Significance
The nature of postnatal cardiomyogenesis in mammals remains in dispute. Here, we provide cell-level evidence for the birth of cardiomyocytes in newborn and adult mice. Our clonal analysis, based on the mosaic analysis with double markers mouse model, shows that cardiomyocytes are the parent cell of origin of cardiomyocytes that are generated postnatally. Our findings confirm that limited, symmetric division of cardiomyocytes is a rare phenomenon in the mouse heart after birth.
Keywords: heart development, cardiovascular progenitors, aging, regeneration
Abstract
The mammalian heart has long been considered a postmitotic organ, implying that the total number of cardiomyocytes is set at birth. Analysis of cell division in the mammalian heart is complicated by cardiomyocyte binucleation shortly after birth, which makes it challenging to interpret traditional assays of cell turnover [Laflamme MA, Murray CE (2011) Nature 473(7347):326–335; Bergmann O, et al. (2009) Science 324(5923):98–102]. An elegant multi-isotope imaging-mass spectrometry technique recently calculated the low, discrete rate of cardiomyocyte generation in mice [Senyo SE, et al. (2013) Nature 493(7432):433–436], yet our cellular-level understanding of postnatal cardiomyogenesis remains limited. Herein, we provide a new line of evidence for the differentiated α-myosin heavy chain-expressing cardiomyocyte as the cell of origin of postnatal cardiomyogenesis using the “mosaic analysis with double markers” mouse model. We show limited, life-long, symmetric division of cardiomyocytes as a rare event that is evident in utero but significantly diminishes after the first month of life in mice; daughter cardiomyocytes divide very seldom, which this study is the first to demonstrate, to our knowledge. Furthermore, ligation of the left anterior descending coronary artery, which causes a myocardial infarction in the mosaic analysis with double-marker mice, did not increase the rate of cardiomyocyte division above the basal level for up to 4 wk after the injury. The clonal analysis described here provides direct evidence of postnatal mammalian cardiomyogenesis.
Although it was widely believed that the adult heart is a quiescent organ, in the past several years reports have argued in favor of the generation of new cardiomyocytes in the mouse and human hearts after birth. The strongest evidence to first incontrovertibly demonstrate this phenomenon “date-stamped” autopsied human hearts by correlating levels of 14C in cardiomyocyte nuclei with atmospheric 14C levels in different years, and revealed that a small percentage of cardiomyocytes is born during adulthood (1). Despite this significant finding, which indirectly correlated nuclear division with cell division, the parent cell of postnatal cardiomyogenesis, as well as the extent of division in the postnatal mammalian hearts, remains vigorously debated. Moreover, the effect of injury on the endogenous rate of mammalian cardiomyocyte generation is unresolved (2–5).
After resection of the ventricular apex, both adult zebrafish and neonatal mice exhibit robust regeneration, which fate-mapping studies suggest occurs through a cardiomyocyte intermediate (6–8). However, the study of cardiomyocyte generation by division postnatally has been controversial (9, 10) in the mammalian heart because it often relies on indirect assays of cell division, which are challenging to interpret in the setting of cardiomyocyte polyploidy (11, 12) as well as potential DNA repair upon injury. Recently, it was shown using a multi-isotope imaging mass spectrometry (MIMS) technique and concomitant fate-mapping that cardiomyocytes renew cardiomyocytes after birth in mice (with, at best, minimal contribution from progenitor cells) (2), but a number of questions about postnatal cardiomyogenesis remains open. For example, it remains unclear whether the daughter cells of cardiomyocytes can also divide (i.e., whether daughter cells can behave as transit-amplifying cells). It is also unknown whether the cell-of-origin of postnatal cardiomyogenesis can generate other cardiovascular cell types at the time of division. Because the majority of studies in this field use indirect assays of cell division that rely on analysis of nuclear division rather than cell division, direct observation of cardiomyocyte generation at the single-cell level has remained elusive. An understanding of postnatal cardiomyocyte generation at the cell level could answer some of the open questions about this controversial phenomenon.
Clonal analysis by lineage tracing is a useful method in cancer biology to trace precursor-progeny relationships of tumorigenic cells (13) and has also revealed the existence of two developmental heart fields (14). Because this technique effectively extends fate-mapping to the single-cell level, we sought to determine whether differentiated cardiomyocytes could generate cardiomyocytes postnatally in mammals using clonal analysis, which could strengthen existing observations on this phenomenon as well as potentially reveal mechanistic details of this property. The results of this study could also inform development of cell therapy for cardiovascular disease.
