromosome region in JWCB018 (ldh−, lane 1), JWCB032 (ldh− adhE+, lane 2), and JWCB033 [ldh− adhE(EA)+, lane 3] amplified by primers DC477 and DC478. M: 1-kb DNA ladder (New England Biolabs). (C) Western blot analysis of C. bescii strains used in this study. The 77 µg of total cell protein lysate isolated from the mid-log–phase cultures grown at various temperatures, either 60 °C, 65 °C, or 70 °C, were electrophoresed and probed with His-tag antibody as described in Methods. Lane 1, ldh−; lane 2, ldh− adhE+; lane 3, ldh− adhE(EA)+; M, MagicMark XP Western Protein Standard (Invitrogen).
Heterologous expression of the AdhE protein was detected in transformants containing the expression cassette by Western hybridization using monoclonal antibodies targeting the His-tag (Fig. 3C). Expression of the wild-type AdhE was readily detected in mid-log–phase cells grown at 60 °C and 65 °C but not at 70 °C. Because the optimal temperature for growth for C. thermocellum is 60 °C (28), the AdhE protein or adhE mRNA may not be stable at 70 °C. All further experiments were performed at 65 °C to ensure production of AdhE and maximize the efficiency of biomass deconstruction by C. bescii. We also expressed a variant of the AdhE protein, AdhE*(EA), from C. thermocellum EA that had been shown to alter cofactor specificity and increase ethanol tolerance in C. thermocellum (26) without losing functional activity for ethanol production, resulting in strain JWCB033 [ldh− adhE(EA)+]. Interestingly, the AdhE*(EA) was detected at 60 °C but not 65 °C or 70 °C, suggesting that either the protein or its mRNA is less thermostable than the wild-type (Fig. 3C). Because the protein was not present at 65 °C, we did not assess its affect on ethanol production or tolerance in C. bescii. Attempts to express the T. saccharolyticum AdhE (Tsac0416) at 60–75 °C were also unsuccessful.
Introduction of a new fermentative pathway for ethanol production in C. bescii could result in poor growth because of a redox imbalance. Therefore, we first examined the growth rate and yield of the adhE-expressing strain relative to the wild-type and parent strains (Fig. 4 A and B). At both 65 °C and 75 °C, the growth rate and growth yield were comparable between wild-type, ldh−, and ldh− adhE+ strains (Fig. 4 A and B and Table S2). Previous work demonstrated that a deletion of the ldh gene in C. bescii led to increased growth yield on cellobiose, likely because of the increased ATP yield concomitant with an increase in acetate production (21). In the present study, there was no obvious difference because cell densities were much higher than those previously reported, likely because of improved growth conditions, including growth with shaking.
Fig. 4.
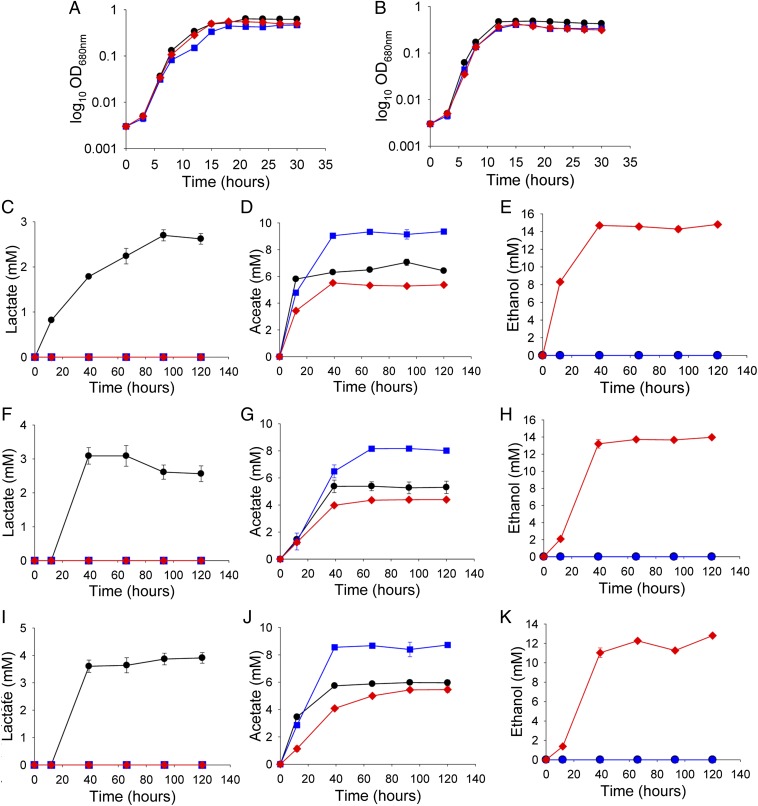
