Congenital heart disease is the most common birth defect. Its genetic component is complex, involving modifier genes, de novo mutations, copy number variants, common variants, and noncoding mutations.
Abstract
Congenital heart disease (CHD) is the most common birth defect. Despite considerable advances in care, CHD remains a major contributor to newborn mortality and is associated with substantial morbidities and premature death. Genetic abnormalities appear to be the primary cause of CHD, but identifying precise defects has proven challenging, principally because CHD is a complex genetic trait. Mainly because of recent advances in genomic technology such as next-generation DNA sequencing, scientists have begun to identify the genetic variants underlying CHD. In this article, the roles of modifier genes, de novo mutations, copy number variants, common variants, and noncoding mutations in the pathogenesis of CHD are reviewed.
Congenital heart disease (CHD) is the most common developmental defect, occurring in almost 3% of neonates when including bicuspid aortic valve (BAV) patients. Numerous studies have shown that the overall incidence of CHD is relatively similar across populations (after accounting for the ascertainment bias associated with improved imaging over the past decades). The distribution of heart lesions is broadly similar with a few notable examples such as the relatively greater incidence of right-sided over left-sided defects among Asians (Sung et al. 1991; Pradat et al. 2003; Wu et al. 2010).
The genetic architecture of CHD is incompletely understood. It has been long appreciated that chromosomal defects and single-gene disorders can cause CHD, often in the context of a multisystem disease. Common examples of aneuploidy with CHD would include trisomy 21, for which ∼50% of individuals will have atrioventricular canal defects and/or tetralogy of Fallot (TOF), and Turner syndrome (45,X), for which aortic coarctation is overrepresented. Examples of Mendelian disorders include autosomal dominant conditions with CHD and pleiomorphic extracardiac abnormalities such as Noonan and Holt–Oram syndromes and rare autosomal dominant isolated CHD such as caused by GATA4 and NKX2.5 mutations. The most common segmental aneuploidy is the 22q11 microdeletion, usually arising de novo, which accounts for a substantial 34% of truncus arteriosus and 16% of TOF (Goldmuntz et al. 1998). Nonetheless, these known genetic causes of CHD are estimated to account for <20% of CHD.
Epidemiologic studies strongly suggest genetic factors as the predominant cause of CHD, although environmental exposures are also relevant. The best population-based study of CHD was performed in Denmark, in which a nationwide medical registry enabled nearly complete ascertainment. In this study, the relative risk for any form of CHD in first-degree relatives was 3.2 (Oyen et al. 2009). The recurrence risks of the same form of CHD varied by lesion. After exclusion of chromosomal defects, the population-associated risk given a positive family history of CHD was 4.2%. Parental consanguinity is associated with a 2- to 3-fold increased offspring risk of CHD, likely because of the shared genetic variants among parents (Nabulsi et al. 2003; Yunis et al. 2006). Finally, family studies of left-sided forms of CHD ranging from BAV to hypoplastic left heart syndrome have indicated a high degree of heritability (Cripe et al. 2004; McBride et al. 2005; Hinton et al. 2007).
In the 1960s, James Nora proposed a multifactorial model for CHD, in which polygenic inheritance combined with environmental factors would underlie cases not readily explained by aneuploidy, Mendelian genetic mutations, or teratogens (Nora 1968). Although broadly popular for complex genetic traits, the multifactorial model has substantial problems when applied to CHD. Specifically, most of the predictions that flow from polygenic inheritance proved untrue for nearly all forms of CHD, patent ductus arteriosus in full-term newborns being the singular exception. In the 1980s, the groundbreaking work of Ruth Whittemore, who studied the progeny of women with CHD, further invalidated the multifactorial model (Whittemore et al. 1982) by documenting a 16% CHD recurrence rate, often with the same heart lesion, much greater than the 2%–3% rate expected for polygenic inheritance. Of interest, the recurrence risk for fathers with CHD is substantially lower. This sex-specific transmission risk remains largely unexplored.
In this article, we will review the role of genetic defects underlying CHD, focusing particularly on recent findings using relatively new genetic and genomic approaches.
MODIFIER GENES
Many monogenes or copy number variants (CNVs) associated with CHD are incompletely penetrant and/or phenotypically heterogeneous. Although complexity can result from nongenetic factors (environmental exposures/epigenetics, stochastic effects), modifier genes are likely to be relevant. Recent efforts, particularly studies with animal models, have begun to address this latter possibility.
