Abstract
Dysregulation of the complement alternative pathway (AP) can cause disease in various organs that may be life-threatening. Severe AP dysregulation can be triggered by autoantibodies to the C3 convertase, termed nephritic factors, which cause pathological stabilisation of the convertase enzyme and confer resistance to innate control mechanisms; unregulated complement consumption followed by deposition of C3 fragments in tissues ensues. The monoclonal antibody, 3E7, and its humanised derivative, H17, have been shown previously to specifically bind activated C3 and prevent binding of both the activating protein, factor B, and the inhibitor, factor H, opposite effects that complicate its potential for therapy. Using ligand binding assays, functional assays and electron microscopy, we show that these antibodies bind C3b via a site which overlaps the binding site on C3 for the Ba domain within factor B, thereby blocking an interaction essential for convertase formation. Both antibodies also bind the preformed convertase, C3bBb, and provide powerful inhibition of complement activation by preventing cleavage of C3. Critically, the antibodies also bound and inhibited C3 cleavage by the nephritic factor-stabilised convertase. We suggest that by preventing enzyme formation and/or cleavage of C3 to its active downstream fragments, H17 may be an effective therapy for conditions caused by severe dysregulation of the C3 convertase, and in particular those involving nephritic factors, such as dense deposit disease.
INTRODUCTION
Complement is part of innate immunity with key roles in defence against pathogens through opsonisation and lysis, clearance of apoptotic cells, handling of immune complexes and modulation of adaptive immune responses (1). Complement can be triggered via three activation pathways: the classical, alternative (AP2) and lectin pathways, all leading to the generation of a C3 cleaving enzyme, or convertase, the central and most important step of the activation cascade. Cleavage of C3 generates C3b which covalently links to target cells, binding factor B (fB) in a Mg2+-dependent manner to form C3bB. This proenzyme is activated by factor D (fD), generating the active C3 convertase, C3bBb. Binding of properdin (P) stabilises this otherwise labile complex. Each C3 convertase cleaves many C3 to C3b, thus providing exponential amplification of the pathway. Complement activation progresses by formation of the C5 cleaving enzyme, resulting in generation of C5a and C5b. C5a is a proinflammatory peptide with anaphylactic and chemotactic properties, while C5b binds the next complement component, C6, marking the start of the terminal pathway which culminates in formation of the cytolytic membrane attack complex (MAC) (2).
The AP ‘ticks over’ constantly in plasma. Spontaneous hydrolysis of C3 generates a C3b-like molecule, C3(H20), that binds fB, which is then processed by fD to form a fluid-phase enzyme, C3(H2O)Bb, that cleaves C3 to C3b, thus ‘priming’ the AP for immediate activation (3). C3b generated in the fluid phase is rapidly inactivated, thus preventing uncontrolled consumption of complement in plasma; however, a proportion binds indiscriminately to any cell in its vicinity and, if not strictly regulated, can drive complement activation and cause damage to host cells. Damage to self is restricted by numerous complement regulatory proteins present in the fluid phase (including factor H; fH) and on cell membranes including CD55, CD35 and CD46. These regulators act by accelerating natural decay of C3bBb or by acting as cofactors for the proteolytic inactivation of C3b by the plasma protease factor I (4, 5).
In health, complement is in homeostatic balance; activation in plasma occurs at a low level and regulation prevents significant deposition of the central component, C3b, and limits further activation except on pathogens. The capacity of complement to initiate quickly and amplify efficiently means that any disturbance in homeostasis can be devastating to health (6). ‘Dysregulation’ of the central components of the amplification loop, C3, fB, fD or the control protein, fH, can cause acute or chronic inflammation and contribute to the pathologies associated with diverse diseases, including rheumatoid arthritis, systemic lupus erythematosus, glomerulonephritis, multiple sclerosis, sepsis, asthma, and ischaemia/reperfusion injuries. In each, complement activation drives a ‘vicious cycle’ of inflammation and tissue damage (7).
It is now established that the prototypic complement dysregulation-associated diseases, dense deposit disease (DDD), atypical hemolytic uremic syndrome (aHUS) and age-related macular degeneration (AMD) are each associated with mutations and/or polymorphisms in the components and regulators of the AP C3 convertase (8, 9). Severe dysregulation is also triggered by autoantibodies against complement components, complexes or regulators. Antibodies which interfere with function of fH are found in some aHUS and DDD patients. Antibodies which bind the AP C3 convertase, C3bBb, known as C3 nephritic factors (C3NeF), are present in over 80% of patients with DDD (10, 11). Once bound to the C3 convertase, C3NeF stabilizes the C3bBb complex, increasing its half-life and preventing regulation by complement regulatory proteins such as fH (12). This stabilised C3 convertase consumes intact C3, thereby generating large amounts of fluid phase activated C3 fragments (C3b, iC3b, C3dg) which locate in the kidney.
Understanding of the mechanisms behind complement-mediated diseases has advanced enormously in recent years, revealing new avenues for development of specific therapies targeting different steps within the complement cascade. These include soluble recombinant complement regulatory proteins, such as TP10 (13), and mAb which block complement proteins, such as Eculizumab™ (anti-C5 (14)) and TA106 (anti-fB Fab; Alexion Pharmaceuticals (15)). Small molecule inhibitors which prevent C3 activation are in trials for disorders such as age-related macular degeneration (compstatin (16, 17)). The most successful anti-complement therapeutic to date is Eculizumab, approved by the FDA for treatment of paroxysmal nocturnal hemoglobinuria (PNH) and aHUS, diseases in which dysregulated alternative pathway triggers terminal pathway activation which plays a critical role in pathology. However, not all complement-mediated diseases occur as a consequence of C5a and membranbe attack complex (MAC) activities; in DDD there is marked activation of C3 and fluid phase C3 activation fragments (primarily iC3b) trapped in the glomerular basement membrane in the kidney are implicated in driving pathology (18).
The mAb, 3E7, and its humanised chimeric counterpart, H17, bind C3b, C3b(H2O) and iC3b but not native C3 or C4/C4b (19, 20). The mAbs compete with fB and fH for binding to C3b, preventing formation of C3 convertase and generation of iC3b respectively; however, their precise mechanism of action is unclear (19). In an in vitro model of PNH, they inhibited deposition of C3b and abolished the haemolysis of PNH erythrocytes (21). Inhibition was due to mAb binding to fluid phase C3(H2O), preventing AP tickover and deposition of C3b on the surface of the erythrocytes. However, it is not clear whether the mAbs bind and influence the AP convertase, important particularly in the context of the pathogenic C3NeF-stabilised AP convertase. We describe here the mechanism by which 3E7/H17 binding influences AP convertase activity, whether unstabilised or stabilised by C3NeF; these data are supported by structural analysis of the mAb in complex with C3b. The data suggest that mAb H17 may be an effective therapy in disorders mediated by dysregulation of the AP C3 convertase and particularly in C3NeF-associated diseases such as DDD.
