Abstract
Germ cells divide and differentiate in a unique local microenvironment under the control of somatic cells. Signals released in this niche instruct oocyte reentry into the meiotic cell cycle. Once initiated, the progression through meiosis and the associated program of maternal mRNA translation are thought to be cell-autonomous. Here we show that translation of a subset of maternal mRNAs critical for embryo development is under the control of somatic cell inputs. Translation of specific maternal transcripts increases in oocytes cultured in association with somatic cells and is sensitive to EGF-like growth factors that act only on the somatic compartment. In mice deficient in amphiregulin, decreased fecundity and oocyte developmental competence is associated with defective translation of a subset of maternal mRNAs. These somatic cell signals that affect translation require activation of the PI3K/AKT/mTOR pathway. Thus, mRNA translation depends on somatic cell cues that are essential to reprogram the oocyte for embryo development.
Germ cell differentiation requires a unique microenvironment created by surrounding somatic cells. In gonads of adult Drosophila and C. elegans, this environment provides the “niche” that is key to the maintenance of the germ stem cell pool1. A similar niche is critical for spermatogonial stem cell replication and differentiation to maintain spermatogenesis in mammals2. In adult mammalian ovaries, the follicle microenvironment in which the oocyte develops is the conduit for bidirectional exchange of signals with surrounding somatic cells3. With the possible exception of transcription in model organisms, the molecular basis for somatic/germ cell interactions is still poorly understood.
Upon signals from the soma in the ovarian follicle, fully grown mammalian oocytes reenter into the meiotic cell cycle, complete the first meiotic division, and progress to metaphase II (MII) of the second division4. Although these transitions still occur if oocytes are freed from surrounding somatic cells and cultured in vitro, it is commonly accepted that, in the absence of somatic cell contacts, oocyte fertilization and embryo development are compromised5-7. These defects probably arise from disruptions in the poorly-defined molecular process by which the oocyte acquires developmental competence, termed cytoplasmic maturation8. Nuclear transfer experiments indeed show that the defects associated with oocyte denudation and in vitro culture reside in the cytoplasm6. Since cytoplasmic maturation of the oocyte and early embryo development proceed in the absence of transcription, competence to develop as an embryo must rely upon a genome-wide program of maternal mRNA translation and degradation.
Oocyte maturation and ovulation induced by the gonadotropin LH requires activation of paracrine/autocrine signals within the follicle. In addition to the release of prostaglandins and steroids, LH induces large increases in amphiregulin (Areg), epiregulin (Ereg), and betacellulin mRNAs within 1-3 hrs of stimulation in mural granulosa cells9, followed by an increase in cumulus cells10. These growth factors bind to the EGF receptor (EGFR) on granulosa cells, and their release mediates the LH-dependent transactivation of EGFR. Genetic and pharmacological data demonstrate that activation of this EGF network is essential to transmit the gonadotropin signal from the mural granulosa cells to the cumulus cells and the oocyte, to induce oocyte maturation, cumulus expansion and ovulation11-13.
Translation of maternal mRNAs during oocyte maturation requires polyadenylation directed by RNA binding proteins, the prototype being the cytoplasmic polyadenylation element binding (CPEB) protein. In Xenopus oocytes, CPEB-mediated translation is under the control of cell cycle regulators which function in a cell-autonomous fashion14. Limited information is available on whether translation during the meiotic cell cycle is affected by somatic cell signals. Here we have tested the hypothesis that the environment in which oocytes complete meiosis and signals from somatic cells control translation in the oocytes. This regulation is critical for mammalian oocyte competence to develop as an embryo.
The accumulation of the spindle component TPX2 is dependent on the environment in which the oocyte matures
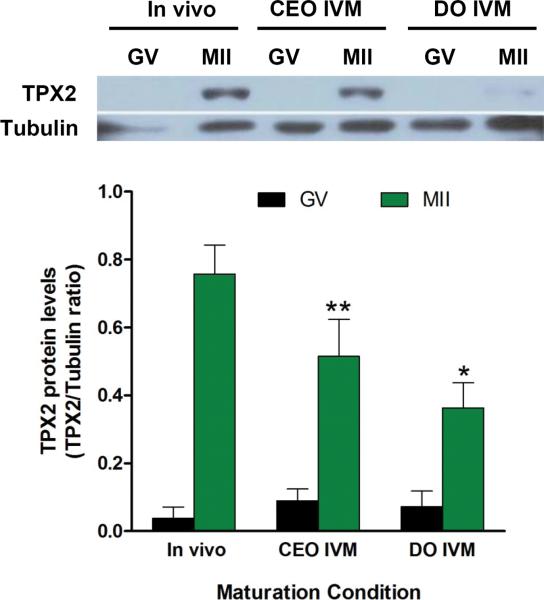
TPX2 (Targeting Protein for the Xenopus kinesin xklp2) is a protein essential for spindle assembly and chromosome interaction with microtubules15, 16. It binds and activates Aurora A by promoting its autophosphorylation17. TPX2 level of expression is critical for spindle function, and altered expression is associated with aneuploidy and cancer18-20. In agreement with a previous report21, we show that TPX2 is undetectable in oocytes in prophase and accumulates during maturation to MII (Fig. 1). It has been proposed that the absence of TPX2 accumulation in prophase is due to protein degradation through APC/Cdh121. Indeed, little change in Tpx2 mRNA translation occurs during the early phases of oocyte maturation, but the late TPX2 accumulation is associated with an increased translation22. Surprisingly, we found that TPX2 protein accumulation is not only dependent on the stage of the meiotic cell cycle. Significant differences in TPX2 protein levels were observed when comparing MII oocytes matured in vivo with those matured in vitro in association with somatic cells, or those matured in vitro after being denuded. This initial finding suggests that TPX2 accumulation is sensitive to the environment in which the oocyte matures.
