Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disease with high prevalence, which imposes a substantial public health problem. The heritability of AD is estimated at 60–80% forecasting the potential use of genetic biomarkers for risk stratification in the future. Several large scale genome-wide association studies using high frequency variants identified 10 loci accountable for only a fraction of the estimated heritability. To find the missing heritability, systematic assessment of various mutational mechanisms needs to be performed. This copy number variation (CNV) genome-wide association study with age at onset (AAO) of AD identified 5 CNV regions that may contribute to the heritability of AAO of AD. Two CNV events are intragenic causing a deletion in CPNE4. In addition, to further study the mutational load at the 10 known susceptibility loci, CNVs overlapping with these loci were also catalogued. We identified rare small events overlapping CR1 and BIN1 in AD and normal controls with opposite CNV dosage. The CR1 events are consistent with previous reports. Larger scale studies with deeper genotyping specifically addressing CNV are needed to evaluate the significance of these findings.
Keywords: Age at onset, Alzheimer’s disease, copy number variation
INTRODUCTION
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder affecting approximately 5.4 million individuals in the US and is the most common cause of dementia in North America and Europe [1–3]. Genetic factors play an important role in the pathogenesis of AD and contribute to the variance of age at onset (AAO) [4, 5]. Rare Mendelian forms of AD have confirmed and elucidated pathways involved in amyloid accumulation, but only account for a small percentage of the cases of AD [6]. The heritability of late-onset AD (LOAD) is estimated at 60–80%. The population attributable risk for the APOE4 allele is calculated at 20–70% in various studies. The recent ADGC study calculated the attributable risk for the ten non-APOE4 loci at 35%; however, it was felt that it might be an overestimate due to the winner’s curse. Additional loci may contribute to the genetic architecture of AD and account for the missing heritability [7]. APOE4 has a strong effect on AAO of AD and an additional 4–5 loci may contribute to the heritability of AAO [8].
Structural variation of submicroscopic DNA segments (deletions, duplications, and inversions) has greatly expanded our understanding of human genetic variation [9]. Copy number variations (CNVs) influence gene expression, phenotypic variation, and adaptation by altering gene dosage and genome organization [10, 11]. CNVs are often multiallelic and have a higher de novo mutation rate, and therefore they are often not adequately tagged by single-nucleotide polymorphism (SNPs) [12]. Because of these attributes, CNVs confer a novel genetic marker map with different properties representing a supplementary approach to SNP association [13]. With the advent of microarray technology allowing genome-wide ascertainment of CNVs, disease associations have been reported in various diseases including neurological diseases [14].
This study of CNV association with AD AAO was designed to examine the role of low-frequency variants with intermediate penetrance in the genetic architecture of AD. AAO serves as a quantitative endophenotype and the cases-only study design eliminates misclassification bias in this common disease with age-dependent penetrance. The mutational load architecture is likely to be locus specific, thus the detection of a given susceptibility locus will be successful by using the marker map that predominantly contributes to the mutational load of that locus, underlining the importance of the systematic assessment of all genetic variants. In addition, we catalogued the CNVs overlapping with the 10 susceptibility loci discovered in the genome-wide association studies (GWAS) to evaluate the role of CNVs contributing to the mutational load of these loci.
Published CNV studies in AD [15–19] have focused mostly on large events (>100 kb) to achieve high confidence calls and performed the tests of association on these events. We previously reported an olfactory receptor cluster association with AAO of AD performing the regression analysis on binned numeric logR ratio data without performing segmentation [20]. An alternative strategy applied here is to use permissive segmentation only to reduce the dataset where events may occur, perform the test of association on the numeric segmented data, and validate the CNV calls if a replicated association signal was detected. This second approach can detect association signals from smaller events that would have been discarded when performing the high confidence calls and overcomes the need to determine exact dosage, which could be problematic at common CNV loci as the reference will deviate from the diploid dosage.