MATERIAL AND METHODS
Monoclonal antibodies and complement components
The mAb 3E7, and its humanised derivative H17, have been described previously (19). Fab fragments of H17 were produced by digestion with papain. C3 was prepared by classical chromatography using an established protocol (22); fB was affinity purified from EDTA plasma on anti-Bb (mAb JC1, in-house) HiTrap column (GE Healthcare). FD was purchased from Complement Technology, Inc (Tyler, USA). C3b was generated by AP activation by co-incubating pure C3, fB and fD as described (23), followed by anion exchange chromatography (Mono Q, GE Healthcare) and size exclusion chromatography (Superdex 200, GE Healthcare). Concentrations of pure proteins were assessed using absorbance at A280 and the following extinction coefficients: fB: 1.43 cm-1(mg/mL)-1; C3: 0.98 cm-1(mg/mL)-1; Immunoglobulin 1.4 cm-1(mg/mL)-1. Soluble recombinant decay accelerating factor (DAF) comprising domains 1-4 (sDAF) was gifted by Professor Susan Lea (University of Oxford). A strongly C3NeF-positive Ig preparation was donated by Dr Margarita López-Trascasa (Hospital Universitário de La Paz, Madrid, Spain).
Surface plasmon resonance (SPR studies)
SPR analyses were conducted at 25°C on a Biacore T100 (GE Healthcare), except for kinetic data which were performed on a Biacore 3000; kinetic/affinity data were collected at 30 μl/min and functional/blocking data at 20 μl/min. Proteins were amine-coupled to a CM5 (carboxymethylated dextran) chip (NHS/EDC coupling kit; GE Healthcare). Interactions were analysed in HEPES-buffered saline, (HBS: 10 mM HEPES pH 7.4, 150 mM NaCl) supplemented with 0.005% surfactant P20 (HBSP) and either 1 mM MgCl2 (HBSPMg) or 1mM Ni2SO4 (HBSPNi). Regeneration was achieved using 10mM sodium acetate pH4, 1M NaCl. Data were evaluated using Biaevaluation 4.1 (Biacore 3000) or Biaevaluation 1.1 software (Biacore T100). Affinity of mAb for C3b was measured by immobilising the mAb (335-400 response units (RU)) and flowing C3b in HBSP at concentrations indicated. Data were analysed using the 1:1 Langmuir binding model. To test avidity effects, C3b was densely immobilised (1200 RU) and mAb flowed.
To test whether mAb 3E7 or H17 blocked fB binding to C3b, mAb (20 μg/ml) was flowed over a C3b surface (1200 RU) in HBSPMg, followed by fB (70 μg/ml). To test whether mAb blocked convertase formation, mAb (varying concentrations as indicated) were flowed over the C3b-coated surface in HBSPMg before flowing fB (140 μg/ml) and fD (1 μg/ml). To investigate whether mAb bound preformed convertase, fB (140 μg/ml) and fD (1 μg/ml) were flowed across the C3b surface in HBSPMg, followed by immediate injection of mAb (10 μg/ml). Convertase on the surface was decayed using sDAF (two injections) and residual mAb binding was measured and compared to that bound to C3b in the absence of convertase formation. To test whether fB prevented mAb binding to C3b, high concentration of fB (350 μg/ml) was flowed in HBSNi before flowing mAb (20 μg/ml); binding of mAb to surface with and without fB was compared.
To test whether mAb affected the capacity of chip-bound convertase to cleave C3 to C3b, C3 convertase was formed by flowing a mix containing fB (as indicated) and fD (1 μg/ml) in HBSPMg in the presence or absence of C3NeF-containing IgG (IND4; 360 μg/ml). When C3NeF Ig was flowed, all normal (non-stabilised) convertase was removed by two injections of sDAF (12). Either mAb (100 μg/ml) or buffer were flowed across the surface, followed by injection of native C3. Cleavage of C3 to nascent C3b resulted in covalent deposition on the chip surface apparent as an increased RU in the sensorgram.
Haemolysis assay
Sheep erythrocytes (E) were sensitized by incubating 1 volume of 4% E (v/v) with 1 volume of 1:4000 Amboceptor (Siemens Healthcare, Marburg-Germany) for 30 min at 37°C. Antibody coated E (EA) were washed twice in complement fixation diluent (CFD; Oxoid, UK) and coated with C3b by incubating with serum depleted of fB and fH and containing the C5 inhibitor OmCI, as previously described (24). C3b-coated EA (C3b-EA) at 2% in AP buffer (5 mM sodium barbitone pH 7.4, 150 mM NaCl, 7 mM MgCl2, 10 mM EGTA pH 7.4) were incubated with factor B (1 μg/ml) and factor D (0.5 μg/ml) for 15 min at 37°C to form C3 convertase. Convertase-coated EA were incubated (20min, 25°C) with increasing concentrations of mAb 3E7 or fH in AP buffer containing 20 mM EDTA. Lysis was developed by adding 50 μl of fB/fH depleted serum diluted 1:25 in PBS/20 mM EDTA. Percent lysis was calculated as described previously.
Electron microscopy of C3b-mAb complexes
C3b (6μg) was incubated with an approximate 3-fold molar excess of H17 Fab fragments (6μg) in 25 mM Tris HCl pH 7.4, 150 mM NaCl, 10 mM DTT for 1 hour at room temperature. C3b-Fab complex was purified by gel filtration chromatography using a Superdex 200 PC 3.2/30 column (GE Healthcare). Fractions were analysed by SDS-PAGE.