Fig. 1. The protein levels of the spindle component TPX2 is dependent on the environment in which the oocyte matures.
Representative and cumulative data of Western blots of TPX2 protein in GV and MII oocytes matured in vivo, or in vitro matured in complex with cumulus cells (CEO), or when denuded prior to maturation in vitro (DO). Tubulin was used as loading control. The bar graph is the summary of densitometric measurements (mean ± SEM) of Western blots done with four different oocyte extracts prepared on different days. Data are presented as ratio of TPX2/ Tubulin intensity (see Supplementary Table S3 for statistics source data). Paired T test: * p=0.013 MII DO versus MII in vivo.** p=0.0022 CEO MII versus MII in vivo. MII DO versus CEO MII not significant (p= 0.316).
Translation of TPX2 and other mRNAs in oocytes is sensitive to somatic cell cues
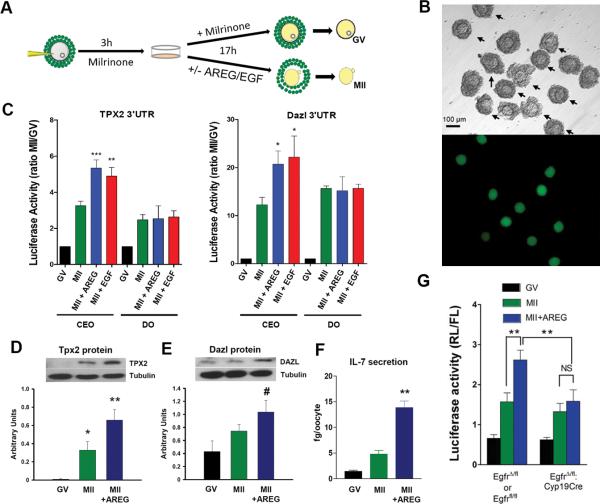
To investigate whether cumulus cells, the somatic cells surrounding the oocyte, play a role in the translation of maternal mRNAs and protein synthesis, we developed an in vitro model that preserves the somatic environment in which the oocyte matures. Translational reporters were constructed and injected into oocytes still surrounded by cumulus cells (cumulus cell - enclosed oocyte, CEO) (Fig. 2A,B). This model enables monitoring translation of selected maternal mRNA in oocytes that maintain contact with cumulus cells. Furthermore, translation rates in CEOs can be compared to those measured in denuded oocytes (DOs), which are no longer exposed to somatic signals.
Fig. 2. EGF-like growth factor stimulation of cumulus/oocyte complexes in vitro increases translation in oocytes.
A. Scheme describing the experimental conditions to measure translation rates in cumulus enclosed oocytes (CEO). Oocytes still in complex with cumulus cells were injected with two reporters, one coding for the renilla luciferase under the control of maternal mRNA (Tpx2 or Dazl) 3’UTR and one coding for the firefly luciferase with polyadenylated 3’ UTR. After three hours of incubation in the presence of milrinone to maintain the meiotic arrest, a group of injected CEO was washed free of inhibitor and incubated with or without EGF-like growth factors. In some experiments, an additional group of CEOs was mechanically denuded and oocytes incubated with growth factors as above. At the end of the incubation, oocytes were dissected free of cumulus cells and luciferase activity was measured in oocyte extracts. B. Expression of GFP in CEOs after microinjection documenting the accuracy of the injection and confinement of the fluorescence to the oocyte. C. Ratio of renilla/firefly luciferase activity in oocytes cultured as CEO or denuded incubated with or without AREG (100nM) or EGF (10nM). Each bar is the ratio of MII/GV activities and mean ± SEM of 6 experiments for TPX2 and 4 experiments for Dazl. T test: * p<0.05; ** p<0.01; *** p<0.001 vs MII. D,E. CEO were incubated as in C and at the end of the incubation, oocytes were denuded, and extracts were used to evaluate endogenous Dazl and TPX2 protein levels by Western Blot. Tubulin was used as loading control. The bar graph is the summary of densitometric measurements (mean ± SEM) of 5 Western blots for TPX2 and 6 for Dazl done with different oocyte extracts. T test: * p<0.05 vs GV; ** p<0.01 vs MII; # p<0.05 vs MII. F. Bars represent the mean ± SEM of 4 experiments where IL-7 accumulation was measured in spent media of CEO cultured as in C with or without AREG. T test: ** p<0.01 vs MII. G CEO were prepared from PMSG-treated Egfrflfl , EgfrΔ/fl, or EgfrΔ/fl:Cyp19 CRE mice. They were then injected with a Tpx2 luciferase reporter and incubated as detailed in panel A in the absence or presence of AREG. At the end of the incubation, oocytes were dissected free of cumulus cells and luciferase activity was measured in oocyte extracts. The data from Egfrflfl and EgfrΔ/fl groups gave similar results and were combined. Data are the mean ± SEM of 4 independent experiments. Paired T test: * p=0.018 MII+AREG vs MII of Egfrflfl /EgfrΔ/fl ; NS = 0.78 MII+AREG vs MII of EgfrΔ/fl:Cyp19 CRE; p=0.033 MII+ AREG of Egfrflfl vs MII+AREG of EgfrΔ/fl:Cyp19 CRE. See Supplementary Table S3 for statistics source data