METHODS
Subject cohorts
781 subjects were enrolled in the study. Probable AD diagnosis was established by NINCDS-ADRDA criteria [21]. The methodology of the Texas Alzheimer Research and Care Consortium project has been described in detail elsewhere [22]. Controls were recruited at each participating site by the same inclusion criteria, including age over 55 years, male and female, unrelated to AD subjects, Clinical Dementia Rating (CDR) global score 0, normal performance on activities of daily living (ADL), and all information was obtained from a surrogate historian. After enrollment all control subjects underwent neuropsychological testing including assessment of Global cognitive functioning/status (Mini-Mental Status Examination and CDR), Attention (Digit Span and Trails A), Executive function (Trails B and Clock Drawing; Texas Card Sorting is optional), Memory (Wechsler Memory Scale (WMS) Logical Memory I and WMS Logical Memory II), Language (Boston Naming and FAS Verbal Fluency), Premorbid IQ (AMNART), Visuospatial Memory (WMS-Visual Reproduction I and II), Psychiatric symptoms (Geriatric Depression Scale; Neuropsychiatric Inventory-Questionnaire) and Functional assessment (Lawton-Brody ADL: PSMS, IADL). Control subjects showing impairment were excluded from the control cohort after consensus review.
Informed consent was obtained from all subjects prior to inclusion. Genomic DNA was isolated from whole blood by the Puregene DNA isolation kit (Qiagen) according to the manufacturer’s instructions.
AAO phenotyping
AAO was determined with two standardized methods in both cohorts: i) caregiver estimate by prompted standard question regarding onset of symptoms and ii) physician estimate of duration of illness using a standardized and validated structured interview with landmark event to facilitate recall [23].
APOE genotyping
Genotyping was performed according to manufacturer‘s instruction with real-time PCR using custom TaqMan probes (Applied Biosystems, Inc) unique for SNPs of rs7412 and rs429358 at nucleotides 112 and 158 of the APOE gene, respectively. All amplifications were carried out in an ABI 7900HT thermal cycler (Applied Biosystems, Inc). APOE genotype was determined from the combination of alleles present at the 112 and 158 polymorphisms.
CNV genotyping by the genome-wide human SNP array 6.0
Array based genotyping was performed on the Genome-Wide Human SNP Array 6.0 (Affymetrix) according to the manufacturer’s instructions. QC measures for the Genome-Wide Human SNP Array 6.0 (Affymetrix) array included contrast QC (>0.4) and Median of the Absolute values of all Pairwise Differences (MAPD) <0.4. Arrays with number of calls more than 2SD from the mean were excluded.
Detection of CNV and test of association
We performed principal component analysis using the Eigenstrat method [24] implemented in Golden-Helix. The Affymetrix intensity data was corrected for 16 principal components as this correction resulted in the QQ plot without evidence of inflation (Supplementary Figure 1; available online: http://www.j-alz.com/issues/33/vol33-2.html#supplementarydata08). The PCA corrected data was segmented by the CNAM algorithm implemented in GoldenHelix (Golden-Helix). The univariate method identifies rare CNVs by scanning the data subject by subject. In the univariate method, we applied a moving window of 20,000 probes: the maximum number of segments per 10,000 markers was set at 1 and we set the minimum number of marker per segment at 1 with maximum pairwise permuted p-value was set at 0.005, applying 2,000 permutations per pair, to have high sensitivity to detect association signals from small events with the expectations that all association signal events will be manually reviewed for validation. The multivariate method compares all samples at each locus and identifies common CNVs. In the multivariate segmentation, we did not apply a moving window and used similar setting as in the univariate segmentation (maximum number of segments per 10,000 markers 10, minimum number of marker per segment 1, maximum pairwise permuted p-value was set at 0.005, applying 2,000 permutations per pair). Both algorithms generate segmentation covariates that results in reduction of the dataset to regions of interests where CNV events occur, thus limiting the number of tests of association.
For test of association, Cox proportional hazard analysis was performed using the covariates from the univariate and multivariate segmentation algorithms. The Cox proportional hazard regression script was created in R and the analysis of the corrected log R ratio data as predictor and AAO as outcome was performed in R. We used the FDR approach and the criteria for significance was set at corrected p-value of <0.05. For the covariates that were associated with AAO of AD, the analysis was repeated incorporating gender and APOE status (number of APOE alleles as categorical variable) in the proportional hazard model.