Immediately after gel filtration, 5-10μl of the peak fraction containing the C3b-H17 Fab complex were diluted to 10μg/ml in the same buffer and adsorbed to glow-discharged carbon-coated copper grids and stained with 0.2 % uranyl formate. Grids were imaged on a JEOL JEM-1230 electron microscope at 100 kV. Micrographs were collected under low-dose conditions (~10 e-/Å2 per exposure) using a 4k × 4k TemCam-F416 camera (TVIPS) at 2.28 Å/pixel. A total of 7227 images of C3b-H17 were selected and binned to 4.56 Å/pixel after CTF correction. Particles were subjected to reference-free alignment and classification and refined using angular refinement methods in EMAN (25). An ab initio model for refinement was obtained using the Random Conical Tilt (RCT) method applied to those averages where the Fab was evident (Supplemental Figure S1). The high correlation between those averages obtained after angular refinement and the reference-free averages supported the correctness of the final structure. The resolution of the structure (30 Å) was estimated using the Fourier Shell Correlation method and a 0.5 correlation coefficient. A pseudo-atomic model was obtained by fitting the atomic structure of C3b (PDB ID 2I07) and a Fab (PDB ID 1H0D; chains A and B) obtained from the Protein Data Bank within the EM density of C3b-H17 using Chimera (26). The top fitting solution showed 0.89 and 0.93 cross-correlation coefficients between the X-ray and the EM structures for C3b and the Fab respectively.
EM-DATA BASE
The structure of the C3b-H17 complex has been deposited in the EM database (http://www.emdatabank.org/index.html) with accession code EMD-2553.
RESULTS
mAbs 3E7 and H17 bind with high affinity to C3b, free and in the AP convertase
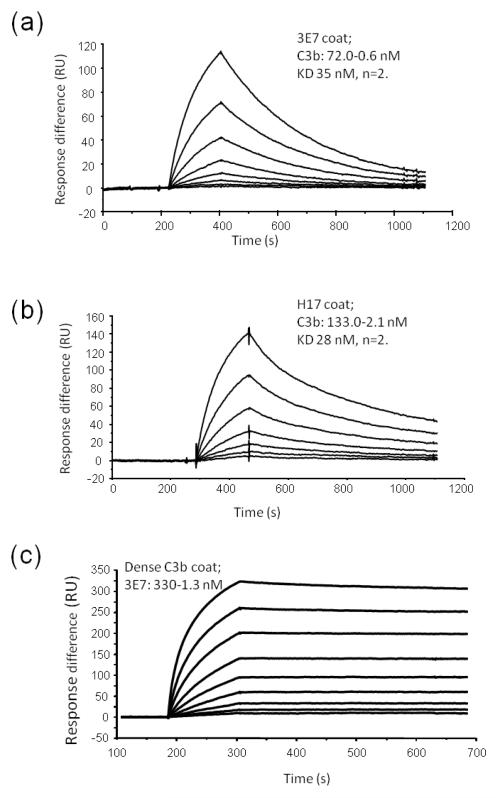
Previous studies have shown that mAb 3E7 binds to C3b and C3(H20). Affinity of the interaction was investigated here using SPR; 3E7 (Figure 1a) was immobilised on the surface of a CM5 sensor chip and C3b was flowed across. Affinity (KD) was 35 nM as determined using the Langmuir 1:1 binding model (χ2 <1.0 indicating a good fit to the model with little heterogeneity). H17 is a humanised, chimeric form of mAb 3E7 in which the mouse Fc region and CH1 domain is replaced with human IgG1 constant regions (19). Affinity of H17for C3b was similar to that of the parent antibody (28nM; figure 1b).The interaction was reversed and 3E7 was flowed across a high density C3b surface (1200RU). Affinity cannot be accurately measured in this latter orientation due to avidity effects and crosslinking of C3b via both Fab arms of 3E7, however, the data illustrate that 3E7 binds tightly to C3b-opsonised surfaces (Figure 1c).
Figure 1. Kinetics of interaction of 3E7/H17 with C3b.
(a) 3E7 or (b) H17 was immobilised on the surface of a CM5 chip and C3b flowed across at indicated concentrations (1:2 serial dilution). Binding was monitored and kinetics evaluated using the 1:1 Langmuir binding model; KD of 3E7 was 35 nM and of H17 was 28 nM (mean of two different experiments using multiple concentrations and global fitting). (c) To investigate how mAb would bind to a C3b-coated surface, C3b (1200 RU) was immobilised on the chip and 3E7 was flowed across; avidity effects markedly alter the binding kinetics, indicating the binding interaction which may take place in vivo on a tissue attacked by complement and coated in C3b.
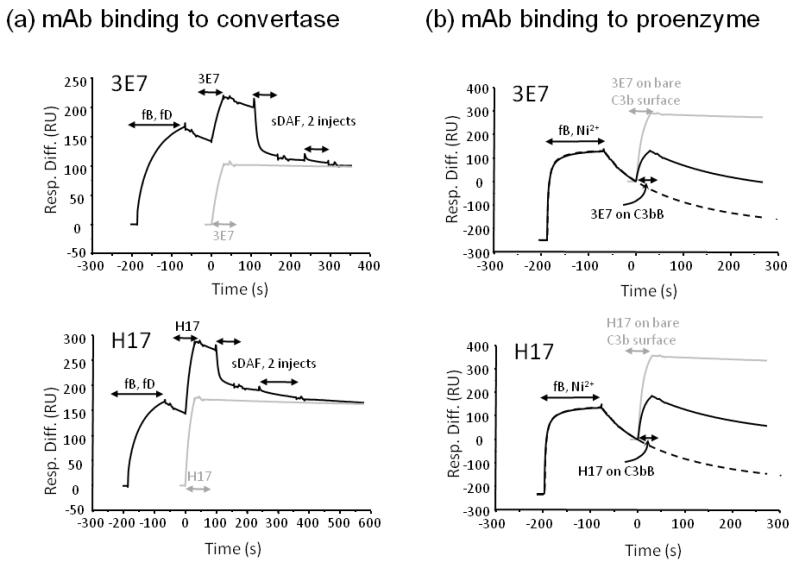
To test whether 3E7/H17 bound C3b in the AP convertase, C3bBb, binding of mAb was compared before and after convertase formation on the surface of the Biacore chip. In order to enable accurate quantitation of mAb binding to C3b in the convertase, Bb bound to C3b was removed after flowing mAb using soluble DAF (sDAF). Both mAb bound equally well to C3b and C3bBb on the surface, demonstrating that mAb binding to C3b is not hampered by the presence of Bb in the complex (Figure 2a). To investigate whether the Ba domain in intact fB interfered with mAb binding, high concentration of fB was flowed over the C3b surface in nickel-containing buffer (to stabilise the C3bB proenzyme complex). The amount of 3E7 or H17 bound to the C3b surface in the absence or presence of fB was compared; data in figure 2b illustrate that intact fB blocked binding of either mAb.
Figure 2. 3E7/H17 bind to the preformed convertase but not the proenzyme.