Reporter constructs with luciferase ORFs under the control of 3’UTRs of Tpx2 or Dazl, an RNA binding protein essential for gametogenesis23, were injected into CEOs. Translation rates of these reporters increased as the oocytes progressed from GV to MII (Fig. 2C), consistent with our report of recruitment of the corresponding endogenous transcripts to the polysomes22. However, translation in CEO is further increased by supplementing the incubation medium with amphiregulin (AREG), an EGF-like growth factor that accumulates physiologically in the follicle during ovulation22, or EGF itself (Fig. 2C). Both ligands signal through EGF receptor (EGFR) on cumulus cells24 and are not expressed by oocytes in culture. Growth factor-induced effects were not detected when meiotic reentry was prevented with the phosphodiesterase inhibitor milrinone (Suppl. Fig 1), when a TPX2 reporter with truncated 3’UTR was injected (Suppl. Fig. 2), or when oocytes were denuded prior to stimulation (Fig. 2C). These findings demonstrate that the 3’ UTR , progression through meiosis, and somatic cells are all required for the growth factor-dependent stimulation. Exposure to AREG did not increase the stability of the reporter, confirming an effect on translation rate (Suppl. Fig. 1B).
Consistent with reporter translation, endogenous TPX2 and DAZL protein levels increased as oocytes mature (Fig. 2D,E). AREG further enhanced these protein levels, confirming that increased translation of the reporter reflects translation of the endogenous mRNA and accumulation of the encoded protein. When incubated with AREG and in contact with somatic cells and AREG, a rise in oocyte protein synthesis was independently confirmed by monitoring accumulation of IL-7, an oocyte secreted chemokine, in the medium (Fig. 2F).
To conclusively demonstrate that AREG does not directly stimulate the oocyte and that the signals are indirect and mediated by somatic cells, we used a genetic mouse model in which EGFR is down-regulated only in somatic cells. Mice carrying a null Egfr allele and a floxed allele (EgfrΔ/fl), and expressing a Cre recombinase under the control of the granulosa cell-specific CYP19A1 promoter25, display a 90% decrease in EGFR expression in granulosa cells13. Although LH-dependent in vivo oocyte maturation is impaired, in vitro spontaneous maturation progresses normally13. We used CEO derived from these mice to test whether Egfr gene inactivation in somatic cells ablates the AREG effects on oocyte translation in vitro. The experiment reported in Fig. 2G demonstrates that AREG does not significantly increase translation rate of TPX2 reporter when CEO from EgfrΔ/fl: CYP19A1-Cre mice are microinjected.
Defective AREG production in vivo is associated with altered translation and compromised oocyte developmental competence
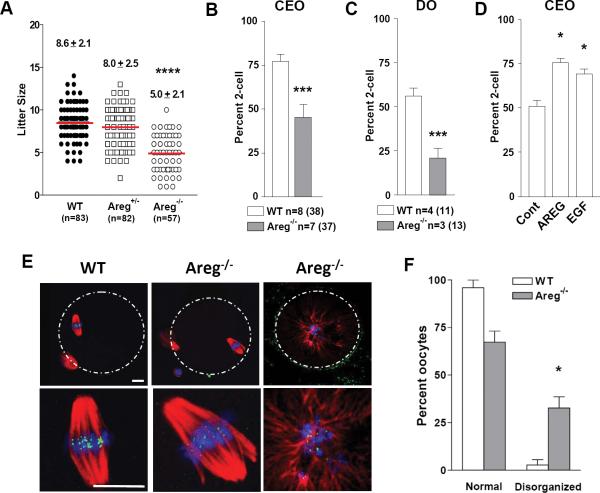
To determine whether this somatic effect on oocyte translation occurs also in vivo and whether global translation is affected, genetic mouse models perturbing the EGF network were investigated. We have reported that the EGF-like growth factors AREG, EREG and BTC are induced by LH at the time of ovulation and that transactivation of EGFR is indispensable for oocyte maturation and ovulation11, 13. Whereas inactivation of EGFR causes a block in oocyte maturation in vivo and defective ovulation11, Areg−/− mice ovulate and are fertile11 but litter size is significantly decreased (Fig. 3A). When fertilization rates were assessed in vitro, CEOs or denuded oocytes derived from Areg−/− mice fertilize at a rate significantly lower than wild type (WT) littermates, suggesting a defect in developmental competence (Fig. 3B and C). Identical to the Areg−/− follicle activated in vivo, isolated CEOs from wild type mice cultured in vitro are not exposed to AREG. Therefore, the role of AREG in promoting cytoplasmic maturation was further tested by adding exogenous AREG during WT CEO in vitro maturation. This treatment improved their fertilization rate (Fig.3D), a finding consistent with other studies demonstrating positive effects of EGF network on developmental competence26-28.
Fig. 3. Compromised developmental competence of oocytes from Areg−/−mice.