NKX2.5 mutations are associated with a range of lesions, including atrial septal defects, (ASDs), conotruncal defects, and hypoplastic left heart syndrome (HLHS), and often cause later-onset atrioventricular conduction block. Similarly, mice heterozygous for a null Nkx2.5 allele on a C57BL/6 background display a range of CHD, most commonly ASDs and ventricular septal defects (VSDs) (Winston et al. 2010). F1 offspring resulting from the outcrossing of C57BL/6 mice to FVB/N or AJ mice have significantly lower incidence of CHD. F2 mice from F1 intercrosses or N2 back-crosses to the parental strains have higher CHD rates (ASD 5%–12% and VSD 12%–15% vs. ASD 7%–8% and VSD 0%–1% in the F1 pups). The analyses suggested the presence of genetic modifiers of Nkx2.5 on ASD and VSD phenotypes. Using a C57BL/6×FVB/N F1×F1 intercross (Winston et al. 2012), linkage analysis for membranous VSDs identified three loci with moderate-sized effects (odd ratios [ORs] of 1.4 to 2.2) (Fig. 1). For muscular VSDs, there was no significant linkage, and two of the three loci for membranous VSDs were excluded. Of interest, a modest maternal age effect (OR = 1.1) was also observed. Future work to identify the precise genetic alterations mediating these modifying effects in mice might be relevant to NKX2.5-related CHD in patients.
Figure 1.
Genetic linkage analyses for loci that modify membranous and muscular ventricular septal defect (VSD) susceptibility in Nkx2-5+/− animals from the C57BL/6×FVB/N F2 population. An example of each VSD type is shown. (A) At least three significant membranous VSD modifier loci exist on chromosomes 6, 8, and 10. Genome-wide significance thresholds are indicated by the dotted lines (α = 0.001 and 0.05). (B) Genetic linkage analysis for muscular VSD modifier loci reveals a significant overlap of the chromosome 6 peak with a membranous VSD locus. The significance thresholds shown were determined by permutation of genotypes on chromosomes 6, 8, and 10, which contain the membranous VSD modifier loci. N = 233 membranous VSDs, 80 muscular VSDs, and 284 structurally normal hearts. (From Winston et al. 2012; reprinted, with permission, from Circulation: Cardiovascular Genetics © 2012.)
A similar strategy was used in mice heterozygous for a null elastin allele (Eln+/−) (Kozel et al. 2011). In humans, the ELN gene resides within the microdeletion associated with Williams syndrome. Haploinsufficiency for elastin is strongly implicated in the aortic pathology in Williams syndrome as individuals with ELN point mutations have the related, but more restricted phenotype, familial supravalvular stenosis. Linkage analyses with intercrossed Eln+/− mice revealed three loci for aortic size and systemic hypertension. Of interest, one of the loci resides near the Eln locus, potentially affecting the Williams syndrome critical region.
A candidate gene resequencing approach of 26 genes in individuals with Down syndrome with either atrioventricular septal defects (AVSDs) or without CHD (Ackerman et al. 2012) identified potentially damaging variants in almost 20% of individuals with Down syndrome with AVSDs but in only 3% of those with Down syndrome without CHD. Six genes were specifically implicated: COL6A1, COL6A2, CRELD1, FBLN2, FRZB, and GATA5. Pathway analysis with these six genes implicated VEGF-A signaling, which was known to have a role in atrioventricular valvuloseptal morphogenesis. The findings in this study provide an initial proof of principal for using individuals with a sensitized genetic background like trisomy 21 that predisposes to CHD to explore additional genetic variants mediating expression of CHD phenotypes.
Further interesting insights into the genetic complexity of CHD were gained through mouse N-ethyl-N-nitrosourea (ENU) mutagenesis screens (Kamp et al. 2010). Three-generation backcrosses were conducted and lines with perinatal lethality were assessed for CHD. This breeding approach favors autosomal recessive mutations. The first CHD gene identified from these ENU screens was Ift172, which encodes an intraflagellar transport protein (Friedland-Little et al. 2011). The mice with the Ift172 mutation showed a phenotype of VACTERL (vertebral defects, anal atresia, cardiac defects, tracheo-esophageal fistula, renal anomalies, and limb abnormalities) with hydrocephalus and CHD. Efforts to identify other genes are currently in progress. The interesting finding, particularly relevant here, is that ∼75% of the lines with CHD had only a single G3-affected pup, despite the assessment of at least 50 animals. Of note, control crosses in which the founders had not been exposed to ENU produced no third-generation offspring with CHD. Possible explanations for this finding include modifier genes because two of the three ENU screens were performed using mice with mixed genetic backgrounds, multigenic inheritance resulting from the ENU-induced mutational load, or fetal demise of some of the CHD-affected animals. Future studies, such as additional breeding to various mouse strains, would distinguish those possibilities and might facilitate identification of modifier genes when present.
DE NOVO MUTATIONS
Another form of genetic complexity is genetic lesions that arise de novo such that the parents do not harbor the mutation and family history is negative for CHD. For highly penetrant mutations underlying severe forms of CHD, this mechanism is more likely a result of low reproductive fitness because the modern era provides strong negative selection against the accumulation of these mutations in the population.