(a) C3 convertase was formed by flowing a mix containing fB and fD over immobilised C3b (1200RU); 3E7/H17 was immediately flowed across the convertase, followed by sDAF (two injections) to remove Bb from C3b (black line). 3E7/H17 was also flowed in the absence of preformed convertase (grey line) and the two mAb injects were aligned. The amount of mAb bound was the same irrespective of whether convertase had been preformed on the surface. (b) Binding to proenzyme was assessed by flowing high concentration of fB in Ni2+-containing buffer (to stabilise) across C3b to form proenzyme followed by mAb. Binding of mAb was compared to that achieved in the absence of fB. Solid black line: convertase followed by mAb; grey line: mAb on bare C3b surface; dashed line convertase only to illustrate proenzyme decay curve.
mAbs 3E7 and H17 prevent AP C3 convertase formation by blocking binding of fB
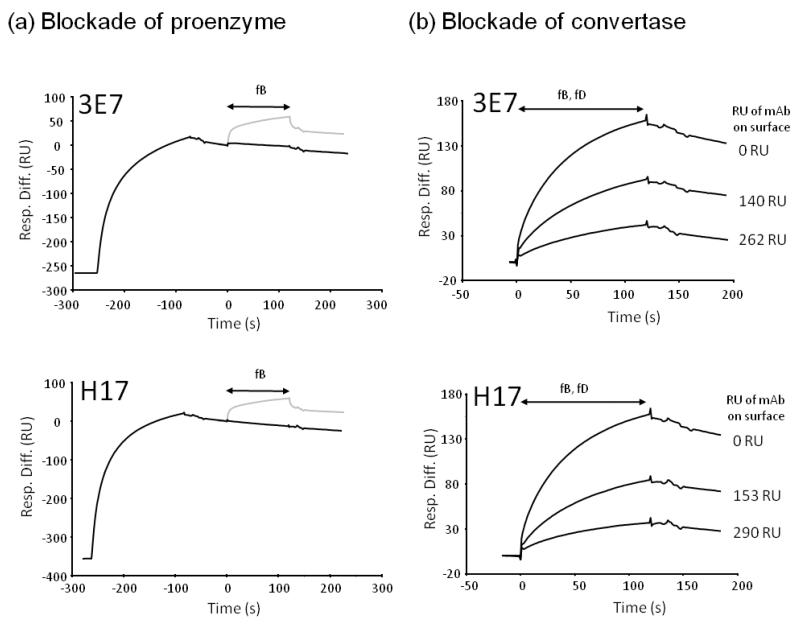
The data above demonstrate that the mAb bound C3b in the preformed C3 convertase, but not the proenzyme, implying that the mAb and the Ba domain share a binding site on C3b. It remained important to assess whether the mAb inhibited convertase formation. To analyse the effect of 3E7/H17 on fB binding to immobilised C3b, the surface was saturated with mAb prior to flowing fB. Binding of mAb to C3b completely prevented subsequent attachment of fB and formation of the proenzyme, C3bB (Figure 3a). Both mAb also prevented convertase formation in a dose-dependent manner (Figure 3b).
Figure 3. 3E7/H17 block proenzyme and convertase formation.
(a) C3b (1200 RU) was immobilised on the chip surface and fB flowed across to form proenzyme C3bB, fB was injected as indicated (grey line). Binding of fB was markedly decreased if 3E7 or H17 was first bound to the surface (black line); the two fB injections were aligned. (b) Various amounts of 3E7/H17 were bound to the C3b surface (as indicated but not shown) prior to flowing fB and fD; the amount of antibody bound to the surface was controlled through multiple injections of mAb at 10μg/ml until the required amount (in RU) was bound. The surface was regenerated to baseline between cycles and identical concentrations of fB and fD were flowed in each case. Convertase (C3bBb) formation was inhibited by the presence of mAb due to blockade of fB binding.
mAB 3E7 inhibits lysis of target cells bearing preformed C3 convertase
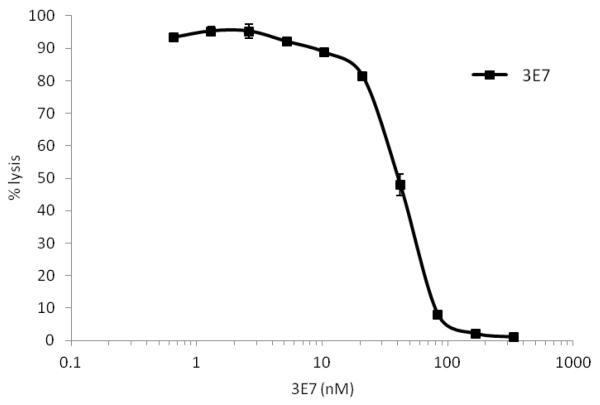
The capacity of 3E7 to inhibit lysis of cells targeted by the AP was investigated by coating sheep erythrocytes (E) with AP convertase, C3bBb. Cells were incubated with increasing concentrations of 3E7 before developing lysis with plasma/EDTA (depleted of fH). Data in Figure 4 demonstrate that 3E7 blocks AP-mediated lysis of E. Due to limited amounts of material, H17 was not tested in this assay, but DiLillo reported that mAb 3E7 and H17 were equally effective in blocking AP-mediated hemolysis of rabbit E (19).
Figure 4. 3E7 inhibits convertase formed on cell surfaces.
C3 convertase was generated on the surface of C3b-coated sheep E by adding fB and fD. The cells were incubated with increasing concentrations of 3E7 in the presence of EDTA (to prevent further convertase formation). Lysis was developed by adding NHS depleted of fB and fH in EDTA to develop MAC. Percentage of lysis was calculated as follows: percentage lysis = 100x (A410 test sample - A410 0% control)/(A410 100% lysis - A4100% control). Means and SD (n=2) are displayed.
