Summary
Nek6 and Nercc1/Nek9 belong to the NIMA family of protein kinases. Nercc1 is activated in mitosis whereupon it binds, phosphorylates and activates Nek6. Interference with Nek6 or Nercc1 in mammalian cells causes prometaphase/metaphase arrest, and depletion of XNercc from Xenopus egg extracts prevents normal spindle assembly. Herein we show that Nek6 is constitutively associated with Eg5, a kinesin necessary for spindle bipolarity. Nek6 phosphorylates Eg5 at several sites in vitro, and one of these sites, Ser1033, is phosphorylated in vivo during mitosis. While Cdk1 phosphorylates nearly all Eg5 during mitosis at Thr926, Nek6 phosphorylates ~3% of Eg5, primarily at the spindle poles. Eg5 depletion arrests cells with a monopolar spindle; this can be rescued by Eg5 wildtype but not by Eg5(Thr926Ala). Eg5(Ser1033Ala) rescues half as well as wildtype whereas an Eg5(Ser1033Asp) mutant is nearly as effective. Thus Nek6 phosphorylates a subset of Eg5 polypeptides during mitosis at a conserved site, whose phosphorylation is critical for the mitotic function of Eg5.
Introduction
The NIMA family of protein kinases are named for NIMA, a kinase of Aspergillus nidulans that participates in a broad array of mitotic processes (O'Connell et al., 2003). The human genome encodes eleven protein kinases whose catalytic domain is evolutionarily related to that of NIMA (O'Connell et al., 2003; Roig and Avruch, 2006). Available evidence indicates that the NIMA family kinases regulate aspects of microtubule function, including these related to cilia and the mitotic spindle (Quarmby and Mahjoub, 2005). Thus Nek2 has a central role in centrosome maturation and disjunction, while Nek1 and Nek8 are proposed to contribute to ciliary function. Nek6, Nek7 (Kandli et al., 2000) and Nek9/Nercc1 (Roig et al., 2002; Holland et al., 2002) are involved in the control of mitotic spindle formation. Except for Nek2 (Hayward and Fry, 2006), few NIMA-family kinase substrates have been described.
Nercc1 is activated in mitosis and microinjection of anti-Nercc1 antibodies in prophase results in prometaphase arrest or abnormalities in chromosome segregation (Roig et al., 2002). In Xenopus mitotic egg extracts, depletion of XNercc impairs spindle assembly as well as Ran-GTP induced aster formation (Roig et al., 2005). Nercc1 is able to bind, phosphorylate and activate Nek6 and Nek7, and Nek6 activity increases in mitosis concomitant with Nercc1 activation (Belham et al., 2003). Induced overexpression of kinase dead Nek6 in HeLa cells results in prometaphase/metaphase arrest (Yin et al., 2003). Similarly, interference with Nek7 (approximately 80% identical to Nek6) results in an increase in multinuclear cells and mitotic cells with a multipolar spindle (Yissachar et al., 2006). Thus the Nercc1/Nek6/Nek7 kinases represent a mitotic signaling cassette; the identification of the targets of this module will be necessary for understanding its function at a molecular level.
Eg5/Kinesin-5 is a plus-end-directed kinesin of the BimC family (Le Guellec et al., 1991) which forms bipolar tetramers capable of moving towards the plus ends of two microtubules simultaneously, thus bundling, sorting, and enabling antiparallel microtubule movement through sliding (Sawin et al., 1992; Kapitein et al., 2005). Eg5 is necessary for premitotic centrosome separation, spindle pole formation and separation, poleward translocation of microtubules, and postmitotic centrosome movement, and is thus one of the motors required for proper spindle organization and function (Walczak et al., 1998). Inhibition or depletion of Eg5 results in mitotic arrest with monopolar microtubule structures and condensed chromosomes surrounding the two unseparated centrosomes (Blangy et al., 1995). The regulation of Eg5 activity is less well defined than its several functions; nevertheless, one clearcut mode of Eg5 regulation is through the control of its binding to microtubules, which requires Cdk1 phosphorylation of a single residue (Thr926 in HsEg5) in the conserved carboxyterminal tail (Blangy et al., 1995; Sawin and Mitchison, 1995). The mechanism by which Cdk1 phosphorylation of some BimC family members induces microtubule binding is unknown; moreover, not all family members have a conserved Cdk1 site motif. Aurora-family kinases have been shown to phosphorylate Eg5 orthologues, modifying the kinesin in the stalk domain in Xenopus laevis (Giet et al., 1999) or in the C-terminal tail domain in Caenorhabditis elegans (Bishop et al., 2005). Although the function of stalk phosphorylation is not clear, in C. elegans, the absence of the Aurora AIR-2 affects kinesin localization to the spindle.
Herein we show that the kinase Nek6 binds to and phosphorylates Eg5 in vitro and in vivo during mitosis at a unique carboxyterminal site. This phosphorylation occurs on a minority of the spindle associated Eg5 and does not alter spindle association, but is required for normal Eg5 function in enabling spindle bipolarity. These results uncover a new mode of Eg5 regulation and identify the first physiologic substrate of the Nercc1/Nek6 signaling module.
Results
Nek6 binds to the tail of Eg5
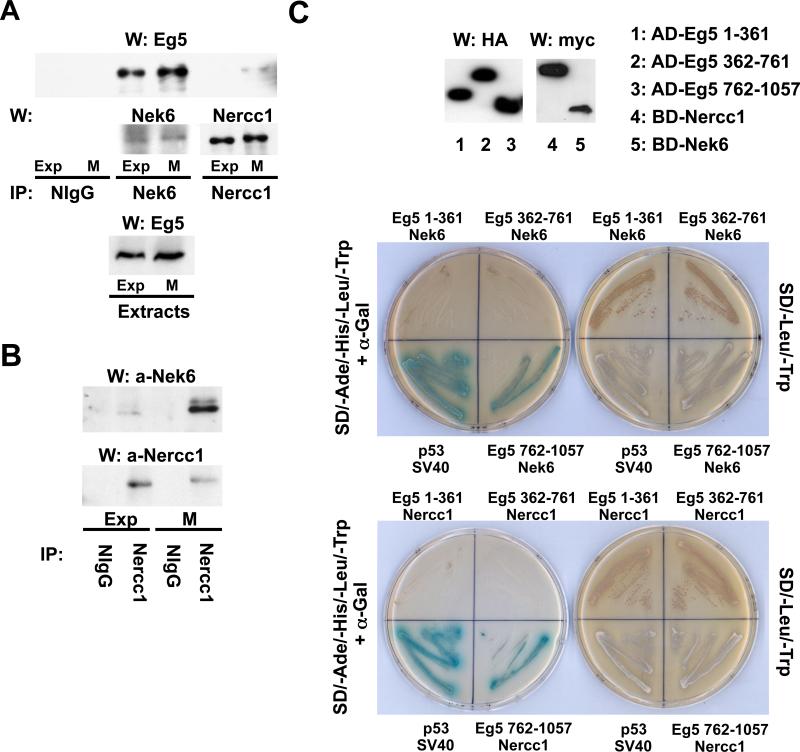
Overexpressed Nek6 retrieves a single ~120 kDa polypeptide band identified, by LC/MS/MS of tryptic peptides, as containing the Nercc1 protein kinase (Roig et al., 2002); further analysis of these tryptic digests indicated the additional presence of the kinesin Eg5. Endogenous Eg5 specifically coprecipitates with endogenous Nek6, using extracts from both exponentially growing and mitotic cells (Figure 1A). In addition, small amounts of Eg5 coprecipitate with Nercc1, but only from extracts of mitotic cells (Figure 1A). The Nercc1-Nek6 interaction also occurs predominantly in mitosis, concomitant with Nercc1 activation (Figure 1B). In a yeast two-hybrid analysis, both Nek6 and Nercc1 exhibit the ability to bind directly to the Eg5 carboxyterminal tail (residues 762-1057) but not to motor head (1-361) or stalk (362-761) domains (Figure 1C). Inasmuch as Eg5 is associated with Nek6 in both interphase and mitosis, but is associated with Nercc1 only in mitosis when the Nercc1/Nek6 interaction is maximal, it is likely that the Eg5/Nercc1 interaction in vivo occurs through the Nercc1-Nek6 interaction.