Results
Mosaic Analysis of Double Markers Transgenesis Unambiguously Labels Progeny Cells.
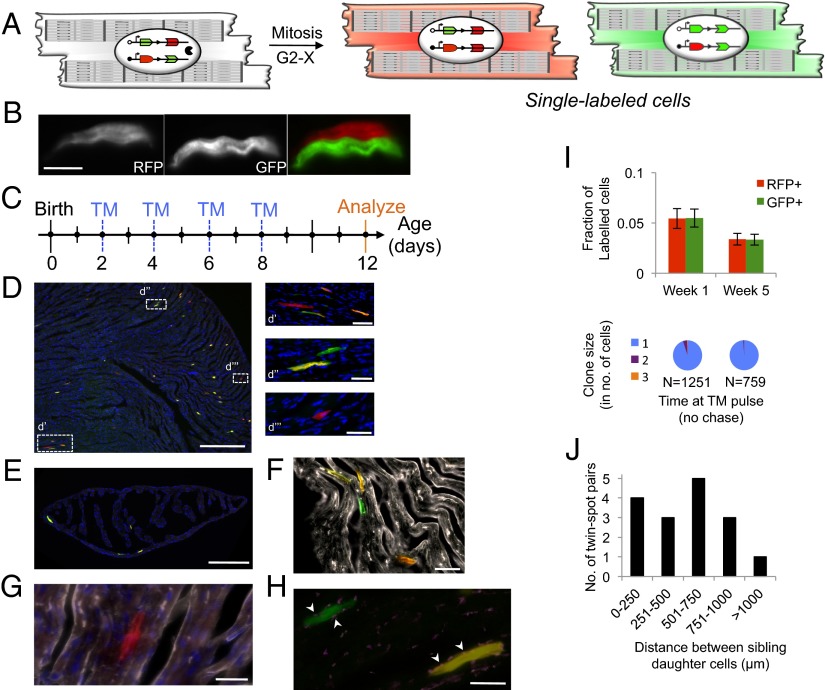
To test our hypothesis, we used the “mosaic analysis with double markers” (MADM) model, in which the two daughter cells of a dividing cell are indelibly and uniquely “single-labeled” either GFP+ or RFP+ because of interchromosomal Cre-loxP recombination after S phase (15, 16) (Fig. 1A and Fig. S1). MADM labeling allows unambiguous identification of progeny cells because cytokinesis is needed to generate individual GFP+ and RFP+ cells (a binucleated cell would be double-labeled as GFP+RFP+, and appear yellow; DNA repair would not cause labeling) (17). Because MADM single-labeling can only be achieved by completion of the cell cycle through cytokinesis, it permits analysis of cell division that is directly related to cytokinesis and uncoupled from karyokinesis, unlike many prior reports that have analyzed division after birth. Thus, MADM is an ideal system in which to test the principles of postnatal division, especially in an organ so prone to controversy as the mammalian heart. Furthermore, asymmetric labeling of the daughter cells enables precise determination of precursor–progeny lineages (e.g., self-renewal and multilineage potential) (Fig. S2) (18). When used with an inducible Cre transgene, MADM permits temporal control of recombination and provides direct evidence for cell division. MADM has been used reliably in the past in different organ systems (19, 20), especially the nervous system, and has revealed key lineage relationships during development as well as identified a cancer cell of origin.
Fig. 1.
Cardiomyocytes divide postnatally in mice. (A) MADM recombination in a parent cardiomyocyte in Myh6-expressing cells leads to RFP+ and GFP+ single-labeled daughter cardiomyocytes (arrowheads). (B) Example of an alternate-label sibling pair. (Scale bar, 15 μm.) (C) TM delivery strategy for D–H. (D) Section through P12 heart reveals double-labeled and sparse single-labeled cells. (Scale bar, 100 µm.) (d′) RFP+ single-labeled cell along with its partner GFP+ single-labeled cell and double-labeled cells. (d′′) GFP+ single-labeled cell (with evident sarcomeric elements) and a double-labeled cell. (d′′′) An RFP+ single-labeled cell. (Scale bars, 10 µm.) (E) MADM labeling in the atria. (Scale bar, 50 µm.) (F) Example of GFP+ cell and double-labeled cell that stain positive for troponin (white). (Scale bar, 10 μm.) (G) α-Actinin staining (white) reveals sarcomeric structures of a RFP+ cardiomyocyte. (Scale bar, 5 μm.) (H) GFP+ cardiomyocyte forms gap junctions (Connexin43, white) with underlying syncytium. (Scale bar, 5 μm.) (I, Upper) Frequency of single-labeled cells at different time points after TM administration. (n = 3, P values calculated with Student t test. Error bars indicate SEM) (Lower) Analysis of same-color clones for the above mice. n = total number of single-labeled cells observed per group. (J) Histogram of the distribution of distance between clonal sibling cells (n = 5 mice).
