Summary
Sickle cell disease (SCD) patients are at high risk of contracting pneumococcal infection. To address this risk, they receive pneumococcal vaccines, and antibiotic prophylaxis and treatment. To assess the impact of SCD and these interventions on pneumococcal genetic architecture, we examined the genomes of over 300 pneumococcal isolates from SCD patients over 20 years. Modern SCD strains retained invasive capacity but shifted away from the serotypes used in vaccines. These strains had specific genetic changes related to antibiotic resistance, capsule biosynthesis, metabolism and metal transport. A murine SCD model coupled with Tn-seq mutagenesis identified 60 non-capsular pneumococcal genes under differential selective pressure in SCD, which correlated with aspects of SCD pathophysiology. Further, virulence determinants in the SCD context were distinct from the general population and protective capacity of potential antigens was lost over time in SCD. This highlights the importance of understanding bacterial pathogenesis in the context of high-risk individuals.
Introduction
Streptococcus pneumoniae (pneumococcus) is a major cause of childhood illness worldwide, causing approximately 14 million episodes of invasive disease and 1 million deaths per year. The first step in invasive pneumococcal disease (IPD) is nasopharyngeal (NP) colonization, but asymptomatic carriage is common, especially in early childhood (~30 - 50%) (Daw et al., 1997). Colonization is usually established by a single pneumococcal strain and persists for 1-2 months before clearance (Ghaffar et al., 1999). Pneumococci lack CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) systems to protect genomic content and are naturally highly transformable, permitting rapid genetic response to evolutionary pressures (Bikard et al., 2012; Croucher et al., 2011). For example, introduction of the 7-valent pneumococcal conjugate vaccine (Prevnar; PCV7) in the USA in 2000 resulted in near-complete elimination of vaccine serotypes and emergence of non-vaccine serotypes (NVT) in colonization and IPD in the general population (Croucher et al., 2013a; Halasa et al., 2013).
Sickle cell disease (SCD), a hemoglobinopathy characterized by chronic hemolysis and sickled red blood cells, is the most common genetic disorder worldwide, with 300,000 affected infants born each year (WHO, 2006). Children with SCD have a 600-fold increased risk of potentially fatal IPD compared with the general population, despite similar colonization rates (Overturf, 1999). The increased risk is related to hyposplenism, complement deficiency, and chronic vascular inflammation promoting upregulation of the ligand for pneumococcal invasion (Miller et al., 2007; Rosch et al., 2010). To address this risk, children with SCD receive long-term penicillin prophylaxis, frequent empiric antibiotic treatment, and pneumococcal vaccines. Pneumococci colonizing children with SCD have been previously shown to develop antibiotic resistance in response to the selective antibiotic pressure and demonstrate capsular serotype switching in response to vaccines (Adamkiewicz et al., 2008; Steele et al., 1996). The SCD host and the pneumococcus represents a unique paradigm for understanding how pathogens adapt to both clinical interventions, including vaccination and antibiotic pressure, as well as unique aspects of host pathophysiology underlying heightened infection risk. Due to the combination of clinical and host factors, we hypothesized that pneumococci found in the SCD population would develop unique genomic adaptations from the selective pressures imparted not only by clinical interventions but also the SCD host environment itself.
To characterize the pneumococcus emerging in the pediatric SCD population between 2004 and 2011 and re-evaluate current disease risk for these vulnerable children, we undertook the largest longitudinal study of pneumococcal colonization in children with SCD. We compared the results with a cohort from 1994-5 to ascertain the impact of deployment of the conjugate PCV7 vaccine in 2000. We also acquired a broad range of IPD isolates from healthy and SCD children from across the United States and obtained sequence data for contemporary NP isolates from the general population (GP) (Croucher et al., 2013a). Whole-genome sequencing of 322 isolates, one of the largest datasets assembled thus far, defined overall gene content and identified genes with differential abundance between isolates from SCD and the GP as well as between historical and contemporary eras. A murine model of SCD coupled to Tn-seq whole-genome mutagenesis and vaccination experiments were used to identify and evaluate pneumococcal gene networks under selective pressure in this host. These data provide a comprehensive analysis of the influence of both clinical interventions and the SCD host environment on the pneumococcus, resulting in unique genetic selection and specialized contribution of genes in virulence in these high-risk patients.
Results
Modern colonizing SCD strains retain invasive characteristics and evade interventions
Pneumococcal strains were obtained from three sources: A) 63 NP SCD isolates from 1994-5 (Daw et al., 1997); B) 186 IPD SCD isolates from the CDC ABC bacterial surveillance core, SJCRH patients, and published collections (McCavit et al., 2011); and C) 98 NP SCD isolates from a longitudinal study spanning 2004-2011 in 195 SCD children followed for up to 4 years with serial NP swabs (Table 1). Children with SCD received PPV23 vaccine after 2 years of age, penicillin prophylaxis from birth to at least 5 years of age (Gaston et al., 1986) and frequent empiric antibiotic therapy. PCV7 vaccine was given only in the contemporary group. Multivariate analysis of the 2004-2011 NP data showed that younger age was significantly associated with increased risk of colonization (OR = 0.783 per year of age increase, p < 0.0001) and that, despite interventions, patients had a colonization rate of 7.1%, a value unchanged from 1994-5 after correction for age (the adjusted colonization rate estimate for median age of 4.3 years was 13.4% in 2004-11 and 13.5% in 1994-5). The rates of acute chest syndrome and pneumonia remained high despite prophylactic interventions (133 episodes/1000 patient years). Penicillin resistance in contemporary SCD NP isolates was unchanged compared with the historical era for both low-level (MIC > 0.06 μg/ml; 54% vs 51%; p = 0.87) and high-level resistance (MIC > 2 μg/ml; 1% vs 5.4%; p = 0.14). Antibiotic prophylaxis appears to have maintained pressure on pneumococci in the SCD population leading to a sustained high rate of penicillin resistance.
Table 1.
Demographic and NP colonization data
| Patient Information | ||
|---|---|---|
| 2004-11 | 1994-5(Daw et al., 1997) | |
| Evaluable patients | 195 | 312 |
| Median months on study (range) | 48 (1 – 59) | N/A |
| Total number of nasopharyngeal swabs | 1372 | 312 |
| Pneumococcal colonization, n (%) | 98 (7.1%)a | 42 (13.5%) |
| Penicillin resistanceb | ||
| High-level: Non-meningitis (PRP) | 1 (1%) | 3 (5.4%) c |
| Low-level: Meningitis (PNSP) | 53 (54.1%) | 29 (51.8%)c |
| Median age at enrollment, years (range) | 5.0 (0.1 – 17.7) |
4.3 (0.3 – 5.4) |
| Median age at time of swab, years | 6.9 | 4.3 |
| Female Gender | 98 (50.3%) | 141 (45.2%) |
| Black Race | 193 (99.0%) | 312 (100%) |
| Sickle Cell Disease Type | ||
| HbS/β-thalassemia | 17 (8.7%) | 24 (7.7%) |
| HbSS Disease | 178 (91.3%) | 195 (62.5%) |
| Hydroxyurea use (ever during study) | 106 (54.4%) | N/A |
| Penicillin prophylaxis (ever during study) | 134 (68.7%) | 208 (67%) |
| Empiric antibiotic (ever during study) | 195 (100%) | N/A |
| Vaccine coverage (ever during study) | ||
| Pneumovax: PPV23 (≥1 dose) | 186 (95.4%) | N/A |
| Prevnar 7: PCV7 (≥1 dose) | 193 (99.0%) | 0 (0.0%) |
|
Infection episodes; n (events/1000 patient
years) |
||
| Invasive pneumococcal disease | 2 (2.8)d | N/A |
| Acute chest syndrome | 95 (133) | N/A |
| Hospital admissions for infection | 200 (280) | N/A |
N/A, not available
Pneumococcal colonization rate in 2004-11 adjusted to median age of 4.3 years = 13.4%.
Includes data for an additional 12 isolates from the same cohort
Clinical and Laboratory Standards Institute susceptibility breakpoints.