Figure 1. Eg5 interacts with Nek6 and Nercc1.
(A) Immunoprecipitates were prepared from exponentially growing (Exp) or nocodazole-arrested mitotic (M) U2OS cell extracts using either normal rabbit IgG (NIgG), anti-Nek6 or anti-Nercc1 antibodies and analyzed by immunoblot with the indicated antibodies. Eg5 in the corresponding extracts is shown in the lower panel. (B) Immunoprecipitates were prepared from exponentially growing (Exp) or nocodazole-arrested mitotic (M) U2OS cells using NIgG or anti-Nercc1 antibody, and immunoblotted for Nek6 and Nercc1. (C) The ability of Eg5 head (Eg5 1-361), stalk (Eg5 362-761) and tail (Eg5 762-1057) domains to interact with Nek6 or Nercc1 was assessed using two hybrid by histidine and adenine prototrophy plus expression of α-galactosidase activity (left plate in both pictures). Fusion protein expression was verified by immunoblot (upper panel). AD, BD: Gal4 activation and binding domains. The interaction between p53 and SV40 is a positive control.
Nek6 phosphorylates Eg5 in vitro specifically and in vivo during mitosis
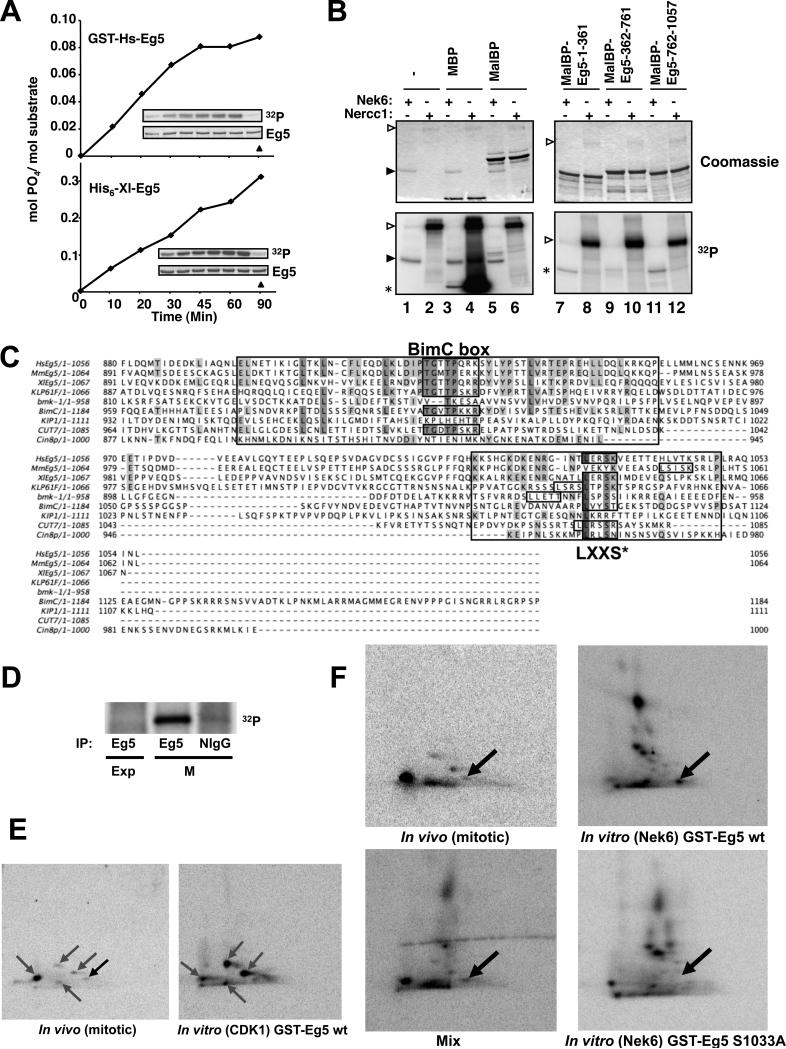
Recombinant FLAG-Nek6 (Belham et al., 2001; Belham et al., 2003), readily phosphorylates GST-HsEg5 or His6-XlEg5 (Figure 2A; but not GST alone, data not shown). Although Nercc1 phosphorylates myelin basic protein (MBP) and GST-Nek7, it is not able to detectably phosphorylate GST-Eg5 in parallel (data not shown). Maltose binding protein (MalBP)-fusions of the Eg5 head (1-361), stalk (362-761) and tail (762-1057) were incubated with active FLAG-Nek6 or FLAG-Nercc1 plus [γ-32P]ATP/Mg2+ (Figure 2B). Whereas the Eg5 stalk is hardly phosphorylated, FLAG-Nek6 phosphorylates the Eg5 head and tail polypeptides, incorporating respectively up to 0.25 and 0.90 mols 32P per mol of protein substrate over a 2 hour reaction. No 32P incorporation occurs with kinase-deficient Nek6[K74M K75M], and mutation of the described Cdk1 phosphorylation site in Eg5 tail (Eg5 762-1057[Thr926Ala]) does not affect the observed phosphorylation (data not shown). As with full length Eg5, Nercc1, although capable of phosphorylating GST-Nek7 or MBP, is unable to phosphorylate the MalBP-Eg5 fusion polypeptides. The identity of the Nek6-catalyzed phosphorylation sites was determined by LC/MS/MS analysis of tryptic peptides derived from MalBP-Eg5[1-361] and MalBP-Eg5[762-1057] phosphorylated to approximately 0.1 and 0.3 moles PO4/mole, respectively. In the head a single phosphorylation at Ser269 was detected, whereas in the tail, phosphorylation was present at Ser776, Thr901 and Ser1033. Notably, each of these sites except Ser776 conforms to the LXXS/T motif defined by (Lizcano et al., 2002) as the preferred Nek6 phosphorylation site.
