Significance
We report the first to our knowledge genetically engineered honeybees, which are important pollinators and interesting biological models for the study of social and complex behaviors as well as caste and sexual development. This genetic manipulation tool will enable systematic studies of biological processes in an organism building complex societies. We demonstrate highly efficient integration and expression of piggyBac-derived cassettes in the honeybee that make this system applicable to colony-based screening approaches and useful for an average beekeeping facility. This cassette was stably and efficiently transmitted and expressed in progeny by two different promoters, offering the prospect for activation or inhibition of gene functions under conditions of stage- and tissue-specific promoters.
Keywords: transposon, transformation, social insect transgene
Abstract
Honeybees (Apis mellifera), which are important pollinators of plants, display remarkable individual behaviors that collectively contribute to the organization of a complex society. Advances in dissecting the complex processes of honeybee behavior have been limited in the recent past due to a lack of genetic manipulation tools. These tools are difficult to apply in honeybees because the unit of reproduction is the colony, and many interesting phenotypes are developmentally specified at later stages. Here, we report highly efficient integration and expression of piggyBac-derived cassettes in the honeybee. We demonstrate that 27 and 20% of queens stably transmitted two different expression cassettes to their offspring, which is a 6- to 30-fold increase in efficiency compared with those generally reported in other insect species. This high efficiency implies that an average beekeeping facility with a limited number of colonies can apply this tool. We demonstrated that the cassette stably and efficiently expressed marker genes in progeny under either an artificial or an endogenous promoter. This evidence of efficient expression encourages the use of this system to inhibit gene functions through RNAi in specific tissues and developmental stages by using various promoters. We also showed that the transgenic marker could be used to select transgenic offspring to be employed to facilitate the building of transgenic colonies via the haploid males. We present here the first to our knowledge genetic engineering tool that will efficiently allow for the systematic detection and better understanding of processes underlying the biology of honeybees.
The honeybee Apis mellifera is an important pollinator of wildflowers and crop plants with great relevance for the global ecosystem. Substantial losses in colonies have been reported in recent years and have been associated with colony collapse disorder, a scenario in which worker bees abruptly disappear from their colony, and with RNA virus infections transmitted via the ectoparasitic mite Varroa destructor (1–5).
Honeybees live in complex societies and display interesting behaviors and developmental processes. The members of a honeybee colony cooperate and produce group phenotypes that allow them to effectively respond to environmental perturbations (6, 7). For instance, honeybees can collectively regulate the temperature of their nest, cooperatively defend against diseases and predators, and exploit food sources efficiently via complex communication systems (8–11).
Research on honeybees has contributed to our understanding of social organization, behavior, physiology, development, and genetics. Important discoveries include the communication of food source locations via waggle dances (8, 12, 13), the identification of neural correlates of cognitive faculties (14), the task specialization of colony members on subsets of tasks performed by the colony (7, 12, 15), the complementary sex determination via heterozygosity at a single gene (16, 17), the caste (queen versus worker) differentiation through differential food and the royalactin protein (18), and the releaser function of pheromones emitted by the queen, which affects social behaviors (19).
Understanding the interesting features of the honeybee has been limited due to a lack of genetic tools to manipulate gene functions. Conditional expression or inhibition of gene functions using different promoters has allowed for the study of the underlying processes in other organisms. Such expression systems have allowed the systematic dissection of the role of gene functions at later developmental stages, at which point many interesting honeybee phenotypes manifest.
Expression cassettes have been introduced into different insect genomes by using transposable elements. The frequencies at which the insects were genetically transformed usually range from below 1 to 5% (20–26). This efficiency would require the screening of at least 100 honeybee colonies to obtain a few transformed queens, making such a system unreasonable. Moreover, the rearing of treated embryos into queens relies on the social environment of a colony, making the development of such procedures difficult.
Honeybee colonies typically consist of thousands of worker bees, a single queen, and hundreds of males (drones). The queen produces all of the eggs. The unfertilized eggs, hemizygous at the complementary sex determiner (csd) gene, differentiate into males. The fertilized eggs that have a heterozygous csd genotype (two different sex-determining alleles) develop into females (17), either a queen or worker depending on the various food provided by the worker bees, such as the royal jelly (18, 27). The worker bees process nectar and pollen collected from plants and rear offspring through repeated feeding.
In this study, we report the highly efficient integration and expression of piggyBac-derived cassettes, which offer the ability to manipulate gene functions throughout development in an average bee facility.