Post-PCV7 rate in healthy children <5 years old = 0.26/1000 patient years; Pre-PCV7 rate in SCD < 5 years old = 24/1000 patient years(Adamkiewicz et al., 2003; Hicks et al., 2007)
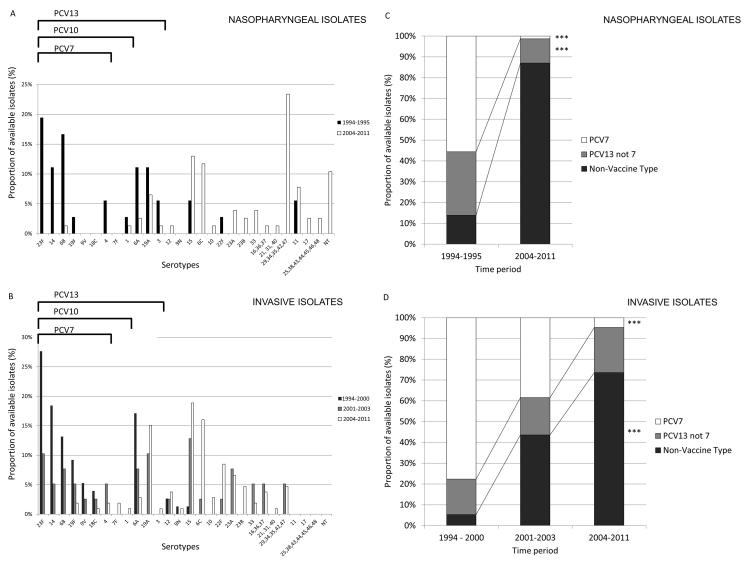
Capsular serogroup was compared between strains from pre (n = 112) and post (n = 225) PCV7 vaccine (Fig 1). After introduction of PCV7, there was the expected marked decrease in prevalence of PCV7 serotypes for both SCD NP and IPD isolates, similar to previous studies in both SCD and normal hosts (Adamkiewicz et al., 2008; Dagan and Klugman, 2008; Halasa et al., 2013) and reflecting the national trend following PCV7 introduction of serotype (Croucher et al., 2013a; Wroe et al., 2012). Unexpectedly, there was also a marked shift away from the additional serotypes in the not yet deployed PCV13 (Fig 1C; p < 0.0001). This indicates that the majority of serotypes found in the pediatric SCD population were outside the coverage of the new PCV10 and PCV13 vaccines prior to their deployment.
Figure 1. Serotype replacement in pneumococcal isolates from children with Sickle Cell Disease.
Serotype distribution and vaccine serotype representation for NP (A,C; n = 113) and IPD isolates (B,D; n = 221) from SCD patients before and after introduction of PCV7 pneumococcal vaccine in 2000. Different color bars in panels A and B represent distinct time periods for which strains were isolated. Different color bars in C and D represent the different vaccine serotypes in relation to inclusion in conjugate vaccines: Included in PCV7 (white bars), additional serotype included in PCV13 (gray bars), or not included in either vaccine (black bars). Data were available for pre- (1994 - 2000) and post-PCV7 (2004 – 2011) time-periods for both IPD and NP isolates, and for the intervening period (2001 – 2003; during rollout of PCV7) for invasive isolates only, as NP isolates were not collected during this period (*** represents p<0.001 by Fisher’s exact test).
The risk for IPD posed by the emerging non-vaccine serotypes (NVT) is a major unanswered question (Syk et al., 2014). Invasiveness depends in part on non-capsular genomic content which could be derived from two general processes with potentially distinctly different invasive potential: capsular switching to NVT while maintaining an invasive, fully virulent genome or serotype replacement via selection of previously rare NVT capsules and potentially less virulent genomes (Wyres et al., 2013). To address this question, we performed whole genome sequencing on 322 SCD strains and determined similarity of all SCD strains in relation to the GP isolates. For the GP cohort, we also compared data from a recently published cohort from pediatric patients in the post-PCV era using the same sequencing technology (Croucher et al., 2013a).
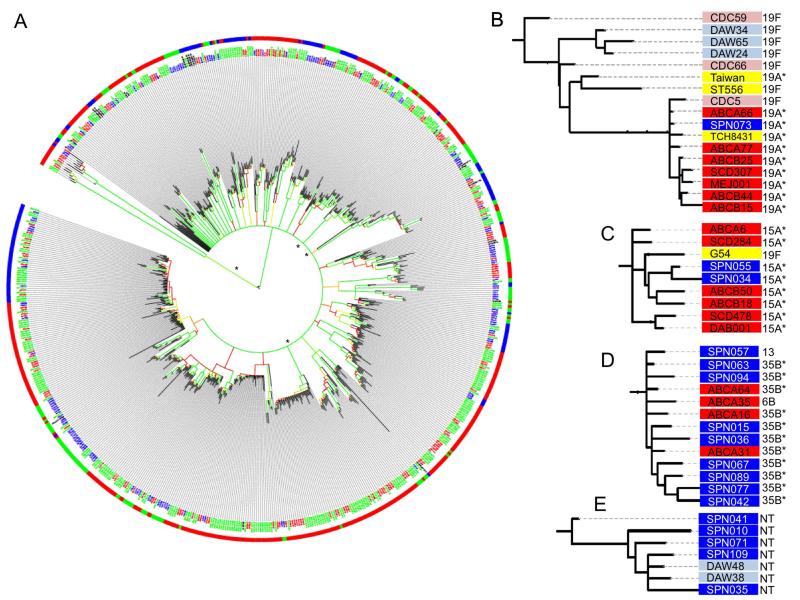
To ascertain the degree of shared protein coding content as well as single nucleotide variants (SNVs) in the core genome between the various isolates, we utilized three approaches: a comparative analysis using either SNVs (Supplementary Figure 1A) or genome composition (Supplementary Figure 1B), and a hybrid approach leveraging both the SNV and gene content (Fig 2). Such pan-genome analysis has been previously shown to provide greater strain resolution compared to methods based on SNV analysis of core genes alone (Hall et al., 2010). An approach using presence/absence of both core and accessory genes has previously been shown to correlate reasonably well with SNV approaches (Donati et al., 2010); hence we employed all approaches to interrogate our strain database. Including the gene content allowed for more accurate assignments than those resolved by SNV alone based on average bootstrap values across all nodes (0.55 for gene content, 0.58 for SNV, 0.65 for combined approach). As expected, in a naturally competent species like S. pneumoniae, low–bootstrap values were typical of deep internal clades, although more recently emerged clades were often highly supported. The geographic distribution of the samples was not a factor in the phylogeny or subsequent genetic analyses as comparison of the SCD NP isolates (Memphis) and the SCD IPD isolates (nationwide) revealed not a single allele found in significantly altered abundance between these geographically distinct groups, similar to previous reports due to wide circulation of pneumococcal strains (Donati et al., 2010). Importantly, all analyses indicated the strains did not segregate based on geography, time period, or disease state (Fig 2A). Strains collected serially from individual patients were also randomly distributed, indicating constant acquisition of new isolates rather than persistent colonization. The effect of recombination can only be incorporated into the individual clades rather than the entire dataset due to the diversity of our sample, similar to previous reports (Croucher et al., 2013a). Hence, analysis was restricted to strains within the same clade or within the hierarchal gene clustering approaches based on gene content.
Figure 2. Phylogeny of pneumococcal isolates derived from gene content and nucleotide variants.
A) Relatedness of the historical SCD (blue text), contemporary SCD (red text), contemporary GP NP cohort (green text), and Genbank strains (black text) as determined by SNVs in core genes and gene content. Colors of the branches reflect bootstrap values (red to green representing 0.65-1.0). Outer circle reflects strain serotype, serotypes included in PCV7 (green), PCV13 (blue), and non-vaccine strains (red) (* represents highly supported clades chosen for further analysis). B-E) Contemporary SCD IPD (dark red), historical SCD IPD (light red), contemporary SCD NP (dark blue), historical SCD NP (light blue), GP NP (yellow). B) Numerous modern vaccine escape 19A SCD strains are closely related to the 19F historical CDC5 isolate. C) Historical GP 19F invasive isolate has undergone capsule switching to contemporary SCD 15A invasive isolates. D) Contemporary SCD IPD and NP strains of several serotypes are highly related at the genetic level. E) An anomalous clade of NVT, non-encapsulated pneumococci with a high prevalence of macrolide resistance was identified in the SCD NP population. Their basal position in the S. pneumoniae tree was determined using nonpneumococcus streptococcal species as outgroups.