MADM Labeling Allows Clonal Identification in the Heart, with No Evidence for Allelic Silencing of the MADM Transgenes.
To first validate the use of MADM in the heart, we generated HprtCre+/−;MADM-11GT/TG mice and analyzed in the embryonic period. Hprt is expressed from the X chromosome and analysis of males permits maximal recombination (because of random X chromosome inactivation in females). Moreover, Hprt can be expressed by any cell, including progenitor cells during development, which would enable observation of discrete clones. Analysis of the male developing heart in embryonic day (E) 16.5 embryos demonstrated clusters of RFP+ cells and GFP+ cells (Fig. S3), as would be expected for proliferation in utero. The cluster sizes were variable, ranging from 6 to 20 cells. Use of Cre recombinase [rather than tamoxifen (TM)-inducible CreER] prevents temporal control of recombination, so the length of the “chase period,” (i.e., the time from induction of MADM recombination to E16.5) is not clear and conclusions about cardiomyocyte proliferation rates at different time points in utero cannot be ascertained from these data. In addition, because Hprt can be expressed in any cell, the identity of the cell type that forms a cluster of cardiomyocytes cannot be revealed by this system. However, this transgenic model (HprtCre;MADM) helped to validate the utility of MADM in the heart, and prompted us to use an inducible cardiomyocyte-specific Cre transgenic mouse model (described below) to investigate clonal analysis of cardiomyocyte proliferation and to identify the lineage relationship between parent and daughter cells.
To ensure that silencing in a single allele of MADM-labeled cells does not occur in the heart, which could lead to false-positive single-labeled cells if it occurs in a double-labeled cell or false-negatives if it occurs in a bona fide single-labeled cell—and would therefore render analysis of cell division inaccurate—a transgenic mouse was generated in which, in each cell, one Hipp11 locus contains the reconstituted GFP gene and the other Hipp11 locus contains the reconstituted RFP gene (MADM-11GG/TT). Tissue sections of the hearts from these mice were analyzed and we observed only GFP+RFP+ double-labeled cells, with no evidence of single-labeled or unlabeled cells, demonstrating that there is no silencing of the Hipp11 locus at either allele (21) (Fig. S4).
In the Postnatal Period, Cardiomyocytes Generate Nondividing Cardiomyocytes That Separate from Each Other.
We created a novel Myh6CreERT2;MADM-11GT/TG mouse, in which MADM recombination (under temporal control by administration of exogenous TM) can only occur in cells that express α-myosin heavy chain (α-MHC or Myh6), a sarcomeric thick filament component commonly used as a marker for differentiated cardiomyocytes (22). We first evaluated the promiscuity of this system by analyzing hearts of 1-mo-old mice without TM exposure and observed no labeling (Fig. S5). Therefore, we next administered a cumulative 2 mg TM dose to newborn pups in the first week of life, and analyzed the hearts at postnatal day (P) 12 (Fig. 1 B–E). We observed a labeling efficiency of 0.78%. Approximately 11% of labeled cells were single-labeled, which can be identified as the progeny of Myh6-expressing cardiomyocytes. All single-labeled cells (n = 500) were α-actinin–positive (Fig. 1 F and H) and sarcomeric elements were evident on longitudinal sections by confocal and epifluorescence imaging (Fig. 1 D, d′′′), which suggests that dividing cardiomyocytes only generate cardiomyocytes. We observed single-labeled cardiomyocytes in all chambers, including the atria, at a similar frequency (Fig. 1E).