Through recent advances in molecular genetic technologies, it is now possible to sequence the ∼2% of the human genome that contains the coding regions for all genes (called the exome), representing ∼180,000 exons and 30 megabases (Mb), in a relatively rapid and affordable manner. Although exome sequencing was initially used to discover mutations underlying Mendelian disorders, current efforts are increasingly focusing on unraveling complex genetic traits.
The Pediatric Cardiac Genomics Consortium (PCGC) (Gelb et al. 2013), a National Heart, Lung, and Blood Institute–funded research collaborative, recently completed a first-of-a-kind study to determine the role of de novo mutations in the etiology of severe forms of CHD (Zaidi et al. 2013). Exome sequencing was performed for 362 parent–offspring trios in which the offspring had a sporadic conotruncal defect, left ventricular outflow track obstructive lesion, or heterotaxy, and was compared with comparable data from 264 control trios. Although the overall rate of de novo point and small insertion/deletion (indel) changes was equivalent between CHD cases and controls, there was an excess burden of protein-altering mutations in genes highly expressed in the heart during heart development (OR of 2.53). These excess mutations accounted for 10% of CHD cases and led to the estimate that approximately 400 genes underlie these cardiac lesions. After filtering to retain only variants most likely to be deleterious (nonsense, splice site, and frameshift defects), the burden among CHD cases increased, attaining an OR of 7.50.
Next, the PCGC investigators asked whether the burden of de novo protein-altering mutations among the CHD cases preferentially targeted particular biologic processes (Zaidi et al. 2013). Indeed, they observed a highly significant enrichment of mutations among genes encoding proteins relevant for chromatin biology, specifically the production, removal, or reading of methylation of Lys4 of histone 3 (H3K4me) (Fig. 2). The phenotypes of the eight subjects harboring H3K4me de novo mutations was diverse, both with respect to the form of CHD and extracardiac manifestations. In addition, two independent de novo mutations were identified in SMAD2, which encodes a protein with relevance for demethylation of Lys27 of histone 3 (H3K27me). SMAD2 contributes to the development of the left–right body axis; both subjects harboring SMAD2 mutations had dextrocardia with unbalanced complete atrioventricular canal defects with pulmonic stenosis. Although the contribution of chromatin remodeling to cardiovascular development generally and to certain rare genetic syndromes with CHD like Kabuki syndrome had been recognized previously, this study exposed a far broader role for H3K4 methylation in CHD pathogenesis. The finding also suggests a fascinating potential link to other birth defects as de novo chromatin-remodeling mutations have also been implicated in autism (O’Roak et al. 2012).
Figure 2.
De novo mutations in the H3K4 and H3K27 methylation pathways. Nucleosome with histone octamer and DNA, with H3K4 methylation bound by CHD7; H3K27 methylation and H2BK120 ubiquitination is shown. Genes mutated in CHD that affect the production, removal, and reading of these histone modifications are shown; genes with damaging mutations are shown in red and those with missense mutations are shown in blue. SMAD2 (2) indicates there are two patients with a mutation in this gene. Genes whose products are found together in a complex are enclosed in a box. (From Zaidi et al. 2013; reprinted, with permission, from the authors.)
COPY NUMBER VARIATION
Copy number variation (CNV), or gains or losses of contiguous DNA ranging in size from 1 kb to several Mbs, affects ∼10% of the human genome (Redon et al. 2006). CNVs are typically detected on a genome-wide basis using single-nucleotide polymorphism (SNP) microarrays or array comparative genomic hybridization. Significant challenges remain in differentiating pathologic CNVs from benign polymorphic ones. Nevertheless, it became clear soon after their discovery that pathologic CNVs contribute significantly to the pathogenesis of certain phenotypes such as autism and schizophrenia.
Thienpont and colleagues (2007) showed the importance of pathologic CNVs for CHD. The studies published to date have often focused on particular subpopulations of individuals with CHD, often based on cardiac anatomy. Certain CNVs are recurrent and at higher frequency in individuals with CHD, often ones that are also associated with extracardiac abnormalities.
The role of CNVs for TOF has been studied in two large studies. In one, 10% of 114 patients with TOF were found to harbor rare de novo CNVs (Greenway et al. 2009). Next, nearly 400 probands with TOF were assessed for CNVs at nine genomic loci and were identified in 5%. Several CNVs were recurrent with both gain and loss at chromosome 1q21.1 being observed in 1% of cases. Of note, 1q21.1 CNVs have previously been associated with other phenotypes such as autism, schizophrenia, and intellectual disabilities.
In the second large study of TOF, more than 400 adults with TOF with or without pulmonary atresia, but without known genetic abnormalities like the 22q11 deletion, were studied (Silversides et al. 2012). Large, rare CNVs (>500 kb) were more prevalent in cases compared with controls (8.8% vs. 4.3%) with mostly genomic gains. Especially when the CNVs contained genes, the subjects were more likely to have extracardiac abnormalities. Several of the CNVs had been previously implicated in diseases including duplications at 1q21.1, which were observed in 1.2% of CHD patients, a 16p11.2 duplication, and a 22q11.21 duplication.