Results
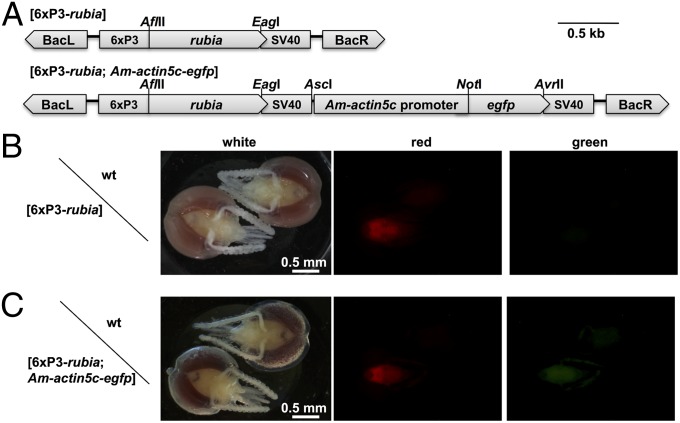
We efficiently introduced two expression cassettes into the honeybee queen’s genome (Fig. 1) using a piggyBac-derived transposon (SI Appendix, Fig. S1; 29, 30). One expression cassette, [6xP3-rubia], contained the transgenic marker rubia under control of the artificial 6xP3 promoter (31). The other, [6xP3-rubia; Am-actin5c-egfp], had an additional expression cassette, the egfp reporter gene coupled with the honeybee promoter Am-actin5c (32; Fig. 1A). We obtained 4 out of 15 (27%) queens from which the offspring possessed the [6xP3-rubia] expression cassette and 2 out of 10 queens (20%) with offspring with the double-expression cassette [6xP3-rubia; Am-actin5c-egfp] (Table 1). The male offspring that possessed the [6xP3-rubia] cassette, as revealed by site-specific DNA amplification (SI Appendix, Fig. S2), showed the red fluorescence signal of the transgenic marker rubia in the transparent head of the pupae in all cases (n = 56; Fig. 1B). The male offspring with the double-expression cassette [6xP3-rubia; Am-actin5c-egfp] exhibited red and green fluorescence signals from the transgenic markers rubia and egfp in the head of pupae (Fig. 1C). We detected no distinct fluorescence signal from background in the area of the compound eyes, possibly due to the fluorescence quenching of the eye pigments (33). These results suggest that we have efficiently introduced and expressed two genes driven by the artificial promoter 6xP3 and the endogenous promoter Am-actin5C in the honeybee genome with a transformation rate equal to or higher than 20%.
Fig. 1.
Genetically transformed honeybees. (A) Structure of the expression cassettes [6xP3-rubia] and [6xP3-rubia; Am-actin5c-egfp] with the piggyBac transposon elements. The boxes indicate the structural elements. BacR, BacL: inverted terminal repeats of the piggyBac transposon, including parts of the transposase coding sequence; 6xP3: six repeating Pax6 response elements upstream of the core promoter of the Tribolium castaneum hsp68 gene (28); rubia and egfp: reporter genes encoding red or green fluorescent proteins, respectively; SV40: the SV40 polyadenylation site; and Am-actin5c promoter: a 1420-bp sequence upstream of the translation start site of the honeybee actin5c gene. The letters above the figure denote the restriction sites that were used for cloning. The scale indicates the size in kilobases (kb). (B) The fluorescent signal of the transgenic marker rubia in the head of pupal drone offspring. A drone with an integrated [6xP3-rubia] expression cassette and a wild-type drone (WT) are shown. (C) The fluorescent signal of the transgenic marker egfp under the control of the Am-actin5c promoter in the pupal drone head. A drone with a [6xP3-rubia; Am-actin5c-egfp] cassette and a WT drone are shown. The heads in B and C are shown under white light and red and green fluorescence detection conditions, as indicated. The drone heads were fixed with paraformaldehyde, cleared with methyl salicylate, which improved the fluorescence detection, and observed using a stereomicroscope.
Table 1.
Rearing of [6xP3-rubia] and [6xP3-rubia; Am-actin5c-egfp] honeybee queens
| Construct | Number of injected embryos | Number of L1 larvae transferred into queen cups | Number of completed queen cells | Number of emerged queens | Number of queens with offspring | Number of queens with [6xP3-rubia] or [6xP3-rubia; Am-actin5c-egfp] offspring* | Transformation rate (%)† |
| [6xP3-rubia] | 440 | 196 | 88 | 58 | 15 | 4 | 27 |
| [6xP3-rubia; Am-actin5c-egfp] | 533 | 190 | 37 | 32 | 10 | 2 | 20 |
Identified using PCR amplifications on genomic DNA and red fluorescence signal of the transgenic marker.
Transformation rate denotes the relative proportion of queens which are inheriting a [6xP3-rubia] or [6xP3-rubia; Am-actin5c-egfp] cassette to the queens with offspring.
To produce these rates, we developed an efficient transformation and rearing method for honeybees. We started the procedure by collecting hundreds of female embryos (0–1.5 h after egg deposition). Queens were confined on a plastic comb in which they lay eggs onto plugs at the bottom of the cells (Jenter Queen Rearing Kit). We removed the plugs with the embryo attached and microinjected the embryos (34) with 30 pg of the pBac plasmid DNA (SI Appendix, Fig. S1) and 60 pg of in vitro synthesized transposase mRNA. The direct transfer of the treated embryos into queen-rearing colonies (colonies which are queenless and which will typically rear new queens from young larvae) was not efficient because all introduced embryos were removed by worker bees within the first 24 h. Hence, we incubated embryos at 34 °C in small plastic boxes until they hatched after 3 d. A mixture of 16% sulfuric acid in water was added to these boxes to prevent mold formation and desiccation of embryos. The acid was replaced with water 3 h before the larvae were expected to hatch.