Of note, the RFP+ and GFP+ single-labeled cells were equivalent in frequency (5.4%, n = 574 cells, P = 0.9894) (Fig. 1I). This parity suggests that the daughter cells are the result of symmetric divisions, as asymmetric proliferation of one daughter cell would lead to an increase in the frequency of that label (Fig. S2). Furthermore, these data also suggest that the two daughter cells of a parent cardiomyocyte have a similar low mitotic potential, which was strengthened by the absence of large GFP+ or RFP+ clones, with rare two-cell clones (Fig. 1I); daughter cardiomyocytes exhibit a very low proliferative capacity. With aging, there was a decline in the percentage of single-labeled cells, despite higher TM doses: when MADM recombination was induced at 1 mo of life, only 6% of all labeled cells were single-labeled, despite a higher labeling index of 2% (Fig. 1I) compared with 11% single-labeled cells observed during the first week of life. Collectively, these data suggest that the size of the mitotically active subset of cardiomyocytes diminishes during aging, especially after the first week.
Unexpectedly, we noticed that the sibling cells were primarily noncontiguous. This finding could be a result of several factors, including: (i) migration of daughter cells away from one another after division, (ii) dispersion of sibling cells among other dividing cell types, or (iii) apoptosis of one daughter cell. To resolve the differences among the different possible explanations, we induced sparse labeling to generate rare division events and analyzed serial sections; rare or sparse labeling allows precise identification of a clone even if its constituents are noncontiguous because of spatial separation and rarity of individual clones. Upon administration of a single dose of 1 mg TM at P7 and analysis at P9, we observed very sparse and spatially distinct single-labeled cells. We noticed a bimodal distribution of distance between alternate-label sibling cells: in a fraction of the pairs, one daughter cell made intimate contact with its sibling cell, but most of the pairs were within 800 µm of each other (Fig. 1J); clonal distinction of sibling pairs could be made unambiguously because they were separated by more than 3,000 µm from other sibling pairs. These results demonstrate the close proximity of single-labeled cardiomyocytes to their sister counterparts, suggestive of cell dispersion, as recently reported in zebrafish hearts (23). The normal distribution of distances between the two closest siblings makes it less likely that these results are a result of one daughter cell surviving and the other undergoing apoptosis. However, we cannot formally exclude this possibility because sparse MADM labeling and an unclear temporal window for potential apoptosis after division renders it technically difficult to stain for apoptotic markers.
We next sought to detect protein-level expression of the gap junction protein Connexin43 to determine whether newborn cardiomyocytes integrate into the existing myocardial network. We consistently observed connexin43 staining between the unlabeled and labeled cells, indicating that daughter cells can migrate (or be dispersed by migration or division of other cell types) yet still form electrochemical coupling with established cardiomyocytes (Fig. 1G). Such postnatal coupling has previously been shown to occur upon transplantation of fetal cardiomyocytes into a syngeneic adult mouse heart (24). Moreover, in the adult zebrafish heart 10% of daughter cells were found remote from their developmental clones in an elegant clonal analysis study, suggesting that dispersion and migration of cardiomyocytes is an evolutionarily conserved mechanism (23).
During mouse development, the predominant ventricular cardiomyocyte population expresses β-MHC and before birth there is an epigenetic switch to the α-MHC isoform, which has greater ATPase activity (25). Given the rare division of Myh6-cardiomyocytes after birth, we sought to determine whether Myh6-expressing cardiomyocytes undergo division during development as well. We induced recombination at E13.5, and observed rare labeling at birth in the pup hearts; this is largely because of the inefficient diffusion of TM across the placenta in addition to the inherently rare MADM recombination events. Although the labeling efficiency (4 × 10−5%) was much lower than during adulthood, reflecting the small number of Myh6-expressing cells present during development, all labeled cells were single-labeled cells and present in one- to four-cell clusters (Fig. S6). Because single-labeled cells are the result of bona fide cell division, whereas double-labeled cells can arise from random recombination in G0 phase, these data indicate that the majority of the small Myh6-expressing cardiomyocyte pool during development is mitotically active. The lack of large clones of single-labeled cells further confirms the limited proliferative capacity of Myh6-expressing cells. Moreover, after birth the majority of the labeled cells are double-labeled, confirming that the mitotically active fraction of Myh6-expressing cardiomyocytes is sharply reduced in the postnatal period. Intriguingly, this data also suggests that division of cardiomyocytes after birth is independent of the MHC isoform. It was previously shown that reexpression of the embryonic β-MHC isoform in adult cardiomyocytes after pressure-overload injury does not lead to division (25). The data above provide an additional line of evidence for this finding by demonstrating that rare α-MHC cardiomyocytes can divide in utero and after birth.