Growth and analysis of fermentation products of C. bescii containing the C. thermocellum adhE gene compared with wild type strains. Growth of C. bescii strains on 1.0% of cellobiose as the carbon source at 65 °C (A) and 75 °C (B) (log10 OD680nm). Analysis of fermentation products lactate (C, F, I), acetate (D, G, J), and ethanol (E, H, K) after growth on cellobiose [1% (wt/vol)] (C, D, E), Avicel [2% (wt/vol)] (F, G, H), and switchgrass [2% (wt/vol)] (I, J, K) at 65 °C. Black circles, JWCB001 wild-type; blue squares, JWCB018 (ldh−); Red diamonds, JWCB032 (ldh− adhE+). Error bars based on two biologically independent experiments.
To determine the functionality of AdhE in C. bescii and its effect on the redirection of flux to ethanol, the fermentation product profiles from C. bescii wild-type and mutant strains were examined via HPLC during growth on a soluble substrate (cellobiose, 1%, equal to 29.2 mM), a model microcrystalline cellulosic substrate [Avicel, 2% (wt/vol)], and a plant biomass substrate [switchgrass, 2% (wt/vol)] (Fig. 4 C–K). We point out that the switchgrass was not even autoclaved, a mild form of pretreatment, before use. The wild-type strain produced lactate (∼3.1 mM) and acetate (∼5.4 mM) but no detectable ethanol on cellobiose, Avicel, or switchgrass. The ldh− strain, JWCB018, made no detectable lactate and increased acetate (7.3 ∼8.2 mM) compared with the wild-type, but there was no detectable ethanol. The ldh− adhE+ strain, JWCB032, produced a lower level of acetate (4.3 mM) and redirected most of the flux to ethanol (14.8 mM) on cellobiose. A similar level of ethanol was produced on 2% (wt/vol) Avicel (14.0 mM), and slightly less (12.8 mM) was produced on switchgrass.
Surprisingly, with no adhE expression optimization or pathway manipulation aside from the use of an ldh mutant background, 70 ∼73% of the detected fermentation products (excluding hydrogen) in the ldh− adhE+ strain was ethanol during growth on cellobiose, Avicel, and switchgrass. By 39 h, cellobiose fermentation was complete. At this point, 14.7 mM ethanol had been produced from 4.4 mM cellobiose (8.8 mM glucose equivalents) that had been removed from the culture, resulting in a molar yield of 1.67 mol ethanol/mole of glucose (Fig. 4). For the overall carbon mass balance, carbon utilization on 1% cellobiose was monitored by HPLC and calculated at 48 h (Fig. S3 and Table S3). The final H2 production was measured by gas chromatography (GC) (Fig. S4). For the wild-type strain, 8% of the cellobiose was used. For the background ldh− strain, 8.1% was used, and ldh− adhE+ used 17%. The overall carbon recovery was 103%, 103%, and 107%, respectively (Table S3). Hydrogen production was measured from all three strains on cellobiose, Avicel, and switchgrass after 48 h of growth. The ldh− strain produced 25% more hydrogen than the wild-type strain on cellobiose, 28%, more on Avicel, and 12% more on switchgrass (Fig. S4). The ldh− adhE+ strain, on the other hand, produced 36% less hydrogen than the ldh− strain on cellobiose, 52% less on Avicel, and 44% less on switchgrass (Fig. S4).