We selected well-formed larvae and grafted them into queen cell cups that were primed with royal jelly. We transferred the cell cups into queen-rearing colonies. With this hatching step in the laboratory, on average, 25% of introduced larvae were accepted by workers in colonies and reared into queens (Table 1). After 10 d, the capped queen cells were removed from the queen-rearing colony and incubated at 34 °C until the queens emerged. Queens emerged from ∼60% ([6xP3-rubia]) or 87% ([6xP3-rubia; Am-actin5c-egfp]) of capped cells (Table 1). Queens were introduced into small, queenless, mini nucleus hives. The nucleus hives were transferred into a closed large flight cage from which no bee can escape into nature. Queens were thereafter repeatedly treated with CO2, which stimulated the laying of eggs. These virgin queens lay exclusively unfertilized eggs, which develop into haploid drones.
The offspring of those transformed queens efficiently expressed the transgenic markers from the 6xP3 promoter (31) and the endogenous promoter Am-actin5c (32). We analyzed at least 34 offspring of each [6xP3-rubia] queen and found that all of the offspring that possessed the transgene were also expressing the transgenic marker (43, 6, 30, and 9% progeny of the different queens). In these drones, we detected the red fluorescence signal of the transgenic marker and amplified the expression cassette from DNA via PCR (Table 2). For queens with the double-expression cassette ([6xP3-rubia; Am-actin5c-egfp]), we studied at least 27 offspring and observed that all progeny possessing the cassette (4 and 6% of progeny) were expressing both transgenic markers (Table 2). Consistently, drones, which did not display the fluorescence signals of the transgenic marker, also possessed no expression cassette, as revealed by PCR amplifications.
Table 2.
Numbers and proportions of offspring that expressed and/or possessed the expression cassettes [6xP3-rubia] or [6xP3-rubia; Am-actin5c-egfp]
| Construct | Queen | Drone offspring |
||
| Drone offspring Number | Exhibiting a red or green fluorescent signal |
|||
| Number (%)* | Possessing [6xP3-rubia] or [6xP3-rubia; Am-actin5c-egfp] cassette (%)† | |||
| [6xP3-rubia] | 11–59 | 93 | 40 (43) | 100 |
| 12–05 | 35 | 2 (6) | 100 | |
| 12–07 | 37 | 11 (30) | 100 | |
| 12–31 | 34 | 3 (9) | 100 | |
| [6xP3-rubia; Am-actin5c-egfp] | actin15 | 27 | 1 (4) | 100 |
| actin22 | 35 | 2 (6) | 100 | |
Denotes the proportion among the drones tested.
Determined by PCR amplifications of expression cassettes on genomic DNA.
We next showed that we can identify living [6xP3-rubia] drones in the comb. We unsealed the brood comb and examined the fluorescence signals of living drone pupae. Drones exhibiting a clearly visible red fluorescence signal possessed the [6xP3-rubia] cassette as revealed later by PCR amplifications, whereas the drones without this signal did not (SI Appendix, Fig. S3; we studied more than 56 drones).
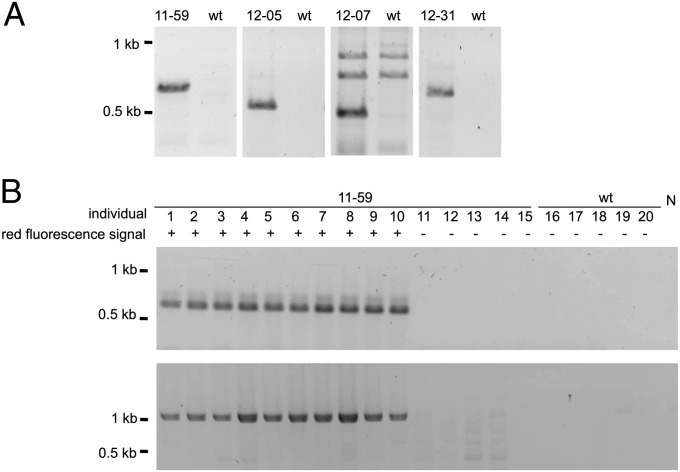
The expression cassette was stably integrated into different genomic loci in the different queens. We used the drone offspring, which derived from haploid, unfertilized eggs, to determine the genomic integration sites of the transgene, which were inherited from the queen. We pooled the DNA of those drones and characterized the integration sites using inverse PCR technique (35). The pooling approach seeks to identify possible distinct integration sites that were inherited from a single queen. We only found a single integration site for the drones of each queen, which we verified by amplifying and sequencing those specific loci (amplicons spanning genomic and transposon sequences; Fig. 2A). DNA from drones derived from other queens did not produce such amplicons. Table 3 lists the integration sites of the expression cassette with its genomic location and linkage group. The cassettes were integrated at different chromosomes and positions on the chromosomes for each population of offspring, which derived from a single queen. Comparison with the reference honeybee genome (honeybee genome assembly version 4.5) showed that the piggyBac-mediated transposition was always accompanied by the duplication of the target sequence TTAA, a finding consistent with integrations of piggyBac-derived vectors in other insect genomes (23). We also studied the location of genomic integration sites in 93 offspring of a single queen. The 40 offspring that showed red fluorescence signals had the [6xP3-rubia] cassette integrated at the same genomic locus and at no other loci, as shown by PCRs amplifying specifically the integration locus or the expression cassette (Fig. 2B). The other 53 offspring of the same queen did not possess the [6xP3-rubia] cassette. The finding of the same genomic integration site in all drones of one queen suggests that the cassette is stably inherited in the honeybee genome.