Limited Cardiomyocyte Division Occurs During Physiological Aging in a MADM Model That Allows Recombination in any Cell Type.
To determine if other cell types can also generate cardiomyocytes postnatally, we created the β-actinCreER;MADM-11GT/TG transgenic mouse (Fig. 2A); β-actin is a ubiquitous housekeeping gene and theoretically should permit MADM recombination in any cell (26). We administered TM to these mice in the first week of life, and the results were similar to the Myh6CreERT2;MADM model: 0.9% labeling efficiency, 17.5% single-labeled cardiomyocytes (Fig. 3A). We observed labeling within vasculature (Fig. 2D), confirming that noncardiomyocyte cell types can also undergo MADM labeling in the β-actin model. The pattern of clone sizes was also similar between the two models (e.g., sibling cells were frequently noncontiguous and the majority of single-labeled cells were without a clonal cluster, in accordance with a model of limited, symmetric division). When we induced recombination at P10 and analyzed after a 3-mo and 18-mo chase, we observed parity of GFP+ and RFP+ single-labeled cells within and between each of the groups, further attesting to the low proliferative capacity of cardiomyocytes born in adulthood (Fig. 3C).
Fig. 2.
Postnatal generation of cardiomyocytes in the actinCreER;MADM model. (A) MADM labeling can occur in any cell type to lead to labeled cardiomyocytes. (B and C) Single-labeled sibling cardiomyocytes exhibit intimate end-on contact. (Scale bars, 10 µm.) (D) Single-labeled RFP cells in a large vessel. [Scale bars, 70 µm (Insets, 5 µm)]. (E and F) Cardiomyocytes and smooth muscle cells do not share MADM labeling. (Scale bars, 20 µm.) (G) Sparse c-kit+ cells (white cells within inset box) did not demonstrate MADM labeling. [Scale bar, 30 µm (Inset, 7.5 µm)]. (H and I) Sections of 10-d-old (H) and 4.5-wk-old mice (I) analyzed after TM pulse reveal a temporal decrease in single-labeling frequency. [Scale bars, 100 µm (H) and 1 mm (I).] (n = 3,3 mice). (J) TM administration timeline for I–H.
Fig. 3.
Clonal analysis reveals limited proliferation underlying postnatal cardiogenesis. (A) (Upper) Frequency of single-labeled cells at different time points after recent TM administration (n = 3 per group). (Lower) Analysis of individual-colored clone size for each time point above. (B, (Left) Two GFP+ cell clones; (Right) two RFP+ cell clones with borders outlined by wheat germ agglutinin staining (orange color). (Scale bars, 5 µm.) (C) Frequency of single-labeled cells 3 and 18 mo after P10 TM pulse reveals no significant proliferation of daughter cells. (Lower) Analysis of clone size for each time point above. N = total number of single-labeled cells observed per group. (P values calculated with Student t test. Error bars indicate SEM).
Furthermore, we did not detect expression of MesP1, Isl1, or Tbx5 in single-labeled cells, which characterize multipotent cardiovascular progenitors that exist during development (27). We did not find c-kit expression in any of the single-labeled cells (Fig. 2G), consistent with several previous studies (5, 28). Additionally, we did not observe any multilineage clusters containing cardiomyocytes, vascular smooth muscle and endothelial cells that shared single- or double-labeling, further arguing against the existence of a multipotent progenitor cell in the adult heart akin to canonical stem cells in other tissues (Fig. 2 E and F). As such, our clonal data support the recent MIMS study in which only cardiomyocytes were shown to generate cardiomyocytes after birth (2).
Limited Cardiomyocyte Proliferation Occurs After Ischemic Cardiac Injury, with No Evidence for Contribution from Postnatal Cardiac Progenitors.