Figure 2. Nek6 phosphorylates Eg5 in vitro at a conserved site that is phosphorylated during mitosis.
(A) Recombinant GST-HsEg5 or His6-XlEg5 were phosphorylated by FLAG-Nek6. The insets show 32P incorporation with time (upper, left to right with no kinase in rightmost lane) and a coomassie blue stain of the substrate (lower). (B) Myelin basic protein (MBP, positive control), Maltose-binding protein (MalBP), or MalBP-fusion proteins containing the Eg5 segments indicated were incubated with [γ-32P]ATP/Mg2+ and either FLAG-Nek6 (lanes 1, 3, 5, 7, 9, 11) or preactivated FLAG-Nercc1 (2, 4, 6, 8, 10, 12) for 30 min followed by SDS-PAGE. The upper panels show Coomassie blue stains of the gels and the lower panels the corresponding autoradiographs. Open arrowheads indicate Nercc1 and filled arrowheads, Nek6. Asterisks indicate the location of MBP (lanes 3,4) or MalBP-fusion proteins (7-12). The left gel is 12% acrylamide, the right gel 7.5%; Nek6 is run off the latter. (C) HsEg5, human Eg5; MmEg5, mouse Eg5; XLEg5, Xenopus laevis Eg5; KLP61F, Drosophila melanogaster KLP61F; bmk-1, Caenorhabditis elegans bmk-1; BimC, Aspergillus nidulans BimC; KIP1, Cin8p, Saccharomyces cerevisiae KIP1 and Cin8p; Cut7, Schizosaccharomyces pombe Cut7. Identical and conserved residues are shaded; regions conserved around the C-terminal Cdk1 site (BimC box) and Nek6 site (LXXS*) are boxed. (D) Using NlIgG and anti-HsEg5 antibodies, immunoprecipitates were prepared from extracts of 32P-labelled U2OS cells growing exponentially (Exp) or incubated overnight in 0.25mM nocodazole (M), and subjected to SDS-PAGE; a PhosphoImage of the gel is shown. (E) Left, two-dimensional (2-D) tryptic phosphopeptide map of endogenous 32P-Eg5 immunoprecipitated from 32P-labeled mitotic U2OS cells, visualized by PhosphorImager. Right, phosphopeptide map of purified recombinant 32P-GST-Eg5 phosphorylated in vitro by immunoprecipitated mitotic Cdk1. The grey arrows mark phosphopeptides attributed to Cdk1; the black arrow indicates a 32P-peptide evident in digests of mitotic Eg5 that is not present in digests of Cdk1-phosphorylated 32P-Eg5. (F) Top left, 2-D phosphopeptide map of endogenous Eg5 immunoprecipitated from 32P-labeled mitotic U2OS cells; top and bottom right, 2-D maps of recombinant GST-Eg5 wt (top right) and GSTEg5[S1033A] (bottom right) phosphorylated in vitro by FLAG-Nek6; note in the lower right map the absence of the most anodally migrating 32P-peptide seen in the upper right map, which encompasses Ser1033-P. Bottom left, a mixture of comparable amounts of the digests of mitotic 32P-Eg5 and FLAG-Nek6 phosphorylated 32P-Eg5. The black arrow marks the phosphopeptide common to the two digests, which contains serine 1033. In the digests of mitotic 32P-Eg5 (E, left; F, upper left), this spot contains ~3% of total 32P, estimated by PhosphoImager.
We next immunoprecipitated endogenous Eg5 from exponentially growing and nocodazole-arrested mitotic U2OS cells, and analyzed Eg5 tryptic peptides by LC/MS/MS; approximately 80% coverage of Eg5 sequence was obtained for both samples. Eg5 from exponentially growing cells exhibited only a single phosphorylation site at Thr926; the detection of this Cdk1-catalyzed phosphorylation site probably reflects the contribution of the cells present in mitosis during unsynchronized growth. Eg5 isolated from mitotic cells exhibited two phosphorylation sites, at Thr926 and Ser1033; the latter corresponds to one of the sites of Eg5 phosphorylated by Nek6 in vitro. Inasmuch as Nek6, like Cdk1, is specifically activated during mitosis (Belham et al., 2003), these results indicate that Eg5 is phosphorylated both by Cdk1 and by Nek6 during mitosis.
A Nek6 motif corresponding to HsEg5[Ser1033] is conserved in the BimC family of kinesins
Alignment of the carboxyterminal aminoacid sequences of several BimC kinesins (Figure 2C) reveals that most, but not all BimC kinesins show a short conserved region at the tail domain surrounding the Cdk1 site (in HsEg5, Thr926) named the “BimC box” (Heck et al., 1993; Sawin and Mitchison, 1995) (Heck et al., 1993; Sawin and Mitchison, 1995). Notably, all but two of the BimC kinesins (MmEg5 and KIP1) have a site containing the LXXS/T motif matching the location of HsEg5[Ser1033]. The nonessential Saccharomyces kinesin KIP1 lacks such a site entirely, as well as a BimC box/Cdk1 site; however the other ScBimC kinesin Cin8 does contain the LXXS/T site. Rodent Eg5s exhibit a LXXS/T site 11 (mouse) or 20 (rat) residues carboxyterminal to the location of HsEg5[Ser1033]. Interestingly, Drosophila melanogaster KLP61F and S. pombe Cut7 have two adjacent or overlapping LXX(S/T) sites, whereas C. elegans bmk-1 has three, with two overlapping. Altogether, it is clear that a residue equivalent to HsEg5[Ser1033] is conserved in the BimC family, indicating that a Nek6 site motif in this region of Eg5 is likely to have an important physiological role, probably related to its phosphorylation by a protein kinase with site-specificity similar to Nek6.
Phosphopeptide maps of mitotic Eg5 confirm Serine 1033 as a relevant mitotic phosphorylation site
We performed two-dimensional tryptic 32P-peptide maps of endogenous Eg5 from 32P-labelled mitotic U2OS cells, to estimate the relative extent of phosphorylation at the two Eg5 sites detected and to determine if other phosphorylation sites exist. Eg5 retrieved from 32P-labelled cycling cells showed no detectable 32P incorporation, whereas Eg5 retrieved from nocodazole-arrested U2OS cells exhibited substantial labeling (Figure 2D). Two-dimensional tryptic 32P-peptide maps (Figures 2E left and 2F top left) of mitotic Eg5 show 5 reproducible 32P-peptides, four of which correspond to 32P-spots present in tryptic digests derived from Eg5 phosphorylated by Cdk1 in vitro (Figure 2E, right), while the most anodally migrating 32P-peptide in the digest (Figure 2E, black arrow) is absent in the maps of Eg5 phosphorylated by Cdk1 in vitro.