Fig. 2.
Stable genomic integration and transmission of the [6xP3-rubia] expression cassette. (A) The different genomic integration sites of the [6xP3-rubia] expression cassette in the four queens as revealed by genomic loci-specific PCRs. The different genomic integration sites were amplified from DNA using a genome- and a cassette-specific oligonucleotide primer in the PCRs. The fragments were separated by agarose gel electrophoresis. The specific oligonucleotide primer pairs used were as follows for the four queens: queen 11–59: #178/#090 (expected size: 586 bp); queen 12–05: #206/#090 (expected size: 428 bp); queen 12–07: #201/#090 (expected size: 529 bp); and queen 12–31: #204/#090 (expected size: 453 bp). The DNA template, representing the queen’s genotype, was obtained by pooling the haploid drone offspring. wt denotes a pool of wild-type honeybee drones. (B) Example of stable integration and transmission for the [6xP3-rubia] cassette in single offspring of queen 11–59. We amplified the sequence from DNA of single offspring in two separate PCRs; one amplifying specifically the integration site (Upper gel image) and one amplifying only the expression cassette alone (Lower gel image). Fragments were separated by agarose gel electrophoresis. Offspring of queen 11–59 that exhibited a red fluorescent signal of the transgenic marker (as indicated by +) had the expression cassette integrated at the same predicted genomic integration site. Offspring that exhibited no red fluorescent signal of the transgenic marker (as indicated by –) possessed no [6xP3-rubia] expression cassette. N denotes the control reaction with no DNA. The gels were stained with ethidium bromide. The identities of the fragments were verified by sequencing.
Table 3.
Genomic integration sites of the [6xP3-rubia] expression cassette
| Queen | Genomic sequence of integration site* |
Linkage group | Position on the chromosome (bp) | ||
| Flanking BacR | Flanking BacL | ||||
| 11–59 | TTCGGTTTGCTTTTT | TTAA | AGGATATGGTTGTAA | LG16 | 4474605 |
| 12–05 | TTTACATAAAATTTA | TTAA | AAATTATATTAAATA | LG10 | 7083925 |
| 12–07 | CCGTCCGTTAATTAA | TTAA | TTCCACGATGAAAGA | LG1 | 21160164 |
| 12–31 | TTATTTTTAGAGTAT | TTAA | AGTTAGGATGATTTT | LG6 | 11531634 |
BLASTn searches against the reference honeybee genome (genome assembly version 4.5).
Discussion
Our results demonstrated an exceptionally high efficiency of integration and expression of piggyBac-derived cassettes in the honeybee. For the [6xP3-rubia] cassette, the transgenic marker was expressed in the offspring of 27% of queens, and the marker for the double cassette, [6xP3-rubia; Am-actin5c-egfp], was expressed in the offspring of 20% of those queens. We showed that the cassette is stably and efficiently transmitted and expressed in the next generation.
This is the first to our knowledge report of directed DNA integrations into the honeybee genome. Integrations into the honeybee genome were mediated through capped and polyadenylated mRNA, encoding transposase proteins. We developed efficient laboratory and colony-rearing methods to produce fertile queens from the DNA/mRNA-injected embryos. For example, by injecting 440 embryos and screening 15 colonies, we were able to produce four [6xP3-rubia] queens (Table 1). A single person in our laboratory can routinely inject 200–300 embryos per day, whereas keeping and maintaining 15 small colonies (nucs) requires only moderate bee facility resources. The demonstration of the efficient integration of active expression cassettes by using modest bee resources encourages the wide use of this system to manipulate gene functions.
We demonstrated that we can identify the [6xP3-rubia] drones derived from living mosaic queens via a transgenic marker in the first generation. We treated the virgin queens with CO2 to obtain solely haploid males from the unfertilized eggs. Because the males are haploid, this method will enable the collection of hemizygous sperm from single drones in which all of the spermatozoa possess the transgene. The collected sperm can be used to instrumentally inseminate queens, which is a routine procedure used in honeybee breeding. With such queens we can build colonies in which the entire worker bee force expresses the cassette, enabling the identification of gene functions in honeybee societies (Fig. 3). The collected sperm can also be used to maintain genetic lines through sperm freezing (36).