We analyzed the NIA-LOAD Familial Study dataset probands deposited in dbGAP (http://www.ncbi.nlm.nih.gov/gap) for replication of the results. 866 AD subjects were included in the analysis from the LOAD dataset selected using the following criteria: probands only, Caucasian, non-Hispanic, and AAO data available. The NIA-LOAD Familial Study genotyping was performed on the Illumina Human610 Quadv1 B array. The logR ratio was calculated in the GenomeStudio (Illumina) software. Candidate locus specific data was extracted and Cox porportinal hazard regression was performed on the logR numeric data because the candidate regions were inadequately covered for segmentation.
RESULTS
756 samples passed the contrast QC and were adequate for copy number analysis. Forty samples failed MAPD cutoff of 0.4 and 143 samples failed due to number of CNV calls more than 2SD of the mean. 573 samples passed QC: 381 AD subjects and 192 normal controls. Six AD subjects had missing AAO or APOE genotype, thus 375 AD subjects entered the cases-only AAO analysis. Demographics of the 375 subjects are depicted in Table 1.
Table 1.
Cohort characteristics
| n | Gender ratio (% F) | Age at Onset (y)
|
Age (y)
|
APOE
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Range | Mean ± SD | Range | N/N n(%) | N/4 n(%) | 4/4 n(%) | |||
| AD | 375 | 59% | (71.4 ± 8.6) | (45–90) | 79.3 ± 8.5 | 56–105 | 143 (38.1%) | 180 (48%) | 52 (13.9%) |
| Control | 180 | 74% | NA | NA | 73.3 ± 8.8 | 57–96 | 130 (67.7%) | 56 (29.1%) | 4 (2.1%)§ |
control (n = 2) missing APOE. F, female.
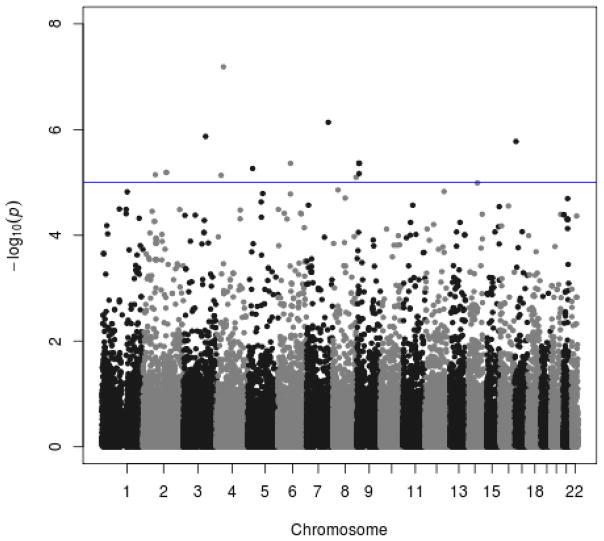
The Cox proportional hazard model identified 14 univariate segments corresponding to 11 chromosomal regions where the association signal survived multiple testing correction depicted in the Manhattan plot (Fig. 1). Five regions remained after manual review of the CNV events and the AAO versus segmented logR ratio scatterplots (Table 2). For all of these probes/regions, the p-value remained comparable after adding APOE and gender into the Cox proportional hazard model (Table 2). The number of events contributing to the association signal is depicted in Supplementary Table 1; consistent with the segmentation method these events were rare, ranging from 1 to 5 instances for the various regions. Three of the 5 regions are known CNV regions; the events on chromosome 3 : 131.9 Mb are intragenic causing a deletion in CPNE4 (Supplementary Figure 2). The detected CNVs were small, ranging from 3.6–24.8 kb. The events contributing to the AAO association signals are detailed in Supplementary Table 3. The multivariate segmented data detected nominal associations only (data not shown); the number of events contributing to these signals were consistent with the segmentation method developed to detect common CNVs. The LOAD familial dataset probands were studied for replication of these signals (Supplementary Table 2). The coverage of the candidate regions was limited on the Illumina 610 array (Supplementary Table 1) precluding conclusive interpretation of the replication study.
Fig. 1.
The Manhattan plot depicts the −log10p for the Cox proportional hazard regression using the array data (univariate segmentation covariates) as predictors and the age at onset (AAO) as outcome (n = 375). The chromosomes are shaded alternately and each segmentation covariate is represented by a dot.
Table 2.