Genomic analysis revealed three important findings regarding the status of pneumococci in the SCD population: First, a number of vaccine escape 19A variants were closely related to the 19F lineage (Fig 2B) and many NVT invasive variants (e.g. 15A) were genetically similar to earlier VT invasive isolates (e.g. 19F; Fig 2C), indicating a high incidence of capsule switching. Contemporary NVT serotypes were found in both NP and IPD classes (such as 35B, 6B and 13 strains; Fig 2D) indicating the NVT strains retained invasive capacity in SCD. Of note is an anomalous clade of non-typeable pneumococci that splits early in the tree suggesting a lineage arising soon after S. pneumoniae diverged from S. mitis (Fig 2E). This clade contains only NP isolates from different subjects across time periods and all isolates were acapsular both serologically and by sequence homology. While penicillin resistance in this group was similar to other modern NP isolates, macrolide resistance was much more frequent (80.0% vs 24.5%; p=0.008) with a higher median erythromycin MIC (>256 vs 0.19 mcg/ml; p=0.002). No significant allelic differences were observed between the NP and IPD SCD isolates, but the limited sample size may not have sufficient power to identify all factors contributing to invasiveness.
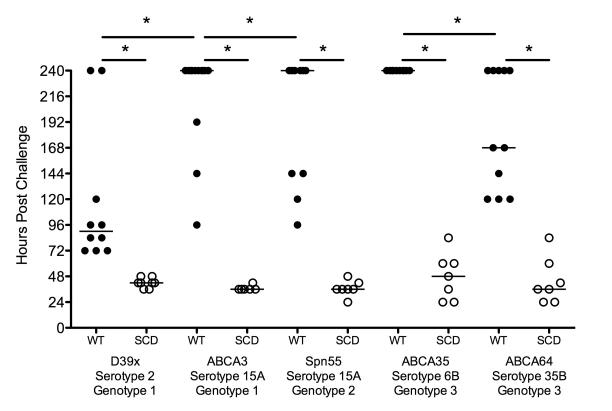
Since genomic analysis suggested that emerging NVT strains retained invasive capacity in SCD, we sought to model the invasive potential of these NVT isolates in a SCD murine system. Using this model, the NVT strains showed varying degrees of pathogenesis in WT mice but all strains were highly virulent in SCD mice (Fig 3). These data indicate that capsule type may not be as critical a requirement for invasive disease in SCD as the GP. Taken together, the data reveal that a high invasive disease risk remains in SCD children outside the scope of available conjugate vaccines. One important caveat is that even though strains were chosen for mouse challenge based on genetic similarity or capsule type, small alterations in genotype can confer strong alterations in virulence (Croucher et al., 2013b). Since the animal model suggested all isolates retained high virulence in the SCD model independent of capsule type or genetic background, we explored other factors that might more accurately define infection risk in this patient population.
Figure 3. Contribution of capsule type and genetic background to IPD.
WT (black circles) and SCD (white circles) animals challenged with the respective pneumococcal strains indicate that regardless of capsule type or genetic background, pneumococcal strains retain high virulence potential in SCD. In WT animals the clinical strains displayed varying degrees of virulence dependent upon both capsule type and genetic background whereas all strain retained equivalent virulence in SCD mice (* represents p<0.05 by log rank test).
SCD isolates show altered genomic content over time
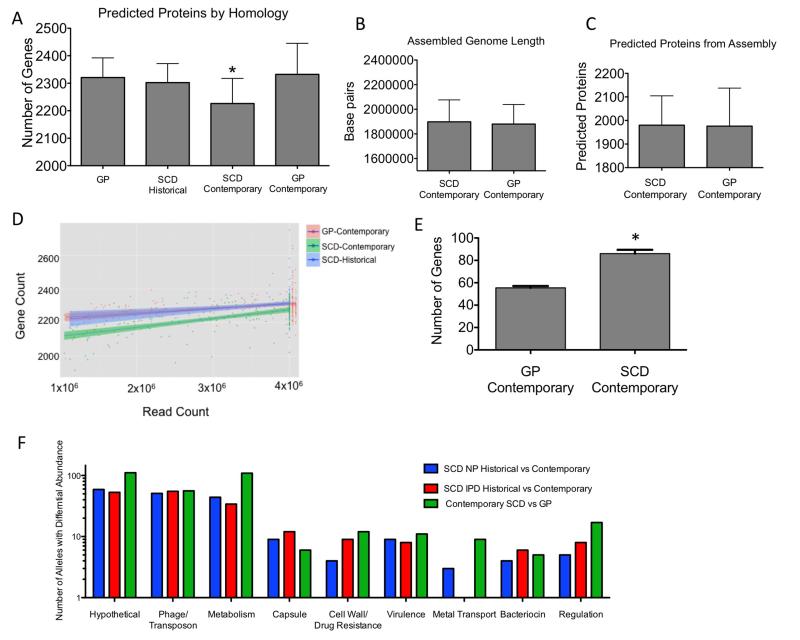
To define any unique features in the SCD genome, contemporary SCD isolates were compared to historical SCD and contemporary GP isolates for total gene content. When genes were identified via mapping of short reads to a reference streptococcal gene set (95% homology cutoff, see methods), the contemporary SCD isolates appeared to have a significantly smaller overall genome size by an average of 76 genes compared to the historical SCD or contemporary GP strains (Fig 4A, p<0.0001). In contrast, when genome length (Fig 4B) and predicted ORFs (Fig 4C) were compared using a genome assembly approach, this was not observed. The difference observed in the contemporary SCD isolates was maintained in comparison to both the contemporary GP isolates as well as the historical SCD isolates regardless of sequence coverage (Fig 4D). Such an apparent decrease in homologous genome content using a mapping approach while preserving physical genome size could represent two processes: actual loss of genetic content or gene chimerism arising from increased rates of recombination. Actual loss of genes was detected in approximately 50% of cases while sequences of the remainder of the apparently missing genes could be matched to parts of more than one divergent pneumococcal sequence indicating novel mosaics arising from intragenic recombination (Fig 4E). Interestingly, the number of loci involved in recombination is very similar to that observed during recombination events between S. mitis and S. pneumoniae under penicillin pressure (Sauerbier et al., 2012). These events were not the result of a single isolated incident, as overall the changes were distributed throughout the chromosome indicating these events were occurring independently. The fact that these events were preferentially observed in the SCD population indicates such strains may be outcompeted or rapidly cleared in the GP.
Figure 4. Genomic comparisons of similarity and allelic content.
A) Average number of genes per genome as determined by homology with known streptococcal genes. # of genomes analyzed: GP (n = 24), SCD historical (n = 141), SCD contemporary (n = 173), GP contemporary (n = 563), (* represents p<0.0001 by Mann-Whitney U test). B,C) Genome length and predicted proteins based on the assemblies of the contemporary GP and SCD isolated showed no significant differences between groups D) Number of coding regions predicted by homology as a function of read counts indicate the contemporary SCD genomes were significantly smaller compared to SCD historical or contemporary GP isolates independent of sequence coverage. E) Putative intragenic recombinant genes that were called as absent by the read mapping approach due to the disjoint mapping of reads to different members of a single gene family (* represents p<0.001 by Mann-Whitney U test). Error bars represent mean and SD. F) Breakdown of annotated function of alleles showing differential abundance or recombination between groups. Differing genes from the total of 3316 genes for each group: Historical vs Contemporary SCD NP (n = 188), Historical vs Contemporary SCD IPD (n = 188), Contemporary SCD vs Contemporary GP (n = 338).