32P-Eg5 generated by phosphorylation with Nek6 in vitro yields a complex 32P-peptide map (Figure 2F upper right), as anticipated from the multiple phosphorylation sites detected on LC/MS/MS of similar digests. Notably, the map of Eg5[Ser1033Ala] phosphorylated by Nek6 in vitro is essentially identical (Figure 2F, lower right) except for the absence of a single 32P-spot, indicated by the arrow. Mixing of the digests of mitotic Eg5 with Nek6-phosphorylated Eg5 confirms that the most anodally migrating 32P-peptide in both digests comigrate (Figure 2F, lower left). These results confirm that Eg5[Ser1033] is phosphorylated in vivo during mitosis and indicate that the relative incorporation of 32PO4 into Ser1033 in nocodazole arrested U2OS cells is approximately 3% that incorporated into Thr926. Moreover, they strongly support the view that in mitotic U2OS cells phosphorylation of Eg5 at sites other than Thr926 and Ser1033, if present, contribute much less than 1% to overall Eg5 phosphorylation.
Phosphospecific antibodies. Nek6 overexpression and activation induces Eg5[Ser1033] phosphorylation in vivo. Eg5[Ser1033P] levels during mitosis
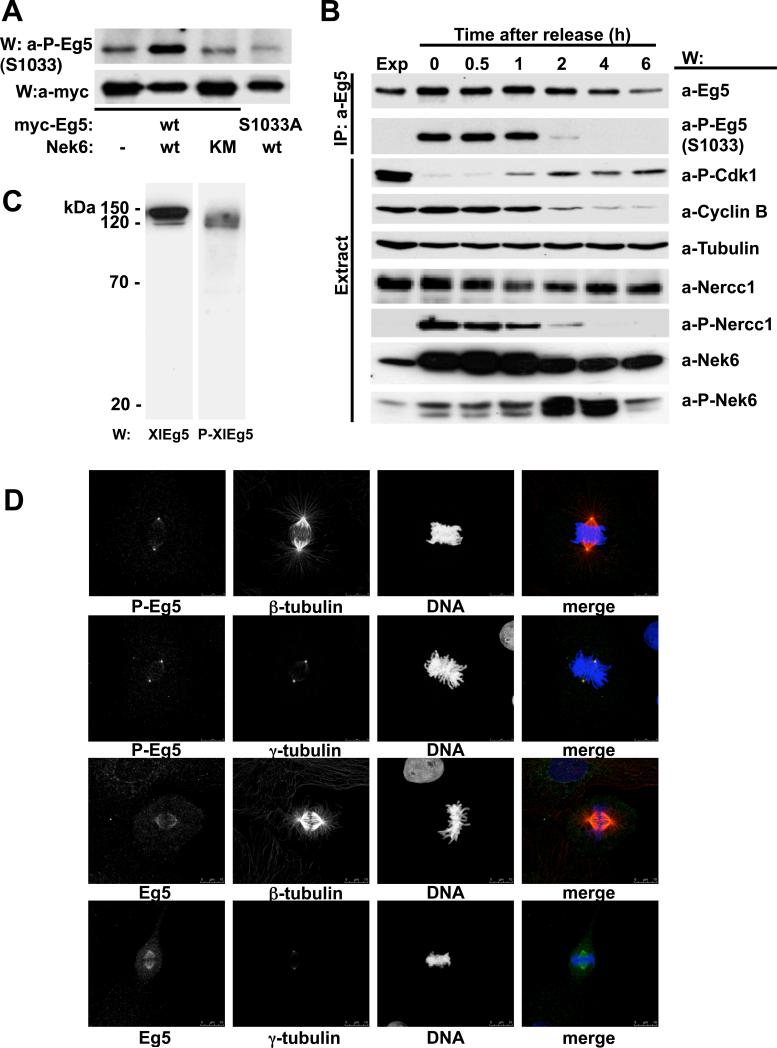
We generated phosphospecific antibodies directed against sequences surrounding HsEg5[Ser1033P] as well as those surrounding the orthologous site on Xenopus laevis Eg5, (XlEg5 serine 1046). The resulting antibodies were totally specific for the Nek6-phosphorylated forms of Eg5 (Supplementary Figure 1), and were used to study the effects of Nek6 overexpression and resulting activation (Belham et al., 2003) on the phosphorylation of Eg5[Ser1033]. Figure 3A shows that coexpression of Nek6 with Eg5 is accompanied by a substantial increase in Eg5[Ser1033P] immunoreactivity, which is not seen with kinase-dead Nek6 or when Nek6 is coexpressed with Eg5[Ser1033Ala] Thus active Nek6 can phosphorylate Ser1033 in vivo as well as in vitro (our attempts to study the effects of Nek6 downregulation on the endogenous levels of Eg5[Ser1033P] were hampered by the incomplete depletion of the kinase obtained using different siRNAs and multiple experimental conditions; in these experiments we did not detect any changes on the levels of active Nek6 (Nek6[Ser206P]) or Eg5[Ser1033P], neither we observed any effect on mitotic progression, data not shown).
Figure 3. Eg5[Ser1033] phosphorylation in vivo.
(A) U2OS cells were cotransfected with either myc-Eg5 wt (lanes 1-3) or myc-Eg5[S1033A] (lane 4) and empty plasmid (lane1), FLAG-Nek6 wt (lane 2 and 4) or FLAG-Nek6 [K74M,K75M] (lane 3). 24 hours later, anti-Myc-immunoprecipitates were subjected to immunoblot with anti-Myc (lower panel) and anti-Eg5[Ser1033-P] antibodies (top panel). (B) U2OS cells growing exponentially were either untreated (first lane, Exp) or arrested with nocodazole (0.25mM overnight; lane 2); arrested cells were allowed to exit mitosis in nocodazole-free medium and extracted at the times indicated (lanes 3 to 7). Eg5 immunoprecipitates (top two rows) and cell extracts (bottom seven rows) were analyzed by immunoblot with the antibodies indicated (a-P-Eg5(S1033), anti-Eg5[serine1033P]; a-P-Cdk1, anti-Cdk1[Tyr15P]; a-P-Nercc1, anti-Nercc1[Thr210P]; a-P-Nek6, anti-Nek6[Ser206P]. (C) Extracts from XL177 Xenopus laevis cells were resolved by SDS-PAGE and immunoblotted with either anti-XlEg5 (left) or anti-XlEg5[Ser1046-P] (right). (D) XL177 cells growing exponentially were fixed and stained with antibodies to XlEg5 (lower two rows), XlEg5[Ser1046P] (upper two rows), and either β- or γ-tubulin as indicated. DNA is stained with DAPI. Representative cells in metaphase are shown; bar, 10 μm.