To our knowledge, this work is the first demonstration of metabolic engineering in an extreme thermophilic organism for the conversion of lignocellulosic biomass to a liquid fuel. Furthermore, ethanol has been produced directly from plant biomass without the use of harsh, expensive, chemical pretreatment. Combining metabolic engineering with the native cellulolytic ability of C. bescii has the potential to transform the biofuels industry by creating a process in which both the pretreatment step and the addition of exogenous cellulases may be eliminated (Fig. 1). The strains developed here will serve as platforms for further metabolic engineering to increase yield and titer to enable cellulosic biofuel production on an industrial scale. Indeed, significant progress has already been made toward process development in C. bescii using high loads of unpretreated plant biomass (31). Deletion of hydrogenase (pathway 1 in Fig. 2), pta and ack genes (pathway 1 in Fig. 2) has been shown to contribute to an increase in ethanol production in T. saccharolyticum (32, 33), and this approach holds promise in C. bescii. Because ∼25% of the carbon fermentation products remain to be redirected to ethanol, improvements in yield will likely come with deletion of the two hydrogenase genes and the acetate kinase gene in C. bescii, and expression of a more thermostable AdhE protein. The relatively high ethanol tolerance of C. bescii (>300 mM) suggests that significant improvements in production will be possible before inhibition becomes an issue.
Although the near-term goal of this work is to generate a cost-effective process for the production of biofuels from lignocellulosic biomass, the strains and conditions developed here may also allow for the production of other classes of products from plant biomass using proteins derived from thermophilic organisms.
Methods
Strains, Media, and Culture Conditions.
C. bescii strains and plasmids used in this study are listed in Table 1. All C. bescii strains were grown anaerobically in liquid or on solid surface in LOD medium (27), final pH 7.0, with maltose [0.5% (wt/vol); catalog no. M5895, Sigma) as the carbon source unless otherwise noted. Liquid cultures were grown from a 0.5% inoculum or a single colony and incubated at 75 °C in anaerobic culture bottles degassed with five cycles of vacuum and argon. For growth of uracil auxotrophic mutants, the LOD medium was supplemented with 40 μM uracil. E. coli strain DH5α was used for plasmid DNA constructions and preparations. Standard techniques for E. coli were performed as described previously (34). E. coli cells were grown in LB broth supplemented with apramycin (50 μg/mL) and plasmid DNA was isolated using a Qiagen Miniprep Kit. Chromosomal DNA from C. bescii strains was extracted using the Quick-gDNA MiniPrep (Zymo) or using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions.
Construction of Vectors for Knock-in of Cthe0423 and Its Derivative into C. bescii.