Cox proportional hazard regression using the univariate segmentation covariates as predictor and age at onset (AAO) as outcome. For the CNVs that showed an association (FDR <0.05) Cox hazard regression was repeated with adding gender and APOE (count of APOE4 alleles as categorical values) as covariates
| Probe | Chr | Start | end | Size bp | ProbChiSq | fdr p | ProbChiSq covariates | Nearest Gene | Carriers AAO mean | Non-carriers AAO mean |
|---|---|---|---|---|---|---|---|---|---|---|
| SNP A-8600234 | 2 | 1.4E + 08 | 1.4E + 08 | 6125 | 6.4626E–06 | 0.0412 | 9.043E–06 | None | 50 | 71.57 |
| SNP A-8329031 | 3 | 1.32E + 08 | 1.32E + 08 | 3571 | 1.3493E–06 | 0.0330 | 1.609E–06 | CPNE4 | 48.5 | 71.63 |
| CN 1082571 | 4 | 42764937 | 42771246 | 6309 | 6.5114E–08 | 0.0051 | 1.992E–07 | ATP8A1 | 56.4 | 71.7 |
| SNP A-8584575 | 8 | 1.4E + 08 | 1.4E + 08 | 4580 | 8.0255E–06 | 0.0421 | 5.911E–06 | COL22A1 | 50 | 71.57 |
| SNP A-8327917 | 9 | 11917137 | 11941926 | 24789 | 4.3441E–06 | 0.0412 | 1.838E–05 | None | 49 | 71.63 |
In order to further assess mutational load, the CNV load of the previously identified 10 susceptibility loci [7] was evaluated in the TARCC dataset. CNVs that are overlapping with genes nearest to the reported associated SNPs are summarized in Table 3. We detected four small events in CR1 at two loci (Supplementary Figure 3) and two events in BIN1. In CD2AP, a small intragenic event was detected in a control.
Table 3.
Mutational load from copy number variation at the ten AD susceptibility loci reported by Naj et al. [7]
| Candidate gene | Chr | Start position | End position | Segment mean | Length (bp) | # Markers | Disease status |
|---|---|---|---|---|---|---|---|
| CR1 | 1 | 205731099 | 205740575 | 0.65599221 | 9477 | 13 | NC |
| 1 | 205733845 | 205740005 | 0.513781071 | 6161 | 9 | NC | |
| 1 | 205779459 | 205797985 | –0.931351781 | 18527 | 3 | NC | |
| 1 | 205779459 | 205797985 | 0.855937362 | 18527 | 3 | AD | |
| BIN1 | 2 | 127505275 | 127585431 | –0.327784389 | 80157 | 50 | AD |
| 2 | 127518845 | 127543003 | 0.608936131 | 24159 | 13 | NC | |
| CD2AP | 6 | 47667873 | 47677623 | –0.586070538 | 9751 | 8 | NC |
DISCUSSION
Five CNV regions were detected with sizes ranging from 3.6–24.8 kb and allele frequencies of 0.001–0.006 suggesting that small events with low frequencies could contribute to the genetic architecture of AD. The previously reported [20] chromosome 14 olfactory receptor cluster association with AAO of AD was confirmed (uncorrected p-value of 0.03) in this analysis workflow, and the size of the effect is consistent with the smaller sample size (the reported association included a sample size of 507 subjects in contrast with the 375 reported here).
Published case-control CNV GWAS in AD applied a fundamentally different workflow by entering only high stringency calls typically in excess of 100 kb in size to the analyses. The CPNE4 deletion has been documented in the database of Genomic Variants (http://projects.tcag.ca/variation/) and was validated in this set (Supplementary Figure 2). CPNE4 encodes copine IV, a calcium-dependent membrane-binding protein highly expressed in the brain. The detected CNV events have a high probability supported by the consistent within event logR ratio values and their presence in the database of Genomic Variants (3 out of 5 loci). The limited availability of probes in the regions of interest in the Illumina Human 610 or 650 arrays confounds the replication of these results in the NIA LOAD study, ADNI study, and the Caribbean Hispanic study, thus their significance remains unknown. Further larger replication studies with adequate coverage of the regions and molecular validation are needed. This emphasizes further challenges in CNV association studies: i) unlike SNPs single probe associations are usually not considered and several consecutive probes are needed to identify an event and ii) due to low allele frequencies, even larger studies are needed to have the power to detect the association than for SNP GWAS. Next generation whole genome sequencing evolved as a tool to detect most genetic variation in a single experiment; however, the cost and the large sample size needed in AD preclude their utility at this point in time.