A. Summary of offspring from mating of wild type, Areg +/−, and Areg−/− mice. The number of litters is reported below the scatterplot. T test: **** p<0.0001 vs. WT. B. Bars represent the mean ± SEM of the two cell-embryo yield after in vitro fertilization (IVF) using CEOs from super-ovulated wild type and Areg−/− mice. T test: *** p<0.0001 vs. WT. The number of IVF experiments performed is reported below the bars. The number in parenthesis indicates the number mice used. C. After superovulation, a group of MII CEOs was stripped of cumulus cells and used for IVF. Bars represent the mean ± SEM of two cell-embryo yield. T test: *** p<0.001 vs. WT. The number of IVF experiment performed is reported below the bars. D. Bars represent the mean ± SEM of two cell embryo yield of wild type CEOs harvested from PMSG treated mice, cultured in vitro in the absence or presence of AREG for 12 hr until they had reached the MII stage and then used for IVF. T test:* p<0.05 vs. WT. E. Representative spindles from wild type and MII Areg−/− oocytes derived from superovulated CEO. Bar corresponds to 10 μm. F. Incidence of aberrant spindle formation n WT and Areg−/− oocytes; Bars represent the mean ± SEM of 3 experiments. paired T test: * p<0.05 vs. WT. See Supplementary Table S3 for statistics source data.
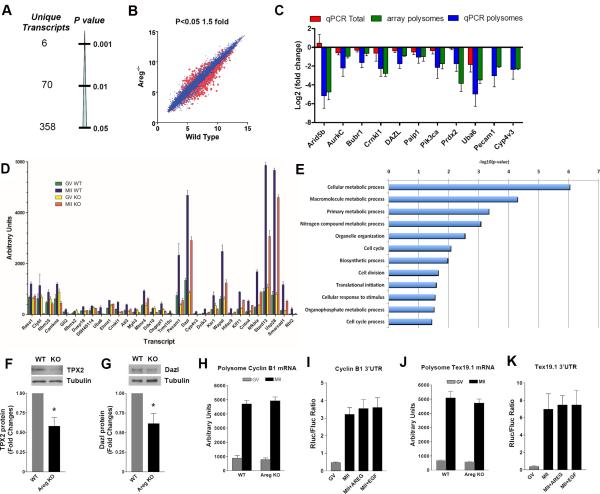
Defective spindle morphology was more frequently detected in MII oocytes derived from Areg−/− mice (Fig. 3E,F). Thus, inactivation of Areg in the somatic cells yields oocytes that mature but with signs of reduced developmental competence. Our model was then used to test whether the translational program executed during oocyte maturation is affected by the absence of this somatic signal. To capture actively translating mRNAs, polysomes were isolated by sucrose density gradient from WT and Areg−/− oocytes. Maternal transcripts associated with the polysomes were isolated and analyzed by microarray hybridization (Fig. 4). Consistent with our previous report, 3208 transcripts were recruited or released from the polysomes during oocyte maturation in wild type oocytes (Suppl. Files l and 2). A similar number of transcripts (3440) moved in and out of the polysome pool also in oocytes from Areg−/− mice, confirming that the translation program is qualitatively intact. However, quantitative analysis revealed significant changes in the level of transcripts associated with polysomes in mutant oocytes. When using a cutoff of p<0.05 (Fig. 4A), polysome association of approximately 200-300 transcripts was altered in the Areg−/− oocytes compared to WT (Fig. 4B). Transcripts normally recruited to the polysome during maturation were decreased in the Areg−/− oocytes (Fig.4C). The affected transcripts included the main functional categories of metabolism, embryonic development, cell cycle and RNA regulators (Fig. 4D). Analysis of the 3’UTRs affected in the Areg-/- oocytes did not reveal the presence of a common signature, with the possible exception of two consensus elements enriched in a subset of transcripts (Suppl. Fig. 3). We confirmed the decrease of selected transcripts in the polysome pool by qPCR (Fig. 4E). Protein levels of DAZL and TPX2 in MII oocytes of the Areg−/− mice were decreased (Fig. 4F,G), which is consistent with the decreased recruitment of these mRNAs to the polysome in vivo. The absence of somatic AREG affected only a subset of transcript translation. Polysome recruitment of Cyclin B1 and Tex19.1, two transcripts known to be highly regulated during oocyte maturation, were not affected in the Areg−/− oocyte and were insensitive to AREG stimulation in the in vitro translation assay (Fig.4H,I,J,K). Thus, somatic cell signals affect translation of a subset of maternal mRNAs in vivo and in vitro.