mAb 3E7 and H17, block C3-cleaving activity of both normal and C3NeF-stabilised AP convertase
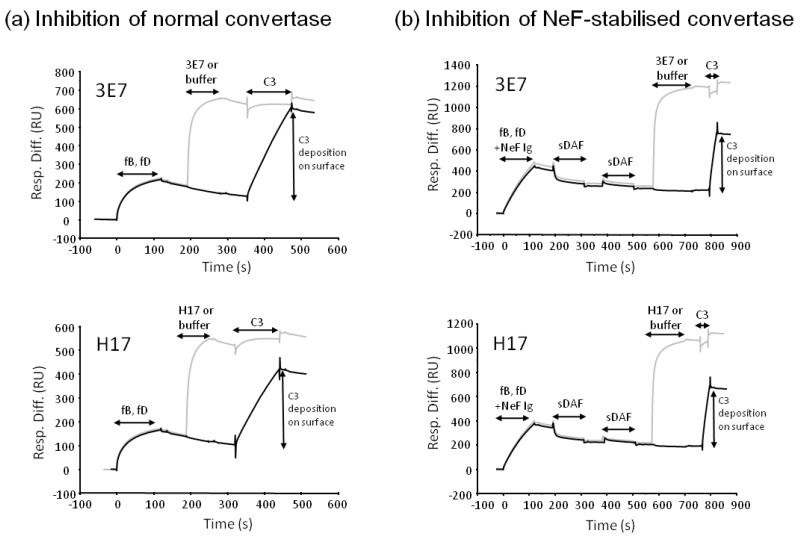
To test whether mAb binding affects C3 convertase activity, cleavage of C3 by the convertase was assessed in real time by SPR. Convertase was formed by flowing fB and fD over immobilised C3b, then mAb was flowed across to bind to free C3b and C3bBb. C3 convertase activity was determined by flowing the substrate, C3, over the surface to generate nascent C3b that binds the chip surface through its exposed thioester. In the absence of mAb, convertase-generated C3b deposition was evident on the chip surface (figure 5a); however, preincubation with mAb prevented C3b deposition, demonstrating that mAb blocked interaction of the convertase with C3. To test the influence of C3NeF, a similar experiment was carried out where C3 convertase was formed in the presence of C3NeF-containing IgG; in this case, all normal (non-stabilised) convertase was removed using sDAF prior to flowing C3. Incubation of the highly active C3NeF-stabilised convertase with 3E7 or H17 blocked C3 cleavage (Figure 5b).
Figure 5. 3E7/H17 inhibit both normal and C3NeF-stabilised convertase by preventing C3 cleavage.
(a) C3 convertase was formed on the surface of a CM5 chip by flowing fB and fD across immobilised C3b. Either 3E7 (grey sensorgram) or buffer (black sensorgram) was flowed across the surface followed by convertase substrate, C3. Binding of mAb blocked C3b deposition by the convertase, C3bBb. (b) C3NeF-containing C3 convertase was formed on the surface of the chip by flowing fB and fD across immobilised C3b in the presence of Ig-containing C3NeF. Two injections of sDAF decayed any normal convertase (non-stabilised) and ensured that only NeF-stabilised convertase remained. Either 3E7/H17 mAb (grey sensorgram) or buffer (black sensorgram) was flowed across the surface followed by convertase substrate, C3. Only a short pulse of C3 was passed over the surface due to the high activity of the C3NeF-stabilised enzyme. Binding of 3E7/H17 blocked C3b deposition.
Structural basis for complement inhibition by mAbs 3E7 and H17
To determine the molecular basis for the observed effects of 3E7 and H17, the C3b-H17 Fab complex was isolated by size-exclusion chromatography (Figure 6a) and analysed by electron microscopy (EM) (Supplemental Figure S1). Images of individual molecules of the complex were collected and reference-free 2D averages of this dataset revealed that complexes were interacting with the support film in several distinct orientations (figure 6b). Strikingly, those views where the triangular shape of C3b was more evident (“side view” from now on) revealed no apparent density that could be assigned to H17. This suggested that H17 projected perpendicular to C3b and thus was not obvious in this orientation (figure 6b). On the other hand, “tilted views” of C3b-H17 revealed a density absent in EM images of C3b alone (27), corresponding to H17 Fab.
Figure 6. Three-dimensional structure of C3b-H17 Fab.
(a) The C3b-H17 complex was purified by gel filtration chromatography. Fractions were analysed by SDS-PAGE (bottom panel) revealing two peaks, the first corresponding to the complex and the second one to the free excess H17. Peak fraction 15 was selected for its observation by EM. (b) Electron microscopy of C3b-H17. The peak fraction was observed in the microscope revealing several views of the complex, including side views with the typical structural features of C3b being evident, and tilted views where density for H17 could be visualised. A 2D average of single molecule images corresponding to each type of view is shown. Scale bar represents 10 nm. (c) Structure of C3b-H17. Two views of the pseudo-atomic model of C3b-H17 with the EM density represented as a white transparent density where the atomic structure of C3b (red colour) (PDB ID 2I07) and Fab (blue colour) have been fitted. The location of H17 on C3b clashes with the binding of the Ba fragment from fB, as determined by the location of the three SCRs in the crystal structure of C3Bb (32). Scale bar represents 3 nm.
A 30 Å resolution structure of the C3b-H17 Fab complex was obtained by angular refinement of the images collected (figure 6c). C3b-H17 revealed two distinct regions of density that could be readily assigned as C3b and H17 after computational fitting of the atomic structures of C3b (PDB ID 2I07) (28) and a Fab (PDB ID 1H0D) within the density of the EM structure (figure 6c). The crystal structures of C3b and Fab perfectly matched within the EM structure (cross-correlation coefficients of 0.89 and 0.93 for C3b and Fab respectively), thus annotating every domain in the complex. The exact epitope mapped by the Fab could not be determined at this resolution, but the pseudo-atomic model of C3b-H17 Fab revealed that H17 bound a site placed within the C3b MG6-MG7 region which is exposed as a consequence of the C3 to C3b conformational change; this site overlapped with the Ba binding site (Fig. 7). As predicted from the single molecule images (Figure 6b, Supplemental Figure S1), H17 projects outwards and perpendicular to C3b (Figure 6c).