We next sought to elucidate specific differences in genomic content between contemporary SCD and GP isolates. This was accomplished by pairwise comparisons of the frequency of the individual alleles utilized for the gene content phylogenetic reconstructions. At the single allele level, a large number of hypothetical genes, transposable elements and genes involved in metabolic networks were differentially abundant or divergent between these two groups (Fig 4F). As this approach is homology based, these differences represent both complete absence and divergent genes, such as the penicillin binding proteins (PBPs) known for their mosaic sequence diversity. Specifically, differences in four key groups were apparent: PBPs, capsular biosynthesis genes, metabolic pathways and metal transport. Differences in genes for capsule biosynthesis and PBPs are expected signatures in a population that routinely receives prophylactic penicillin and has very high PCV7 vaccination rates (Croucher et al., 2011; Zapun et al., 2008). However, the emergence of differences in metabolic networks and metal uptake loci suggests that absence or recombination within these loci in the SCD host environment may be advantageous or neutral whereas in the GP such disruptions may prove detrimental.
Analysis of SCD host selective pressure by Tn-seq
Identification of genes associated with SCD or IPD via abundance in the strain collection can be problematic, due to the fact that very slight changes can impact virulence and the likely multifactorial contribution of genes to bacterial fitness. Hence, we opted for an experimental approach using Tn-Seq to model selective pressures of SCD physiology on the contribution of pneumococcal genes to bacterial fitness in this host (van Opijnen and Camilli, 2013). Pools of thousands of tagged, transposon-induced loss-of-function pneumococcal mutants were created in a GP invasive isolate (TIGR4) and injected into SCD and WT mice. Bacterial mutants recovered from the blood of septic mice were sequenced and compared to the input inoculum to generate a fitness value for each gene. If a gene product contributes to pathogenesis, then its loss-of-function mutant replicates less well in the host. Conversely, if deletion of a gene is favorable, the mutant replicates more rapidly or escapes immune clearance and is over-represented in the output pool.
The fitness of each pneumococcal gene was calculated independently for infection in either WT or SCD mice (Supplementary Table S1). Comparison of the datasets revealed 60 genes whose transposon disruption resulted in significantly different fitness between the two hosts (Supplementary Table S2), indicating distinct requirements for SCD infection. Transposon-induced disruption of 41 genes significantly lowered fitness selectively in WT relative to SCD mice, while disruption of 19 other genes significantly lowered fitness in SCD relative to WT mice (Supplementary Fig. S2). Moreover, the population bottleneck from indiscriminant bacterial clearance was significantly greater in WT animals compared to SCD (Supplementary Fig. S3). Thus the bloodstream of normal mice is not only a more hostile environment for S. pneumoniae, but also is a significantly distinct environment compared to SCD.
The differential fitness of the gene knockouts predicted by Tn-seq was validated by creating individual deletion mutations for six genes: five which had shown reduced fitness in WT but not SCD hosts and one with reduced fitness only in SCD. Compared to the parental TIGR4 strain in a competitive index model of pneumococcal bacteremia, the mutant fitness predicted by Tn-Seq proved robust (Fig 5A,D, Supplementary Fig. S4). These data indicate that pneumococci utilize distinct virulence gene networks in a host-specific manner. This also predicts that there could be a unique and defined set of critical genes differentially selected by SCD host pathophysiology.
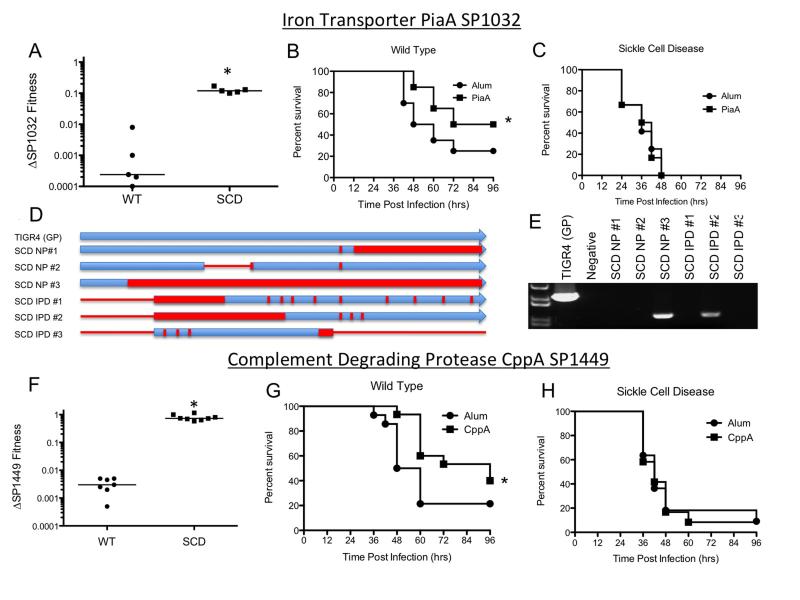
Figure 5. Host-specific differences in contribution of two pneumococcal genes to virulence and protection by vaccination.
A-E: Iron transporter SP1032: A) Equivalent CFUs of TIGR4 and the SP1032 deletion mutant were co-inoculated into wild type (WT) and SCD mice and the respective amounts of each strain recovered from blood at 24 hours was enumerated. A fitness value of 1 indicates that the mutant and TIGR4 parent have equal virulence. Each data point represents an individual mouse and medians are shown by lines (* represents p<0.01 by Mann-Whitney U test). B,C) Vaccination with Sp1032/PiaA confers protection against subsequent challenge in WT but not SCD mice (* represents p<0.05 by log rank test). D,E) Alterations of the SP1032 gene in SCD isolates shown by genomic sequencing (D) and PCR (E) reveal significant divergence/point mutations (red bars) and deletions/truncations (red lines). F-H): Complement degrading proteinase SP1449: F) Attenuation of the SP1449 mutant in WT but not SCD mice (experiment performed as indicated in panel A). G,H) Wild type (G) and SCD mice (H) were vaccinated with recombinant CppA with alum or alum alone and subsequently challenged with TIGR4. Vaccinated wild type mice showed significantly improved survival (p=0.03 by Log rank test) but the vaccine conferred no protection for SCD mice.
Human SCD host physiology correlates with Tn-seq and genomic content
To relate the putative SCD genes identified in the Tn-seq analysis to the human context, two analyses were performed: the function of the pneumococcal gene product was compared to known SCD pathophysiology and the frequency of mutation of these genes in clinical SCD isolates was determined, hypothesizing that genes dispensable in the SCD host may not be maintained or may be significantly divergent in the altered contemporary SCD genome.
The functions of genes under selective pressure in SCD mice correlated strongly with aspects of SCD pathophysiology in humans, including abnormal iron homeostasis, purine metabolism, and complement function (Table 2). Secondly, six genes predicted by Tn-seq to be under SCD pressure were either a distinct allelic variant, altered or absent in contemporary clinical SCD isolates. One important feature to note is that while Tn-seq represents an IPD model of bacterial fitness, pneumococcal prevalence in the clinical cohort is also related to transmissibility and colonization potential. We opted for this model because gene content was indistinguishable between the NP and IPD SCD isolates, transmission is not well modeled in a murine system, and fulminant sepsis is a classic presentation of pneumococcal disease in SCD. Genes identified to be under selective pressure in the murine model by Tn-seq and found in significantly (p<0.05 by Fishers exact test) altered abundance in the SCD clinical isolates were SP511, SP0946, SP1032, SP1449, SP1483, and SP1835. For Sp0511, SP0946, SP1032, SP1483, and SP1835, alterations were the result of truncations at the N or C terminus, recombination events, or outright deletions. In contrast, alterations in SP1449 consisted of point mutations only.
Table 2.