Fig. 4. Altered mRNA translation of a subgroup of maternal mRNAs in Areg−/−.
oocytes. A,B. Analysis of polysome-associated transcripts in wild type and Areg−/− MII oocytes. Three distinct pools of 500-750 oocytes from wild type and Areg−/− mice were used for the analysis. The actual data are reported in Supplemental Tables1 and 2. C. Comparison of hybridization data and qPCR of total and polysome-bound distribution of selected transcripts; T test p <0.05 from three distinct mRNA preparations. D. Comparison of recruitment to the polysome of most downregulated transcripts in Areg-/-oocytes. The ratio MII/GV was calculated for the top 31 transcripts whose levels were most significantly different in polysomes from MII Areg−/− oocytes compared to WT MII. Data are reported as Mean ± SE intensity for three biological replicates. Actual data and statistics are reported in Table 1 and 2. Of note, many of the transcripts downregulated in the Areg−/− are recruited to the polysome during the GV-to-MII transition. E. Gene Ontology analysis of transcripts significantly different in WT and Areg−/− oocytes. Transcripts recovered at levels significantly different in the polysomes (p<0.05) and with WT/Areg−/− ratio of at least 1.5 were included in the analysis. Enriched Gene Ontology terms were discovered using DAVID (see methods for details). F,G. comparison of TPX2 and Dazl levels in wild type and Areg-/- super-ovulated MII oocytes. The bars are the mean ± SEM of densitometric analysis of 5 Western blots for TPX2 and 4 Western blots for Dazl proteins don on different groups of oocytes. T test: * p<0.05 vs. WT. H. Bars represent the mean ± SEM of polysome associated Cyclin B1 mRNA in wild type and Areg−/− oocytes. G. In vitro translation of cyclin B1 3’ UTR reporter in CEO cultured with or without AREG. Bars represent the mean ± SEM of 3 distinct experiments with different pools of CEOs. J. Bars represent the mean ± SEM of polysome associated Tex19.1 mRNA in wild type and Areg−/− oocytes. K. In vitro translation of the Tex19.1 3’ UTR reporter in CEO cultured with or without AREG. Bars are the mean ± SEM of 3 separate experiments. In experiments reported in panel H-K, no statistical difference could be observed between the WT and Areg−/− groups. See Supplementary Table S3 for statistics source data.
PI3K/AKT/mTOR signaling is involved in the somatic regulation of oocyte mRNA translation
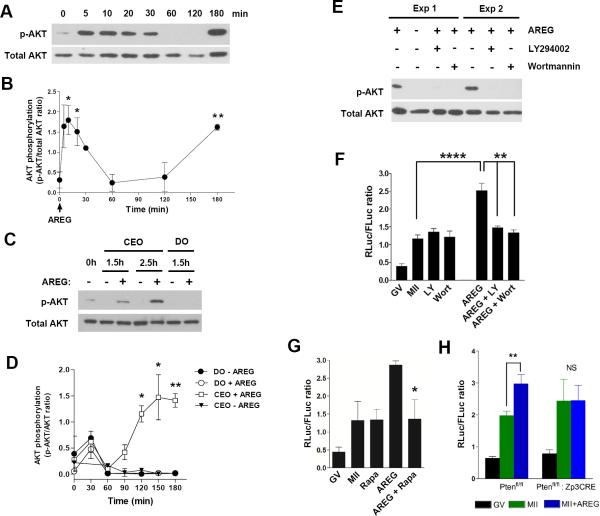
Incubation of CEO with AREG caused a rapid increase in AKT phosphorylation indicative of activation of the PI3K pathway (Fig. 5A,B). Surprisingly, we found that the time course of CEO activation is biphasic with a secondary increase in AKT phosphorylation detected at 120-180 min of incubation with AREG (Fig. 5A,B). Because the delayed increase in AKT phosphorylation was reminiscent of the AKT phosphorylation detected in the oocyte 2-3 hrs after LH/hCG stimulation29, 30, we investigated whether this secondary increase takes place in the oocyte. CEOs were incubated in the presence of AREG; at the end of the incubation, the oocytes were denuded and used for Western Blot analysis (Fig. 5C,D) or in situ detection by immunofluorescence (Suppl. Fig. 5). A delayed AKT phosphorylation was observed in oocytes exposed to AREG while still in complex with cumulus cells; conversely, this delayed phosphorylation could not be detected in denuded oocytes exposed to AREG (Fig. 5C,D and Suppl. Fig. 5). Variable AKT phosphorylation was observed in oocytes within the first 30 min whether cultured as CEO or denuded (Fig. 5D), a signal independent of AREG and likely caused by the mechanical denudation of the oocytes. These data suggest that AREG stimulation of CEOs causes a delayed transient increase in AKT phosphorylation in the oocyte. Pharmacological inhibition of PI3K with Wortmannin or LY 294002 blocked the AKT phosphorylation in oocytes when cultured in complex with cumulus cells in the presence of AREG for 2 hrs (Fig. 5E). Under these conditions, the AREG-dependent increased translation of the TPX2 reporter was abolished, whereas the cell cycle-dependent increase in reporter translation was not affected (Fig. 5F). Similar results were obtained with rapamycin (Fig. 5G and Suppl. Fig. 6), an mTOR inhibitor31.
Fig. 5. PI3K/AKT/mTOR signaling is involved in the somatic regulation of oocyte mRNA translation.