Fig. 3.
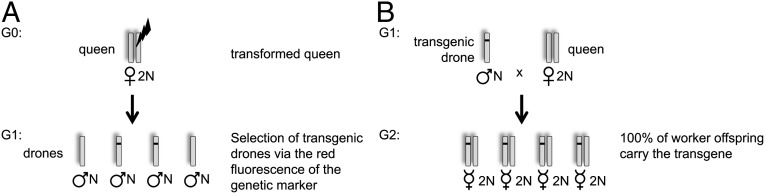
Generation of transgenic colonies using haploid drones. (A) Generation 1 (G1) drones with the expression cassette are selected via the fluorescent signal of the transgenic marker at pupal stage. The drones are haploid (denoted by N); hence, each sperm cell will possess the expression cassette. (B) The semen of the selected drone is used to instrumentally inseminate a wild-type diploid (2N) queen. All of the diploid progeny (workers and queens) of this cross will possess the expression cassette.
Efficient integration of expression cassettes enables systematic studies of gene functions and processes that are underlying features or causes of the interesting biological traits in honeybees. Utilization of different promoters enables the transcription of tissue- and stage-specific hairpin RNA from the cassette, directing conditional inhibition of gene functions by RNAi. Additionally, we can direct the transcription of mRNA and translation of the ORF, resulting in the conditional activation of gene functions.
Additional work is needed to determine the conditions under which different promoters are activated. A set of promoter sequences of interest in honeybees that can be used to conditionally manipulate gene functions has been tested in insect cells, including the Am-actin5c promoter (also used in this study), the heat shock inducible promoters Am-hsp83 and Am-hsp70, and the brain-specific promoter elp2l (32). Conditional inhibition of gene function is of great importance in honeybees because many genes have multiple functions throughout development, and many important traits manifest relatively late in development in the pupa or in the adult stage. For example, genes that are essential in early honeybee development, and which are functionally affected by permanent DNA lesions, will preclude the understanding of their function in the pupa or at adult stages.
Permanent lesions at specific target sites of genes have been induced in recent years through the use of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9)-associated systems (reviewed in ref. 37). Molecular components of these systems, which mediate small insertions or deletions at specific target sites in the genome, are typically injected into the embryos of the relevant organism. The highly efficient rearing and mRNA injection methods described in this study also encourage the use of these genome-editing systems in honeybees.
We suggest that the richness of A and T nucleotides in the genome, the weakly activated transposon defense system in the embryos, and the provision of transposase activity through capped and polyadenylated mRNA have together allowed us to achieve these high integration rates in honeybees. The integration rates of transposons in other insect species typically range from less than 1 to 5% (the number of fertile generation 0 individuals producing transgenic offspring), suggesting that we have increased the rate in honeybees by 6- to 30-fold (20–26). Previous studies have shown that by providing the transposase activity by injecting mRNA instead of a helper plasmid can increase the integration rate of the Minos transposon for the medfly (Ceratitis capitata) by 6-fold and for the fruit fly (Drosophila melanogaster) by 11-fold (38). The honeybee genome is more rich in the nucleotides A and T than other sequenced insect genomes (67% A+T in honeybee, compared with 58% in D. melanogaster and 56% in Anopheles gambiae) (39). Hence, the honeybee genome has a relatively high abundance of TTAA sequences, the target site of the piggyBac transposon, which will facilitate genomic integrations. Finally, the honeybee genome is low in abundance of transposon sequences compared with other sequenced genomes (39), suggesting that there is no requirement of a highly activated endogenous defense system that silences the activity of transposons in the embryos.
The efficient integration and expression of piggyBac-derived cassettes in the honeybee genome encourage the use of this system for the conditional manipulation of gene functions. The system offers the prospect of identifying underlying processes involved in the fascinating features of honeybees, including those required for social organization.
Materials and Methods
Microinjection and Rearing.