Clinical correlation with SCD pathophysiology of selected genes with significant differential fitness between wild type and SCD mice. (WT: Virulence attenuated in wild type mice compared with SCD mice; SCD: Virulence attenuated in SCD mice compared to wild type mice).
| Locus | Virulence Attenuated in: |
Name | Correlation with Sickle Cell Disease Pathophysiology |
|---|---|---|---|
| SP0045 | WT | phosphoribosylformylglycinamidine synthase (putative) |
These genes are involved in the purine biosynthetic pathway. Serum levels of purines, which are essential for bacterial DNA and RNA synthesis, are elevated in SCD, possibly due to chronic hemolysis, inflammation or renal disease. (Cerqueira et al., 2011) |
| SP0046 | WT | amidophosphoribosyltransferase | |
| SP0050 | WT | formyltransferase / IMP cyclohydrolase |
|
| SP0051 | WT | phosphoribosylamine-glycine ligase | |
| SP1032 | WT | iron-compound ABC transporter | These genes are involved in homeostasis of metal ions, especially iron. Iron metabolism in SCD is complex with increased serum hemin due to hemolysis, but depressed free iron due to increased renal losses. (Koduri, 2003) |
| SP1638 | WT | iron-dependent transcriptional regulator |
|
| SP0963 | SCD | dihydroorotate dehydrogenase, electron transfer subunit |
These genes are involved in nucleotide and DNA synthesis. Purines and pyrimidines are more abundant in the SCD host.(Cerqueira et al., 2011) |
| SP1018 | SCD | thymidine kinase | |
| SP0768 | WT | ribosomal RNA large subunit methyltransferase N |
These genes are involved in the RNA synthesis and processing. Availability of nucleotide precursors is generally increased in the SCD host. (Cerqueira et al., 2011) |
| SP1483 | WT | ATP-dependent RNA helicase, DEAD/DEAH box family |
|
| SP2016 | SCD | nicotinate-nucleotide pyrophosphorylase |
This gene is involved in the process of NAD biosynthesis by catalyzing the production of nicotinic acid mononucleotide. NAD+ plays an important role in bacterial DNA biosynthesis. Erythrocyte total NAD and NAD/NADH ratio are both increased in the SCD host. (Zerez et al., 1988) |
| SP0823 | SCD | amino acid ABC transporter, permease protein |
These genes are involved in arginine transport and regulation. Arginine is a critical source of nitric oxide for the host and a required nutrient for pneumococci. Availability of arginine in SCD hosts is known to be decreased compared with healthy controls due to reduced plasma levels. (Sullivan et al., 2010) |
| SP0893 | WT | putative transcriptional repressor | |
| SP1203 | WT | putative transcriptional repressor | |
| SP1665 | WT | ylmE protein | These genes are involved in amino acid transport. Serum concentrations of many amino acids, including leucine, isoleucine and valine, are significantly lower in SCD, due in part to elevated urinary excretion. (VanderJagt et al., 1997) |
| SP0751 | WT | branched-chain amino acid ABC transporter protein |
|
| SP0668 | SCD | glucokinase | This gene is involved in glucose metabolism and regulation of other metabolic pathways. Glucose metabolism and availability of other sugars appears to be altered in SCD, with higher insulin levels and higher mean HOMA-IR scores, attributable to oxidative damage (Alsultan et al., 2010) |
| SP1619 | SCD | PTS system, IIA component | This gene is involved in fructose transport and metabolism. Plasma fructose concentration is markedly decreased in the SCD host. (Osuagwu and Mbeyi, 2007) |
| SP1121 | SCD | 1,4-alpha-glucan branching enzyme | This gene is involved in conversion of glucose to glycogen, the primary storage carbohydrate for bacteria. Availability of sugars, including glucose and fructose, is altered in the SCD host.(Osuagwu and Mbeyi, 2007) (Alsultan et al., 2010) |
| SP0715 | SCD | lactate oxidase | This gene is involved in metabolism of lactate to pyruvate. In the SCD host environment, systemic lactate clearance is impaired, erythrocyte lactate uptake is increased, and serum lactate dehydrogenase levels are increased. (Freund et al., 1992; Pattillo and Gladden, 2005) |
| SP1157 | WT | voltage-gated chloride channel family protein |
Function of these gene products appears to be dependent on chloride and sodium concentrations. SCD hosts have frequent episodic hyponatremia during acute illness, related to renal salt-wasting.(Radel et al., 1976) |
| SP1318 | WT | v-type sodium ATP synthase, subunit G |
|
| SP1449 | WT | C3-degrading proteinase | This gene is involved in bacterial evasion of complement- mediated immune killing. Complement function is severely impaired in SCD. (Battersby et al., 2010) |
| SP1916 | WT | PAP2 family protein | This gene is involved in synthesis of triacylglycerol and glycerophospholipid. Serum triglycerides are frequently elevated in SCD, whereas total and HDL cholesterol are decreased (Seixas et al., 2010) |
The most significant difference was observed in the SP1032 iron transporter gene, found to be dispensable in the SCD host by Tn-seq and found in significantly reduced abundance or recombined in the modern SCD population (p<0.001). The divergence of the SP1032 sequences ranged from large deletions to SNPs (Fig 5B-C). Furthermore, additional genes in this operon that did not meet the required fitness-difference threshold in the Tn-seq analysis were also found to be absent or divergent in the modern SCD isolates, including the SP1034 permease component of the ABC transporter and SP1035 ATP binding protein (p<0.001). One potential explanation for this locus to be under particularly strong negative selection could arise from both the lack of necessity for iron acquisition in the iron-rich SCD host and the fact that this protein complex is highly immunogenic (Whalan et al., 2005). Such factors could allow for more permissive recombination events at this locus in the SCD population that would be disadvantageous in the GP. These data indicate the potential for the selective pressures observed in murine models of SCD to be reflected in the population genetics of isolates circulating and causing invasive disease in these patients.
Efficacy of a vaccine antigen differed between hosts
An important implication of these analyses relates to vaccine design. Gene products required for full virulence are attractive vaccine candidates, but are usually identified primarily in otherwise healthy individuals. The most obvious shortcoming of a potential vaccine would be if the potential vaccine target was not present in strains circulating amongst a target population, as was observed with the SP1032 gene in SCD. This has also been observed both in our study and in previous reports whereby a number of genes encoding antigenic surface proteins are easily lost and reacquired due to selective pressure driven by the host immune response (Donati et al., 2010; Muzzi et al., 2008). Further, our demonstration that even universally conserved genes, essential for IPD in normal hosts, are not required in SCD suggests that these antigens would be paradoxically inefficacious for immunizing children with SCD. We tested this hypothesis using two genes which were required for WT but not SCD infection: Sp1032 (PiaA) which was extremely divergent in SCD isolates and SP1449 (CppA) which was highly conserved in all isolates but shown by Tn-Seq and deletion analyses to be superfluous in the SCD setting (Fig 5D). Following vaccination with either recombinant CppA or PiaA, challenge with TIGR4 showed strong vaccine-induced protection in WT but not SCD mice (Fig 5E,F) despite similar antibody titers (Supplementary Figure S5). These data indicate that vaccines derived from virulence determinants in normal hosts may not be efficacious in high-risk patient populations with a differing host environment.
Discussion
Identification of the underlying factors that contribute to heightened infection risk is a fundamental challenge in infectious disease. This is underscored by the observation that the heritability factor for dying of infectious disease is greater than any other condition including cardiovascular disease and cancer (Sorensen et al., 1988). Heightened risk is associated with rare defects in host immune defenses (Ram et al., 2010) or bacterial adaptation to clinical interventions (Ding et al., 2009). The pneumococcus is particularly adept at rapidly acquiring antibiotic resistance and evading capsule-based vaccines (Adamkiewicz et al., 2003; Halasa et al., 2013). Pneumococcal isolates from children with SCD were previously known to be highly antibiotic resistant as an adaptation to the high rate of antimicrobial exposure in this population. It had been less clear whether the pathogen could tailor itself to an altered host metabolic environment resulting in strains capable of causing disease in high-risk individuals but not the GP. Using strains from one of the largest longitudinal study of pneumococcal colonization in children with SCD coupled to extensive pneumococcal genomic sequencing, the signatures of three distinct selective pressures were clearly evident in the changes in the pneumococcal genome over the last 20 years: penicillin prophylaxis, pneumococcal vaccines, and the SCD host environment.