A. Representative Western blot of the time course of AKT Ser473 phosphorylation in CEOs incubated with AREG for different times. Extracts from total CEOs were used for the Western blot. B. Densitometric analysis of AKT Ser473 phosphorylation at different times after AREG exposure. Total CEO extracts were used for these measurements. Each point is the mean ± SEM of 3 different experiments. * T test: p<0.05 and ** p<0.01 vs zero time point. C. Representative Western blot of AKT Ser473 phosphorylation measured in oocyte extracts after stimulation when still in complex with cumulus cells (CEOs) or after denudation (DO). D. Densitometric analysis of AKT Ser473 phosphorylation measured in oocytes incubated with or without AREG when still in complex with cumulus cells (CEO) or after denudation. Each point is the mean ± SEM of 3 different experiments. T test: * p<0.05 and ** p<0.01 vs DO groups. Of note, phosphorylation of AKT at 0 and 30 min was highly variable. Although not significant, this trend to an increase in S473 phosphorylation at 30 min is likely due to mechanical stress due to manipulation of the oocytes during denudation. This increase is observed whether oocytes are derived from CEO or are incubated as denuded oocytes in the absence or presence of AREG. E,F. The AREG dependent activation of translation of the TPX2 reporter is prevented by PI3K inhibitors (LY294002 and Wortmannin) which block AKT phosphorylation in the oocyte. After microinjection of the TPX2 reporter, CEO were preincubated with inhibitors for 30 min and then incubated with or without AREG for 2 additional hrs. At the end of the incubation, oocytes were freed of cumulus cells and used for Western blot analysis (E). For luciferase assays (F), CEOs were incubated overnight. All oocytes used in the luciferase assays had reached the MII stage as indicated by the presence of polar body. Data are the mean ± SEM of 4 independent experiments. T test: **** p<0.0001 and ** p<0.01 . G. Effect of the mTOR inhibitor rapamycin on the AREG-dependent increase in TPX2 reporter translation. Data are the mean ± SEM of 4 independent experiments. T test:* p<0.05 vs. AREG. H. Intact PI3K signaling in the oocyte is necessary for the AREG-dependent increase in Tpx2 reporter translation. CEOs were derived from Ptenfl/fl: Zp3-CRE mice, where Pten gene is deleted only in the oocytes, and Ptenfl/fl littermates which behave as wild type mice. The incubation and the luciferase assay were conducted as detailed in Fig 2. Each point is the mean ± SEM of 3 experiments conducted in different days. T test: ** p=0.009 Ptenfl/fl MII+AREG vs MII; NS (not significant) Ptenfl/fl: Zp3-CRE MII+AREG vs MII. See Supplementary Table S3 for statistics source data.
It should be noted that, in the experiments reported above, the pharmacological inhibitors used block the PI3K/AKT/mTOR pathway both in somatic cells and in the oocyte. To determine whether the PI3K pathway function is required in the oocyte, we used a genetic model to test whether disruption of this pathway exclusively in the oocyte is sufficient to block the AREG-dependent increase in translation. To this aim, we used cumulus/oocyte complexes from Ptenfl/fl: ZP3-CRE mice. In this genetic model, the Cre recombinase is under the control of an oocyte-specific promoter and the Pten gene is ablated only in oocytes but not in cumulus cells. Previous data with this model show that Pten inactivation causes a constitutive increase in AKT phosphorylation32. In these oocytes, translation of the Tpx2 reporter is no longer dependent on AREG (Fig. 5H). This latter finding confirms that an intact PI3K pathway in the oocyte is necessary to translate the AREG signal from the soma.
Discussion
Our findings demonstrate that translation of a subset of oocyte maternal mRNAs is under the control of somatic cell inputs acting through the PI3K/AKT/mTOR pathway. This regulation functions in concert with the translational control by meiotic cell cycle regulators and is involved in establishing the oocyte competence to successfully develop as embryos.
Our in vivo and in vitro data document that exposure of somatic cell/oocyte complexes to the EGF-like growth factor AREG causes an increase translation of a subset of maternal mRNAs in the oocyte. This activation requires cell-to-cell communication, as it is lost in denuded oocytes. The use of alleles affecting Egfr and PI3K signaling in the somatic and germ cell compartments respectively confirms that AREG action requires Egfr expression in the soma and an intact PI3K signaling in the oocyte. Thus, Egfr signaling in the somatic cellular compartment is translated into PI3K activation in the contiguous cell compartment, the oocyte. Given the critical role of EGFR signaling in tumor growth33, 34, it is possible that the signaling across cells described here is also necessary for communication of transformed cell with the surrounding cellular environment.
Growth factor regulation of translation via the PI3K/AKT/mTOR pathway is established for somatic cells35, 36. However, the molecular mechanism underlying the specific regulation of a subset of transcripts in growth factor-target cells remains controversial. Through target of rapamycin complex 1 (mTORC) the PI3K pathway converges on regulation of S6 ribosomal protein and eukaryotic initiation factor 4E (eIF4E), a key rate-limiting initiation factor for cap-dependent translation35, 37. 4E-binding protein 1 (4EBP1), one of the three 4EBP binding proteins expressed in mammals, is phosphorylated by mTORC1 causing the dissociation from eIF4E and the formation of an active initiation complex38. Transcripts sensitive to mTOR regulation have signature sequences at the 5’ UTR, including a polypyrimide (5’TOP)39 or PRTE sequence40, but it is not clear whether these sequences are indispensable. The mechanism of translational regulation we describe in the oocyte is clearly distinct. 5’TOP or PRTE sequences are not readily discernible in the 5’ UTR of the prototype transcripts we have investigated in the oocyte. A selective activation is retained when different luciferase reporters are injected in oocytes, even though they have an identical 5’ UTR sequence. Conversely, the 3’ UTR of these mRNAs is essential because TPX2 3’UTR reporter recapitulates this regulation whereas the 3’UTR of, for instance, cyclin B does not support this regulation. Elements present in the 3’ UTR are necessary because a truncated TPX2 3’UTR is not sensitive to somatic cell signals. An additional feature that distinguishes the effects we describe in the oocyte from those in somatic cells is that none of the oocyte targets of regulation identified are ribosomal proteins, whereas many of the growth factor targets in somatic cells are ribosomal proteins41. On the contrary, the regulation we describe takes place on a background of widespread destabilization and decreased translation of mRNAs coding for ribosomal proteins22, 42. The pharmacological and genetic analyses performed indicate that the AKT/mTOR pathway is required for the AREG-depended translational regulation in the oocyte. We propose that mTOR signaling in oocytes controls translation via unique mechanisms that target the 3’UTR (Fig. 6). The possible role of the 5’UTR in this growth factor-dependent regulation remains to be determined.