Several small events overlapping genes in the 10 known susceptibility loci were identified suggesting that CNVs may contribute to the genetic architecture of these loci in AD. The CNV involving the CR1 gene at the low copy repeat region (Supplementary Figure 3, pink and blue bars) is consistent with previous reports and the direction of the CNV dosage in cases and controls consistent with the previously reported association [16]. Further work is needed to delineate the effect of these CNVs in AD.
Challenges to identifying the loci contributing to the heritability of AD are many fold. The disease affects the elderly, is common, and has an age-dependent penetrance, all contributing to misclassification bias which decreases power. The cases-only design attempts to eliminate the misclassification bias. Identification of genes that regulate AAO in AD may result in valid therapeutic targets as projections suggest that by postponing AAO by only 5 years may decrease the prevalence of AD by half [2]. CNVs are thought to contribute to the genetics of various diseases by intermediate frequency alleles with intermediate penetrance, which is confirmed in this study. Due to the lower allele frequencies compared to SNPs, CNV association studies need even larger sample sizes. Additional work is needed to validate and confirm these associations.
There have been several genome-wide scans involving over 10,000 cases and 10,000 controls which have identified replicable loci [7]. However, these loci together account for only a small fraction of the heritability of AD [7]. In order to find the missing heritability, systematic assessment of structural variants, rare variants, DNA methylation, and common variants need to be performed.
Supplementary Material
Acknowledgments
This study was supported by the Texas Alzheimer’s Research and Care Consortium (TARCC) funded by the state of Texas through the Texas Council on Alzheimer’s Disease and Related Disorders and an Alzheimer Association New Investigator Research Grant to KS.
Authors’ disclosures available online (http://www.j-alz.com/disclosures/view.php?id=1495).
Investigators from the Texas Alzheimer’s Research and Care Consortium: Baylor College of Medicine: Susan Rountree MD, Valory Pavlik PhD, Wen Chan PhD, Paul Massman PhD, Eveleen Darby, Tracy Evans RN, Aisha Khaleeq; Texas Tech University Health Science Center: Benjamin Williams, MD, Gregory Schrimsher, PhD, Andrew Dentino, MD, Ronnie Orozco; University of North Texas Health Science Center: Thomas Fairchild, PhD, Janice Knebl, DO, Sid E. O’Bryant, PhD, James R. Hall, PhD, Robert C. Barber, PhD, Douglas Mains, Lisa Alvarez; University of Texas Southwestern Medical Center: Perrie Adams, PhD, Roger Rosenberg, MD, Myron Weiner, MD, Mary Quiceno, MD, Joan Reisch, PhD, Ryan Huebinger, PhD, Guanghua Xiao, PhD, Doris Svetlik, Amy Werry, Janet Smith; University of Texas Health Science Center – San Antonio: Donald Royall, MD, Raymond Palmer, PhD, Marsha Polk.