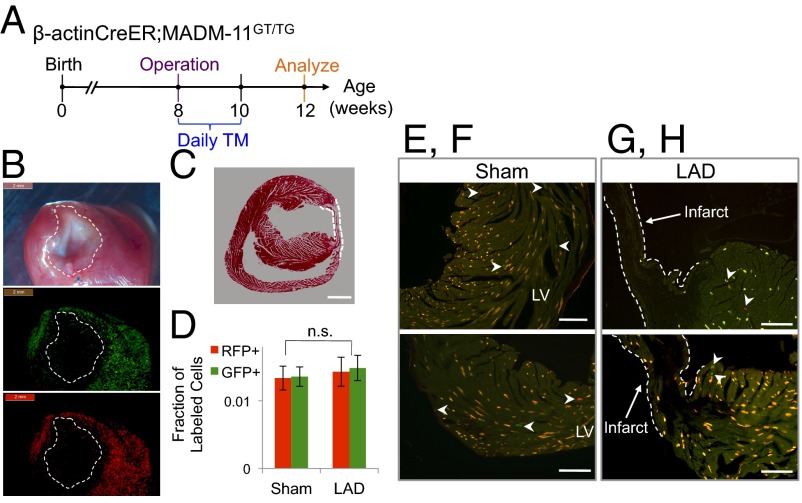
Recent reports using a dilution of indelible labeling strategy have provided indirect evidence for generation of new cardiomyocytes from a progenitor cell after an infarction injury (29). However, other studies have failed to find evidence of cardiomyocyte renewal from progenitor cells (2). We sought to find clonal evidence for progenitor cells leading to cardiomyocytes in the β-actin–CreER;MADM-11GT/TG mice that underwent left anterior descending artery (LAD) ligation (n = 5) or sham (n = 3) operation at 8 wk of age and then given TM for 2 wk starting at the time of the operation (Fig. 4A); no recombination occurred in the absence of TM administration. In the sham mice, we found sparse single-labeled cells (2.7% single-labeled cells within the 6.9% of all labeled cells), but did not see clones (Fig. 4 C–E). Contrary to expectation, the global frequency of single-labeled cells in the LAD group was equivalent to the sham group (2.9%, P = 0.6300) and no single-labeled cardiomyocyte clones were observed (Fig. 4 F and G), consistent with a pattern of limited, symmetric cardiomyocyte division that occurs during physiological aging in the sham model. On gross specimens there was sparse labeling in the infarct and peri-infarct region (Fig. 4B), with rare single-labeled cells but no clonal expansion; single-labeled cells were c-kit−. Per our results, a myocardial infarction (MI) does not stimulate cardiomyocyte division above the basal level, nor does it appear to activate any putative latent progenitor cells. Interestingly, we did not observe an increase in the pool of labeled fibroblasts, which may be a result of apoptosis of these cells before analysis 1 mo after the operation (30).
Fig. 4.
Limited birth of new cardiomyocytes after MI in adult mice. (A) Timeline for injury, TM delivery, and analysis of β-actinCreER;MADMGT/TG transgenic mouse model. (B) Gross images of the heart after LAD ligation reveals fibrosis (dashed circle) in the infarct region (Top, phase) and limited infarct and peri-infarct labeling (Middle and Bottom: GFP and RFP channel, respectively). (Scale bars, 2 mm.) (C) H&E section of a MI heart demonstrates an area of fibrosis (within dashed lines). (Scale bar, 150 µm.) (D) Frequency of single-labeled cells is similar between the sham (n = 3) and LAD mice (n = 5). (P values calculated with Student t test. Error bars indicate SEM) (E–H) There are few single-labeled cells (white arrowheads) in the left ventricle (LV) after an MI or sham operation. (Scale bars, 120 µm.)
Cardiomyocytes Are the Source of Cardiomyocytes Generated After Ischemic Cardiac Injury.
To confirm the above findings directly in cardiomyocytes, we similarly administered TM for 2 wk [starting several hours before sham (n = 3) or LAD ligation (n = 5) operation] to 2-mo-old Myh6CreERT2;MADM mice (Fig. 5A); there was no recombination postinjury in mice that were not administered TM. Both sham- and LAD-operated hearts exhibited a slightly decreased labeling index of 4.2–4.5% compared with the β-actin models (likely as a result fewer cells capable of undergoing MADM recombination), but the percentage of labeled cells that were single-labeled was 2.0–2.3%. In both sham and LAD mice, the numbers of RFP+ and GFP+ cells were in parity within each group as well as across both injury models (Fig. 5B). Moreover, we did not observe any clusters—of cardiomyocytes or of multiple cell types—in either operation arm. We also did not observe any robust labeling in the border zone compared with the rest of the heart in the LAD mice.
Fig. 5.