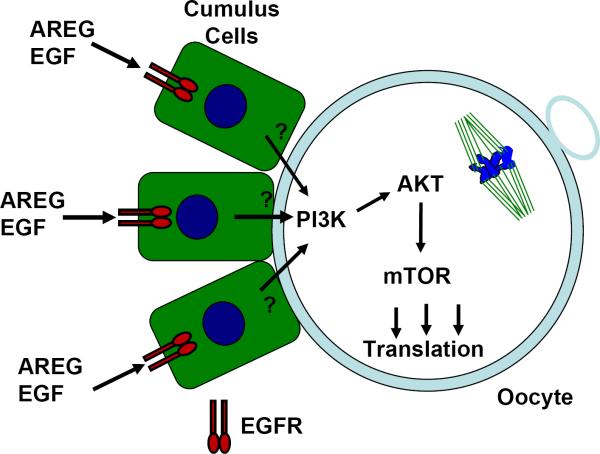
Fig. 6. Model of the signaling pathways involved in somatic cell control of oocyte translation.
Cumulus cells are represented as green squares with a blue nucleus. An MII oocyte is schematically represented with a spindle and a polar body. EGFR dimers are represented by green/red symbols. EGF-like growth factors released from mural granulosa cells interact with EGFRs expressed on cumulus cells and cause activation on undefined signaling pathway/s across the cumulus/oocyte plasma membrane. These signals lead to PI3K activation in the oocyte. The resulting phosphorylation of AKT and mTOR activation in oocytes leads to an increase in translation of a subset of maternal mRNAs.
Correct mRNA translation and accumulation of corresponding proteins in the oocyte is critical for efficient meiotic cell cycle progression, fertilization, and embryo development and necessary to attain full developmental potential. Our findings provide a molecular explanation for the widely reported observation that fully grown oocytes dissociated from the surrounding somatic cells may complete nuclear maturation but are defective in fertilization and embryo development4, 6, 26. Our findings support the idea that components required for maturation, fertilization and embryo development reach optimal levels when oocyte-somatic cell communication is preserved. In the absence of somatic cells, these components accumulate at levels sufficient for cell cycle progression but robustness and stability of the oocyte machinery is compromised, with meiosis becoming prone to errors. This conclusion is supported by our observation that TPX2, a key component in spindle assembly21, accumulates significantly less in the absence of somatic input, and spindle defects become more frequent in MII oocytes. In the same vein, decreased accumulation of Dazl, which may function upstream of TPX222, likely destabilizes the network of RNA binding protein required for maturation and embryo development. Thus, correct execution of the program of maternal mRNA translation and somatic inputs are essential for oocyte developmental competence. As an extension of this concept, biomarkers monitoring this program, such as secreted proteins, should be of prognostic value for the “fitness” of an oocyte to develop into an embryo that successfully implants and sustains pregnancy to term.
Supplementary Material
Acknowledgments
We thank Drs. Diana Laird, Davide Ruggero, Todd Nystul, and Robert Belloch at UCSF for advice during the studies and for critical reading of the manuscript, and Andrej Susor for assisting with the oocyte confocal analysis. This work was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH cooperative agreement 1U54HD055764-06, as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and RO1-GM097165 to MC.