Medical interventions of penicillin prophylaxis and vaccination are universally deployed in SCD children to decrease invasive pneumococcal disease. Over 20 years, there has been no change in overall pneumococcal colonization rate in SCD children, and rates of IPD and pneumonia/acute chest syndrome remain markedly higher in SCD than in the general population (Halasa et al., 2007; Payne et al., 2013). Penicillin non-susceptible pneumococcal colonization is much more common in SCD children than in the GP (Daw et al., 1997), but rates have not changed over time despite PCV7 vaccination. Detection of non-typeable pneumococcal isolates in this study, especially those with high-level macrolide resistance, is particularly concerning as non-encapsulated pneumococci offer opportunities for accelerated gene transfer between organisms due to their increased transformability. A finding with major clinical impact is that contemporary SCD colonizing strains have almost all been replaced with serotypes not included in PCV13 (p<0.0001), despite lack of exposure to this new vaccine (only PCV7 had been deployed during the study period). Emergent colonization with non-PCV7 serotypes and the erosion of PCV13 serotypes even before introduction of the PCV13 vaccine, is particularly alarming in tandem with the observation that SCD isolates retain virulence potential in SCD but not necessarily the GP. This was reflected in murine models of infection whereby pneumococcal strains varied in lethality based on capsular type or genetic background in WT but not SCD mice, where they were all lethal. The invasive potential of strains emerging by capsular serotype switching in response to vaccine pressure has been recognized as a pressing question (Syk et al., 2014). We conclude that circulating pneumococci are not only excluded from conjugate vaccine coverage by capsule switching but also have retained invasive capacity in these vulnerable hosts. This represents an unrecognized, persistent risk for invasive infection in vaccinated SCD children who are uniquely susceptible, even to strains that might rarely be pathogenic in the GP.
One of the most challenging aspects of genomic analyses in a naturally competent organism is the high rate of recombination. Lateral gene transfer can obscure accurate phylogenetic reconstruction as genes are transferred both within and outside of a particular bacterial species. In the case of homologous recombination, different homologous positions in a SNV-based phylogenetic reconstruction can support different topologies. This will result in low bootstrap support of phylogeny bipartitions. A gene presence/absence approach can mitigate this problem because it is not affected by the presence of SNVs that have different evolutionary histories, although it is still prone to additive transfer events. The only assumption is that genes are gained or lost along a phylogeny at a constant rate. Consequently, we used a combined SNV-gene presence/absence approach since recombination between related streptococci occurs frequently. We believe this approach is more comprehensive than using only SNV data when comparing strains that frequently recombine with related species. Indeed, this approach provides the best overall bootstrap support for the phylogenetic reconstruction compared to either dataset alone.
In addition to effects of penicillin and vaccine exposure, whole genome sequencing revealed a noticeable impact of the SCD host environment on pneumococcal genome content. One of the most striking features was the decreased homologous coding content of the contemporary SCD strains and the extensive intragenic recombination leading to the creation of chimeric genes from divergent sources. Potential explanations for increased frequency of recombination in SCD isolates include unique selection by the combination of a novel SCD host environment, the global population bottleneck created by the introduction of PCV7, and constant antimicrobial pressure. The distinct population bottleneck effect imposed by penicillin prophylaxis, vaccine coverage, and altered host physiology could also result in a predominance of otherwise rare variants in this population. Such a different bottleneck constraint agrees with the breakthrough of infections in SCD mice by strains of significantly low virulence for WT mice, a finding reflected in the clinical isolates whereby atypical capsule types continue to cause invasive disease in SCD. A lower Tn-seq bottleneck observed during experimental SCD infection indicates that the SCD environment is less hostile for pneumococcal survival and suggests that the environment could result in variants that cause breakthrough infection in SCD but not the GP. This was supported by the large number of alleles showing differential abundance between the contemporary SCD and GP isolates, a number similar to that found between isolates before and after introduction of PCV which completely reshaped the circulating pneumococcal population.
Previous studies have showed high rates of antibiotic resistance in pneumococci colonizing SCD hosts (Steele et al., 1996). This occurred without decreased carriage rates, implying that the organisms had developed unique features specific to SCD penicillin prophylaxis. Our data expands and supports this notion in two important ways. For one, the increased proportion of mosaic genes in the SCD but not GP isolates indicates that distinct selective pressures have had a measurable effect on the composition of the pneumococcal genome within the individual patients. Secondly, strains that might otherwise be non-invasive in the GP appear to retain full pathogenicity in SCD, a prediction in line with our observations that all isolates tested retained full virulence in SCD but not WT mice and that strains with various targeted deletions predicted by Tn-seq were defective in invasive disease in WT but not SCD mice. These findings indicate that rapid recombination events may be more frequent in strains colonizing SCD patients resulting in strains with decreased fitness capacity when transmitted back into the GP. The mechanism by which the sickle-cell host influences the pneumococcal genome remains unknown. It is possible that individual mutations occur during periods of carriage, that strains which are rapidly cleared in the GP may colonize and cause IPD in SCD patients, or that transmission to SCD patients from family members or other patients at clinic visits occurs. A combination of these factors or other unrecognized factors may also be responsible.
We identified 60 genes that had differential effects on bacterial fitness in the SCD and WT host, and showed correlation of the function of these genes with SCD pathophysiology. For example, the SCD host has defects in complement function (Test and Woolworth, 1994) and altered plasma levels of zinc, iron, purines, amino acids, and carbohydrates (Darghouth et al., 2011; Prasad et al., 1976) , which in turn correlate with the dispensability of complement degrading proteinase, a transcriptional regulator of zinc/manganese influx, purine biosynthesis enzymes, arginine utilization, and iron acquisition genes in this population. These findings are underscored by the genomic analyses of clinical SCD isolates that showed high sequence divergence and even deletion of a subset of genes that correlated with the lack of selective pressure to maintain these functions. In the SCD host, the constraints of pathogenesis are clearly different and hence genetic selection may proceed differently than in the GP. These studies establish that host environments place recognizable, specific selective pressures on bacterial evolution. This process in pneumococci is rapid, likely facilitated by the plasticity of the genome, allowing the possibility of emergence of a pneumococcus more tailored to the SCD host than the GP.
We conclude that, although pneumococcal colonization rates are similar in SCD and the general population, the environment of the SCD host is more permissive to invasive pneumococcal disease and, in contrast to the GP, risk of infection appears to be driven less by capsule type than by non-capsular genomic content. This pressure appears to have created a unique evolutionary bottleneck that has resulted in a shift of dozens of genes from essential to nonessential for SCD virulence. The identification of strains lacking important virulence determinants has important implications for vaccine design. Gene products that are normally required for disease progression in healthy hosts may not be required for pathogenesis in highly susceptible individuals, and may even be absent from strains causing disease. Both CppA (SP1449) and PiaA (SP1032) are putative future vaccine candidate proteins which no appear to be dispensable for SCD disease. Both vaccines protect wild type but not SCD mice, and PiaA is frequently absent or modified in pneumococcal strains colonizing SCD patients. Inclusion of these antigens in vaccines might be efficacious in the general population but provide inadequate protection in high risk individuals with predisposing medical conditions such as SCD. Extending Tn-seq and genome sequencing to other disorders as a probe for differences in the genes required for disease progression could aid in the development of vaccines and therapeutics for high-risk patients. To avoid excluding uniquely vulnerable groups, such as children with SCD, from the benefits of new vaccines, these factors should be considered in vaccine design.
Experimental Procedures
2004-2011 Patient cohort
All human studies were prospectively approved by the St. Jude Institutional Review Board. Children (<18 years) with confirmed HbSS or HbS/β thalassemia disease attending the St Jude MidSouth Sickle Cell Clinic were eligible for enrollment. Enrollment was stratified by age (≤24 months, >2 to 5 years, >5 – 18 years). Visits were scheduled 3 monthly for children under 2 years, and six monthly for all patients 2 years and over. For up to 4 years from enrollment, data collected at each routine visit included: antibiotic use, vaccinations, hydroxyurea treatment, surgical splenectomy, comorbidities, intercurrent illnesses and hospitalization for infection, acute chest syndrome/pneumonia, or serious bacterial infection (SBI). NP samples were collected at each visit on sterile rayon swabs, plated directly onto blood agar and cultured at 37°C for 48 hours in 5% CO2. S. pneumoniae isolates were identified by colony morphology, Gram-stain, and catalase and optochin testing. Serogrouping was performed by latex agglutination [Pneumotest-Latex, Statens Serum Institut, Denmark]. Non-typeable or PCV7, PCV10 or PCV13 vaccine serogroup isolates were serotyped by Quellung reaction.