Limited division of cardiomyocytes after MI in adult mice. (A) Timeline for injury, TM delivery, and analysis of Myh6CreER; MADMGT/TG transgenic mouse model. (B) Rare single-labeled cells in the sham heart. (Scale bar, 100 µm.) (Inset) Magnified image of dashed box. (Scale bar, 10 µm). (C, Left) Analysis of clone size for each surgery group; (Right) frequency of single-labeled cells is similar between the sham (n = 3) and LAD mice (n = 5). (P values calculated with Student t test. Error bars indicate SEM) (D) There are few single-labeled cells (white arrowheads) in the left ventricle (LV) after an MI. Dashed lines depict the borders of the infarcted area. [Scale bar, 20 μm (Top), 100 μm (Middle), and 10 μm (Bottom).]
Discussion
In this study we use several transgenic mouse models that enable clonal analysis of postnatal cardiomyogenesis. Experiments with the MADM system demonstrate at the cellular level that cardiomyocytes generate cardiomyocytes after birth in mice, in the setting of aging and injury, albeit in a very limited way, given the rarity of such division events, as well as limited evidence that the generated daughter cells can further divide. Indeed, we show that most examples of postnatal cardiomyocyte division are single iteration events. We did not observe any clusters of newly formed cardiomyocytes (i.e., a group containing greater than four cells) in our analyses of postnatal development or after injury, whereas during embryonic development using the HprtCre model, we clearly observed clusters of more than 20 cells in the heart. Although it has been reported that Myh6 is expressed in certain postnatal cardiac stem cells (31), we do not find clonal evidence of asymmetric division or multipotentiality among Myh6-expressing cells that undergo division, despite using a model that is optimal for detecting asymmetric self-renewal. Rather, our observations argue against the existence of robust putative stem cells. The capacity to divide postnatally appears to be relegated to a small fraction of cardiomyocytes, and this property diminishes over time and appears insensitive to stimulation by an infarction injury within a 4-wk time period after the infarct.
The concordance between the two MADM models presented here further argues against the contribution to postnatal cardiogenesis by a stem or progenitor cell, although we cannot formally exclude their existence. It is possible that cardiac stem/progenitors were, by chance, excluded from a MADM recombination event and therefore no clonal expansion from a single stem/progenitor cell could be observed. However, the rarity of cardiac tumors in mammals, as well as our identification herein of cardiomyocytes as the parent cell of postnatal cardiomyocytes, suggests that precursor cells are, at best, latent. Our data corroborate the findings of the MIMS report at the single-cell level and further reveal that the progeny of postnatally dividing cardiomyocytes mostly do not divide again. Senyo et al. also show that in the 6 mo after an infarction there is increased cardiomyogenesis in the border zone (2), which we did not observe in the 4 wk period after injury: the several months longer chase period between injury and analysis in the former study may explain the difference between our findings. It is also possible that recombination inefficiency of the MADM system led to underestimation of cardiomyocyte generation at the infarct border zone.
It has recently been shown that deletion of a single gene, Meis1, can cause cardiomyocytes to divide after birth (32), which further suggests that limited postnatal cardiomyogenesis in mammals is the result of a Meis1-imposed limit on the division of cardiomyocytes, as well as near-absence of cardiac stem cells (as cardiomyocytes were the only source of new cardiomyocytes). It would be interesting to determine whether the rare number of dividing cardiomyocytes in the wild-type mouse heart have intrinsically low or absent levels of Meis1.
Our data provide direct evidence that cardiomyocytes can be born after birth at very low rates and that they appear to form electrochemical contacts with cardiomyocytes already present. Our clonal analysis lends cell-level insights into this controversial topic, and identifies the cell of origin of the postnatal cardiomyogenesis, as well as characterizes the behavior of the postnatally generated daughter cardiomyocytes; our work provides fundamental insight into the postnatal cell growth of the heart. The findings presented herein support the potential utility of cell transplantation therapy in the heart by demonstrating the existence of an innate mechanism that permits the electrochemical coupling of new cardiomyocytes into the existing syncytium (33).
Materials and Methods
See SI Materials and Methods for full materials and methods and Fig. S7 for the genetic breeding strategy to generate experimental MADM mice.
Mice.
Mice were bred and maintained at the Stanford University Research Animal Facility in accordance with Stanford University guidelines. All of the animals were housed in sterile microinsulators and given water and rodent chow ad libitum. Myh6CreERT2, MADM-GT/GT, and TG/TG strains were obtained from The Jackson Laboratory; the β-actinCreER mice were a gift from Corrinne G. Lobe (Sunnybrook and Women’s College Health Sciences Centre, Tornonto, ON, Canada).