Endogenous Eg5 was immunoprecipitated from freely cycling and nocodazole arrested U2OS cells, as well as from cells harvested at intervals after nocodazole washout (Figure 3B); no Ser1033P immunoreactivity is detected in Eg5 from freely cycling U2OS cells, whereas a strong signal is evident in mitotic Eg5 which fades over the first two hours after nocodazole washout. The pattern of Eg5[Ser1033P] immunoreactivity parallels that of Nercc1 phosphorylation at the activating site on the T-loop. Surprisingly, whereas Nek6 T-loop phosphorylation increases in parallel with Eg5[Ser1033P] immunoreativity during mitotic arrest, it exhibits a further increase at 2-4 hours after nocodazole washout; the mechanism of this late-mitotic and/or early G1 phosphorylation of Nek6 is not known and its timing suggests a previously unappreciated function of Nek6 in mitotic exit or early G1. Nevertheless, these results further confirm the mitotic phosphorylation of Eg5[Ser1033] coincident with Nercc1 activation and likely mediated by the Nercc1/Nek6 complex.
Subcellular localization of Nek6-phosphorylated Eg5
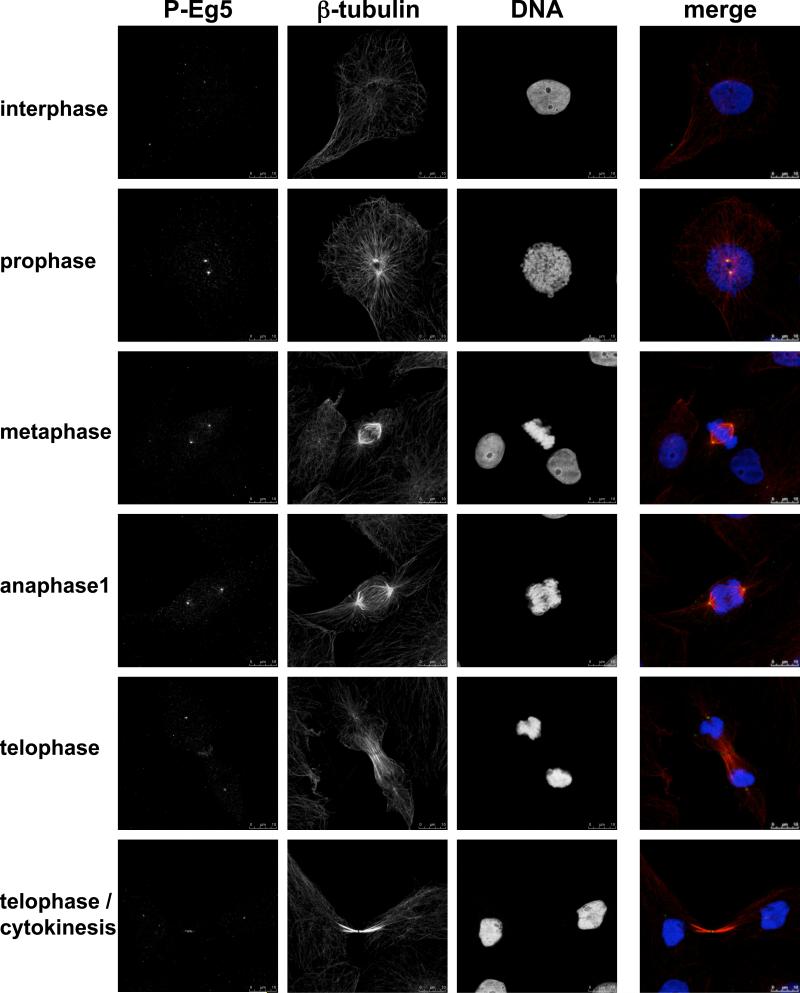
We sought next to define the subcellular localization of Nek6 phosphorylated Eg5. Our anti-HsEg5[Ser1033P] antibodies recognize several bands of lower molecular weight on immunoblots (identity unknown), rendering them unsuitable for immunocytochemistry. We therefore employed the anti-XlEg5[Ser1046P] antibodies, which show comparable phosphospecificity (Supplementary Figure 1) but exhibit only a single band on immunoblot of extracts from Xenopus XL177 cells (Figure 3C). In mitotic XL177 cells, XlEg5 polypeptide (Figure 3D, bottom two rows) resides in the cytoplasm and along the length of the metaphase spindle, somewhat more prominently near the poles but extending to the spindle midzone without covering it; XlEg5 is not detectable on astral microtubules. As expected anti-XlEg5[Ser1046P] antibodies label only mitotic XL177 cells (Figure 4), with a maximal signal detectable in metaphase (Figure 3D, Figure 4). Eg5[Ser1046P] immunoreactivity is highly concentrated at spindle poles, as can be clearly seen by costaining with γ-tubulin (Figure 3D, second row). A minor amount of XlEg5Ser1046P] is detectable on spindle microtubules, however no signal is observed at astral microtubules. Interestingly, whereas XlEg5[Ser1046P] reactivity is evident almost exclusively at the spindle poles from prophase through anaphase (Figure 4), a clear signal is seen abutting the cytokinetic furrow at telophase. The highly localized concentration of XlEg5[Ser1046P] at the spindle poles is consistent with the view that Ser1046 phosphorylation occurs on a minor subset of Cdk1 phosphorylated Eg5; this in turn is in agreement with our phosphopeptide maps, which show that the extent of Eg5 phosphorylation by Nek6 is much less than that catalyzed by Cdk1. Moreover, the Nercc1/Nek6 module is also activated at spindle poles (Roig et al., 2005), suggesting that Eg5 phosphorylation by Nek6 occurs at poles.
Figure 4. Phosphoserine 1046 immunolocalization in XL177 cells in interphase and different phases of mitosis.
As in Figure 3D; bar, 10 μm.