Honeybee embryos were collected using the Jenter egg collector system (Jenter Queen Rearing Kit) and were microinjected 0–1.5 h after egg deposition (34, 40). Honeybee queens were confined to a plastic comb box that contained removable cell plugs at the bottom of the worker cells. The queen usually lays single fertilized eggs at the bottom of each cell. We removed the single cell plugs with the attached eggs and affixed the plugs on Petri dishes in rows using plasticine; this setup allowed us to microinject hundreds of embryos. We used a microinjection device (PLI-100, Medical Systems Corporation) and an Oxford micromanipulator (Singer Instruments Co.) to inject the embryos under a stereomicroscope. We injected 30 pg of pBac[3xP3-rubia] or pBac[3xP3-rubia; Am-actin5c-egfp] plasmid DNA and 60 pg of the transposase mRNA into each honeybee embryo using 53-mm injection pipettes, which were made from borosilicate capillary tubes (Hilgenberg). The tips of the pipettes were rigid and beveled at a 37° angle. The inner diameter of the pipette tip was 5 µm. The injection time was 120 ms, the injection pressure was 60 kPa, and the balance pressure was 5 kPa. Using these settings, we injected an average volume of ∼400 pL into each embryo (40). Improved transformation rates were obtained when the embryos were injected in the dorsal posterior region. The embryos were incubated in plastic boxes at 34 °C with 0.5 mL of 16% (vol/vol) sulfuric acid to prevent mold formation. We replaced the acid with water 4 h before the larvae hatched. The hatched larvae (∼72 h after egg deposition) were grafted into queen cell cups that were primed with royal jelly. The royal jelly-primed cups were created by transferring young wild-type larvae into the queen cups and by transferring these cups into a queenless colony on the day before the larvae hatched. We replaced these wild-type larvae with our manipulated larvae and transferred the manipulated larvae to the queenless colony. After 10 d, the completed queen cells were removed from the colony and incubated at 34 °C in queen-banking cages. The emerged queens were supplied with young worker bees, and the queens were then placed into small queenless mini mating nucleus hives (Kirchhain nucs; Holtermann). We used different containment procedures so that no transformed bee could escape into nature. This includes a large closed flight cage in which the nucleus hives with the transformed queens were kept. When the queens were 8 d old, they were treated with CO2 for 7 min on two successive days. This treatment induced the laying of unfertilized eggs, which developed into drones (41).
Bee Sources.
The bees were feral colonies of the A. mellifera carnica strain.
DNA Preparation, PCR, Nucleotide Sequence Analysis, and mRNA Synthesis.
Genomic DNA were isolated by phenol/chloroform extraction (42) from honeybee larvae in the 2–5 instar stages. The plasmids were prepared for microinjection using the Plasmid Midi Kit (Qiagen). The isolated genomic or plasmid DNA were diluted in double distilled H2O. The restriction enzymes and DNA modifying enzymes were obtained from Thermo Scientific. PCR reactions were performed using standard conditions (43). The sequences of the synthesized oligonucleotide primers (MWG Eurofins) are listed in SI Appendix, Table S1. The fragments used for cloning were amplified using Phusion High-Fidelity polymerase (Finnzymes) and were inserted into the pGEM-T T-overhang vector (Promega). Nucleotide sequences were analyzed using Sanger sequencing (MWG Eurofins), and BLASTn searches of those sequences were performed against the honeybee genome (genome assembly version 4.5; http://blast.ncbi.nlm.nih.gov/).
We created four adapter-ligated libraries of genomic DNA derived from pools of drones (queen’s offspring) to identify the genomic integration sites of the transgenes using inverse PCR technique (35). We used the restriction enzymes AluI, DpnI, DraI, or SspI to restrict the genomic DNA. Adapters were ligated to the restricted DNA fragments (35). To produce the adapters, 10 µM of each oligonucleotide (sequence described in ref. 35) were heated at 98 °C for 5 min and subsequently cooled to room temperature. Nested PCRs were performed using oligonucleotide primers that bind to the adapter and the transposon sequence; in the outer PCR, we amplified the 5′ integration site using the oligonucleotide primers #96 and #105 and the 3′ integration site using the oligonucleotide primers #101 and #105. The resulting PCR products were diluted 1:50 and were used as the template for the inner PCRs. The 5′ integration site was amplified using the oligonucleotide primer pair #102/#103, and the 3′ integration site was amplified using the primer pair #104/#102. The final verification of the genomic integration requires a second PCR, which uses primers matching the sequence of the specific genomic locus and the transgene.
Transposase mRNA were synthesized following the instructions of the mMESSAGE mMACHINE Kit (Ambion). We cloned the coding sequence of the transposase gene from the phspBac plasmid, which was kindly provided by Gregor Bucher [Georg August University, Göttingen, Germany (31)], into the pGEM-T plasmid (Promega, Mannheim, Germany) downstream of the T7 promoter site. We linearized the plasmid using the SalI restriction enzyme and synthesized the transposase mRNA via the T7 promoter using the mMESSAGE mMACHINE Kit, which also introduces a 5′ capping structure. We polyadenylated the mRNA using the Poly(A) Tailing Kit and purified them with the MEGAclear Kit (both from Ambion).
Cloning of the Bac[6xP3-rubia] and Bac[6xP3-rubia; Am-actin5c-egfp] Genes.