Footnotes
References
- 1.Ebly EM, Parhad IM, Hogan DB, Fung TS. Prevalence and types of dementia in the very old: Results from the Canadian Study of Health and Aging. Neurology. 1994;44:1593–1600. doi: 10.1212/wnl.44.9.1593. [DOI] [PubMed] [Google Scholar]
- 2.Kukull WA, Higdon R, Bowen JD, McCormick WC, Teri L, Schellenberg GD, van Belle G, Jolley L, Larson EB. Dementia and Alzheimer disease incidence: A prospective cohort study. Arch Neurol. 2002;59:1737–1746. doi: 10.1001/archneur.59.11.1737. [DOI] [PubMed] [Google Scholar]
- 3.Rocca WA, Hofman A, Brayne C, Breteler MM, Clarke M, Copeland JR, Dartigues JF, Engedal K, Hagnell O, Heeren TJ, et al. Frequency and distribution of Alzheimer’s disease in Europe: A collaborative study of 1980–1990 prevalence findings. The EURODEM-Prevalence Research Group. Ann Neurol. 1991;30:381–390. doi: 10.1002/ana.410300310. [DOI] [PubMed] [Google Scholar]
- 4.Daw EW, Heath SC, Wijsman EM. Multipoint oligogenic analysis of age-at-onset data with applications to Alzheimer disease pedigrees. Am J Hum Genet. 1999;64:839–851. doi: 10.1086/302276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66:196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, Thomas-Anterion C, Michon A, Martin C, Charbonnier F, Raux G, Camuzat A, Penet C, Mesnage V, Martinez M, Clerget-Darpoux F, Brice A, Frebourg T. Early-onset autosomal dominant Alzheimer disease: Prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad DF, Hurles ME. The population genetics of structural variation. Nat Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarroll SA. Extending genome-wide association studies to copy-number variation. Hum Mol Genet. 2008;17:R135–R142. doi: 10.1093/hmg/ddn282. [DOI] [PubMed] [Google Scholar]
- 14.Boone PM, Wiszniewski W, Lupski JR. Genomic medicine and neurological disease. Hum Genet. 2011;130:103–121. doi: 10.1007/s00439-011-1001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzen EL, Need AC, Hayden KM, Chiba-Falek O, Roses AD, Strittmatter WJ, Burke JR, Hulette CM, Welsh-Bohmer KA, Goldstein DB. Genome-wide scan of copy number variation in late-onset Alzheimer’s disease. J Alzheimers Dis. 2010;19:69–77. doi: 10.3233/JAD-2010-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouwers N, Cauwenberghe CV, Engelborghs S, Lambert JC, Bettens K, Bastard NL, Pasquier F, Montoya AG, Peeters K, Mattheijssens M, Vandenberghe R, Deyn PP, Cruts M, Amouyel P, Sleegers K, Broeckhoven CV. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol Psychiatry. 2012;17:223–233. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, Foroud T, Mayeux R, Saykin AJ. Analysis of copy number variation in Alzheimer’s disease: The NIA-LOAD/NCRAD Family Study. Curr Alzheimer Res. 2012 doi: 10.2174/156720512802455331. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghani M, Pinto D, Lee JH, Grinberg Y, Sato C, Moreno D, Scherer SW, Mayeux R, St George-Hyslop P, Rogaeva E. Genome-wide survey of large rare copy number variants in Alzheimer’s disease among Caribbean hispanics. G3 (Bethesda) 2012;2:71–78. doi: 10.1534/g3.111.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, Kim S, Shen L, Risacher SL, Foroud T, Pankratz N, Potkin SG, Huentelman MJ, Craig DW, Weiner MW, Saykin AJ The Alzheimer’s Disease Neuroimaging Initiative A. Genomic copy number analysis in Alzheimer’s disease and mild cognitive impairment: An ADNI study. Int J Alzheimers Dis. 2011;2011:729478. doi: 10.4061/2011/729478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw CA, Li Y, Wiszniewska J, Chasse S, Zaidi SN, Jin W, Dawson B, Wilhelmsen K, Lupski JR, Belmont JW, Doody RS, Szigeti K. Olfactory copy number association with age at onset of Alzheimer disease. Neurology. 2011;76:1302–1309. doi: 10.1212/WNL.0b013e3182166df5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blacker D, Albert MS, Bassett SS, Go RC, Harrell LE, Folstein MF. Reliability and validity of NINCDS-ADRDA criteria for Alzheimer’s disease. The National Institute of Mental Health Genetics Initiative. Arch Neurol. 1994;51:1198–1204. doi: 10.1001/archneur.1994.00540240042014. [DOI] [PubMed] [Google Scholar]
- 22.O’Bryant SE, Waring SC, Cullum CM, Hall J, Lacritz L, Massman PJ, Lupo PJ, Reisch JS, Doody R. Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: A Texas Alzheimer’s research consortium study. Arch Neurol. 2008;65:1091–1095. doi: 10.1001/archneur.65.8.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doody RS, Dunn JK, Huang E, Azher S, Kataki M. A method for estimating duration of illness in Alzheimer’s disease. Dement Geriatr Cogn Disord. 2004;17:1–4. doi: 10.1159/000074078. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.