The plasmids described below were generated using high-fidelity pfu AD DNA polymerase (Agilent Technologies) for PCR reactions, restriction enzymes (New England Biolabs), and Fast-link DNA Ligase kit (Epicentre Biotechnologies) according to the manufacturers’ instructions. Plasmid pDCW144 (Fig. 3A and Fig. S2) was constructed in four cloning steps. First, a 2.31-kb DNA fragment containing the targeted insertion region sequences (intergenic space between convergent genes Cbes0863-Cbes0864) in the C. bescii chromosome was amplified using primers DC456 (with KpnI site) and DC457 (with EcoRI site) using C. bescii genomic DNA as a template. The 4.0-kb DNA fragments containing an apramycin resistance-gene cassette, pyrF cassette (30), and the pSC101 replication origin, were amplified from pDCW88 (19) using primers DC081and DC356. The DC081 and DC356 primers were engineered to contain KpnI and EcoRI sites, respectively. These two linear DNA fragments were digested with KpnI and EcoRI, and ligated to construct the 6.33 kb pDCW139 (Fig. S2). Plasmid pDCW140 (Fig. S2) was constructed by inserting the 3.28 kb DNA fragment which contains the 134 bp of upstream sequences of Cbes2303 (S-layer protein), 3,507 bp of Cbes2303 coding sequences, and 86 bp of its downstream sequences, into pDCW139. This DNA fragment was amplified using primers DC460 (with PvuI site) and DC461 (with NotI site) using C. bescii genomic DNA as a template. The 6.1-kb DNA fragment was amplified from pDCW139 using primers DC458 (with PvuI site) and DC459 (with NotI site) to be used as a back-bone fragment. These two linear DNA fragments were digested with PvuI and NotI, and ligated to a construct pDCW140. The resulting plasmid contains the 5′ flanking region (1,013 bp) and the 3′ flanking region (1,012 bp) of targeted insertion site in the C. bescii genome in addition to the S-layer protein expression cassette (Fig. S2). Plasmid pDCW142, the back-bone vector for knock-in plasmid, was constructed by adding restriction sites for cloning and C-terminal 6x Histidine-tag in front of a stop codon, in addition to removing the Cbes2303 coding sequences in pDCW140. The 6.3-kb DNA fragment was amplified from pDCW139 using primers DC464 (with BamHI site) and DC466 (with SphI site, 6x Histidine-tag, and stop codon) using pDCW140 as a template. This DNA fragment was blunt-end ligated after treatment with T4 PNK (New England Biolabs) to construct pDCW142 (Fig. S2). In the last step, a 2.62-kb DNA fragment containing the coding sequence of Cthe0423 was amplified by PCR using DC469 (with BamHI site) and DC470 (with SphI site) using C. thermocellum ATCC 27405 genomic DNA as a template. This DNA fragment was digested with BamHI and SphI, and then cloned into pDCW142 that had been digested with BamHI and SphI (Fig. S2). Plasmid pDCW145 is identical to pDCW144 except for the cloning of Cthe0423 adhE*(EA) (26), which contains two point mutations in coding sequences, into pDCW142. To make this change, a 2.62-kb DNA fragment containing the coding sequence of Cthe0423* was amplified by PCR using DC469 (with BamHI site) and DC470 (with SphI site) using C. thermocellum EtOH (26) genomic DNA as a template. DNA sequences of the primers are shown in Table S4. E. coli strain DH5α cells were transformed by electroporation in a 2-mm gap cuvette at 2.5 V and transformants were selected for apramycin resistance. The sequences of all plasmids were verified by Automatic sequencing (Macrogen). All plasmids are available on request. Primers used for plasmid constructions are listed in Table S4.
Transformation, Screening, Purification, and Sequence Verification of Engineered C. bescii Mutants.
To construct strain JWCB032, 1 μg of pDCW144 DNA was used to electrotransform JWCB018 (ΔpyrFA Δcbe1) as described previously (19). Cells were then plated onto solid LOD medium and uracil prototrophic transformant colonies were inoculated into liquid medium for genomic DNA extraction and subsequent PCR screening of the targeted region to confirm the knock-in event of pDCW144 into the chromosome. Confirmed transformants were inoculated into nonselective liquid-defined medium, with 40 μM uracil, and incubated overnight at 75 °C to allow loop-out of the plasmid DNA leaving the adhE expression cassette (Fig. 3A and Fig. S2). The cultures were then plated onto 5-FOA (8 mM) containing solid medium. After initial PCR screening, 5-FOA–resistant colonies containing the expected knock-in were further purified by one additional passage on solid medium and screened a second time by PCR to check for segregation of the PS-layer-adhE insertion. The location of the insertion was verified by PCR amplification and sequence analysis. A PCR product was generated from genomic DNA using primers (DC477 and DC478) outside the homologous regions used to construct the knock-in, and internal primers (DC456, DC457, DC462, and DC463). PCR products were sequenced to confirm. Construction of JWCB033 was the same as JWCB032 except that pDCW145 (Table 1) was used to electrotransform JWCB018. All primers used are listed in Table S4.