Genome sequencing and analysis
All 322 SCD pneumococcal genomes were sequenced using the Hi-Seq with 100-base paired-end reads. Paired-end sequence data was acquired for a sample of 327 GP strains from the European Nucleotide Archive (Croucher et al. 2013; accession: ERP000809). Reads were randomly selected from each sample of the GP data to give a similar range of genome coverage as those of the SCD isolates. Adapters and low-quality terminal bases were removed using Cutadapt (Martin, 2011) and the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), respectively. Each sample had a minimum of 106 reads (~46x coverage for genome of 2.16 MB). Twenty four complete reference genomes were in silico sequenced using DWGsim (https://github.com/nh13/DWGSIM) to reduce potential biases in the comparison of Illumina-sequenced and fully assembled reference genomes. Three million paired-end 100-base sequences were randomly sampled from the reference genomes with linearly increasing error rates across each read from 0.001 to 0.01. All fully-assembled genomes and plasmids from the genus Streptococcus (n = 109) were downloaded as Genbank files and the coding sequences were extracted using PERL. 155,257 coding sequences (hereafter referred to as genes) were reverse complemented when necessary, translated, and used to construct a genome index for short-read alignment using SHRiMP2 (David et al., 2011). OrthoMCL was used to partition the reference set of genes into 8668 orthologous groups ranging from 1 to 233 orthologs and inparalogs across the reference genomes using an inflation parameter of 3 (Li et al., 2003). All reads from both the Illumina-sequenced and the in silico-sequenced genomes were aligned to the full set of streptococcal reference genes by a paired-end, exhaustive mapping approach using SHRiMP2 (David et al., 2011).
A gene was considered present n a sample using the mapping-based approach if reads aligned to >95% of any member of an orthologous group. Genes were also identified using the RAST server(Aziz et al., 2008) to annotate genome assemblies. Genomes were assembled using Edena (Hernandez et al., 2008)and PAGIT (Swain et al., 2012). The reference genome used for contig scaffolding of each sample was determined by mapping sample reads to each reference genome independently and selecting the one with the highest coverage. Further details regarding sequence analysis is detailed in the Supplementary material.
Tn-seq calculation of gene fitness
Tn-seq was performed as described previously (van Opijnen et al., 2009; van Opijnen and Camilli, 2010, 2012). After fitness was calculated for each insertion, values were normalized against a set of neutral genes with no fitness effect to express all fitness values relative to the wild type background. 3 requirements were established to determine that a gene had a differential effect on fitness in wild type and SCD mice: i) fitness was composed of at least 3 data points, ii) fitness deviated by at least 20%, and iii) fitness was significantly different in a one-sample t-test with p < 0.05 after Bonferroni correction for multiple testing.
Statistical analysis
Generalized estimating equation (GEE) model analysis was used to determine independent risk factors for colonization and to adjust colonization rate for age. GEE was used to account for possible correlation between repeated measures from the same participants. Fisher’s exact test was used to determine significance of difference between proportions and the Mann-Whitney U test was used to determine significance of difference between medians for continuous variables.
Supplementary Material
Over 600 pneumococcal isolates from sickle cell disease (SCD) patients sequenced
SCD isolates retain invasive capacity but diverge from vaccine strains
Pneumococcal genes under selective pressure in SCD correlated with SCD pathophysiology
Due to host-specificity, high-risk patients require unique vaccine composition
Acknowledgements
JWR was supported by grant U54 HL-070590 NHLBI Sickle Scholar Award and by the American Lebanese Syrian Associated Charities (ALSAC). EIT was supported by R01-AI27913, NHLBI U54 HL-070590 and ALSAC. TvO is supported the Charles H. Hood Foundation; AC is an investigator of the Howard Hughes Medical Institute. Funding agencies had no role in the conduct of the study or interpretation of the data. We thank the ABC surveillance core of the Centers for Disease Control and Prevention for graciously providing many of the isolates. We thank Drs. Bernard Beall and Tim McCavit for providing SCD IPD strains and Dr. Beall for pneumococcal serotyping assistance. We thank all members of the St Jude Mid-South Sickle Cell Clinic for assistance with handling of patient samples and Dr Najat C. Daw for providing SCD pneumococcal isolates from 1994-5.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamkiewicz TV, Sarnaik S, Buchanan GR, Iyer RV, Miller ST, Pegelow CH, Rogers ZR, Vichinsky E, Elliott J, Facklam RR, et al. Invasive pneumococcal infections in children with sickle cell disease in the era of penicillin prophylaxis, antibiotic resistance, and 23-valent pneumococcal polysaccharide vaccination. J Pediatr. 2003;143:438–444. doi: 10.1067/S0022-3476(03)00331-7. [DOI] [PubMed] [Google Scholar]
- Adamkiewicz TV, Silk BJ, Howgate J, Baughman W, Strayhorn G, Sullivan K, Farley MM. Effectiveness of the 7-valent pneumococcal conjugate vaccine in children with sickle cell disease in the first decade of life. Pediatrics. 2008;121:562–569. doi: 10.1542/peds.2007-0018. [DOI] [PubMed] [Google Scholar]
- Alsultan AI, Seif MA, Amin TT, Naboli M, Alsuliman AM. Relationship between oxidative stress, ferritin and insulin resistance in sickle cell disease. Eur Rev Med Pharmacol Sci. 2010;14:527–538. [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby AJ, Knox-Macaulay HH, Carrol ED. Susceptibility to invasive bacterial infections in children with sickle cell disease. Pediatr Blood Cancer. 2010;55:401–406. doi: 10.1002/pbc.22461. [DOI] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Cerqueira BA, Boas WV, Zanette AD, Reis MG, Goncalves MS. Increased concentrations of IL-18 and uric acid in sickle cell anemia: contribution of hemolysis, endothelial activation and the inflammasome. Cytokine. 2011;56:471–476. doi: 10.1016/j.cyto.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013a;45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Mitchell AM, Gould KA, Inverarity D, Barquist L, Feltwell T, Fookes MC, Harris SR, Dordel J, Salter SJ, et al. Dominant role of nucleotide substitution in the diversification of serotype 3 pneumococci over decades and during a single infection. PLoS Genet. 2013b;9:e1003868. doi: 10.1371/journal.pgen.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan R, Klugman KP. Impact of conjugate pneumococcal vaccines on antibiotic resistance. Lancet Infect Dis. 2008;8:785–795. doi: 10.1016/S1473-3099(08)70281-0. [DOI] [PubMed] [Google Scholar]
- Darghouth D, Koehl B, Madalinski G, Heilier JF, Bovee P, Xu Y, Olivier MF, Bartolucci P, Benkerrou M, Pissard S, et al. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood. 2011;117:e57–66. doi: 10.1182/blood-2010-07-299636. [DOI] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical Short Read Mapping. Bioinformatics. 2011;27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- Daw NC, Wilimas JA, Wang WC, Presbury GJ, Joyner RE, Harris SC, Davis Y, Chen G, Chesney PJ. Nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae in children with sickle cell disease. Pediatrics. 1997;99:E7. doi: 10.1542/peds.99.4.e7. [DOI] [PubMed] [Google Scholar]