Two studies focused on CNVs in individuals with defects of the left ventricular outflow tract (Hitz et al. 2012; Payne et al. 2012). In a small study of children with HLHS, CNVs, predominantly small lesions, were more common among subjects than controls (Payne et al. 2012). The frequency of rare CNVs, however, did not vary between the groups. In a study that included individuals with a broad range of left-sided cardiac lesions with a focus on those from families with more than one affected individual, the overall burden of CNVs was not increased in cases over controls, but a number of rare CNVs were identified in cases only, none of which was recurrent in the study (Hitz et al. 2012). Of interest, these rare CNVs were found to harbor genes relevant for angiogenesis. After filtering, the investigators identified 25 putative candidate genes for left-sided heart defects.
The role of CNVs in AVSDs, both in patients with and without Down syndrome, was investigated (Priest et al. 2012). None of the 29 individuals with AVSD without Down syndrome had a pathologic CNV on chromosome 21 and only two harbored rare CNVs elsewhere in the genome, deletions of 1–1.5 Mb that were deemed of uncertain significance. None of the 50 individuals with Down syndrome with AVSD had a CNV of clear significance. The investigators concluded that larger rare CNVs altering chromosome 21 do not contribute significantly to the pathogenesis of AVSDs in patients without Down syndrome nor do large CNVs contribute to the AVSDs in Down syndrome.
The role of rare CNVs was investigated in the etiology of heterotaxy, resulting from asymmetric development along the left–right body axis, leading to thoracic and abdominal organ defects (Fakhro et al. 2011). More than 250 patients with heterotaxy and nearly 1000 controls were genotyped with SNP arrays. There were nearly twofold more rare CNVs among the heterotaxy cohort compared with the controls (14.5% vs. 7.4%, respectively). In the 38 smaller CNVs identified, 14 of 61 genes altered were relevant to biological processes of relevance for left–right axis development: ciliary function, zinc finger transcription factors, and TGF-β signaling. Five genes contained within CNV areas were shown to be significant by examining conserved genes located within these intervals and functionally evaluating the effect of knockdown of these genes in the frog Xenopus tropicalis. Specifically, knockdown of NEK2, ROCK2, TCGBR2, GALNT11, and NUP188 disrupted left–right development (Fig. 3); none had been previously implicated in left–right patterning.
Figure 3.
Left–right (LR) abnormalities from morpholino oligonucleotide (MO) knockdown in X. tropicalis. MOs were injected at the one-cell stage and heart and gut looping were assayed in tadpoles at stage 45/46. Views are from the ventral aspect, shown in schematic form in F. (A) Heart (area outlined in red box as in schematic in F) showing normal D-looping. The inflow (red arrow) is on the tadpole’s left; the outflow tract (yellow arrow) is on the tadpole’s right. (B) Heart showing abnormal, anterior, A-looping. The inflow (red) and outflow (yellow) are both at the midline, with no discernible LR orientation. (C) Heart showing abnormal, reversed L-looping. The inflow (red) is on the tadpole’s right; the outflow (yellow) is on the tadpole’s left. (D) Normal clockwise rotation of the gut. (E) Abnormal gut rotation. (F). Schematic of Xenopus tadpole at stage 45/46; ventral view with anterior to the top; arrows indicate heart and gut. (G) Heart looping in MO knockdown tadpoles. Both dnah9 and ift88 are positive controls; standard control MO (StdCtrl), uninjected control (UiC), and dye-injected (Dye) are negative controls. Bars show the total percentage of abnormally looped hearts: divided into A-loop (blue) and L-loop (red). (H) Gut looping in MO knockdown tadpoles. Red bars show the percentage of abnormal gut loops. Heart and gut looping were analyzed by two independent readers blinded to group status with 95% concordance. *P < 10−4 versus standard control. (From Fakhro et al. 2011; reprinted, with permission, from the National Academy of Sciences © 2011.)
A genome-wide CNV study of various types of CHD compared more than 2000 CHD cases including approximately 800 with TOF to nearly 900 controls (Soemedi et al. 2012b). They found a significant burden of rare loss CNVs >100 kb containing genes (OR 1.8; population-attributable risk of nearly 3.5%). There was a trend toward greater ORs with increasing CNV deletion size and gene content. Enrichment analysis with the rare loss CNVs revealed a significant association with WNT signaling, which was altered in a broad range of CHD. Examining CNVs that were recurrent, the investigators confirmed prior observations of gain and loss CNVs including GATA4 at 8p23.1 and deletions at 15q11.2 in 0.5% of cases, an OR of 8.2 compared with controls. Finally, the parent of origin for de novo CNVs was assessed in 11 cases, of which 10 were paternal.