Preparation of Cell Lysates and Western Blotting.
Cell-free extracts of C. bescii were prepared from 500-mL cultures grown to mid-log phase at various temperatures (60 °C, 65 °C, 70 °C, and 75 °C), harvested by centrifugation at 6,000 × g at 4 °C for 15 min, and resuspended in Cel-Lytic B-cell lysis reagent (Sigma). Cells were lysed by a combination of 4× freeze-thawing and sonication on ice. Protein concentrations were determined using the Bio-Rad protein assay kit with BSA as the standard. Next 77-μg protein samples were electrophoresed in a 4–15% gradient Mini-Protean TGX gels (Bio-Rad) and electrotransferred to PVDF membranes (ImmobilonTM-P; Millipore) using a Bio-Rad Mini-Protean 3 electrophoretic apparatus. The membranes were then probed with His-tag (6xHis) monoclonal antibody (1:5,000 dilution; Invitrogen) using the ECL Western Blotting substrate Kit (Thermo Scientific), as specified by the manufacturer.
Growth Curve Analysis, Measurement of Ethanol Tolerance, and Fermentation Conditions.
Analysis of growth and ethanol tolerance was conducted in stoppered 125-mL serum bottles containing 50 mL LOD medium supplemented with 10 g/L cellobiose (catalog no. M5895; Sigma) and 1 mM uracil. Duplicate bottles were inoculated with a fresh 2% (vol/vol) inoculum and incubated at both 65 °C and 75 °C with shaking at 150 rpm. Optical cell density was monitored using a Jenway Genova spectrophotometer, measuring absorbance at 680 nm. Batch fermentations were performed for 5 d, at 65 °C in the same culture conditions, except using 10 g/L cellobiose, 20 g/L Avicel (catalog no.11365, Fluka), or 10 g/L unpretreated switchgrass [sieved −20/+80-mesh fraction; washed with warm water but no additional pretreatment (note that the biomass samples were not autoclaved before use); provided by Brian Davison, Oak Ridge National Laboratory, Oak Ridge, TN] as carbon sources.
Analytical Techniques for Determining Fermentation End Products.
Fermentation products, acetate, lactate, and ethanol, were analyzed on an Agilent 1200 infinity HPLCy (HPLC) system (Agilent Technologies). Metabolites were separated on an Aminex HPX-87H column (Bio-Rad Laboratories) under isocratic temperature (50 °C) and a flow (0.6 mL/min) condition in 5.0 mM H2SO4, and then passed through a refractive index detector (Agilent 1200 Infinity Refractive Index Detector). Identification was performed by comparison of retention times with standards, and total peak areas were integrated and compared against peak areas and retention times of known standards for each compound of interest. H2 was measured using a GC model 8610C (SRI Instruments) equipped with a thermal conductivity detector at 80 °C with a N2 reference flow and a 1.83 m × 3.18 mm (inside diameter) stainless-steel column packed with Porapak-Q (80/100 mesh).
Supplementary Material
Acknowledgments
We thank Jennifer Copeland for outstanding technical assistance; Brian Davison for providing the switchgrass used in this study; Sidney Kushner for expert advice in the execution of the experiments; and Joe Groom and Jenna Young for critical review of the manuscript. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402210111/-/DCSupplemental.