- Ding F, Tang P, Hsu MH, Cui P, Hu S, Yu J, Chiu CH. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics. 2009;10:158. doi: 10.1186/1471-2164-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, et al. Structure and dynamics of the pan genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund H, Lonsdorfer J, Oyono-Enguelle S, Lonsdorfer A, Bogui P. Lactate exchange and removal abilities in sickle cell patients and in untrained and trained healthy humans. J Appl Physiol. 1992;73:2580–2587. doi: 10.1152/jappl.1992.73.6.2580. [DOI] [PubMed] [Google Scholar]
- Gaston MH, Verter JI, Woods G, Pegelow C, Kelleher J, Presbury G, Zarkowsky H, Vichinsky E, Iyer R, Lobel JS, et al. Prophylaxis with oral penicillin in children with sickle cell anemia. A randomized trial. N Engl J Med. 1986;314:1593–1599. doi: 10.1056/NEJM198606193142501. [DOI] [PubMed] [Google Scholar]
- Ghaffar F, Friedland IR, McCracken GH., Jr. Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J. 1999;18:638–646. doi: 10.1097/00006454-199907000-00016. [DOI] [PubMed] [Google Scholar]
- Halasa NB, Grijalva CG, Arbogast PG, Talbot TR, Craig AS, Griffin MR, Schaffner W. Near Complete Elimination of the Seven Valent Pneumococcal Conjugate Vaccine Serotypes in Tennessee. Pediatr Infect Dis J. 2013 doi: 10.1097/INF.0b013e318287fe0d. [DOI] [PubMed] [Google Scholar]
- Halasa NB, Shankar SM, Talbot TR, Arbogast PG, Mitchel EF, Wang WC, Schaffner W, Craig AS, Griffin MR. Incidence of invasive pneumococcal disease among individuals with sickle cell disease before and after the introduction of the pneumococcal conjugate vaccine. Clin Infect Dis. 2007;44:1428–1433. doi: 10.1086/516781. [DOI] [PubMed] [Google Scholar]
- Hall BG, Ehrlich GD, Hu FZ. Pan-genome analysis provides much higher strain typing resolution than multi-locus sequence typing. Microbiology. 2010;156:1060–1068. doi: 10.1099/mic.0.035188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduri PR. Iron in sickle cell disease: a review why less is better. Am J Hematol. 2003;73:59–63. doi: 10.1002/ajh.10313. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17 [Google Scholar]
- McCavit TL, Quinn CT, Techasaensiri C, Rogers ZR. Increase in Invasive Streptococcus Pneumoniae Infections in Children with Sickle Cell Disease since Pneumococcal Conjugate Vaccine Licensure. J Pediatr. 2011;158:505–507. doi: 10.1016/j.jpeds.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ML, Gao G, Pestina T, Persons D, Tuomanen E. Hypersusceptibility to invasive pneumococcal infection in experimental sickle cell disease involves platelet-activating factor receptor. J Infect Dis. 2007;195:581–584. doi: 10.1086/510626. [DOI] [PubMed] [Google Scholar]
- Muzzi A, Moschioni M, Covacci A, Rappuoli R, Donati C. Pilus operon evolution in Streptococcus pneumoniae is driven by positive selection and recombination. PLoS One. 2008;3:e3660. doi: 10.1371/journal.pone.0003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuagwu CG, Mbeyi CU. Altered plasma hexose sugar metabolism in sickle cell anaemia. Afr J Biochem Res. 2007;1:37–40. [Google Scholar]
- Overturf GD. Infections and immunizations of children with sickle cell disease. Adv Pediatr Infect Dis. 1999;14:191–218. [PubMed] [Google Scholar]
- Pattillo RE, Gladden LB. Red blood cell lactate transport in sickle disease and sickle cell trait. J Appl Physiol. 2005;99:822–827. doi: 10.1152/japplphysiol.00235.2005. [DOI] [PubMed] [Google Scholar]
- Payne AB, Link-Gelles R, Azonobi I, Hooper WC, Beall BW, Jorgensen JH, Juni B, Moore M. Invasive Pneumococcal Disease among Children with and without Sickle Cell Disease in the United States, 1998-2009. Pediatr Infect Dis J. 2013 doi: 10.1097/INF.0b013e3182a11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AS, Ortega J, Brewer GJ, Oberleas D, Schoomaker EB. Trace elements in sickle cell disease. JAMA. 1976;235:2396–2398. [PubMed] [Google Scholar]
- Radel EG, Kochen JA, Finberg L. Hyponatremia in sickle cell disease. A renal salt-losing state. J Pediatr. 1976;88:800–805. doi: 10.1016/s0022-3476(76)81118-3. [DOI] [PubMed] [Google Scholar]
- Ram S, Lewis LA, Rice PA. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev. 2010;23:740–780. doi: 10.1128/CMR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch JW, Boyd AR, Hinojosa E, Pestina T, Hu Y, Persons DA, Orihuela CJ, Tuomanen EI. Statins protect against fulminant pneumococcal infection and cytolysin toxicity in a mouse model of sickle cell disease. J Clin Invest. 2010;120:627–635. doi: 10.1172/JCI39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbier J, Maurer P, Rieger M, Hakenbeck R. Streptococcus pneumoniae R6 interspecies transformation: genetic analysis of penicillin resistance determinants and genome-wide recombination events. Mol Microbiol. 2012;86:692–706. doi: 10.1111/mmi.12009. [DOI] [PubMed] [Google Scholar]
- Seixas MO, Rocha LC, Carvalho MB, Menezes JF, Lyra IM, Nascimento VM, Couto RD, Atta AM, Reis MG, Goncalves MS. Levels of high-density lipoprotein cholesterol (HDL-C) among children with steady-state sickle cell disease. Lipids Health Dis. 2010;9:91. doi: 10.1186/1476-511X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- Steele RW, Warrier R, Unkel PJ, Foch BJ, Howes RF, Shah S, Williams K, Moore S, Jue SJ. Colonization with antibiotic-resistant Streptococcus pneumoniae in children with sickle cell disease. J Pediatr. 1996;128:531–535. doi: 10.1016/s0022-3476(96)70365-7. [DOI] [PubMed] [Google Scholar]
- Sullivan KJ, Kissoon N, Sandler E, Gauger C, Froyen M, Duckworth L, Brown M, Murphy S. Effect of oral arginine supplementation on exhaled nitric oxide concentration in sickle cell anemia and acute chest syndrome. J Pediatr Hematol Oncol. 2010;32:e249–258. doi: 10.1097/MPH.0b013e3181ec0ae5. [DOI] [PubMed] [Google Scholar]
- Swain MT, Tsai IJ, Assefa SA, Newbold C, Berriman M, Otto TD. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc. 2012;7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syk A, Norman M, Fernebro J, Gallotta M, Farmand S, Sandgren A, Normark S, Henriques-Normark B. Emergence of hyper-virulent mutants resistant to early clearance during systemic serotype 1 pneumococcal infection in mouse and man. J Infect Dis. 2014 doi: 10.1093/infdis/jiu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Test ST, Woolworth VS. Defective regulation of complement by the sickle erythrocyte: evidence for a defect in control of membrane attack complex formation. Blood. 1994;83:842–852. [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Camilli A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol Chapter. 2010;1 doi: 10.1002/9780471729259.mc01e03s19. Unit1E 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature reviews Microbiology. 2013;11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderJagt DJ, Kanellis GJ, Isichei C, Patuszyn A, Glew RH. Serum and urinary amino acid levels in sickle cell disease. J Trop Pediatr. 1997;43:220–225. doi: 10.1093/tropej/43.4.220. [DOI] [PubMed] [Google Scholar]
- Whalan RH, Funnell SG, Bowler LD, Hudson MJ, Robinson A, Dowson CG. PiuA and PiaA, iron uptake lipoproteins of Streptococcus pneumoniae, elicit serotype independent antibody responses following human pneumococcal septicaemia. FEMS Immunol Med Microbiol. 2005;43:73–80. doi: 10.1016/j.femsim.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Wroe PC, Lee GM, Finkelstein JA, Pelton SI, Hanage WP, Lipsitch M, Stevenson AE, Rifas-Shiman SL, Kleinman K, Dutta-Linn MM, et al. Pneumococcal carriage and antibiotic resistance in young children before 13-valent conjugate vaccine. Pediatr Infect Dis J. 2012;31:249–254. doi: 10.1097/INF.0b013e31824214ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Linares J, Jacobs MR, Kristinsson KG, Beall BW, Klugman KP, et al. Pneumococcal capsular switching: a historical perspective. J Infect Dis. 2013;207:439–449. doi: 10.1093/infdis/jis703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- Zerez CR, Lachant NA, Lee SJ, Tanaka KR. Decreased erythrocyte nicotinamide adenine dinucleotide redox potential and abnormal pyridine nucleotide content in sickle cell disease. Blood. 1988;71:512–515. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.