Restricting the search for CNVs to genes previously implicated in CHD, another study compared more than 800 CHD cases without known genomic defects (e.g., Down syndrome, 22q11del) to 3000 controls (Tomita-Mitchell et al. 2012). Large, rare CNVs (>100 kb for losses and >200 kb for gains) were observed in 4.3% of cases compared with 1.8% of controls, a significant difference. The previously noted gain at 1q21.1 and loss at 8p23.1 that includes GATA4 were detected in this study as well. Of interest, two CHD subjects without a clear syndrome were found to harbor gains of HRAS, the gene with gain-of-function mutations underlying Costello syndrome.
The role of 1q21.1 CNVs in CHD was investigated in a mixed CHD population, although enriched for TOF (Soemedi et al. 2012a). Gain CNVs were observed in 8/948 cases compared with 3/10,910 controls, an OR of 30.9. Smaller gain CNVs (100–200 kb) including GJA5 were also increased among the TOF cases with an OR of 10.7. Although no 1q21.1 losses were identified in this TOF cohort, three losses (but no gains) were observed among nearly 1500 cases of assorted other forms of CHD. Taken as a whole, this study suggested that 1q21.1 gains account for ∼1% of TOF, possibly a result of altered expression of GJA5.
COMMON VARIANTS
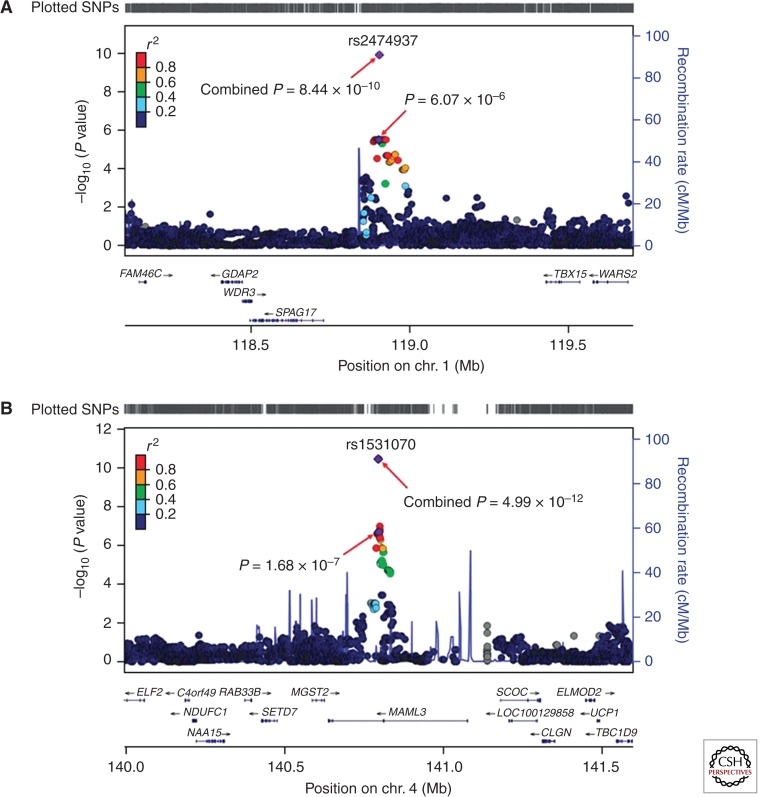
To examine the possibility that common genetic variants contribute to the etiology of CHD, two groups have recently completed genome-wide association studies (GWASs). A GWAS including nearly 1000 Han Chinese with ASD and/or VSD and more than 1200 racially matched controls in the discovery group and (Hu et al. 2013) nearly 1600 ASD/VSD subjects and 2300 controls in the replication cohort identified two SNPs, at 1p12 and 4q31.1 that were replicated in the second stage as well as within an additional second replication cohort (582 cases and 1565 controls). In aggregate, the associations of these two SNPs with CHD were highly significant (P < 1 × 10−9 and 5 × 10−12, respectively) and both had ORs of 1.4 (Fig. 4). For the 1p12 locus, the nearest relevant gene is TBX15, which encodes a T-box transcription factor. Although mutations in several TBX genes cause CHD in humans and mice, loss of Tbx15 in mice is not associated with heart abnormalities and no TBX15 mutation has been found in a human. The gene MAML3, which encodes mastermind-like 3, resides in the 4q31.1 locus. Mastermind proteins bind Notch, a key developmental regulator, and have transcription activation domains. Loss of Maml3 in mice is not associated with an observable phenotype, and no human MAML3 mutation has been identified to date. Finally, this GWAS did not confirm associations with any of the previously published SNPs. It is not clear whether the failure to replicate is attributable to prior published false positives, as most of the previously published studies were based on modest-sized cohorts, or to differences in race and ethnicity of the subjects.
Figure 4.