References
- 1.John UM. Contribution of the Ethanol Industry to the Economy of the United States. Washington, DC: Renewable Fuels Association; 2013. [Google Scholar]
- 2.Lynd LR, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26(2):169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 3.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 4.Kerr RA. Climate change. Global warming is changing the world. Science. 2007;316(5822):188–190. doi: 10.1126/science.316.5822.188. [DOI] [PubMed] [Google Scholar]
- 5.Solomon BD. Biofuels and sustainability. Ann N Y Acad Sci. 2010;1185:119–134. doi: 10.1111/j.1749-6632.2009.05279.x. [DOI] [PubMed] [Google Scholar]
- 6.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488(7411):320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 7.Tilman D, et al. Energy. Beneficial biofuels—The food, energy, and environment trilemma. Science. 2009;325(5938):270–271. doi: 10.1126/science.1177970. [DOI] [PubMed] [Google Scholar]
- 8.Keshwani DR, Cheng JJ. Switchgrass for bioethanol and other value-added applications: A review. Bioresour Technol. 2009;100(4):1515–1523. doi: 10.1016/j.biortech.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 9.Thomason WE, et al. Switchgrass response to harvest frequency and time and rate of applied nitrogen. J Plant Nutr. 2004;27(7):1199–1226. [Google Scholar]
- 10.Schmer MR, Vogel KP, Mitchell RB, Perrin RK. Net energy of cellulosic ethanol from switchgrass. Proc Natl Acad Sci USA. 2008;105(2):464–469. doi: 10.1073/pnas.0704767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gronenberg LS, Marcheschi RJ, Liao JC. Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol. 2013;17(3):462–471. doi: 10.1016/j.cbpa.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan DB, et al. Plant cell walls to ethanol. Biochem J. 2012;442(2):241–252. doi: 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- 13.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23(3):396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Bokinsky G, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci USA. 2011;108(50):19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linger JG, Adney WS, Darzins A. Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Appl Environ Microbiol. 2010;76(19):6360–6369. doi: 10.1128/AEM.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argyros DA, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77(23):8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: Status and prospects. Curr Opin Biotechnol. 2008;19(3):210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Yang SJ, et al. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75(14):4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung D, Farkas J, Westpheling J. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol Biofuels. 2013;6(1):82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cha M, Wang H, Chung D, Bennetzen JL, Westpheling J. Isolation and bioinformatic analysis of a novel transposable element, ISCbe4, from the hyperthermophilic bacterium, Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol. 2013;40(12):1443–1448. doi: 10.1007/s10295-013-1345-8. [DOI] [PubMed] [Google Scholar]
- 21.Cha M, Chung D, Elkins J, Guss A, Westpheling J. Metabolic engineering of Caldicellulosiruptor bescii yields increased hydrogen production from lignocellulosic biomass. Biotechnol Biofuels. 2013;6(1):85. doi: 10.1186/1754-6834-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. Methylation by a unique α-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PLoS ONE. 2012;7(8):e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carere CR, et al. Linking genome content to biofuel production yields: Ameta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol. 2012;12:295. doi: 10.1186/1471-2180-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riederer A, et al. Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose. Appl Environ Microbiol. 2011;77(4):1243–1253. doi: 10.1128/AEM.02008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rydzak T, et al. Proteomic analysis of Clostridium thermocellum core metabolism: Relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 2012;12:214. doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown SD, et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci USA. 2011;108(33):13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farkas J, et al. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol. 2013;40(1):41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrero AA, Gomez RF. Development of ethanol tolerance in Clostridium thermocellum: Effect of growth temperature. Appl Environ Microbiol. 1980;40(3):571–577. doi: 10.1128/aem.40.3.571-577.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dam P, et al. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 2011;39(8):3240–3254. doi: 10.1093/nar/gkq1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung D, Cha M, Farkas J, Westpheling J. Construction of a stable replicating shuttle vector for Caldicellulosiruptor species: Use for extending genetic methodologies to other members of this genus. PLoS ONE. 2013;8(5):e62881. doi: 10.1371/journal.pone.0062881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basen M, et al. Degradation of high loads of crystalline cellulose and of unpretreated plant biomass by the thermophilic bacterium Caldicellulosiruptor bescii. Bioresour Technol. 2014;152:384–392. doi: 10.1016/j.biortech.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Shaw AJ, Hogsett DA, Lynd LR. Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J Bacteriol. 2009;191(20):6457–6464. doi: 10.1128/JB.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw AJ, et al. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;105(37):13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.