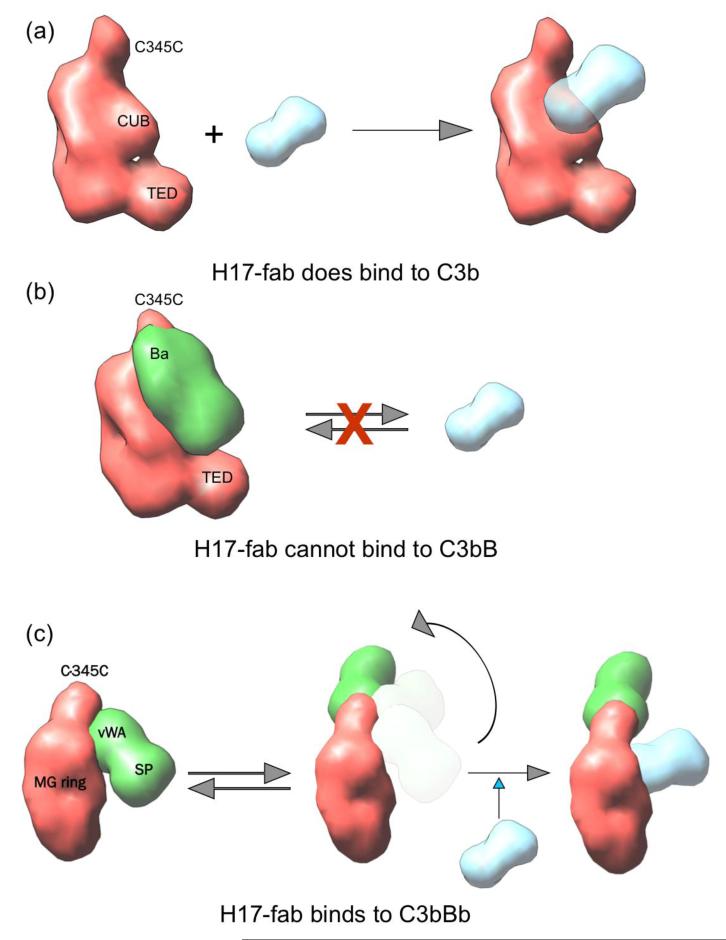
Figure 7. Model for H17 activity.
(a) A cartoon representing a side view of C3b shows that H17 can access C3b. (b) H17 cannot bind to C3bB due to steric clashes with the Ba domain. (c) H17 partially clashes with Bb in the structure of the C3bBb convertase (PDB ID 2WIN) (41) which would impede H17 binding to the AP C3 convertase. We hypothesise that the flexibility of the C345C-Bb module in the C3bBb convertase could allow H17 binding by displacing the Bb region (27). In (c), C3bBb is shown with the MG ring facing the viewer for clarity. The locations of C345C, the MG ring, and the TED domain in each cartoon model are labelled.
DISCUSSION
The AP C3 convertase, C3bBb, is the key enzyme of the complement system, delivering tickover activation and amplification that are essential to roles in immune defence and waste disposal. AP activation and dysregulation also contribute to many disease processes (7), this can be caused by polymorphisms and mutations in AP proteins or by autoantibodies against AP components (6). C3NeF, autoantibodies that bind the AP C3 convertase, cause AP dysregulation by inhibiting both spontaneous and accelerated decay (12). To date, no specific therapies to counter the effects of C3Nef have been developed.
The mAb 3E7 and the humanised derivative, H17, were previously shown to bind C3b, iC3b and C3(H2O) but not native C3 (19). Here we confirmed that binding to C3b was high affinity and stable. Binding of the mAb to C3b prevented the binding of fB to form the proenzyme, C3bB, and thus stopped further AP activation. The mAb also efficiently bound pre-formed C3bBb, implying that the mAb binding site overlapped the Ba-binding region on C3b. The importance of Ba binding in formation of the proenzyme has been emphasised in several earlier studies (27, 29, 30). Importantly, we also found that 3E7/H17 binding to the C3 convertase abrogated its C3 cleaving capacity and prevented further complement activation. The mAbs therefore have multiple effects as inhibitors of AP convertase formation and blockers of AP convertase C3-cleaving capacity. The compound effect is powerful inhibition of AP convertase formation and function, raising the possibility that the mAbs could play roles in therapy of AP-driven diseases. While binding of 3E7/H17 prevents fH binding (data not shown), this will have no pathogenic effect as we show that the mAb-bound convertase is non-functional.
C3NeF stabilise the AP convertase and are likely pathogenic because the convertase continues to cleave C3, causing unregulated activation of the AP. To test whether 3E7/H17 could be used as therapy in diseases associated with C3NeF, the C3NeF-stabilised convertase was generated using IgG isolated from the serum of a patient with a proven high titre pathological C3NeF. C3 was added as substrate to test whether bound 3E7/H17 blocked C3-cleaving activity; the mAb completely blocked C3 cleavage by the C3NeF-stabilised enzyme. The mAb will therefore inhibit the activity of C3NeF in two ways; first by preventing the formation of C3bBb by binding nascent C3b and second by binding to the preformed NeF-stabilised convertase and inhibiting C3-cleaving activity. It should be noted that, although all C3NeF stabilise the convertase and most prevent accelerated decay, they are heterogeneous between, and possibly within, patients (12). So far, the inhibitory effects of 3E7/H17 have only been demonstrated for one C3NeF. It is entirely possible that some C3NeF-stabilised C3 convertases will be resistant to mAb inhibition; however, even in this eventuality, the mAb will still inhibit AP activation by preventing fB binding to C3b and thus convertase formation.
In order to shed further light on the mechanism by which 3E7/H17 affected convertase formation and activity, complexes of C3b and a Fab fragment of H17 were analysed using high resolution EM. The components of the complex were obvious in the images obtained with the Fab binding in the vicinity of the MG6 and MG7 domains of C3b. The transition from C3 to C3b is accompanied by major structural rearrangements that include a substantial shift of the MG7 domain, likely explaining the finding that the mAb binds C3b, iC3b and C3(H2O), but not native C3 (28). The structure of the proenzyme C3bB (PDB 2XWJ) shows that the three SCRs that comprise Ba interact with the α’NT and CUB domains of C3b and occlude binding sites for decay accelerators, fH and DAF (31, 32). These Ba-binding domains are immediately adjacent to MG6 and MG7; indeed, α’NT and MG6 form one continuous exposed region that includes binding sites for fB, fH and CR1 (28, 32). Binding of the Fab thus likely: i) impedes subsequent binding of fB by occluding the Ba binding site; and ii) when bound to the convertase, directly interferes with cleavage of substrate. Binding of the mAb could cause distortion of the MG ring in C3b thus preventing C3 binding; alternatively, it might displace the Bb serine protease domain away from the substrate (C3) thus preventing cleavage (Figure 7). It is also possible that, through binding to MG7, the antibody prevents the substrate, C3, from binding to the convertase. The crystal structure of CVF with C5 implicates this domain in inter-molecular interactions and a disease-associated two amino acid deletion within the MG7 domain has been shown to prevent substrate binding (33, 34). The binding site(s) of C3NeF on the C3 convertase have yet to be mapped; our data demonstrate that, at least for the C3NeF tested, these are distinct from the mAb and substrate binding domains described above.