We replaced the reporter gene DsRed with the rubia gene (nucleotide sequence reported in SI Appendix, Fig. S4) using the AflII/EagI restriction sites in the pBac [6xP3 Tc-hsp core DsRed Express SV40] plasmid, which was kindly provided by Gregor Bucher (29). The resulting plasmid was named pBac[6xP3-rubia] (SI Appendix, Fig. S1). The artificial 3xP3 promoter, which contains Pax6-responsive enhancer elements (P3) in combination with a core promoter sequence, can drive the expression of fluorescent marker proteins in the compound eyes or in parts of the central nervous system in a variety of insects (31, 44, 45). We used the Rubia protein, which is a variant of the monomeric Mars-like red fluorescent protein that differs at four amino acid residues (46). Rubia is thought to be more resistant to photobleaching and to provide a more intense fluorescence signal. To construct the pBac[6xP3-rubia; Am-actin5c-egfp] plasmid, we inserted a multiple cloning site via the EagI/MssI restriction sites downstream of the rubia gene in the pBac[6xP3-rubia] plasmid. We inserted an Am-actin5c promoter sequence (32) into the multiple cloning site via the AscI/NotI restriction sites and inserted the egfp coding sequence via the NotI/AvrII restriction sites (Fig. 1A). We inserted the SV40 polyA site downstream of the rubia coding sequence using the EagI/AscI restriction sites (Fig. 1A). We introduced the new restriction sites into the fragment via oligonucleotide primers that were used in the PCRs.
Microscopic Analysis.
We used the DsRed filter set [excitation filter: 546/10 nm; barrier filter: 565 nm long pass (LP)] and the GFP2 filter set (excitation filter: 480/40 nm; barrier filter: 510 nm LP) on a Leica MZ FLIII microscope to detect the red and green fluorescence signals, respectively, that were derived from the Rubia and EGFP proteins. For improved detection of the fluorescence signal, we cleared the head tissue. We fixed the heads overnight in 4% paraformaldehyde in PBS buffer, followed by three 10-min washes in PBS buffer. We dehydrated the heads in an ethanol series of 30, 50, 70, 90, 96, and 100% ethanol. The heads were cleared in 100% methyl salicylate for 2 d and examined in a 100% methyl salicylate solution.
Supplementary Material
Acknowledgments
We thank Dr. Gregor Bucher (Georg August University, Göttingen) for providing the piggyBac and helper plasmids. We would also like to thank Nina Rossié and Jenni Matheis for their technical support. We thank Michael Griese for beekeeping support. This work was supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8708.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402341111/-/DCSupplemental.
References
- 1.Bowen-Walker PL, Martin SJ, Gunn A. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite varroa jacobsoni Oud. J Invertebr Pathol. 1999;73(1):101–106. doi: 10.1006/jipa.1998.4807. [DOI] [PubMed] [Google Scholar]
- 2.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Cox-Foster D. Effects of parasitization by Varroa destructor on survivorship and physiological traits of Apis mellifera in correlation with viral incidence and microbial challenge. Parasitology. 2007;134(Pt 3):405–412. doi: 10.1017/S0031182006000710. [DOI] [PubMed] [Google Scholar]
- 4.Genersch E, et al. The German bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Apidologie. 2010;41(3):332–352. [Google Scholar]
- 5.Guzman-Novoa E, et al. Varroa destructor is the main culprit for the death and reduced populations of overwintered honey bee (Apis mellifera) colonies in Ontario, Canada. Apidologie. 2010;41(4):443–450. [Google Scholar]
- 6.Page RE, Jr, Erber J. Levels of behavioral organization and the evolution of division of labor. Naturwissenschaften. 2002;89(3):91–106. doi: 10.1007/s00114-002-0299-x. [DOI] [PubMed] [Google Scholar]
- 7.Robinson GE. Regulation of division of labor in insect societies. Annu Rev Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. [DOI] [PubMed] [Google Scholar]
- 8.von Frisch K. 1967. The Dance Language and Orientation Of Bees, trans Chadwick LE (Harvard Univ Press, Cambridge, MA) [Google Scholar]
- 9.Wilson EO. The Insect Societies. Cambridge, MA: Belknap Press of Harvard Univ Press; 1971. [Google Scholar]
- 10.Michener CD. The Social Behavior of the Bees. Cambridge, MA: Harvard Univ Press; 1974. [Google Scholar]
- 11.Seeley TD. The Wisdom of the Hive. Cambridge, MA: Harvard Univ Press; 1995. [Google Scholar]
- 12.Robinson GE. Genomics and integrative analyses of division of labor in honeybee colonies. Am Nat. 2002;160(Suppl 6):S160–S172. doi: 10.1086/342901. [DOI] [PubMed] [Google Scholar]
- 13.Liang ZS, et al. Molecular determinants of scouting behavior in honey bees. Science. 2012;335(6073):1225–1228. doi: 10.1126/science.1213962. [DOI] [PubMed] [Google Scholar]
- 14.Menzel R. The honeybee as a model for understanding the basis of cognition. Nat Rev Neurosci. 2012;13(11):758–768. doi: 10.1038/nrn3357. [DOI] [PubMed] [Google Scholar]
- 15.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322(5903):896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dzierzon J. 1845. Gutachten über die von Herrn Direktor Stöhr im ersten und zweiten Kapitel des General-Gutachtens aufgestellten Fragen. Eichstädter Bienenzeitung 1:109–113, 119–121.