Labeling and Clonal Analysis.
To determine percent labeling, the total number of cells in a section was determined using ImageJ software (National Institutes of Health); specifically, the total number of nuclei was counted in the DAPI channel and the total number of cells was calculated by adjusting for the age-specific fraction of polyploid cardiomyocytes based on previous reports. For clonal analysis, the frequency of single-labeled cells was determined as a percentage of all labeled cells: for each specimen, the total number of labeled cells (i.e., either single- or double-labeled) and the numbers of RFP+ and GFP+ single-labeled cells were manually identified in every fourth section (in 7-μm-thick serial sections). The fraction of RFP+ and GFP+ single-labeled cells for each section was calculated and averaged with all other sections in a single specimen. The mean and SEM for each group of mice was calculated using the average data from each specimen in the group. Cluster/clone size analysis was performed by manually identifying single-labeled cells and categorizing them by clone size (using wheat germ agglutinin staining to demarcate cell borders).
Supplementary Material
Acknowledgments
We thank T. Naik, T. Storm, and L. Jerabek for laboratory management; A. Mosley, C. Manalac, and A. McCarty for assistance with animal care; W. Jiang for assistance with confocal imaging; Y. Gong for performing the operations; and M. Krasnow and K. Red-Horse for critical discussions. This work was supported in part by Grant RC1-00354-1 from the California Institute of Regenerative Medicine (to R.A. and I.L.W.); American Heart Association Grant AHA-BGA 12BGIA8960008 (to R.A.); a Howard Hughes Medical Institute Medical Research Fellowship (to S.R.A.); the Stanford Medical Scholars Program (to S.R.A.); the Paul and Daisy Soros Fellowship (to S.R.A.); and an American Heart Association Student Scholarship in Cardiovascular Disease (to S.R.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408233111/-/DCSupplemental.
References
- 1.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol. 1997;272(1 Pt 2):H220–H226. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- 4.Beltrami AP, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344(23):1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 5.Jesty SA, et al. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci USA. 2012;109(33):13380–13385. doi: 10.1073/pnas.1208114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann O, et al. Cardiomyocyte renewal in humans. Circ Res. 2012;110(1):e17–e18. doi: 10.1161/CIRCRESAHA.111.259598. author reply e19–e21. [DOI] [PubMed] [Google Scholar]
- 10.Kajstura J, et al. Cardiomyogenesis in the aging and failing human heart. Circulation. 2012;126(15):1869–1881. doi: 10.1161/CIRCULATIONAHA.112.118380. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271(5 Pt 2):H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 13.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488(7412):527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meilhac SM, et al. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130(16):3877–3889. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 15.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121(3):479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Hippenmeyer S, et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68(4):695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83(1):15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 18.Weissman IL. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100(1):157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 19.Hippenmeyer S, Johnson RL, Luo L. Mosaic analysis with double markers reveals cell-type-specific paternal growth dominance. Cell Reports. 2013;3(3):960–967. doi: 10.1016/j.celrep.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasic B, et al. Extensions of MADM (mosaic analysis with double markers) in mice. PLoS ONE. 2012;7(3):e33332. doi: 10.1371/journal.pone.0033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89(1):20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484(7395):479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soonpaa MH, Koh GY, Klug MG, Field LJ. Formation of nascent intercalated disks between grafted fetal cardiomyocytes and host myocardium. Science. 1994;264(5155):98–101. doi: 10.1126/science.8140423. [DOI] [PubMed] [Google Scholar]
- 25.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32(1):8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126(6):1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121(18):1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8(4):389–398. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virag JI, Murry CE. Myofibroblast and endothelial cell proliferation during murine myocardial infarct repair. Am J Pathol. 2003;163(6):2433–2440. doi: 10.1016/S0002-9440(10)63598-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey B, et al. Cardiac stem cell genetic engineering using the alphaMHC promoter. Regen Med. 2009;4(6):823–833. doi: 10.2217/rme.09.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497(7448):249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardehali R, et al. Prospective isolation of human embryonic stem cell-derived cardiovascular progenitors that integrate into human fetal heart tissue. Proc Natl Acad Sci USA. 2013;110(9):3405–3410. doi: 10.1073/pnas.1220832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.