Plots are shown for the two loci associated with congenital heart malformations. 1p12 (A) and 4q31.1 (B). Imputation was performed for each region using The 1000 Genomes Project CHB (Han Chinese in Beijing, China) and JPT (Japanese in Tokyo, Japan) data (November 2010 release) as a reference. Results [−log10 (P values)] are shown for SNPs in the 1.6-Mb regions centered on the proxy SNPs. Proxy SNPs are shown in purple, and the r2 values of the other SNPs are indicated by color. The genes within the regions of interest are annotated, and arrows represent the direction of transcription. The right y-axis shows the recombination rate estimated from the HapMap samples. chr., chromosome. (From Hu et al. 2013; reprinted, with permission, from Nature Publishing Group © 2013.)
A GWAS of more than 1800 European Caucasians with various forms of CHD with more than 5100 controls failed to identify any SNPs with genome-wide significance (Cordell et al. 2013a). Subgroup analyses based on CHD anatomy produced suggestive P values for ASD and VSD. Replication cohorts (more than 400 ASD and 200 VSD cases compared with more than 2500 controls) genotyped for 10 SNPs from six regions showed association of ASD to a locus at chromosome 4p16 (combined P < 3 × 10−10 and OR of 1.5). Within this region, the best candidate gene was MSX1, which encodes a transcription factor relevant for atrial septal development. Loss-of-function MSX1 mutations in humans alter craniofacial and dental development, but have not been associated with CHD.
In a separate GWAS publication, this group studied 800 cases of TOF and more than 5100 controls in the discovery phase and 800 cases and 3000 controls for replication (Cordell et al. 2013b) and identified two regions of interest at 12q24 (P < 8 × 10−11; OR 1.3) and 13q32 (P = 3.0 × 10−11; OR 1.3). The 12q24 locus has been implicated in GWASs for other complex genetic traits such as type 1 diabetes and coronary artery disease, but no other developmental disorder. The best candidate gene at 12q24 is PTPN11, which encodes SHP-2, a protein tyrosine phosphatase. PTPN11 mutations cause Noonan and LEOPARD syndromes, which can include pulmonary valve stenosis. The best candidate gene from the 13q32 locus is GPC5, which encodes the heparan sulphate proteoglycan glypican 5. Glypicans serve as regulators of signaling pathways during development, and mutations in genes for other glypican family members are associated with CHD.
Of interest, both major CHD GWAS studies identified associations for ASDs, albeit different loci. Because this heart lesion would seem to have the smallest effect on reproductive fitness, common variants not subject to strong purifying selection is more likely than for other lesions. Future work will need to confirm these findings and then establish the biological effects of the relevant common variant in the region.
NONCODING MUTATIONS
Mutations in gene promoters, enhancers, locus control regions, microRNAs, and even intergenic regions are likely to be relevant for CHD causality. Although ongoing large-scale genomics efforts such as ENCODE are facilitating such work (Dunham et al. 2012), the task of identifying disease-causing variation in noncoding regions of the human genome remains daunting.
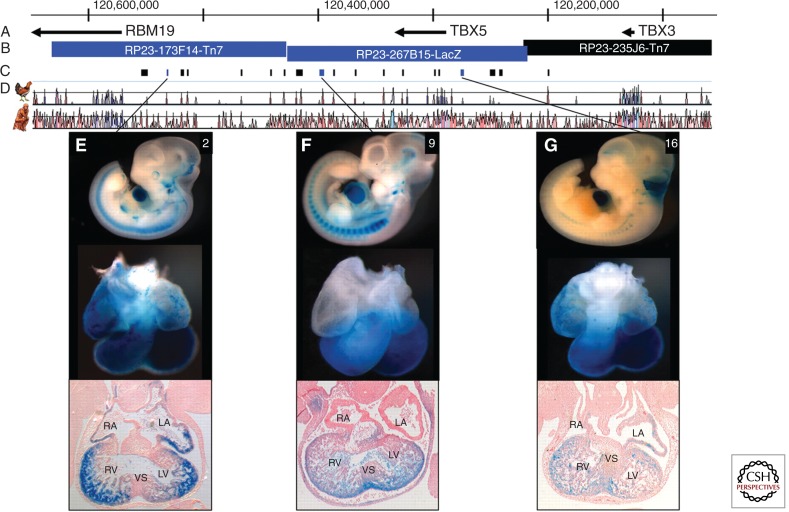
A recent study focused on the identification of cis-regulatory elements for TBX5, the gene mutated in Holt–Oram syndrome (Smemo et al. 2012). Using a series of constructs with LacZ reporters in transgenic mice, a 700-kb region around TBX5 was scanned and three enhancer elements residing 380 kb, 140 kb, and 9 kb downstream of TBX5 that drove cardiac expression were identified (Fig. 5). The three enhancer regions were sequenced in 260 unrelated individuals with ASDs, VSDs, or AVSDs. Among the nine rare or novel variants identified, one altering a highly conserved nucleotide was homozygous for the variant allele in a subject with a VSD. When tested in vitro, the oligonucleotide with the variant bound the transcription factor less avidly. Strikingly, use of the variant sequence with reporters in transgenic mice or zebrafish embryos resulted in dramatically reduced expression in the heart. Thus, the investigators provided strong evidence that this single nucleotide variant in a TBX5 enhancer alters expression of the gene during heart development. The challenges, well illuminated through this study, are how to prove causality definitively and how to scale such work as large numbers of candidate variants are identified through whole-genome sequencing.