Antibody based therapeutics represent one of the fastest growing sectors of the pharmaceutical industry and, with the rise of the anti-C5 mAb Eculizumab™, are now impacting on complement-driven diseases (35, 36). Different diseases may require that different steps of the complement cascade are blocked by targeting a specific component (37, 38). Eculizumab abolishes the generation of C5a and MAC formation, thereby reducing tissue damage caused by activation of the terminal pathway. Additional strategies for treatment include upstream inhibition of C3 activation, which can be mediated by 3E7/H17, as well as the newly developed CR2-fH chimeric construct, TT30 (21, 39). Several other mAb targetting AP components (fD, fB) are in development (37), although none have yet been tested in C3NeF-driven pathologies. Inhibiting the AP convertase carries the risk of increasing susceptibility to bacterial infection; however, judicious use of prophylactic antibiotics, or administration during acute flares of disease, should minimise this risk. The humanised mAb, H17, shown here to be effective in controlling the AP C3 convertase, is an exciting new candidate for therapy in many diseases driven by dysregulation of the C3 convertase, including aHUS and PNH. The amount of therapeutic antibody needed to block a neoepitope on C3b will be far less than that required to block native C3, although the mAb can also bind to C3(H20) (19), which could act as a decoy and raise the effective dose required for blocking pathologic complement activation. The exact dosing of antibody will depend on the nature of the disease, fluid phase versus surface, and the extent of complement dysregulation at the level of C3. The demonstration that the mAb bind a highly active C3NeF-stabilised convertase and inhibit its capacity to cleave C3 opens the possibility that the mAb could provide the first specific therapy for DDD, a disease that currently lacks effective treatment options; 80% DDD cases are associated with presence of C3NeF. The mAb are specific for the human C3 convertase and thus cannot be tested in rodent models of DDD. They do, however, inhibit AP activation in primate serum (21), raising the possibility that a primate model of DDD could be developed to test the therapeutic effects of the mAb (40).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Leslie Casey and EluSys Therapeutics for generously providing mAb H17. This work was presented in abstract form at the 23rd International complement Workshop, New York, 2010.
Footnotes
This work was supported by Kidneeds, USA (to CLH and BPM), Medical Research Council (G0701298; CLH and BPM), Ramón Areces Foundation (to O.L.), the Autonomous Region of Madrid (S2010/BMD-2316 to SRdC and OL) and the Spanish Ministry of Science and Innovation (SAF2011-22988 to OL, SAF2011-26583 to SRdC). DPC was a Kidney Research UK Post-Doctoral Fellow (PDF5/2010). O.L. is additionally supported by the “Red Temática de Investigación Cooperativa en Cáncer (RTICC)” from the “Instituto de Salud Carlos III” (RD06/0020/1001). SRdeC is also supported by the Fundación Renal Iñigo Alvarez de Toledo, the Ciber de Enfermedades Raras (CIBERER) and the 7FP European Union project EURenOmics.
ABBREVIATIONS: AP, alternative pathway; fB, factor B; fD, factor D; fH, factor H; DDD, dense deposit disease; aHUS, atypical hemolytic uremic syndrome; PNH, paroxysmal nocturnal hemoglobinurea; C3NeF, C3 nephritic factor; SPR, surface plasmon resonance; EM, electron microscopy; RU, resonance units
REFERENCES
- 1.Walport MJ. The Roche Rheumatology Prize Lecture. Complement deficiency and disease. British Journal of Rheumatology. 1993;32:269–273. doi: 10.1093/rheumatology/32.4.269. [DOI] [PubMed] [Google Scholar]
- 2.Morgan BP. The complement system: an overview. Methods Mol Biol. 2000;150:1–13. doi: 10.1385/1-59259-056-X:1. [DOI] [PubMed] [Google Scholar]
- 3.Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Formation of the initial C3 convertase of the alternative complement pathway. Acquisition of C3b-like activities by spontaneous hydrolysis of the putative thioester in native C3. J Exp Med. 1981;154:856–867. doi: 10.1084/jem.154.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan BP, Harris CL. Complement Regulatory Proteins. Academic Press; London: 1999. [Google Scholar]
- 5.Morgan BP, Meri S. Membrane proteins that protect against complement lysis. Springer Semin Immunopathol. 1994;15:369–396. doi: 10.1007/BF01837366. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez de Cordoba S, Tortajada A, Harris CL, Morgan BP. Complement dysregulation and disease: from genes and proteins to diagnostics and drugs. Immunobiology. 2012;217:1034–1046. doi: 10.1016/j.imbio.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 8.Pickering MC, Cook HT. Translational mini-review series on complement factor H: renal diseases associated with complement factor H: novel insights from humans and animals. Clin Exp Immunol. 2008;151:210–230. doi: 10.1111/j.1365-2249.2007.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris CL, Heurich M, Rodriguez de Cordoba S, Morgan BP. The complotype: dictating risk for inflammation and infection. Trends Immunol. 2012;33:513–521. doi: 10.1016/j.it.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitzer RE, Vallota EH, Forristal J, Sudora E, Stitzel A, Davis NC, West CD. Serum C’3 lytic system in patients with glomerulonephritis. Science. 1969;164:436–437. doi: 10.1126/science.164.3878.436. [DOI] [PubMed] [Google Scholar]
- 11.Smith RJH, Harris CL, Pickering MC. Dense deposit disease. Mol Immunol. 2011;48:1604–1610. doi: 10.1016/j.molimm.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paixao-Cavalcante D, Lopez-Trascasa M, Skattum L, Giclas PC, Goodship TH, de Cordoba SR, Truedsson L, Morgan BP, Harris CL. Sensitive and specific assays for C3 nephritic factors clarify mechanisms underlying complement dysregulation. Kidney Int. 2012;82:1084–1092. doi: 10.1038/ki.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisman HF, Bartow T, Leppo MK, Boyle MP, Marsh HC, Jr., Carson GR, Roux KH, Weisfeldt ML, Fearon DT. Recombinant soluble CR1 suppressed complement activation, inflammation, and necrosis associated with reperfusion of ischemic myocardium. Transactions of the Association of American Physicians. 1990;103:64–72. [PubMed] [Google Scholar]