Footnotes
AUTHOR CONTRIBUTIONS
JC developed the COC translation assay and performed the microarray experiments and some of the AKT assays; ST performed the characterization of the Areg null phenotype; FX contributed with Western blot studies and oocyte isolation for microinjection; CJL helped with immunostaining experiments and the microinjections in cumulus oocyte complexes; HC performed the experiments on protein secretion and contributed to the writing of the manuscript. FF performed microinjections in CEO, KH contributed with the preparation of the translational luciferase reporters, CO and JS performed the bioinformatic analysis of the microarray data. MIC advised on data analysis and discussed results; MRS provided reagents and constructs and advised in the interpretation of the data; MC conceived the project, designed the experiments, analyzed the data and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests
References
- 1.Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida S. Spermatogenic stem cell system in the mouse testis. Cold Spring Harb Symp Quant Biol. 2008;73:25–32. doi: 10.1101/sqb.2008.73.046. [DOI] [PubMed] [Google Scholar]
- 3.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 4.Gosden R, Lee B. Portrait of an oocyte: our obscure origin. J Clin Invest. 2010;120:973–983. doi: 10.1172/JCI41294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chian RC, Lim JH, Tan SL. State of the art in in-vitro oocyte maturation. Curr Opin Obstet Gynecol. 2004;16:211–219. doi: 10.1097/00001703-200406000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Chang HC, Liu H, Zhang J, Grifo J, Krey LC. Developmental incompetency of denuded mouse oocytes undergoing maturation in vitro is ooplasmic in nature and is associated with aberrant Oct-4 expression. Human reproduction. 2005;20:1958–1968. doi: 10.1093/humrep/dei003. [DOI] [PubMed] [Google Scholar]
- 7.Luciano AM, et al. Developmental capability of denuded bovine oocyte in a co-culture system with intact cumulus-oocyte complexes: role of cumulus cells, cyclic adenosine 3′,5′-monophosphate, and glutathione. Molecular reproduction and development. 2005;71:389–397. doi: 10.1002/mrd.20304. [DOI] [PubMed] [Google Scholar]
- 8.Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev. 1996;8:485–489. doi: 10.1071/rd9960485. [DOI] [PubMed] [Google Scholar]
- 9.Park JY, et al. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- 10.Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and Autocrine Regulation of Epidermal Growth Factor-Like Factors in Cumulus Oocyte Complexes and Granulosa Cells: Key Roles for Prostaglandin Synthase 2 and Progesterone Receptor. Mol Endocrinol. 2006;20:1352–1365. doi: 10.1210/me.2005-0504. M. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh M, et al. Luteinizing hormone-dependent activation of the epidermal growth factor network is essential for ovulation. Mol Cell Biol. 2007;27:1914–1924. doi: 10.1128/MCB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panigone S, Hsieh M, Fu M, Persani L, Conti M. Luteinizing hormone signaling in preovulatory follicles involves early activation of the epidermal growth factor receptor pathway. Mol Endocrinol. 2008;22:924–936. doi: 10.1210/me.2007-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh M, Thao K, Conti M. Genetic dissection of epidermal growth factor receptor signaling during luteinizing hormone-induced oocyte maturation. PloS one. 2011;6:e21574. doi: 10.1371/journal.pone.0021574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Gruss OJ, et al. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- 16.De Luca M, Lavia P, Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 2006;5:296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 17.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang H, et al. The TPX2 gene is a promising diagnostic and therapeutic target for cervical cancer. Oncol Rep. 2012;27:1353–1359. doi: 10.3892/or.2012.1668. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y, et al. Expression of targeting protein for xklp2 associated with both malignant transformation of respiratory epithelium and progression of squamous cell lung cancer. Clin Cancer Res. 2006;12:1121–1127. doi: 10.1158/1078-0432.CCR-05-1766. [DOI] [PubMed] [Google Scholar]
- 20.Shigeishi H, et al. Expression of TPX2 in salivary gland carcinomas. Oncol Rep. 2009;21:341–344. [PubMed] [Google Scholar]
- 21.Brunet S, et al. Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PloS one. 2008;3:e3338. doi: 10.1371/journal.pone.0003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, et al. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brook M, Smith JW, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 24.Conti M, Hsieh M, Park JY, Su YQ. Role of the epidermal growth factor network in ovarian follicles. Mol Endocrinol. 2006;20:715–723. doi: 10.1210/me.2005-0185. [DOI] [PubMed] [Google Scholar]
- 25.Fan HY, et al. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development. 2008;135:2127–2137. doi: 10.1242/dev.020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De La Fuente R, O'Brien MJ, Eppig JJ. Epidermal growth factor enhances preimplantation developmental competence of maturing mouse oocytesHum. Reprod. 1999;14:3060–3068. doi: 10.1093/humrep/14.12.3060. [DOI] [PubMed] [Google Scholar]
- 27.Prochazka R, Kalab P, Nagyova E. Epidermal Growth Factor-Receptor Tyrosine Kinase Activity Regulates Expansion of Porcine Oocyte-Cumulus Cell Complexes. VitroBiol Reprod. 2003;68:797–803. doi: 10.1095/biolreprod.102.005520. [DOI] [PubMed] [Google Scholar]
- 28.Richani D, Ritter LJ, Thompson JG, Gilchrist RB. Mode of oocyte maturation affects EGF-like peptide function and oocyte competence. Molecular human reproduction. 2013;19:500–509. doi: 10.1093/molehr/gat028. [DOI] [PubMed] [Google Scholar]
- 29.Han SJ, et al. Protein kinase B/Akt phosphorylation of PDE3A and its role in mammalian oocyte maturation. Embo J. 2006;25:5716–5725. doi: 10.1038/sj.emboj.7601431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalous J, et al. PKB/AKT is involved in resumption of meiosis in mouse oocytes. Biol Cell. 2006;98:111–123. doi: 10.1042/BC20050020. [DOI] [PubMed] [Google Scholar]
- 31.Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer Res. 2012;72:3891–3900. doi: 10.1158/0008-5472.CAN-12-0026. [DOI] [PubMed] [Google Scholar]
- 32.Jagarlamudi K, et al. Oocyte-specific deletion of Pten in mice reveals a stage-specific function of PTEN/PI3K signaling in oocytes in controlling follicular activation. PloS one. 2009;4:e6186. doi: 10.1371/journal.pone.0006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busser B, Sancey L, Brambilla E, Coll JL, Hurbin A. The multiple roles of amphiregulin in human cancer. Biochimica et biophysica acta. 2011;1816:119–131. doi: 10.1016/j.bbcan.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature reviews. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 35.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 36.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 37.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 38.Ruggero D, Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- 39.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su YQ, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.