- 17.Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114(4):419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 18.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473(7348):478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 19.Slessor KN, Winston ML, Le Conte Y. Pheromone communication in the honeybee (Apis mellifera L.) J Chem Ecol. 2005;31(11):2731–2745. doi: 10.1007/s10886-005-7623-9. [DOI] [PubMed] [Google Scholar]
- 20.Lozovskaya ER, Nurminsky DI, Hartl DL, Sullivan DT. Germline transformation of Drosophila virilis mediated by the transposable element hobo. Genetics. 1996;142(1):173–177. doi: 10.1093/genetics/142.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catteruccia F, et al. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405(6789):959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 22.Tamura T, et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18(1):81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- 23.Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem Mol Biol. 2002;32(10):1211–1220. doi: 10.1016/s0965-1748(02)00084-x. [DOI] [PubMed] [Google Scholar]
- 24.Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito Anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. J Biol Chem. 2002;277(11):8759–8762. doi: 10.1074/jbc.C100766200. [DOI] [PubMed] [Google Scholar]
- 25.Uhlírová M, Asahina M, Riddiford LM, Jindra M. Heat-inducible transgenic expression in the silkmoth Bombyx mori. Dev Genes Evol. 2002;212(3):145–151. doi: 10.1007/s00427-002-0221-8. [DOI] [PubMed] [Google Scholar]
- 26.Sumitani M, Yamamoto DS, Oishi K, Lee JM, Hatakeyama M. Germline transformation of the sawfly, Athalia rosae (Hymenoptera: Symphyta), mediated by a piggyBac-derived vector. Insect Biochem Mol Biol. 2003;33(4):449–458. doi: 10.1016/s0965-1748(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 27.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 28.Schinko JB, et al. Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev Biol. 2010;10:53. doi: 10.1186/1471-213X-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posnien N, Koniszewski NDB, Hein HJ, Bucher G. Candidate gene screen in the red flour beetle Tribolium reveals six3 as ancient regulator of anterior median head and central complex development. PLoS Genet. 2011;7(12):e1002416. doi: 10.1371/journal.pgen.1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handler AM, McCombs SD, Fraser MJ, Saul SH. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc Natl Acad Sci USA. 1998;95(13):7520–7525. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402(6760):370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 32.Schulte C, et al. Honey bee promoter sequences for targeted gene expression. Insect Mol Biol. 2013;22(4):399–410. doi: 10.1111/imb.12031. [DOI] [PubMed] [Google Scholar]
- 33.Hediger M, Niessen M, Wimmer EA, Dübendorfer A, Bopp D. Genetic transformation of the housefly Musca domestica with the lepidopteran derived transposon piggyBac. Insect Mol Biol. 2001;10(2):113–119. doi: 10.1046/j.1365-2583.2001.00243.x. [DOI] [PubMed] [Google Scholar]
- 34.Gempe T, et al. Sex determination in honeybees: Two separate mechanisms induce and maintain the female pathway. PLoS Biol. 2009;7(10):e1000222. doi: 10.1371/journal.pbio.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chenchik A, Moqadam F, Siebert PD. A new method for full-length cDNA cloning by PCR. In: Krieg PA, editor. A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis. New York: Wiley-Liss; 1996. pp. 273–321. [Google Scholar]
- 36.Kaftanoglu O, Peng YS. Preservation of honeybee spermatozoa in liquid nitrogen. J Apic Res. 1984;23(3):157–163. [Google Scholar]
- 37.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapetanaki MG, Loukeris TG, Livadaras I, Savakis C. High frequencies of Minos transposon mobilization are obtained in insects by using in vitro synthesized mRNA as a source of transposase. Nucleic Acids Res. 2002;30(15):3333–3340. doi: 10.1093/nar/gkf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beye M, Härtel S, Hagen A, Hasselmann M, Omholt SW. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Mol Biol. 2002;11(6):527–532. doi: 10.1046/j.1365-2583.2002.00361.x. [DOI] [PubMed] [Google Scholar]
- 41.Laidlaw HH, Page RE. Queen Rearing and Bee Breeding. Kalamazoo, MI: Wicwas Press; 1997. [Google Scholar]
- 42.Hunt GJ, Page RE., Jr Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 1995;139(3):1371–1382. doi: 10.1093/genetics/139.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasselmann M, Beye M. Signatures of selection among sex-determining alleles of the honey bee. Proc Natl Acad Sci USA. 2004;101(14):4888–4893. doi: 10.1073/pnas.0307147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210(12):630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 45.Horn C, Schmid BGM, Pogoda FS, Wimmer EA. Fluorescent transformation markers for insect transgenesis. Insect Biochem Mol Biol. 2002;32(10):1221–1235. doi: 10.1016/s0965-1748(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 46.Fischer M, Haase I, Simmeth E, Gerisch G, Müller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 2004;577(1-2):227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.