Figure 5.
Regulatory landscape of the TBX5 locus. Whole mount and histological sections through the heart of embryonic day 11.5 (E11.5) embryos. Top row: In situ hybridizations showing endogenous expression (purple) of (A) RBM19, (B) TBX5, and (C) TBX3. Bottom row: β-Galactosidase staining (blue) captures the regulatory landscape of enhancers within bacterial artifical chromosomes (BACs): (D) RP23-173F14-Tn7, (E) RP23-267B15-LacZ, and (F) RP23-235J6-Tn7. The genes probed for in A, B, and C are contained, respectively, within the BACs tested in D, E, and F. In B and D–F, the forelimb is outlined. A–E, transverse sections; F, sagittal section. RA, right atrium; LA, left atrium; AS, atrial septum; RV, right ventricle; LV, left ventricle; VS, ventricular septum; AVC, atrioventricular canal. (From Smemo et al. 2012; reprinted, with permission, from Oxford University Press © 2012.)
CONCLUDING REMARKS
Since the earliest ideas about the genetics of CHD as a complex trait were proposed 35 years ago, substantial progress has been made in understanding that complexity. Following an era of gene discovery based on highly penetrant mutations with Mendelian segregation with CHD, we have now entered an exciting period in which the landscape of the genetic complexity is being explored robustly. This has been enabled by advances in genotyping and sequencing technology and new methods for detecting CNVs. The most recent large-scale efforts to understand the noncoding portions of the human genome, particularly through ENCODE (Dunham et al. 2012), will allow CHD researchers to expand their search for causative variation.
As exemplified by the finding that de novo mutations underlying CHD often alter chromatin remodeling, epigenetic modifications are probably important for CHD pathogenesis. Because epigenetic states can be tissue-specific, efforts to procure and study cardiac tissues will be needed. Moreover, these discoveries underscore the likelihood that interactions of genetic variation with environmental exposures will prove relevant for understanding the complexity of CHD. Research methodologies for addressing gene×environment interactions are still emerging and may require international efforts to assemble CHD cohorts of sufficient size to power studies.
Finally, researchers and physicians caring for individuals with CHD and their families will need to creatively and cooperatively translate genetic findings into meaningful medical advances. Low-hanging fruit is likely to include better prediction of recurrence risks for couples with a prior child with CHD and offspring risk for adults with CHD as well as improved prognostication of extracardiac involvement for infants with CHD. More challenging, but potentially more impactful, would be the development of therapies to reduce its incidence.
ACKNOWLEDGMENTS
This work is funded in part with support of National Heart, Lung and Blood Institute grants HL098123 to B.D.G. and HL098163 to W.K.C.
REFERENCES
- Ackerman C, Locke AE, Feingold E, Reshey B, Espana K, Thusberg J, Mooney S, Bean LJ, Dooley KJ, Cua CL, et al. 2012. An excess of deleterious variants in VEGF-A pathway genes in Down-syndrome-associated atrioventricular septal defects. Am J Hum Genet 91: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, Cosgrove C, Blue G, Granados-Riveron J, Setchfield K, et al. 2013a. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet 45: 822–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Topf A, Mamasoula C, Postma AV, Bentham J, Zelenika D, Heath S, Blue G, Cosgrove C, Granados Riveron J, et al. 2013b. Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Hum Mol Genet 22: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW 2004. Bicuspid aortic valve is heritable. J Am Coll Cardiol 44: 138–143 [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M 2011. Rare copy number variations in congenital heart disease patients identify unique genes in left–right patterning. Proc Natl Acad Sci 108: 2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland-Little JM, Hoffmann AD, Ocbina PJ, Peterson MA, Bosman JD, Chen Y, Cheng SY, Anderson KV, Moskowitz IP 2011. A novel murine allele of Intraflagellar Transport Protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Hum Mol Genet 20: 3725–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Kaski JP, Kim R, Kline J, Mercer-Rosa L, Porter G, et al. 2013. The Congenital Heart Disease Genetic Network Study: Rationale, design, and early results. Circ Res 112: 698–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmuntz E, Clark BJ, Mitchell LE, Jawad AF, Cuneo BF, Reed L, McDonald-McGinn D, Chien P, Feuer J, Zackai EH, et al. 1998. Frequency of 22q11 deletions in patients with conotruncal defects. J Am Coll Cardiol 32: 492–498 [DOI] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, et al. 2009. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 41: 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton RB Jr, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson DW 2007. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol 50: 1590–1595 [DOI] [PubMed] [Google Scholar]
- Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D’Amours G, Lemyre E, Cullum R, et al. 2012. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet 8: e1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shi Y, Mo X, Xu J, Zhao B, Lin Y, Yang S, Xu Z, Dai J, Pan S, et al. 2013. A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet 45: 818–821 [DOI] [PubMed] [Google Scholar]
- Kamp A, Peterson MA, Svenson KL, Bjork BC, Hentges KE, Rajapaksha TW, Moran J, Justice MJ, Seidman JG, Seidman CE, et al. 2010. Genome-wide identification of mouse congenital heart disease loci. Hum Mol Genet 19: 3105–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel BA, Knutsen RH, Ye L, Ciliberto CH, Broekelmann TJ, Mecham RP 2011. Genetic modifiers of cardiovascular phenotype caused by elastin haploinsufficiency act by extrinsic noncomplementation. J Biol Chem 286: 44926–44936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KL, Pignatelli R, Lewin M, Ho T, Fernbach S, Menesses A, Lam W, Leal SM, Kaplan N, Schliekelman P, et al. 2005. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am J Med Genet A 134A: 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabulsi MM, Tamim H, Sabbagh M, Obeid MY, Yunis KA, Bitar FF 2003. Parental consanguinity and congenital heart malformations in a developing country. Am J Med Genet A 116A: 342–347 [DOI] [PubMed] [Google Scholar]
- Nora JJ 1968. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases. The genetic–environmental interaction. Circulation 38: 604–617 [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M 2009. Recurrence of congenital heart defects in families. Circulation 120: 295–301 [DOI] [PubMed] [Google Scholar]
- Payne AR, Chang SW, Koenig SN, Zinn AR, Garg V 2012. Submicroscopic chromosomal copy number variations identified in children with hypoplastic left heart syndrome. Pediatr Cardiol 33: 757–763 [DOI] [PubMed] [Google Scholar]
- Pradat P, Francannet C, Harris JA, Robert E 2003. The epidemiology of cardiovascular defects: Part I. A study based on data from three large registries of congenital malformations. Pediatr Cardiol 24: 195–221 [DOI] [PubMed] [Google Scholar]
- Priest JR, Girirajan S, Vu TH, Olson A, Eichler EE, Portman MA 2012. Rare copy number variants in isolated sporadic and syndromic atrioventricular septal defects. Am J Med Genet A 158A: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, Yuen T, Rickaby J, Thiruvahindrapuram B, Marshall CR, et al. 2012. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet 8: e1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA 2012. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet 21: 3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Topf A, Wilson IJ, Darlay R, Rahman T, Glen E, Hall D, Huang N, Bentham J, Bhattacharya S, et al. 2012a. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet 21: 1513–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Wilson IJ, Bentham J, Darlay R, Topf A, Zelenika D, Cosgrove C, Setchfield K, Thornborough C, Granados-Riveron J, et al. 2012b. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet 91: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung RY, So LY, Ng HK, Ho JK, Fok TF 1991. Echocardiography as a tool for determining the incidence of congenital heart disease in newborn babies: A pilot study in Hong Kong. Int J Cardiol 30: 43–47 [DOI] [PubMed] [Google Scholar]
- Thienpont B, Mertens L, de Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K 2007. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J 28: 2778–2784 [DOI] [PubMed] [Google Scholar]
- Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, Hidestrand M, Harris SE, Goetsch MA, Simpson PM, Bick DP, et al. 2012. Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics 44: 518–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore R, Hobbins JC, Engle MA 1982. Pregnancy and its outcome in women with and without surgical treatment of congenital heart disease. Am J Cardiol 50: 641–651 [DOI] [PubMed] [Google Scholar]
- Winston JB, Erlich JM, Green CA, Aluko A, Kaiser KA, Takematsu M, Barlow RS, Sureka AO, LaPage MJ, Janss LL, et al. 2010. Heterogeneity of genetic modifiers ensures normal cardiac development. Circulation 121: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JB, Schulkey CE, Chen IB, Regmi SD, Efimova M, Erlich JM, Green CA, Aluko A, Jay PY 2012. Complex trait analysis of ventricular septal defects caused by Nkx2-5 mutation. Circ Cardiovasc Genet 5: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Chen HC, Lu CW, Wang JK, Huang SC, Huang SK 2010. Prevalence of congenital heart disease at live birth in Taiwan. J Pediatr 156: 782–785 [DOI] [PubMed] [Google Scholar]
- Yunis K, Mumtaz G, Bitar F, Chamseddine F, Kassar M, Rashkidi J, Makhoul G, Tamim H 2006. Consanguineous marriage and congenital heart defects: A case-control study in the neonatal period. Am J Med Genet A 140: 1524–1530 [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498: 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]