Mutation of HsEg5[Ser1033] to Ala disrupts normal Eg5 function but not mitotic localization
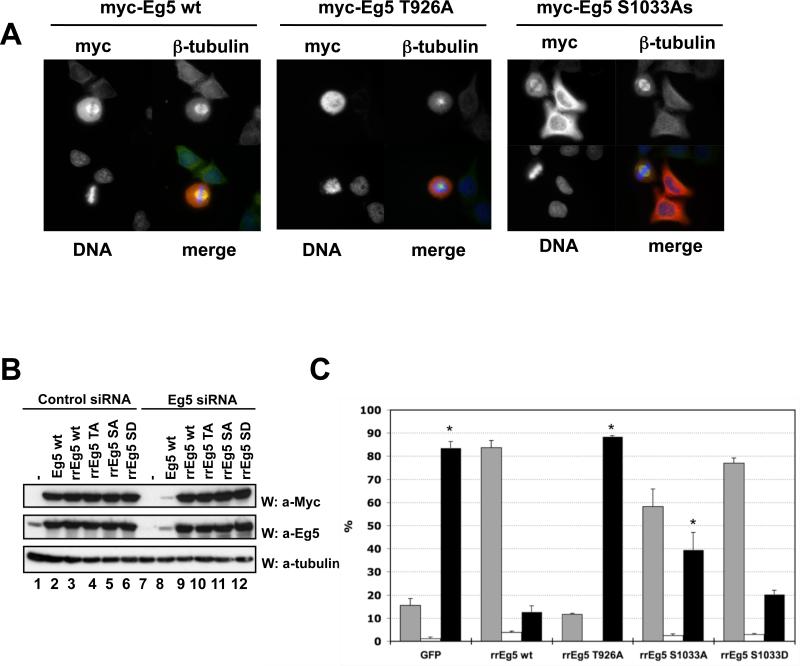
Different Eg5 forms (wildtype, Thr926Ala; Ser1033Ala; Ser1033Asp) were purified from baculovirus-infected insect cells and characterized in vitro. On gel filtration, the oligomeric state of the wildtype and mutant Eg5 polypeptides were indistinguishable. We assessed the ability of these Eg5 variants to bind microtubules by addition of equal amounts to assembled microtubules, followed by pelleting. If the amount of wildtype Eg5 in the pellet is set at 100%, then Eg5[Thr926Ala] is recovered at approximately 28%, the Eg5[Ser1033Ala] mutants at approximately 63% and the Eg5[Ser1033Asp] 42% (Supplementary Figure 2). Despite these differences in microtubule binding, each Eg5[Ser1033] variant exhibited rapid movement on microtubules, performing indistinguishably by visual inspection on microtubule gliding assays. The ability of these variants to bind to the mitotic spindle in vivo was evaluated during transient expression (Figure 5A). Myc-Eg5 wildtype binds along the length of the mitotic spindle (left) whereas mitotic cells expressing Myc-Eg5[Thr926Ala] invariably showed a diffuse localization usually associated with a monopolar spindle (middle). The Myc-Eg5[Ser1033Ala] mutant showed a diffuse cytoplasmic localization in interphase and a strong association with a normal appearing spindle (right), similar to Eg5 wildtype. Thus at these levels of expression, Myc-Eg5[Ser1033Ala] binds normally to the spindle and does not appear to alter mitotic progression.
Figure 5. Eg5[Ser1033] phosphorylation is required for normal Eg5 function in mitotic spindle assembly.
(A) U2OS cells transiently expressing Myc-Eg5 wt, Thr926Ala or Ser1033Ala were stained for Myc, β-tubulin and DNA. A representative transfected cell in mitosis is shown in each case, except for Myc-Eg5 S1033A, where a mitotic plus two interphase transfected cells are shown. (B). Hela cells were co-transfected with either control oligonucleotides (lanes 1-6) or siRNA oligonucleotides directed against HsEg5 (lanes 7-12), together with empty plasmid (lanes 1,7) or plasmids encoding Myc- Eg5 wt, unmodified (lanes 2 and 8) or Myc-Eg5 variants rendered RNAi-resistant (Eg5 wild type, rrwt; Eg5[Thr926Ala], rrTA; Eg5[Ser1033Ala], rrSA; Eg5[Ser1033Asp], rrSD). Cell lysates were immunoblotted with anti-Myc (upper panel) or anti-Eg5 (middle panel) to determine recombinant and total Eg5 expression, respectively, and with anti-α-tubulin antibodies to evaluate loading (lowest panel). (C) HeLa cells were co-transfected with siRNA oligonucleotides directed against HsEg5 and plasmids encoding Myc-GFP (control) or several RNAi-resistant Myc-Eg5 (rrEg5) variants. Forty hours after transfection, cells were fixed and stained with anti-Myc and anti-β-tubulin antibodies and with DAPI. Myc-positive cells were scored as being in interphase (grey columns), in normal mitosis (white columns), or in an abnormal mitosis (mostly monopolar spindles; black columns). The figure represents the mean of three independent experiments, wherein >100 cells were scored for each point in each of the experiments; error bars indicate S.E.M.; The fraction of abnormal mitoses (black columns) in cells expressing rrEg5 wildtype was compared to each of the other conditions by Student's t test; p values < 0.05 are marked with an asterisk. The comparison of S1033A with 1033D gave a p value of 0.068.
Nevertheless, higher level expression of Myc-Eg5[Ser1033Ala] does interfere with normal spindle assembly, in comparison with a similar amount of Myc-Eg5 wildtype (not shown). To evaluate the functional efficacy of this mutant in a systematic and quantitative manner, we used siRNAs capable of depleting endogenous Eg5 (Weil et al., 2002) and concomitantly introduced the Myc-Eg5 variant cDNAs, rendered resistant to the siRNA by several silent point mutations, achieving comparable levels of expression of each Eg5 polypeptide (Figure 5B). GFP was used as a negative control. Transfected cells were identified by Myc positivity. Figure 5C shows that, as expected, Eg5 siRNA cotransfected with GFP resulted in approximately 80% of transfected cells exhibiting a mitotic arrest with condensed chromosomes arranged around a monopolar spindle focused on two non-separated centrosomes (black columns). Cells expressing Myc-Eg5 wildtype were predominantly in interphase (grey columns) and the fraction of transfected cells exhibiting a monopolar spindle was greatly reduced as compared with the GFP transfected cells; in contrast Myc-Eg5[Thr926Ala], despite expression comparable to wildtype Myc-Eg5, does not ameliorate (and may increase further) the siRNA-induced mitotic arrest. Notably, Myc-Eg5[Ser1033Ala] is substantially less effective in rescue than Myc-Eg5 wildtype. Whereas Myc-Eg5 wildtype reduces the fraction of cells with monopolar spindles from ~80% to ~10%, Myc-Eg5[Ser1033Ala] reduces this proportion to ~40%, significantly less than that achieved by Eg5 wildtype. Notably, the Myc-Eg5[Ser1033Asp] mutant is more efficacious, reducing the fraction of cells with monopolar spindles to ~20%, significantly better than Myc-Eg5[Ser1033Ala] and quite close to Myc-Eg5 wildtype.
Thus although the Ser1033Ala mutation does not interfere with the ability of Eg5 to bind to the mitotic spindle, it does impair one or more of the several functions of Eg5 necessary for normal mitotic progression.
Discussion
These results demonstrate that during mitosis, HsEg5 is phosphorylated at both Thr926 by Cdk1 (Blangy et al., 1995), and at the newly identified site Ser1033 by Nek6. Ser1033 phosphorylation occurs on a subset of the spindle associated Eg5 polypeptides. Comparison of peak intensities in our MS analysis of Eg5 indicates that ~90% of mitotic Eg5 is phosphorylated at Thr926, probably explaining the detection of this site in samples of Eg5 from exponentially growing cells. Comparison of the 32P content of peptides on our 2D maps indicates that 32P incorporation into Eg5[Ser1033] is approximately 3% that of Thr926. Whereas the Thr926 phosphorylation is necessary for Eg5 spindle association, elimination of Ser1033 does not detectably affect Eg5 spindle binding. Nevertheless, Eg5[Ser1033Ala] is clearly impaired for one or more functions required for normal spindle assembly, which can be better supplied by an Eg5[Ser1033Asp] mutant. Thus, cells have spatially localized and differentially modified pools of Eg5; this may help to explain how the kinesin can perform the multiple cellular roles that have been assigned to it. The specific functions of Eg5[Ser1033] phosphorylation are not known. The baculoviral Eg5 Ser1033 mutants do not differ dramatically from wildtype Eg5 in microtubule binding or motility on microtubular monolayers, suggesting that major effects on motor function are unlikely. Our efforts to characterize the effect of Eg5 phosphorylation in vitro on the function of Eg5 is severely hampered by the lability of the motor function. Consistent with its centrosomal localization, serine 1033 phosphorylation may be related to centrosome separation before and/or during spindle formation. Although further speculation is unwarranted, the conservation of at least one LXXS/T motif at or near that of HsEg5[Ser1033] in practically all the members of the BimC family of kinesins strongly points to a conserved function for this phosphorylation in many organisms.