- 14.Kaplan M. Eculizumab (Alexion) Curr Opin Investig Drugs. 2002;3:1017–1023. [PubMed] [Google Scholar]
- 15.Thurman JM, Kraus DM, Girardi G, Hourcade D, Kang HJ, Royer PA, Mitchell LM, Giclas PC, Salmon J, Gilkeson G, Holers VM. A novel inhibitor of the alternative complement pathway prevents antiphospholipid antibody-induced pregnancy loss in mice. Mol Immunol. 2005;42:87–97. doi: 10.1016/j.molimm.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 16.Sahu A, Kay BK, Lambris JD. Inhibition of human complement by a C3-binding peptide isolated from a phage-displayed random peptide library. J Immunol. 1996;157:884–891. [PubMed] [Google Scholar]
- 17.Ricklin D, Lambris JD. Compstatin: a complement inhibitor on its way to clinical application. Adv Exp Med Biol. 2008;632:273–292. doi: 10.1007/978-0-387-78952-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paixao-Cavalcante D, Hanson S, Botto M, Cook HT, Pickering MC. Factor H facilitates the clearance of GBM bound iC3b by controlling C3 activation in fluid phase. Mol Immunol. 2009;46:1942–1950. doi: 10.1016/j.molimm.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiLillo DJ, Pawluczkowycz AW, Peng W, Kennedy AD, Beum PV, Lindorfer MA, Taylor RP. Selective and efficient inhibition of the alternative pathway of complement by a mAb that recognizes C3b/iC3b. Mol Immunol. 2006;43:1010–1019. doi: 10.1016/j.molimm.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Pawluczkowycz AW, Lindorfer MA, Waitumbi JN, Taylor RP. Hematin promotes complement alternative pathway-mediated deposition of C3 activation fragments on human erythrocytes: potential implications for the pathogenesis of anemia in malaria. J Immunol. 2007;179:5543–5552. doi: 10.4049/jimmunol.179.8.5543. [DOI] [PubMed] [Google Scholar]
- 21.Lindorfer MA, Pawluczkowycz AW, Peek EM, Hickman K, Taylor RP, Parker CJ. A novel approach to preventing the hemolysis of paroxysmal nocturnal hemoglobinuria: both complement-mediated cytolysis and C3 deposition are blocked by a monoclonal antibody specific for the alternative pathway of complement. Blood. 2010;115:2283–2291. doi: 10.1182/blood-2009-09-244285. [DOI] [PubMed] [Google Scholar]
- 22.Harrison RA, Herzenberg LA, Blackwell C. Purification, assay and characterization of complement proteins from plasma. In: Herzenberg LA, Weir DM, editors. Weir’s handbook of experimental immunology. II, Cell surface and messenger molecules of the immune system. Blackwell Science; Cambridge, MA: 1996. pp. 75.71–75.50. [Google Scholar]
- 23.Heurich M, Martinez-Barricarte R, Francis NJ, Roberts DL, Rodriguez de Cordoba S, Morgan BP, Harris CL. Common polymorphisms in C3, factor B, and factor H collaborate to determine systemic complement activity and disease risk. Proc Natl Acad Sci U S A. 2011;108:8761–8766. doi: 10.1073/pnas.1019338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tortajada A, Montes T, Martinez-Barricarte R, Morgan BP, Harris CL, de Cordoba SR. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum Mol Genet. 2009;18:3452–3461. doi: 10.1093/hmg/ddp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludtke SJ. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol Biol. 2010;673:157–173. doi: 10.1007/978-1-60761-842-3_9. [DOI] [PubMed] [Google Scholar]
- 26.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Torreira E, Tortajada A, Montes T, Rodriguez de Cordoba S, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc Natl Acad Sci U S A. 2009;106:882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 29.Hourcade DE, Wagner LM, Oglesby TJ. Analysis of the short consensus repeats of human complement factor B by site-directed mutagenesis. J Biol Chem. 1995;270:19716–19722. doi: 10.1074/jbc.270.34.19716. [DOI] [PubMed] [Google Scholar]
- 30.Pryzdial EL, Isenman DE. Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J Biol Chem. 1987;262:1519–1525. [PubMed] [Google Scholar]
- 31.Torreira E, Tortajada A, Montes T, Rodriguez de Cordoba S, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc Natl Acad Sci U S A. 2009;106:882–887. doi: 10.1073/pnas.0810860106. doi: 810.1073/pnas.0810860106. Epub 0810862009 Jan 08108601091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Barricarte R, Heurich M, Valdes-Canedo F, Vazquez-Martul E, Torreira E, Montes T, Tortajada A, Pinto S, Lopez-Trascasa M, Morgan BP, Llorca O, Harris CL, Rodriguez de Cordoba S. Human C3 mutation reveals a mechanism of dense deposit disease pathogenesis and provides insights into complement activation and regulation. J Clin Invest. 2010;120:3702–3712. doi: 10.1172/JCI43343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laursen NS, Andersen KR, Braren I, Spillner E, Sottrup-Jensen L, Andersen GR. Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 2011;30:606–616. doi: 10.1038/emboj.2010.341. doi: 610.1038/emboj.2010.1341. Epub 2011 Jan 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber J, Fakhouri F, Roumenina LT, Loirat C, Fremeaux-Bacchi V. Use of eculizumab for atypical haemolytic uraemic syndrome and C3 glomerulopathies. Nat Rev Nephrol. 2012;8:643–657. doi: 10.1038/nrneph.2012.214. [DOI] [PubMed] [Google Scholar]
- 36.Hill A, Hillmen P, Richards SJ, Elebute D, Marsh JC, Chan J, Mojcik CF, Rother RP. Sustained response and long-term safety of eculizumab in paroxysmal nocturnal hemoglobinuria. Blood. 2005;106:2559–2565. doi: 10.1182/blood-2005-02-0564. Epub 2005 Jun 2528. [DOI] [PubMed] [Google Scholar]
- 37.Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: therapeutic interventions. J Immunol. 2013;190:3839–3847. doi: 10.4049/jimmunol.1203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risitano AM, Notaro R, Pascariello C, Sica M, del Vecchio L, Horvath CJ, Fridkis-Hareli M, Selleri C, Lindorfer MA, Taylor RP, Luzzatto L, Holers VM. The complement receptor 2/factor H fusion protein TT30 protects paroxysmal nocturnal hemoglobinuria erythrocytes from complement-mediated hemolysis and C3 fragment. Blood. 2012;119:6307–6316. doi: 10.1182/blood-2011-12-398792. [DOI] [PubMed] [Google Scholar]
- 40.Gozalo AS, Cheng LI, St Claire ME, Ward JM, Elkins WR. Pathology of captive moustached tamarins (Saguinus mystax) Comp Med. 2008;58:188–195. [PMC free article] [PubMed] [Google Scholar]
- 41.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.