The present data establish the Nercc1/Nek6 cassette as one of the regulators of Eg5 function in mitosis and provide a plausible mechanism for at least part of the observed phenotypes resulting from interference with Nek6 (Yin et al., 2003), its activator kinase Nercc1/Nek9 (Roig et al., 2002; Roig et al., 2005), and possibly Nek7 (Yissachar et al., 2006). Nek6 thus joins Cdk1 and the Aurora-family kinase pEg2 as kinases capable of phosphorylating and regulating Eg5 kinesins. The present data is, to our knowledge, the first example of regulation of a kinesin by a NIMA-family kinase.
Materials and methods
Cell culture and transfection
293T, U2OS and HeLa cells were grown at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% heat inactivated fetal calf serum (FCS) and 50IU of penicillin per ml and 50mg of streptomycin per ml (Invitrogen). XL177 cells were grown in 70% Leibovitz-15 media supplemented with L-glutamine, 10% heat inactivated FCS and 50IU of penicillin per ml and 50mg of streptomycin per ml at 25°C. 293T and HeLa cells were transfected with the indicated expression plasmids using Lipofectamine and Lipofectamine 2000 respectively according to the manufacturers instructions (Invitrogen). siRNA and DNA cotransfection was performed using Lipofectamine 2000 according to manufacturers instructions.
Antibodies and Immunotechniques
Polyclonal anti-Nek6, anti-Nercc1, anti-P-Nek6/7 and anti-P-Nercc1 antibodies were produced as described in (Belham et al., 2003) and (Roig et al., 2002; Roig et al., 2005). Anti-Xl Eg5 antibodies were produced as described in (Miyamoto et al., 2004). Monoclonal anti-Eg5 antibodies and anti-cyclin B antibodies were purchased from BD Biosciences (San Jose, CA, USA), anti-CDK1 Y15 from Cell Signaling (Danvers, MA, USA), normal IgG from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA), anti-myc, anti-β-tubulin, anti-α-tubulin and anti-γ-tubulin from Sigma (St Louis, MO, USA). Secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Cell lysis, immunoprecipitation, and immunoblotting methods have been described previously (Roig et al., 2002) unless stated.
Two-hybrid analysis
cDNAs coding for the HsEg5 fragments indicated in Fig.1 were subcloned into pGBKT7 and transformed into S. cerevesiae AH109 strain by the lithium acetate method. Nercc1 and Nek6 cDNAs were subloned into pGADT7 and transformed into S. cerevesiae Y187 strain. Mating was carried out according to manufacturer procedures (Clontech Laboratories, Inc. Paol Alto, CA, USA) in 2X YPD medium for 16 hours. Selection was carried out in SD/-Leu/-Trp/-His/-Ade and interactions were verified using α-Gal.
Protein expression and purification
MalBP-Eg5 fusion protein expression in DH5α was induced with 0.4mM IPTG for 5h at 37°C and purified using amylose resin according to the manufacturers instructions (New England Biolabs, Ipswich, MA, USA). GST-Eg5 was expressed in 293T cells by transient transfection. Forty eight hours later cells were lysed in buffer A (150mM NaCl, 1% Triton-X 100, 0.5% sodium deoxycholate, 0.1% SDS, 1mM DTT, 1mM EDTA, 1mM EGTA, 50mM Tris pH8.0, protease inhibitor tablet (Roche Applied Science, Penzberg, Germany) and adsorbed to glutathione sepharose (Amersham Biosciences, Buckinghamshire, UK) and eluted with 25mM reduced glutathione after washing. Flag-Nek6 wildtype, Flag-Nek6(K74/75M) and Flag-Nercc1/Nek9 were transiently expressed in 293T cells. Forty eight hours after transfection cells were lysed in buffer B (150mM NaCl, 1% NP-40, 50mM Tris pH8.0, protease inhibitor tablet (Roche)), the recombinant polypeptides adsorbed to Flag-Agarose and eluted with Flag peptide (0.1 μg/μl, Sigma) after washing. His6-Eg5 polypeptides were generated in SF9 cells using recombinant baculovirus according to the manufacturers instructions (Invitrogen). His6-Eg5 polypeptides were extracted and purified using Nickel agarose according to the manufacturers instructions (Qiagen, Hilden, Germany) followed by FPLC gel filtration.
In vitro kinase assays
Maltose binding Protein (MalBP), MalBP-Eg5 fusion proteins and myelin basic protein (2μg each) were phosphorylated by Flag-Nek6 wildtype or preactivated (with 100μM ATP for 30 min at 30°C) Flag-Nercc1/Nek9 wild-type in kinase buffer (50mM MOPS pH7.4, 10mM MgCl2, 2mM EGTA, 20mM β-glycerophosphate, 5μM ATP, 0.83pmol [γ-32P] ATP) in a total volume of 50μl for 30 min at 30°C. GST-Eg5 and His6-XlEg5 (2μg each) were phosphorylated as above except that kinase buffer was supplemented with 20μM ATP. Reactions were stopped by the addition of 20μl 5X Laemmli buffer. Samples were separated by SDS-PAGE, Coomassie blue stained, exposed to a phosphorImager and sometimes quantitated by liquid scintillation counting.
Mass Spectrometry (MS)
Endogenous Eg5 was immunoprecipitated from extracts of cycling or nocodazole-arrested (0.5μg/ml, 24h) U2OS cells. The lysis buffer contained 150mM NaCl, 1% Triton-X 100, 0.5% sodium deoxychlate, 0.1% SDS, 1mM DTT, 1mM EGTA, 1mM EDTA, 10mM β-glycerophosphate, 1mM Na3VO4, 100nM calyculin, 0.1M NaF, 50mM Tris pH8.0. Immunoprecipitation utilized 2mg extract protein and 8μg anti-Eg5 (BD Biosciences) conjugated to 120μl protein G Dynal beads (Invitrgen) in lysis buffer, incubated with rotation for 1h at 4°C. After three washes in lysis buffer, bound proteins were eluted into 70μl 3X Laemmli buffer. Recombinant MalBP-Eg5[1-361] and MalBP-Eg5[762-1057] were phosphorylated by Flag-Nek6 as described above using 20μM ATP; parallel assays were performed with [γ-32P] ATP to define extent of phosphorylation. After SDS-PAGE, fixation and staining, in situ tryptic digestion of gel slices and LC/MS/MS analysis of phosphorylation sites was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA, USA) as described previously (Roig et al., 2005). Note that the HsEg5 sequence used here, published in the NCBI database as accession number P52732 and confirmed by our MS analysis, differs by one residue from that used by (Blangy et al., 1995; Blangy et al., 1997) and (Nousiainen et al., 2006); we find HsEg5 residues 674/675 to be EL, whereas the latter authors report these as RNS, resulting in one additional residue.
In vivo labeling and two-dimensional tryptic phosphopeptide mapping
U2OS cells either growing exponentially or treated with nocodazole (250 ng/ml) for 16 hours, were washed with TBS, resuspended in phosphate-free Dulbecco's modified Eagle's medium containing 10% PBS-dialyzed FCS, without or with nocodazole. After 30 minutes at 37 °C, 32Pi (1mCi/ml) was added for an additional 90 minutes. The 32P-labelled cells were extracted and Eg5 immunoprecipiates were subjected to SDS-PAGE, fixation, and staining. The gel slices containing Eg5 were equilibrated in 50 mM ammonium bicarbonate (pH 8), homogenized, and subjected to several rounds of tryptic digestion. The dried, salt-free digest was separated by thin layer electrophoresis at pH 1.9 followed by TLC as described previously (Belham et al., 2003). 32P was visualized and quantified using a PhosphorImager.
Phosphospecific antibodies
Rabbits were immunized with synthetic peptides corresponding to HsEg5[1028-1038] or XlEg5[1041-1051] containing a phosphorylated Serine at position 1033 and 1046 respectively, coupled to KLH through an aminoterminal cysteine. Phosphopeptide-specific antibodies were affinity purified from sera depleted of antibodies reactive with the nonphosphorylated peptides.
Immunohistochemistry
XL177 cells grown on coverslips were rinsed with 70% phosphate-buffered saline (PBS), and fixed by immersion in methanol at −20°C for 5 min, rinsed with PBS, immersed in solution A (3% bovine serum albumin in PBS plus 0.1% Triton X-100 and 0.02% azide) for 30 min and subsequently in solution A containing specific antibodies for as follows (final concentration): rabbit anti-P-XlEg5 (1:100); rabbit anti-Eg5 (1:2000); mouse anti-β-tubulin (1:1000); anti-γ-tubulin (1:400). Primary antibodies were visualized with either Cy2-conjugated donkey anti-rabbit or rhodamine X-conjugated donkey anti-mouse. DNA was visualized with 4,6-diamidino-2-phenylindole (DAPI, 0.01 mg/ml). Rinsed coverslips were mounted on a microscope slide. Images were taken using a Leica TCS SPE confocal system with a DM2500 CSQ upright microscopy and a 63× 1.30 ACS Apo lens, and edited using Leica LAS AF 1.6.3 software (Leica Microsystems, Mannheim, Germany).
Supplementary Material
Acknowledgements
We acknowledge the seminal research of Christopher Belham demonstrating the binding of Nek6 to Eg5. We thank Dr. M. Kress (CNRS, Villejuif, France) for Eg5 cDNAs and Dr. Isabelle Vernos (Centre de Regulació Genòmica, Barcelona, Spain) for XL177 cells and insightful comments. JR acknowledges support from the Ramón y Cajal Program and the Plan Nacional I+D grant BFU2005-05812 (MEC, Spain), the European Commission through the Marie Curie IRG MIRGCT-2005-031088, and institutional funds. JA from NIH grant DK17776 and institutional funds.
Footnotes
Supplemental Material
Supplemental Materials include Supplemental Experimental Procedures and two figures.
References
- Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem. 2003;278:34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- Belham C, Comb MJ, Avruch J. Identification of the NIMA family kinases NEK6/7 as regulators of the p70 ribosomal S6 kinase. Curr Biol. 2001;11:1155–1167. doi: 10.1016/s0960-9822(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272:19418–19424. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PM, Milne A, Garka K, Johnson RS, Willis C, Sims JE, Rauch CT, Bird TA, Virca GD. Purification, cloning, and characterization of Nek8, a novel NIMA-related kinase, and its candidate substrate Bicd2. J Biol Chem. 2002;277:16229–16240. doi: 10.1074/jbc.M108662200. [DOI] [PubMed] [Google Scholar]
- Kandli M, Feige E, Chen A, Kilfin G, Motro B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics. 2000;68:187–196. doi: 10.1006/geno.2000.6293. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Le Guellec R, Paris J, Couturier A, Roghi C, Philippe M. Cloning by differential screening of a Xenopus cDNA that encodes a kinesin-related protein. Mol Cell Biol. 1991;11:3395–3398. doi: 10.1128/mcb.11.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Deak M, Morrice N, Kieloch A, Hastie CJ, Dong L, Schutkowski M, Reimer U, Alessi DR. Molecular basis for the substrate specificity of NIMA-related kinase-6 (NEK6). Evidence that NEK6 does not phosphorylate the hydrophobic motif of ribosomal S6 protein kinase and serum- and glucocorticoid-induced protein kinase in vivo. J Biol Chem. 2002;277:27839–27849. doi: 10.1074/jbc.M202042200. [DOI] [PubMed] [Google Scholar]
- Miyamoto DT, Perlman ZE, Burbank KS, Groen AC, Mitchison TJ. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MJ, Krien MJ, Hunter T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003;13:221–228. doi: 10.1016/s0962-8924(03)00056-4. [DOI] [PubMed] [Google Scholar]
- Quarmby LM, Mahjoub MR. Caught Nek-ing: cilia and centrioles. J Cell Sci. 2005;118:5161–5169. doi: 10.1242/jcs.02681. [DOI] [PubMed] [Google Scholar]
- Roig J, Groen A, Caldwell J, Avruch J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol Biol Cell. 2005;16:4827–4840. doi: 10.1091/mbc.E05-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J, Mikhailov A, Belham C, Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J, Avruch J. Nek protein kinases. In: Watling KJ, editor. SIGMA-RBI Handbook of Receptor Classification and Signal Transduction. 5th Edition Sigma-RBI; Natick: 2006. pp. 286–290. [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Weil D, Garçon L, Harper M, Duménil D, Dautry F, Kress M. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques. 2002;33:1244–1248. doi: 10.2144/02336st01. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Shao L, Voehringer D, Smeal T, Jallal B. The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J Biol Chem. 2003;278:52454–52460. doi: 10.1074/jbc.M308080200. [DOI] [PubMed] [Google Scholar]
- Yissachar N, Salem H, Tennenbaum T, Motro B. Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 2006;580:6489–6495. doi: 10.1016/j.febslet.2006.10.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.